Abstract
The significance of the major immediate-early gene ie3 of mouse cytomegalovirus (MCMV) and that of the corresponding ie2 gene of human cytomegalovirus to viral replication are not known. To investigate the function of the MCMV IE3 regulatory protein, we generated two different MCMV recombinants that contained a large deletion in the IE3 open reading frame (ORF). The mutant genomes were constructed by the bacterial artificial chromosome mutagenesis technique, and MCMV ie3 deletion mutants were reconstituted on a mouse fibroblast cell line that expresses the MCMV major immediate-early genes. The ie3 deletion mutants failed to replicate on normal mouse fibroblasts even when a high multiplicity of infection was used. The replication defect was rescued when the IE3 protein was provided in trans by a complementing cell line. A revertant virus in which the IE3 ORF was restored was able to replicate with wild-type kinetics in normal mouse fibroblasts, providing evidence that the defective growth phenotype of the ie3 mutants was due to disruption of the ie3 gene. To characterize the point of restriction in viral replication that is controlled by ie3, we analyzed the pattern of expression of selective early (β) and late (γ) genes. While we could detect transcripts for the immediate-early gene ie1 in cells infected with the ie3 mutants, we failed to detect transcripts for representative β and γ genes. These data demonstrate that the MCMV transactivator IE3 plays an indispensable role during viral replication in tissue culture, implicating a similar role for the human CMV ie2 gene product. To our knowledge, the ie3 deletion mutants represent the first MCMV recombinants isolated that contain a disruption of an essential gene.
Gene expression during the lytic replication cycle of cytomegalovirus (CMV) is, as in all herpesviruses, regulated in a cascade fashion (27). Viral gene expression starts with the transcription of immediate-early (IE or α) genes immediately after infection. Transcription of IE genes is carried out by the cellular RNA polymerase II and is not dependent on de novo synthesis of viral proteins. Viral transactivator proteins that are synthesized during the IE phase activate transcription of early (β) genes and give rise to a more extended gene expression program during the early phase of the replication cycle. Expression of late (γ) genes occurs after the onset of the viral DNA replication.
The structural organizations of the major IE gene regions of mouse and human CMV (MCMV and HCMV, respectively) show remarkable similarity (31). A complex regulatory sequence, the major IE enhancer promoter (MIEP), controls transcription of the IE genes. Five exons are encoded downstream of the MIEP. The first three exons are spliced to either exon 4, generating the ie1 transcript, or to exon 5, generating the ie2 transcript. In HCMV, the ie1 transcript is translated into the acidic 72-kDa IE1 phosphoprotein. The HCMV ie2 transcript gives rise to the 86-kDa IE2 phosphoprotein. The corresponding IE transcripts of MCMV encode the 89-kDa acidic IE1 phosphoprotein pp89 (15, 16) and the 88-kDa IE3 protein (24).
The functions of the HCMV IE proteins have been well analyzed during recent years. Both of the major HCMV IE proteins are involved in regulation of viral gene expression. It has been suggested that the IE1 protein augments its own expression by positive autoregulation of the MIEP (6, 32, 33). IE1 also has a costimulatory function in the activation of viral early promoters (reviewed in 31 and 27). More recently, it has been shown that IE1 mediates the disruption of nuclear structures, the promyelocytic leukemia protein (PML)-associated nuclear bodies or nuclear domains 10 (ND10), probably in order to generate a favorable environment for replication of the HCMV genome (1, 2, 13, 17). The HCMV IE2-p86 protein is a potent transactivator of HCMV early promoters and of heterologous viral as well as cellular promoters (reviewed in 31 and 27). It is believed that the IE2-p86 protein is the key regulatory protein that governs early and most likely also late gene expression of HCMV. In addition, IE2 down-regulates transcription from its own promoter by binding to the cis-repression signal (crs) target site near the transcription start site of ie1/ie2, thereby mediating autoregulation of its own expression (21, 22, 36). Recent studies suggest that the IE2 protein is also involved in blocking the cell cycle of infected cells (35). In contrast to the thorough functional characterization of the isolated HCMV IE2 protein that was mostly done by transient transfection assays, little is known about its role in the context of the viral infection. An HCMV mutant virus with a deletion of the ie2 gene is not available yet.
Although the functions of the MCMV IE3 protein are not as well analyzed as those of its HCMV counterpart, it is nevertheless clear that the MCMV IE3 protein plays a similar role for replication of MCMV as the HCMV IE2 protein does for HCMV. Namely, it activates MCMV early promoters and is able to repress transcription from the MCMV MIEP (5, 24). This functional equivalence is also reflected in the conservation of the amino acid sequences between the MCMV IE3 and the HCMV IE2 proteins. It is assumed that CMV IE proteins have an important role not only for initiation of the lytic replication cycle but probably also during reactivation of CMV from latency. This aspect will presumably be studied best with MCMV mutants in the mouse model. There is indeed evidence for episodes of ie1 transcription during latency of MCMV (12, 18, 19). It is more interesting, however, that the occurrence of ie3 transcripts during induced reactivation was often associated with more extended gene transcription of MCMV and with virus recurrence (20). The availability of an MCMV ie3 mutant offers the possibility to study the function of the IE3 protein for growth of MCMV in tissue culture as well as its role in pathogenesis of the MCMV infection in vivo.
Here we report on the generation and characterization of MCMV ie3 mutants. The ie3-deficient mutants did not replicate in normal mouse fibroblasts, but growth could be restored by a complementing cell line that provided the IE3 protein in trans. Our data show an essential regulatory function of the IE3 protein during the lytic replication cycle of MCMV.
MATERIALS AND METHODS
Cells and viruses.
Mouse NIH 3T3 cells (ATCC CRL1658) were grown in Dulbecco's modified Eagle medium supplemented with 10% newborn calf serum. Primary mouse embryonic fibroblasts were prepared from BALB/c.ByJ mice and grown in Dulbecco's modified Eagle medium with 10% fetal calf serum. The bacterial artificial chromosome (BAC)-derived MCMV strain MW97.01 (34), which we refer to as parental MCMV in this study, was propagated on NIH 3T3 cells. The ie3-deficient mutants were grown on the complementing cell line NIH 3T3-Bam25.
Construction of NIH 3T3-Bam25 cells.
NIH 3T3-Bam25 cells were derived from NIH 3T3 cells by cotransfecting pBam25 (14) and pPUR (Clontech, Palo Alto, Calif.), a plasmid containing the puromycin resistance gene, using the calcium phosphate technique (10) and selecting cells in medium containing puromycin (Sigma) at 5 μg/ml. Plasmid pBam25 contains a 10.6-kbp BamHI fragment of the MCMV genome (nucleotides [nt] 176,441 to 187,035 [30]) and encodes the MCMV ie1 and ie3 genes under control of the authentic MCMV enhancer ie1/ie3 promoter (14). Cultures were re-fed every 3 to 5 days. Single colonies were picked using cloning cylinders and analyzed for IE1 expression by indirect immunofluorescence using monoclonal antibody Croma 101 (kindly provided by S. Jonjic). Reverse transcription-PCR (RT-PCR) using ie1- and ie3-specific primers was carried out to confirm the presence of the ie1 and ie3 transcripts in NIH 3T3-Bam25 cells. Several cell lines were obtained. For the purpose of this study, we primarily used clone 23 and confirmed our results with clone 18.
Viral growth curves.
Monolayers of NIH 3T3 cells or NIH 3T3-Bam25 cells in 24-well dishes were infected at a multiplicity of infection (MOI) of 2 (for single-cycle growth curves) or 0.05 PFU/cell (for multicycle growth curves) with the different MCMV recombinants. After a 1-h adsorption period, cells were washed three times with phosphate-buffered saline and fed with fresh medium. At different time points after infection, the supernatants of three separate cultures were harvested, cleared of cellular debris, frozen, and thawed. Viral titers were determined by standard plaque assays on NIH 3T3-Bam25 cells.
Plasmid construction.
The recombination plasmid pSTKSie3 was constructed to delete the ie3 gene from the BAC plasmid pSM3fr. Briefly, plasmid pSL301 (Invitrogen, Carlsbad, Calif.) was modified by insertion of an oligonucleotide adapter providing MunI, HindIII, and NsiI sites (forward, 5′-agc tgc aat tgc gaa gct tgg atg cat cc-3′; reverse; 5′-aat tgg atg cat cca agc ttc gca att gc-3′) into the MunI/HindIII digested vector. A 3.1-kbp NsiI/PstI fragment (nt 175,044 to 178,117 of MCMV [30]) was isolated from plasmid HindIII K (7) and cloned into the NsiI site of the vector resulting in plasmid pCBie3. A 3.2-kbp HindIII/MunI fragment (equivalent to MCMV nt 179,510 to 182,682 [30]) was excised from pp89UC (24) and integrated between the HindIII and MunI sites of pCBie3, resulting in plasmid pp89.4. The complete insert was then transferred as a 6.3-kbp NsiI/MunI fragment into the shuttle plasmid pST76KS11, a derivative of pST76KSacB (4) that encodes the negative selection marker sacB (9).
For insertion of the green fluorescent protein (GFP) marker into the MCMV BAC plasmid, the recombination plasmid pST76KS-GFP was generated. Plasmid pUCH3L, comprising the MCMV HindIII L fragment (7), was digested with HpaI, which released a 79-bp fragment (nt 184,236 to 184,315 [30]). An oligonucleotide adapter (forward, 5′-ggg atg cat tag ttt aaa cgg cgc gcc-3′; reverse, 5′-ggc gcg ccg ttt aaa cta atg cat ccc-3′) was inserted that provided the restriction enzyme sites NsiI and AscI, resulting in plasmid pHMM5. The polylinker of plasmid pEGFP-C1 (Clontech) was removed by digestion with BamHI and BglII followed by religation. Then a 1.6-kbp NsiI/MluI fragment comprising the HCMV MIEP, the GFP open reading frame (ORF), and the simian virus 40 polyadenylation signal was excised from the modified pEGFP-C1 plasmid and inserted between the NsiI and AscI sites of pHMM5. A 7.2-kbp MscI/BamHI fragment was excised and transferred to shuttle plasmid pST76KSacB (4).
For construction of a revertant virus genome, recombination plasmid pST76KSie3rev was made as follows; plasmid pp89.4 was digested with NsiI and HindIII, and a 5.7-kbp NsiI/HindIII K fragment (MCMV nt 175,044 to 180,728 [30]) representing the genomic MCMV ie3 sequence was inserted. Then the complete NsiI/MunI insert (equivalent to MCMV nt 175,044 to 182,682 [30]) was transferred to the shuttle vector pST76KS11.
BAC mutagenesis and reconstitution of MCMV mutants.
Recombination between the shuttle plasmids and the MCMV BAC plasmid pSM3fr (34) was performed by a two-step replacement procedure in the Escherichia coli strain CBTS as first described by O'Connor et al. (28) utilizing the recently described modifications (3, 4, 34). Recombinant viruses were reconstituted by transfection of the BAC plasmids into murine embryonic fibroblasts, NIH 3T3, or the complementing cell lines using the calcium phosphate transfection method.
Viral nucleic acid isolation and analysis.
Preparation of total DNA from infected cells, restriction enzyme analysis, and gel electrophoresis was essentially done as described previously (3). MCMV BAC plasmids were isolated from 400-ml E. coli cultures by using Nucleobond PC 500 columns (Macherey-Nagel, Düren, Germany) according to the instructions of the manufacturer.
RT-PCR.
NIH 3T3 cells or NIH 3T3-Bam25 cells were infected with the different recombination viruses at an MOI of 0.5 PFU/cell. For selective expression of IE transcripts, the cultures were incubated from 30 min prior to infection to 13 h postinfection (p.i.) in the presence of cycloheximide (100 μg/ml; Sigma). Total RNA was isolated at the indicated time points after infection by using the RNAzol B method (Tel-Test, Inc., Friendswood, Tex.) according to the manufacturer's protocol. RNA samples were treated with RNase-free DNase I for 15 min at room temperature, and the DNase was inactivated at 65°C for 15 min. The RNA was reverse transcribed using oligo(dT) primers at 42°C for 50 min, and reactions were terminated by heating at 70°C for 15 min. The reverse transcribed products were treated with RNase H for 20 min at 37°C and amplified using specific primers. Primers ie1-R (5′-tac agg aca aca gaa cgc tc-3') and ie1/ie3-F (5′-cct cga gtc tgg aac cga aa-3′) were used to amplify a 188-bp product within the ie1 gene, primers ie3-R (5′-tgt gag gca gta gtt ata cc-3′) and ie1/ie3-F were used to amplify a 299-bp fragment within the ie3 gene, primers gB-R (5′-aga atg tca cgt gcg act gg-3′) and gB-F (5′-gca cgt cgt agg taa att gc-3′) were used to amplify a 509-bp region within the gB gene, and primers HPRT-R (5′-aga ttc aac ttg cgc tca tct tag gc-3′) and HPRT-F (5′-ttg gat aac agg cca gaa ctt tgt tgg-3′) were used to amplify a 163-bp product within the hypoxanthine phosphoribosyltransferase (HPRT) cellular gene. These primer sets have been previously described (18, 19). Primers M54C (5′-cga gtt cgt tca cgt ttc cac ag-3′) and M54NC (5′-gat atg caa gaa gag gta tat cg-3′) were designed to amplify a 660-bp product within the MCMV M54 gene. Primers M115C (5′-atc ttg atc tgg tcg ctg act ga-3′) and M115NC (5′-gac ctc acc acc gta tac gtg tta-3′) were designed to amplify a 679-bp product within the M115 gene. PCRs were performed under the following conditions: 1 cycle at 94°C for 3 min; 30 cycles of 1 min at 94°C, 1 min at the corresponding annealing temperature, and 1 min at 72°C; and 1 cycle at 72°C for 10 min. Annealing temperatures were as follows: 51°C for the M115-specific primers, 58°C for the ie1, ie3, gB, and HPRT primers, and 60°C for the M54-specific primers. The presence of introns in the viral ie1 and ie3 genes and the cellular HPRT gene made it possible to distinguish the correct amplified RNA from contaminant viral or cellular DNA by its size. In the case of the gB, M54, and M115 genes lacking introns, amplificates derived from RNA and DNA could not be distinguished by size. Control reactions carried out in the absence of reverse transcriptase were used to assess the specific detection of RNA. Amplified products were separated on a 1% agarose gel and visualized by ethidium bromide staining.
RESULTS
Construction of MCMV genomes with a deletion in the IE3 ORF.
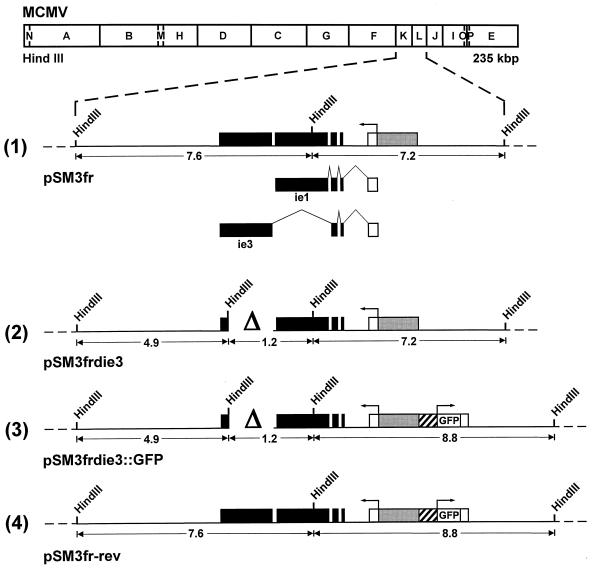
All functions described so far for the MCMV IE3 protein have been deduced from data that were obtained with transient transfection experiments (24). To examine the function of IE3 during the replication cycle of MCMV, we generated MCMV mutants with a deletion in the IE3 ORF. Construction of the mutant genomes was performed by using the recently established BAC mutagenesis procedure (25, 34). The MCMV BAC plasmid pSM3fr (34; Fig. 1, line 1) represents the parental genome that was used to construct the ie3 deletion genomes. pSM3fr contains the complete MCMV genome cloned into a BAC vector. After transfection into permissive cells, it gives rise to recombinant MCMV whose growth properties are indistinguishable from wild-type (wt) MCMV (34). In order to disrupt the IE3 ORF, a 1.4-kbp deletion was introduced into the cloned MCMV genome by making use of the recombination procedures in E. coli as described in Materials and Methods. The deletion (nt 178,117 to 179,510 of the MCMV genome [30]) removed almost entirely the fifth exon of the MCMV ie1/ie3 transcription unit (see Fig. 1, line 2). Thus, the MCMV genome of BAC plasmid pSM3frdie3 was unable to encode the IE3 protein. The position of the deletion in the BAC plasmid pSM3frdie3 was tagged with a HindIII restriction enzyme site (Fig. 1, line 2) in order to facilitate the characterization of the mutant genome. In a second step, the green fluorescent protein (GFP) reporter gene under control of the HCMV MIEP was introduced into the ie3-deficient genome, resulting in BAC plasmid pSM3frdie3::GFP (Fig. 1, line 3). The GFP expression cassette was inserted in front of the ie2 gene (26) since it was previously shown that foreign genes can be inserted at this location without affecting the growth of the recombinant MCMV (23). Insertion of the GFP gene was performed with the intention to follow replication of the ie3-deficient genome in transfected cells by monitoring GFP expression. Finally, we generated a revertant genome by restoring the IE3 ORF. The revertant genome pSM3fr-rev was made to test whether the phenotype of the ie3 knock-out mutants was caused solely by disruption of the ie3 gene. The revertant genome also contained the GFP gene and could therefore be distinguished from the genome of the parental virus (Fig. 1, compare lines 1 and 4).
FIG. 1.
Construction of ie3-deficient MCMV BAC genomes. The HindIII map of the MCMV genome is shown at the top. The expanded map of the HindIII K and L fragments represents the major IE gene region of MCMV. Coding exons are shown in black, and the first noncoding exon of the ie1/ie3 transcription unit is depicted as an open rectangle. The gray box marks the MCMV enhancer ie1/ie3 promoter. The structure of the ie1 and ie3 transcripts is indicated below line 1 of the expanded map. Starting with the parental MCMV BAC plasmid pSM3fr (line 1), the other BAC plasmids pSM3frdie3 (line 2), pSM3frdie3::GFP (line 3), and pSM3fr-rev (line 4) were generated by successive rounds of homologous recombination in E. coli as described in Materials and Methods. The deletion in the fifth exon of the ie3 gene is marked by the delta (Δ). The cross-hatched box in front of the GFP ORF represents the HCMV MIEP.
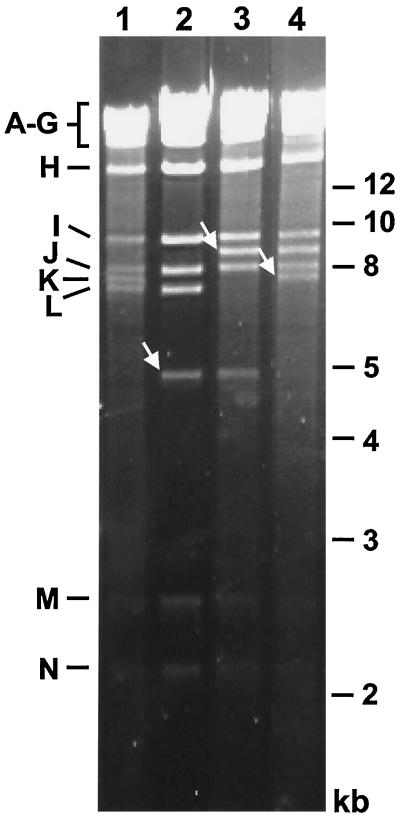
The structure of the BAC plasmids was analyzed by digestion of plasmid DNA with restriction enzyme HindIII followed by agarose gel electrophoresis (Fig. 2). The 7.6-kbp HindIII K fragment of the parental BAC plasmid pSM3fr was missing in the BAC plasmid pSM3frdie3 and was replaced by two new fragments of 1.2 and 4.9 kbp (Fig. 1, lines 1 and 2; Fig. 2, compare lanes 1 and 2). Insertion of the GFP expression cassette in BAC plasmid pSM3frdie3::GFP resulted in a shift of the 7.2-kbp HindIII L fragment to a new fragment of 8.8 kbp (Fig. 1, lines 2 and 3). Hence, the 7.2-kbp HindIII fragment disappeared in the DNA of BAC plasmid pSM3frdie3::GFP, and a new band of 8.8 kbp was observed (Fig. 2, lane 3). Restoration of the ie3 gene led to the reappearance of the 7.6-kbp HindIII K fragment in the revertant BAC plasmid pSM3fr-rev (Fig. 2, lane 4). Additional characterization of the ie3-deficient BAC plasmids was performed by digestion with restriction enzymes EcoRI and NsiI. The observed DNA patterns of the BAC plasmids were as expected (data not shown). These results show that the intended modifications were introduced in the MCMV BAC plasmids and that no adventitious deletions or rearrangements could be detected anywhere else in the cloned genomes.
FIG. 2.
Structural analysis of the ie3-deficient MCMV BAC genomes. Ethidium bromide-stained agarose gel of HindIII-digested BAC plasmids pSM3fr (lane 1), pSM3frdie3 (lane 2), pSM3frdie3::GFP (lane 3), and pSM3fr-rev (lane 4) after separation on a 0.7% agarose gel. The names of the MCMV HindIII fragments (7) and the sizes of the molecular-weight markers are shown in the left and right margin, respectively. New fragments in the BAC plasmids are marked with white arrows.
The ie3 gene is essential for viral DNA infectivity.
To test whether the ie3 gene is essential for infectivity, the MCMV BAC plasmids were transfected into NIH 3T3 cells that are permissive for MCMV infection. The results of the experiments are shown in Table 1. Transfection of the parental BAC plasmid pSM3fr and of the revertant BAC plasmid pSM3fr-rev reproducibly resulted in the formation of plaques. Plaques occurred usually around day 4 or 5 posttransfection, and the infection spread rapidly throughout the monolayers. Cells harboring the revertant virus genome displayed a green fluorescence due to expression of GFP. Transfection of the ie3-deficient genomes pSM3frdie3 and pSM3frdie3::GFP into NIH 3T3 cells did not lead to plaque formation. Identical results were obtained after transfection of the BAC plasmids into mouse embryonic fibroblasts (data not shown). These results suggested that the ie3 gene is essential for the lytic replication cycle of MCMV.
TABLE 1.
Plaque formation on NIH 3T3 cells after transfection of the MCMV BAC plasmids in the absence or presence of pBam25a
| Transfection condition | Plaque formation after transfection of BAC plasmid:
|
|||
|---|---|---|---|---|
| pSM3fr | pSM3frdie3 | pSM3frdie3::GFP | pSM3fr-rev | |
| BAC plasmid only | +++ | − | − | +++ |
| Cotransfection of BAC plasmid and pBam25b | +++ | + | + | ++ |
Scoring of the transfections was as follows: +++, 10–20 plaques; ++, 5–10 plaques; +, 1–5 plaques; −, no plaques.
Plasmid pBam25 spans the deletion in the ie3-deficient BAC plasmids and encodes the MCMV IE genes ie1 and ie3.
In a first attempt to prove that the failure of the ie3-deficient BAC plasmids to form plaques was due to the disrupted ie3 gene, we performed rescue experiments by cotransfection of plasmid pBam25. pBam25 spans the deleted region and encodes the MCMV IE proteins IE1 and IE3 (15, 24). After cotransfection, pBam25 provides the missing IE3 protein in trans, to initiate the MCMV replication cycle. Recombination between the plasmid and the ie3-deficient genomes may eventually result in the reconstitution of replication-proficient genomes. The results of this experiment are shown in Table 1 (second line). The infectivity of BAC plasmids pSM3fr and pSM3fr-rev was not influenced by cotransfection of plasmid pBam25. A few plaques appeared after cotransfection of the ie3-deficient BAC plasmids and pBam25. Typically, the plaques were first seen at 7 to 8 days posttransfection. After occurrence of the plaques, the infection spread rapidly throughout the tissue culture. Infected cells in the culture transfected with BAC plasmid pSM3frdie3::GFP displayed a green fluorescence. The reduced number of plaques as well as the delayed kinetics in plaque formation was consistent with the expectation that reconstitution of replication-proficient genomes by recombination with the complementing plasmid had to occur prior to plaque formation. Analysis of viral DNA obtained from these cultures showed that the reconstituted viruses had indeed acquired the ie3 gene from the cotransfected plasmid (data not shown). Altogether, these experiments indicated that the ie3-deficient MCMV genomes cannot give rise to infectious virus in normal murine fibroblasts and that the replication-deficient genomes can be rescued by cotransfection of a complementing plasmid.
trans-complementation of viral DNA infectivity and reconstitution of MCMV ie3 mutants.
Since the ie3 gene seemed to be essential for replication of MCMV, a complementing cell line that provided the missing IE3 protein in trans was needed in order to reconstitute mutant viruses from the recombinant BAC plasmids. To this end, NIH 3T3 cells were transfected with plasmid pBam25 that encodes the MCMV IE genes ie1 and ie3, and stable NIH 3T3-Bam25 cell lines were isolated as described in Materials and Methods.
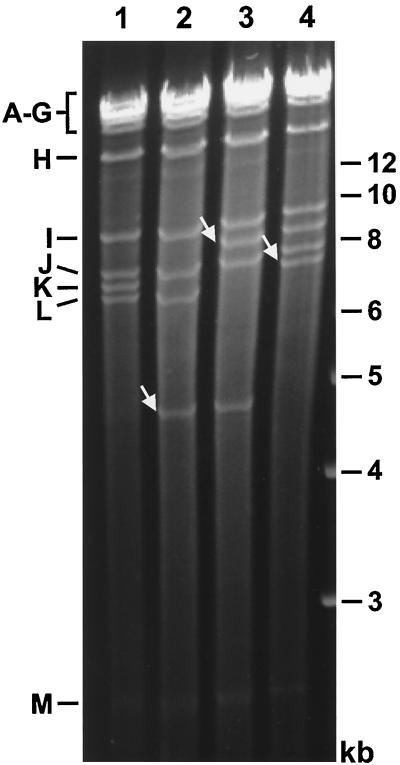
The four different BAC plasmids were then transfected into one of the NIH 3T3-Bam25 cell lines. Plaques appeared around 5 to 7 days posttransfection, and the infection spread throughout the culture. To analyze the genome structure of the reconstituted mutants, viral DNA was isolated from infected NIH 3T3-Bam25 cells and subjected to restriction enzyme digestion. Fig. 3 shows the DNA fragment profiles after HindIII digestion of the four different MCMV genomes. The DNA patterns of the viral genomes were identical to those of the corresponding BAC plasmids (compare Fig. 2 and 3), confirming that the viral mutants were reconstituted from these BAC plasmids and that the viral genomes did not change during replication in the complementing cell line. These data demonstrate that the cell line was able to support replication of the ie3-deleted genomes and growth of the ie3-deficient MCMV mutants.
FIG. 3.
Structural analysis of the genomes of the MCMV ie3 mutants. DNA isolated from NIH 3T3-Bam25 cells infected with the parental MCMV (lane 1), the ie3-deficient mutants MCMVdie3 (lane 2), and MCMVdie3::GFP (lane 3), or the revertant virus MCMVrev (lane 4) was subjected to HindIII digestion, separated on a 0.7% agarose gel, and stained with ethidium bromide. Size markers are shown in the right margin, and the names of the HindIII fragments (7) are indicated in the left margin. New fragments in the genomes of the MCMV mutants are marked by white arrows.
Growth analysis of the ie3 mutants.
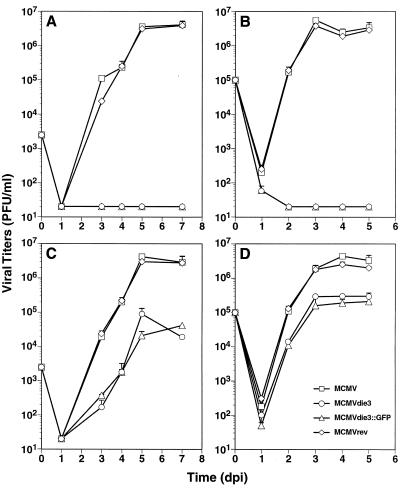
The possibility to propagate the ie3 mutants on the complementing cell line allowed us to prepare viral stocks. Thus, we could then perform infection experiments with the mutant viruses and test whether the ie3 gene is definitely required for growth of MCMV in normal mouse fibroblasts. When NIH 3T3 cells were infected with a low MOI of 0.05 PFU/cell, the amount of virus that could be found in the supernatant of cultures infected with the ie3 mutant dropped below the detection level by 1 day p.i. Even 1 week p.i., viral progeny was not obtained in these cultures (Fig. 4A). Cultures that were infected with the parental MCMV strain displayed a rapid increase in the viral titers. The growth kinetics of the revertant virus MCMVrev were comparable to those of the parental virus, demonstrating that reinsertion of the ie3 gene led to a complete rescue of the growth phenotype (Fig. 4A). This experiment indicated that the ie3 gene is essential for replication of the MCMV when a low-input dose is used. Still, it was possible that the requirement for ie3 could be overcome by using a high MOI. To examine the growth dependence of the mutant viruses on the input dose, infection experiments were performed by using an MOI of 2. Again, no growth of the ie3 mutants was observed when cells were infected under these conditions. In contrast, the viruses that encode the IE3 protein rapidly grew to high titers (Fig. 4B). We concluded from these experiments that the ie3 gene is absolutely essential for growth of MCMV in normal fibroblast cells, regardless whether a low- or a high-input dose is used.
FIG. 4.
Growth curve analysis of the MCMV ie3 mutants. NIH 3T3 (A, B) or NIH 3T3-Bam25 (C, D) cells were infected at an MOI of 0.05 (A, C) or 2 PFU per cell (B, D) with the parental MCMV, MCMVdie3, and MCMVdie3::GFP and the revertant MCMVrev. At the indicated time points after infection (days p.i. [dpi]), supernatants from the infected cultures were harvested and titered on monolayers of NIH 3T3-Bam25 cells. The limit of detection was 20 PFU/ml. Error bars indicate the standard deviation from three separate cultures.
Growth analyses were next performed on the NIH 3T3-Bam25 cell line to examine the growth behavior of the ie3 mutants and the capability of the complementing cell line to support replication of the mutants. When NIH 3T3-Bam25 cells were infected with the ie3 mutants at a low MOI of 0.05 PFU/cell, virus production could be detected at 3 days p.i., and a rise of the virus titers was observed on days 4 to 5 p.i. (Fig. 4C). The increase in the titers of the ie3 mutants was reduced in comparison to the titers of the parental virus. Maximal titers were obtained at day 5 p.i. with values of about 4 × 104 to 1 × 105 PFU/ml, while the ie3-expressing viruses achieved titers of about 4.2 × 106 PFU/ml. Similar observations were made when the NIH 3T3-Bam25 cells were infected at an MOI of 2 (Fig. 4D). Virus progeny was found 2 and 3 days p.i., but there was little further increase of the titers after day 3 p.i. The titers that were achieved with the parental and revertant viruses were about 1 to 1.5 orders of magnitude higher than those obtained with the ie3 mutants. The difference between the titers of parental MCMV and ie3 viruses was already seen on days 2 and 3 p.i. (Fig. 4D). Altogether, these data clearly indicate that the complementing cell line was able to support growth of the ie3 deletion mutants, although the growth behavior of the mutants was altered in comparison to wt virus.
The ie3 gene product is essential for early gene expression.
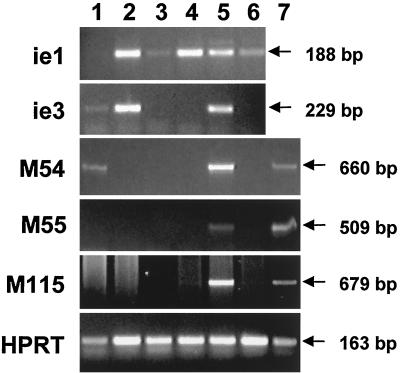
Next, we asked at which stage the viral gene expression was blocked when cells were infected with the ie3 mutants that are unable to express the regulatory protein IE3. Expression of representative viral genes was analyzed by RT-PCR using RNA that was isolated from cells infected with the ie3 deletion mutant MCMVdie3::GFP and, for comparison, RNA that was with the parental MCMV. First, we tested whether the IE genes were transcribed when cells were infected in the presence of cycloheximide, i.e., in the absence of de novo protein synthesis. Transcripts arising from the ie1 and ie3 genes were detected in cells infected with the parental virus (Fig. 5, lane 2). In cells infected with the ie3 mutant MCMVdie3::GFP, only ie1 transcripts could be detected (Fig. 5, lane 4). As expected, transcripts of IE genes accumulated under this condition of infection because transcription of IE genes was performed by the transcription machinery of the cell, and de novo synthesis of viral proteins was not required. Transcription of early and late genes did not occur since viral transactivator proteins that were required for early and late gene expression were not synthesized in the presence of cycloheximide. The data clearly indicate that the ie3 mutant did not synthesize an ie3 transcript (Fig. 5, lane 4). This result confirmed that the ie3 gene had been disrupted in the ie3 mutant and that the mutant was therefore unable to encode the regulatory IE3 protein.
FIG. 5.
Detection of viral transcripts after infection with an MCMV ie3 mutant. NIH 3T3 (lanes 1 through 6) or NIH 3T3-Bam25 (lane 7) cells were infected at an MOI of 0.5 PFU per cell with parental MCMV (lanes 1, 2, and 5) or MCMVdie3::GFP (lanes 3, 4, 6, and 7) in the presence (lanes 2 and 4) or absence (lanes 1, 3, and 5 to 7) of cycloheximide. Whole-cell RNA was harvested at 13 h p.i. (lanes 1 through 4) or 20 h p.i. (lanes 5 through 7), treated with DNase, and reverse transcribed using oligo(dT). PCRs were performed using primer sets specific for ie1, ie3, M54, M55, M115, and HPRT as described in Materials and Methods. Amplified products were separated on 1.5% agarose gels and visualized by ethidium bromide staining. Sizes of the amplified products are indicated by arrows. Specific PCR-amplified products were not detected in control reactions in which the reverse transcriptase was not added during the RNA reverse transcription reaction (data not shown).
Next, we examined the viral gene expression at 13 h p.i., a time point during the early phase of the infection cycle well before the onset of viral DNA replication (14; Fig. 5, lanes 1 and 3). In addition to the ie1 and ie3 transcripts, we found transcripts of the early gene M54, encoding the viral DNA polymerase (8) and a small amount of the transcript encoding the glycoprotein B (M55 [29, 30]) in MCMV-infected cells (Fig. 5, lane 1). In cells infected with the ie3 mutant, viral gene expression was restricted to the ie1 gene (Fig. 5, lane 3). The level of ie1 transcripts in cells infected with the ie3 mutant seemed higher than in cells infected with the parental virus (Fig. 5, compare lanes 1 and 3). This might indicate that feedback regulation of IE gene expression by the IE3 protein that leads to reduced levels of ie1 transcripts in MCMV-infected cells during the early phase (Fig. 5, lane 1) cannot occur in cells infected with the ie3 mutant and results in enhanced expression of the ie1 gene (Fig. 5, lane 3). Transcripts of early genes could not be detected in MCMVdie3::GFP-infected cells by 13 h p.i. (Fig. 5, lane 3). This result indicated that either activation of early gene transcription was completely impossible in cells infected with the ie3 mutants or the time course of the viral gene expression program was delayed.
To distinguish between these possibilities, RNA isolated in the late phase of the infection cycle (20 h p.i.) was analyzed. RNA from MCMV-infected cells contained transcripts of the late gene M115 encoding glycoprotein L (37), in addition to the early and IE transcripts that were already detected at the earlier time point. Again, in RNA isolated from cells infected with the ie3 mutant, only transcripts arising from the ie1 gene could be detected (Fig. 5, lane 6). Thus, gene expression in MCMVdie3::GFP-infected cells was always restricted to the IE gene ie1, irrespective of whether the cells were infected in the presence or absence of CH and at which time point p.i. the infected cells were analyzed (Fig. 5, compare lanes 3, 4, and 6). Accordingly, the protein encoded by the ie3 gene must exert a key function in activation of early gene expression.
To test directly whether ie3 is important for activating early genes, we analyzed RNA isolated from the complementing NIH 3T3-Bam25 cell line that had been infected with the ie3 mutant. Since the complementing cell line encodes the missing protein, the expression of early and late genes should be restored if the protein mediates the proposed regulatory function. The results of the experiment revealed the same profile of early and late viral transcripts in MCMVdie3::GFP-infected NIH 3T3-Bam25 cells as in MCMV-infected NIH 3T3 cells (Fig. 5, compare lanes 7 and 5). We concluded from these experiments that the complementing cell line provided a sufficient amount of the transactivating protein to achieve activation of early and late gene expression and to substitute for the lack of ie3 expression by the ie3 mutant. In summary, the data indicate that the protein encoded by the ie3 gene plays an essential role for the activation of the viral gene expression program.
DISCUSSION
In this study, we report on the generation of MCMV ie3-deficient mutants. Disruption of the ie3 gene on the cloned MCMV genome was achieved by utilizing the recently established BAC mutagenesis procedure. Transfection of ie3-deficient genomes into permissive cells did not result in plaque formation, indicating that the genomes were replication deficient. Infectious viruses could be reconstituted by transfection of the ie3-deficient genomes into a cell line that provided the missing IE protein in trans. The ie3 mutants could not grow on normal non-complementing cells, indicating the essential function of the ie3-encoded protein. Transcript analysis in cells infected with an ie3 mutant showed that early and late genes were not activated. Altogether, these data provided direct evidence for an essential regulatory role of ie3 for replication of MCMV.
Construction of MCMV ie3-deficient mutants by the BAC technique.
The mutant MCMV genomes were constructed by site-directed mutagenesis of the cloned MCMV genomes in E. coli (25, 34). This technique might be especially useful for mutagenesis of essential genes since construction of the mutant genome is completely independent of the ability of the corresponding mutant virus to grow in cell culture. Thus, we can first manipulate any gene of interest on the cloned genome, and, in a second separate step, we can examine the phenotypic consequences of the manipulation, e.g., whether the deleted gene is essential or nonessential.
The mutant BAC plasmids isolated from bacterial cultures were of clonal origin. Therefore, after transfection of the BAC plasmids into the complementing cell line, we got mutant viruses only, and no selection against parental virus was required. We consider this a major advantage of our technique in comparison to conventional recombination techniques in complementing cell lines, because selection and isolation of mutant viruses might be quite cumbersome if the mutant has an impaired growth potential, in comparison to the wt virus.
Furthermore, we demonstrated that consecutive rounds of mutagenesis can be performed on the cloned MCMV genome without the need to reconstitute viral intermediates. We showed that intermediate steps can lead to replication-deficient genomes and that eventually a revertant replication-proficient genome can be reconstituted. The GFP marker inserted into the revertant genome allowed us to differentiate between the parental and the revertant viruses. Rescue of the growth potential by reinsertion of the ie3 gene in the revertant genome confirmed that the observed growth deficit of the ie3 mutants was indeed due to the disruption of the ie3 gene and excluded the possibility that any other mutation that may have been accidentally introduced somewhere else in the genome might have been responsible for the phenotype. To our knowledge, this is the first report on the generation and complementation of an MCMV mutant with a disruption of an essential gene.
Properties of the complementing cell line.
The MCMV ie3 mutants could be reconstituted and propagated on NIH 3T3-Bam25 cell lines. Neither the successful generation of the complementing cell line nor the fact that the cell line was able to support growth of the ie3 mutants seems to be trivial. For example, several IE proteins of other herpesviruses turned out to be toxic for cells (11). Accordingly, construction of cell lines expressing such IE proteins was difficult. Since it has been reported that the HCMV IE2 protein, which is homologous to the MCMV IE3 protein, is able to block the cell cycle in transfected cells (35), one could expect that generation of MCMV IE3-expressing cell lines may be rather complicated. As is observed with coexpressing ie1 and ie2 of HCMV (A. Angulo and P. Ghazal, unpublished results), we did not encounter any problems in generating the NIH 3T3-Bam25 cell lines that express both of the MCMV major IE genes. Also, we reported before on the successful generation of a similar cell line that encodes the MCMV IE1 and IE3 proteins (5). However, this particular cell line failed to complement the ie3 mutants. The reason for this is unclear at present but may be the result of inappropriate expression or modification of the MCMV major IE proteins.
The IE3 protein expressed by the complementing cell line was sufficient to allow growth of the ie3 mutants. However, the titers of the ie3 mutants obtained on the cell line did not reach those levels which were achieved with the parental and revertant viruses. The titers of the different viruses were determined on the complementing cell lines, and we have to consider that the efficiency of plaque formation of the ie3 mutants might be lower than that of the parental virus. Although we do not have any indication for a reduced efficiency of plaque formation, we cannot completely rule out that the titers of the ie3 mutants were underestimated. But even if the input titers were underestimated, the results indicate that the ie3 mutants grow to lower final titers. There are several possible explanations for the altered growth kinetics of the ie3 mutants on the complementing cell line. First, the amount of the IE3 protein in the cell line might not be as high as that during infection with wt viruses. Second, the amount of IE3 required may vary during the infection cycle. There is indeed evidence that the IE3 protein autoregulates expression from its own promoter (24). Though the IE genes in the complementing cell line were expressed from their authentic promoter, it is not clear whether correct transcriptional regulation is maintained when the viral genes are integrated into the cellular chromatin. There is also evidence for posttranslational modification of the MCMV IE3 protein during the replication cycle, most likely by phosphorylation (24). We do not know whether appropriate modification of IE3 occurs in the complementing cell line. Finally, the copy number of the viral genomes goes up during replication, whereas the number of integrated IE genes in the cellular genomes remains constant. The lower titers of the ie3 mutants in the complementing cell line can be easily explained if a certain amount of IE3 protein is required per viral genome. In this case, IE3 will become limiting in the cell line when the viral copy number increases. Accordingly, virus production will already cease at lower titers. This will not happen when IE3 is expressed from the wt genomes since the copy number of the ie3 gene increases coordinately with the increase of the viral genomes. Additional experiments are required to explain the limited growth of the ie3 mutants in the complementing cell line.
The ie3 gene is essential for viral growth, and the ie3 encoded protein is a key regulator for early gene expression.
We provide several lines of evidence that the MCMV ie3 gene is essential for viral growth. (i) MCMV BAC genomes with a large deletion in the ORF encoding the IE3 protein were unable to generate viral progeny. (ii) Viral infectivity could be restored in cis by cotransfection of a plasmid spanning the deleted region and in trans by transfection of the BAC plasmids into a complementing cell line that provided the missing IE protein. (iii) The ie3 mutants that were reconstituted on the complementing cell line were unable to grow on normal fibroblasts either at low or high MOI.
During the IE phase of the infection cycle MCMV expresses at least two proteins that are encoded by the major IE region, namely the 89-kDa protein pp89 and the 88-kDa protein IE3. Due to disruption of exon 5 of the ie1/ie3 transcription unit, it is clear that the ie3 mutants are unable to express the IE3 protein. The ie1 gene is not affected by the deletion, and the RNA analyses indicated that the ie3 mutants express ie1 transcripts in infected cells irrespective of whether infection occurred in the presence or absence of cycloheximide. Though enhanced expression of the IE1 protein might occur in cells infected with the ie3 mutants because the lack of IE3 protein might lead to a failure in autoregulation of IE transcription (24), we consider it unlikely that the observed growth phenotype is due to altered IE1 expression. This belief is supported by the fact that disruption of the ie1 gene does not result in a lethal phenotype (25 and unpublished data). The most likely explanation for the growth defect of the ie3 mutants is their inability to synthesize the IE3 protein. This conclusion is further supported (i) by the observation that the complementing cell line that provides the IE3 protein in trans allows growth of the ie3 mutants and (ii) by the fact that repair of the IE3 ORF in the revertant virus rescued the growth potential.
The restricted viral gene expression profile displayed by the ie3 mutants is explained best by the absence of the regulatory function of the IE3 protein. Transcription of viral genes was confined to ie1, which can occur in the absence of viral regulatory proteins. Though our RNA analyses were limited to the important early genes encoding the viral DNA polymerase and the glycoprotein B and we cannot completely rule out that some early genes might be activated in the absence of the IE3 protein, it seems that gene expression by the ie3 mutants is blocked at the IE stage of the infection cycle. Accordingly, the IE3 protein has a key regulatory function for activation of viral gene expression, i.e., for the switch from α to β gene expression. Further studies are required to investigate whether the IE3 protein is just needed to initiate early gene activation or is required throughout the replication cycle in order to maintain viral gene expression.
The observation of the important regulatory function of the IE3 protein has two implications. If the counterpart of the MCMV IE3 protein in HCMV, IE2, possesses a similar key regulatory function, it may be possible to combat the HCMV infection by developing and using therapeutic compounds that interfere with this function of the HCMV IE2 protein. Second, gene activation by CMV IE proteins might represent a bottleneck not only during initiation of the lytic replication cycle but also during reactivation of CMV from latency. Indeed, data from Reddehase et al. suggest that the regulatory function of the IE3 protein is also pivotal during reactivation of MCMV (20). Again, this promises to offer a point of intervention at which to inhibit recurrence of CMV and to control the CMV infection at a very early stage.
In conclusion, we have shown that the ie3 gene plays a key role for activation of MCMV gene expression. Given the many similarities between the MCMV IE3 and the HCMV IE2 protein, the data of our experiments predict a comparable essential role of the IE2 protein for gene expression of HCMV during the lytic replication cycle. The precise mechanism(s) and functional significance of the major IE transactivator of CMV in promoting lytic replication in vitro and in vivo are topics that remain to be explored.
ACKNOWLEDGMENTS
We thank Andrea Reus for technical assistance.
This work was in part supported by grants from the Bundesministerium für Bildung and Forschung (projects 01GE96140 and 01GE9918) and the Deutsche Forschungsgemeinschaft (project A2 of the Sonderforschungsbereich 455) to M.M. and the National Institutes of Health to P.G. (AI-30627). A.A. is supported by a fellowship from the University of California Universitywide AIDS Research Program.
Footnotes
Publication 13327-IMM from The Scripps Research Institute.
REFERENCES
- 1.Ahn J H, Brignole E J, Hayward G S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angulo A, Messerle M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst E M, Hahn G, Koszinowski U H, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J Virol. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buhler B, Keil G M, Weiland F, Koszinowski U H. Characterization of the murine cytomegalovirus early transcription unit e1 that is induced by immediate-early proteins. J Virol. 1990;64:1907–1919. doi: 10.1128/jvi.64.5.1907-1919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebeling A, Keil G M, Knust E, Koszinowski U H. Molecular cloning and physical mapping of murine cytomegalovirus DNA. J Virol. 1983;47:421–433. doi: 10.1128/jvi.47.3.421-433.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott R, Clark C, Jaquish D, Spector D H. Transcription analysis and sequence of the putative murine cytomegalovirus DNA polymerase gene. Virology. 1991;185:169–186. doi: 10.1016/0042-6822(91)90765-4. [DOI] [PubMed] [Google Scholar]
- 9.Gay P, Le C D, Steinmetz M, Ferrari E, Hoch J A. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J Bacteriol. 1983;153:1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghazal P, Nelson J A. Enhancement of RNA polymerase II initiation complexes by a novel DNA control domain downstream from the cap site of the cytomegalovirus major immediate-early promoter. J Virol. 1991;65:2299–2307. doi: 10.1128/jvi.65.5.2299-2307.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glorioso J C, DeLuca N A, Fink D J. Development and application of herpes simplex virus vectors for human gene therapy. Annu Rev Microbiol. 1995;49:675–710. doi: 10.1146/annurev.mi.49.100195.003331. [DOI] [PubMed] [Google Scholar]
- 12.Henry S C, Hamilton J D. Detection of murine cytomegalovirus immediate early 1 transcripts in the spleens of latently infected mice. J Infect Dis. 1993;167:950–954. doi: 10.1093/infdis/167.4.950. [DOI] [PubMed] [Google Scholar]
- 13.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keil G M, Ebeling-Keil A, Koszinowski U H. Temporal regulation of murine cytomegalovirus transcription and mapping of viral RNA synthesized at immediate early times after infection. J Virol. 1984;50:784–795. doi: 10.1128/jvi.50.3.784-795.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keil G M, Ebeling-Keil A, Koszinowski U H. Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J Virol. 1987;61:526–533. doi: 10.1128/jvi.61.2.526-533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keil G M, Ebeling-Keil A, Koszinowski U H. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J Virol. 1987;61:1901–1908. doi: 10.1128/jvi.61.6.1901-1908.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 18.Kurz S, Steffens H P, Mayer A, Harris J R, Reddehase M J. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71:2980–2987. doi: 10.1128/jvi.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurz S K, Rapp M, Steffens H P, Grzimek N K, Schmalz S, Reddehase M J. Focal transcriptional activity of murine cytomegalovirus during latency in the lungs. J Virol. 1999;73:482–494. doi: 10.1128/jvi.73.1.482-494.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurz S K, Reddehase M J. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J Virol. 1999;73:8612–8622. doi: 10.1128/jvi.73.10.8612-8622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang D, Stamminger T. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J Virol. 1993;67:323–331. doi: 10.1128/jvi.67.1.323-331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macias M P, Stinski M F. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc Natl Acad Sci USA. 1993;90:707–711. doi: 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning W C, Mocarski E S. Insertional mutagenesis of the murine cytomegalovirus genome: one prominent alpha gene (ie2) is dispensable for growth. Virology. 1988;167:477–484. [PubMed] [Google Scholar]
- 24.Messerle M, Buhler B, Keil G M, Koszinowski U H. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J Virol. 1992;66:27–36. doi: 10.1128/jvi.66.1.27-36.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messerle M, Keil G M, Koszinowski U H. Structure and expression of murine cytomegalovirus immediate-early gene 2. J Virol. 1991;65:1638–1643. doi: 10.1128/jvi.65.3.1638-1643.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mocarski, E. S. 1996. Cytomegaloviruses and their replication, p. 2447–2492. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Lippincott-Raven Publishers, Philadelphia, Pa.
- 28.O'Connor M, Peifer M, Bender W. Construction of large DNA segments in Escherichia coli. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 29.Rapp M, Messerle M, Buhler B, Tannheimer M, Keil G M, Koszinowski U H. Identification of the murine cytomegalovirus glycoprotein B gene and its expression by recombinant vaccinia virus. J Virol. 1992;66:4399–4406. doi: 10.1128/jvi.66.7.4399-4406.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stenberg R M. The human cytomegalovirus major immediate-early gene. Intervirology. 1996;39:343–349. doi: 10.1159/000150505. [DOI] [PubMed] [Google Scholar]
- 32.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stenberg R M, Stinski M F. Autoregulation of the human cytomegalovirus major immediate-early gene. J Virol. 1985;56:676–682. doi: 10.1128/jvi.56.3.676-682.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner M, Jonjic S, Koszinowski U H, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiebusch L, Hagemeier C. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J Virol. 1999;73:9274–9283. doi: 10.1128/jvi.73.11.9274-9283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J, Jupp R, Stenberg R M, Nelson J A, Ghazal P. Site-specific inhibition of RNA polymerase II preinitiation complex assembly by human cytomegalovirus IE86 protein. J Virol. 1993;67:7547–7555. doi: 10.1128/jvi.67.12.7547-7555.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, Scalzo A A, Lyons P A, Farrell H E, Rawlinson W D, Shellam G R. Identification, sequencing and expression of the glycoprotein L gene of murine cytomegalovirus. J Gen Virol. 1994;75:3235–3240. doi: 10.1099/0022-1317-75-11-3235. [DOI] [PubMed] [Google Scholar]