Abstract
Candida is a polyphyletic genus of asexually reproducing yeasts in the Saccharomycotina with more than 400 species that occur in almost all families of the subclass and its name is strongly connected with the infectious disease candidiasis. During the last two decades, approximately half of the Candida species have been reassigned into more than 36 already existing genera and 14 newly proposed genera, but the polyphyletic feature of the genus largely remained. Candida auris is an important, globally emerging opportunistic pathogen that has caused life-threatening outbreaks in healthcare facilities worldwide. This species belongs to the Candida auris-Candida haemuli (CAH) clade in the Metschnikowiaceae, a clade that contains multidrug-resistant clinically relevant species, but also species isolated from natural environments. The clade is phylogenetically positioned remotely from the type species of the genus Candida that is Candida vulgaris (currently interpreted as a synonym of Candida tropicalis) and belongs to the family Debaryomycetaceae. Although previous phylogenetic and phylogenomic studies confirmed the position of C. auris in the Metschnikowiaceae, these analyses failed to resolve the position of the CAH clade within the family and its delimitation from the genera Clavispora and Metschnikowia. To resolve the position of the CAH clade, phylogenomic and comparative genomics analyses were carried out to address the phylogenetic position of C. auris and related species in the Metschnikowiaceae using several metrics, such as the average amino acid identity (AAI) values, the percentage of conserved proteins (POCP) and the presence-absence patterns of orthologs (PAPO). Based on those approaches, 13 new genera are proposed for various Candida and Hyphopichia species, including members of the CAH clade in the Metschnikowiaceae. As a result, C. auris and related species are reassigned to the genus Candidozyma. Fifty-five new combinations and nine new species are introduced and this will reduce the polyphyly of the genus Candida.
Citation: Liu F, Hu Z-D, Zhao X-M, et al. 2024. Phylogenomic analysis of the Candida auris-Candida haemuli clade and related taxa in the Metschnikowiaceae, and proposal of thirteen new genera, fifty-five new combinations and nine new species. Persoonia 52: 22–43. https://doi.org/10.3767/persoonia.2024.52.02 .
Keywords: AAI, Candida, Metschnikowiaceae, new taxa, nomenclature, PAPO, phylogenomics, POCP, statistics, taxonomy
INTRODUCTION
The genus Candida contains ascomycetous yeasts that belong to Saccharomycotina without a known sexual state and that reproduce asexually by budding and that may form pseudo hyphae or true hyphae and lack distinctive morphological features that distinguish it from other asexually (and sexually) reproducing ascomycetous yeast genera ( Lachance et al. 2011). For a long time, the genus served as a dustbin genus for many asexual ascomycetous yeast species that did not show any distinct properties that could be used for their placement in a specific genus. This broad definition of the genus and the past classification system with dual naming for sexual and asexual morphs has made this genus large and phylogenetically heterogeneous. In the fifth edition of The Yeasts, a taxonomic study ( Lachance et al. 2011) and Daniel et al. (2014), 365 and 434 species were recognized in the genus Candida, respectively. Many molecular phylogenetic studies (e.g., Kurtzman & Robnett 1998, Kurtzman 2011a, Lachance et al. 2011, Daniel et al. 2014) indicated that Candida is a highly polyphyletic genus with its members distributed in almost all families of Saccharomycotina. Some species of Candida are of major importance in the medical field, and among the most important opportunistic pathogens ( Lachance et al. 2011, Stavrou et al. 2019, Takashima & Sugita 2022), e.g., Candida albicans, Candida dublinenesis, Candida glabrata (also known as Nakaseomyces glabratus, see below), Candida parapsilosis and Candida tropicalis. Besides, many emerging species have been identified ( Stavrou et al. 2019), such as Candida auris. Following the implementation of the ‘One Fungus, one name’ principle by the International Code of Nomenclature for algae, fungi, and plants (ICNafp; McNeill et al. 2012), C. glabrata has recently been transferred to the genus Nakaseomyces in the Saccharomycetaceae (as N. glabratus) ( Takashima & Sugita 2022). Fortunately, the clinically most relevant species C. albicans, C. dubliniensis, C. parapsilosis and C. tropicalis belong to the core of the genus that is represented by its nomenclatural type species, Candida vulgaris (a synonym of C. tropicalis) in the family Debaryomycetaceae ( Lachance et al. 2011, Daniel et al. 2014).
Candida auris, an emerging fungal opportunist, was firstly isolated from the external ear canal of a Japanese patient in 2009 and placed in the Candida haemuli clade (also known as the Candida haemulonii clade, CAH) in the family Metschnikowiaceae ( Satoh et al. 2009, Cendejas-Bueno et al. 2012), which from a phylogenetic perspective is distantly related to C. tropicalis. This species has been isolated around the world and causes a threat to global health due to its high mortality and resistance to multiple antifungal drugs ( Clancy & Nguyen 2017, Lockhart et al. 2017, Rabaan et al. 2023). A few other species closely related to C. auris also may cause infections, i.e., Candida khanbhai, C. haemuli (also known as C. haemulonii), Candida duobushaemuli (also known as Candida duobushaemulonii), Candida pseudohaemuli (also known as Candida pseudohaemulonii) and Candida vulturna were found to be resistant to multiple antifungal drugs, mainly amphotericin B and with a reduced susceptibility to various azoles ( Cendejas-Bueno et al. 2012, Sipiczki & Tap 2016, Muthusamy et al. 2022, De Jong et al. 2023). Other species belonging to the clade, such as Candida chanthaburiensis, Candida konsanensis, Candida heveicola and Candida ruelliae, were isolated from natural habitats, i.e., flowers and tree bark ( Saluja & Prasad 2008, Wang et al. 2008, Limtong & Yongmanitchai 2010, Sarawan et al. 2013). Although C. auris, C. haemuli and C. vulturna are mostly obtained prominent from humans and animals, some isolates originated from plants or marine substrates ( Van Uden & Kolipinski 1962, Sipiczki & Tap 2016, Arora et al. 2021, Yadav et al. 2022). Sipiczki & Tap (2016) and Klaps et al. (2020) published three new species as C. vulturna pro tempore, Candida ohialehuae pro tempore and Candida metrosideri pro tempore in the CAH clade. The authors indicated that the placement in the genus Candida was provisional considering the distant relationships of those new species with the core Candida species in the Lodderomyces clade, where the type species of the genus Candida is placed. However, this made the names formally invalid according to Art. 36.1(a) (ICNafp Shenzhen code; Turland et al. 2018) as indicated in Index Fungorum and MycoBank. Recently, De Jong et al. (2023) validated the names C. chanthaburiensis, C. konsanensis, C. metrosideri, C. ohialehuae and C. vulturna, and described a new species Candida khanbhai in the CAH clade, but they did not revise the taxonomy of the CAH clade.
Recently, genome-based metrics, i.e., the average amino acid identity (AAI) values and the percentage of conserved proteins (POCP), have been used to characterize genera in prokaryotes ( Luo et al. 2014, Varghese et al. 2015, Parks et al. 2018, 2022, Hayashi Sant’Anna et al. 2019, Meier-Kolthoff & Göker 2019, Barco et al. 2020, Nouioui & Sangal 2022). Such approaches and the presence-absence patterns of orthologs (PAPO) were also employed to delimit yeast genera using presently well-recognized genera in the Saccharomycetaceae as an example ( Liu et al. 2024). From the above study, a range of 80–92 % POCP values and a range of 60–70 % AAI values might be estimated thresholds to discriminate genera in Saccharomycetaceae ( Liu et al. 2024).
The recent addition of so-called Candida species in the CAH clade by Sipiczki & Tap (2016) and Klaps et al. (2020), prompted us to carry out a phylogenomic and comparative genome analysis of the CAH clade and related species using some genome-based metrics that have been used in the studies of Takashima et al. (2019) and Liu et al. (2024), such as the AAI, the POCP and the PAPO. The genomes of 150 species including 154 strains in the Metschnikowiaceae have been analyzed to resolve the taxonomy of the CAH clade. A new genus, Candidozyma, is proposed to accommodate the members of the CAH clade. Furthermore, our analyses revealed 12 other lineages in the Metschnikowiaceae for which new generic names are proposed.
MATERIALS AND METHODS
Ribosomal DNA (rDNA) phylogenetic analysis
The sequences of the ITS (including 5.8S) and the D1/D2 domains of the large subunit (LSU) (Table S1) were downloaded from the NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide/) and aligned using the MAFFT program G-INS-i ( Katoh & Standley 2013). Three datasets, the ITS, the D1/D2 and the combined ITS + D1/D2 sequences were used to construct Maximum Likelihood (ML) trees with the GTR+G+I model using the software of RAxML v. 8.2.12 ( Stamatakis 2014) with 1 000 bootstrap replicates.
Genome sequencing, assemblies and annotation
DNA from yeast colonies of 14 strains, viz., C. chanthaburiensis NBRC 102176T, Candida eppingiae JCM 17241T, C. haemuli SLLAear13-1, C. heveicola SLLAear14-10, C. khanbhai AFear10, CBS 16213T, CBS 16555, C. konsanensis NBRC 109082T, Candida linzhiensis AS 2.3073T, Candida melibiosica JCM 9558T, C. ruelliae CBS 10815T, Candida sp. XZY238F3, Danielozyma pruni NYUN 218101T and Metahyphopichia laotica CBS 13022T, was extracted using the method described by Wang & Bai (2008). Genomic libraries (150 bp paired-end) were constructed following the manufacturer’s protocols of TruSeq DNA Nano library prep kit (Illumina) and sequenced on an Illumina HiSeq 2000 platform using TruSeq SBS Kit (Illumina). The adapter sequence and low-quality reads were removed with default parameters using Fastp v. 0.20.1 ( Chen et al. 2018). SPAdes v. 3.15.0 ( Bankevich et al. 2012) was used to assemble the genomes of the above 14 yeast strains with the following parameters: “--memory 800 -k 21,33,55,77,99 --careful --cov-cutoff auto”. Quast v. 5.0.2 ( Gurevich et al. 2013) was assessed for genome quality. Gene prediction was done using GeneMark-ES ( Ter-Hovhannisyan et al. 2008).
Phylogenomic analysis and comparative genomics
Phylogenetic relationships of members of the CAH clade and related taxa in Metschnikowiaceae were evaluated by identifying single-copy homologs. BUSCO v. 5.3.2 ( Manni et al. 2021) was used to evaluate the integrity and obtain a single copy of the BUSCO sequence. Genomes with less than 60 % BUSCO completeness were eliminated and 155 genomes (154 strains in Metschnikowiaceae and 1 strain in Debaryomycetaceae as outgroup) were retained (Table 1). Alignment of single-copy BUSCO sequences was done using MAFFT v. 7.475 ( Katoh & Standley 2013) with L-INS-I model. The maximum Likelihood (ML) tree was constructed using IQ-TREE v. 2.1.2 ( Minh et al. 2020) with MFP as the model and 1 000 ultrafast bootstrap repeats (-m MFP -B 1000 -redo -mredo -nt AUTO). The phylogenomic tree and the final alignment are saved in the TreeBASE (www.treebase.org, No. 31145).
Table 1.
List of yeast strains and genomes used in this study.
| Species | Strain | Assembly | Complete BUSCOs | Complete and single-copy BUSCOs (S) | No. proteins | Total length | GC ( %) | Clade | Source |
|---|---|---|---|---|---|---|---|---|---|
| Candida berkhoutiae | CBS 11722T | GCA_030578995.1 | 93.20 % | 93.10 % | 5390 | 12617291 | 44.99 | C. blattae clade | NCBI |
| Candida blattae | NRRL Y-27698T | GCA_003706955.2 | 93.80 % | 93.70 % | 5661 | 12022277 | 49.77 | C. blattae clade | NCBI |
| Candida dosseyi | NRRL Y-27950T | GCA_030573325.1 | 93.60 % | 93.50 % | 5642 | 11984081 | 49.87 | C. blattae clade | NCBI |
| Candida ecuadorensis | CBS 12653T | GCA_030579155.1 | 90.40 % | 90.30 % | 5610 | 12617072 | 48.07 | C. blattae clade | NCBI |
| Candida ezoensis | CBS 11753T | GCA_030569115.1 | 92.70 % | 92.60 % | 5346 | 12574817 | 45.14 | C. blattae clade | NCBI |
| Candida flosculorum | NRRL Y-48731T | GCA_030568875.1 | 93.70 % | 93.60 % | 5422 | 12064463 | 48.2 | C. blattae clade | NCBI |
| Candida intermedia | CBS 572T | GCA_900106115.1 | 95.70 % | 95.60 % | 5931 | 13162108 | 43.53 | C. blattae clade | NCBI |
| Candida inulinophila | CBS 11725T | GCA_030562885.1 | 93.60 % | 93.50 % | 5405 | 13306216 | 46.04 | C. blattae clade | NCBI |
| Candida middelhoveniana | CBS 12306T | GCA_030557965.1 | 93.50 % | 93.30 % | 5495 | 12832034 | 44.96 | C. blattae clade | NCBI |
| Candida pseudoflosculorum | CBS 8584T | SRR16989025 | 93.30 % | 93.20 % | 5422 | 12171906 | 48.07 | C. blattae clade | NCBI |
| Candida pseudointermedia | NRRL Y-10939T | GCA_030557285.1 | 94.60 % | 94.50 % | 5658 | 13085940 | 43.47 | C. blattae clade | NCBI |
| Candida sharkensis | NRRL Y-48380T | GCA_030567015.1 | 94.10 % | 94.00 % | 5998 | 14008157 | 43.46 | C. blattae clade | NCBI |
| Candida suratensis | CBS 10928T | GCA_030566815.1 | 89.70 % | 89.60 % | 5803 | 13760921 | 47.91 | C. blattae clade | NCBI |
| Candida thailandica | CBS 10610T | GCA_022023595.1 | 90.70 % | 90.50 % | 5491 | 16310147 | 45.96 | C. blattae clade | NCBI |
| Candida tsuchiyae | NRRL Y-17840T | GCA_030566995.1 | 93.40 % | 93.30 % | 5288 | 12514726 | 46.31 | C. blattae clade | NCBI |
| Clavispora xylosa | NYNU 174173T | GCA_023158955.1 | 71.30 % | 71.20 % | 5045 | 15305418 | 50.87 | C. blattae clade | NCBI |
| Candida citri | CBS 11858T | GCA_030571295.1 | 92.70 % | 92.60 % | 5323 | 13045557 | 43.4 | C. citri clade | NCBI |
| Candida danieliae | CBS 8533T | GCA_030579135.1 | 90.80 % | 90.70 % | 5344 | 11795114 | 50 | C. danieliae clade | NCBI |
| Candida entomophila | NRRL Y-7783T | GCA_030555945.1 | 92.10 % | 91.70 % | 5261 | 10209960 | 54.43 | C. entomophila clade | NCBI |
| Candida sp. | CBS 14106 | SRR16974439 | 95.90 % | 95.60 % | 5658 | 11453389 | 51.23 | C. entomophila clade | NCBI |
| Candida eppingiae | JCM 17241T | NMDC60137102 | 85.50 % | 85.40 % | 5217 | 11274499 | 50.9 | C. eppingiae clade | this study |
| Candida kutaoensis | CBS 11388T | GCA_030562905.1 | 78.00 % | 77.80 % | 4631 | 9654364 | 55.76 | C. kutaoensis clade | NCBI |
| Candida baotianmanensis | CBS 11898T | GCA_030556145.1 | 85.00 % | 84.90 % | 5066 | 11060496 | 55.16 | C. melibiosica clade | NCBI |
| Candida melibiosica | JCM 9558T | NMDC60137103 | 85.40 % | 85.30 % | 5049 | 11018273 | 55.36 | C. melibiosica clade | this study |
| Candida rhizophorensis | NRRL Y-48382T | GCA_030573355.1 | 86.40 % | 86.30 % | 5165 | 11723898 | 51.63 | C. melibiosica clade | NCBI |
| Candida oregonensis | NRRL Y-5850T | GCA_003707785.2 | 95.30 % | 95.10 % | 5471 | 10887972 | 47.46 | C. oregonensis clade | NCBI |
| Clavispora reshetovae | CBS 11556T | GCA_030558395.1 | 92.80 % | 92.30 % | 6224 | 13479832 | 46.44 | C. oregonensis clade | NCBI |
| Candida bambusicola | CBS 11723T | GCA_030563705.1 | 91.40 % | 91.30 % | 5105 | 12107016 | 42.59 | C. succicola clade | NCBI |
| Candida nongkhaiensis | CBS 11724T | GCA_030563825.1 | 91.60 % | 91.40 % | 5266 | 12680896 | 39.96 | C. succicola clade | NCBI |
| Candida picinguabensis | NRRL Y-27814T | GCA_030582875.1 | 92.10 % | 92.00 % | 5146 | 11996333 | 44.14 | C. succicola clade | NCBI |
| Candida robnettiae | CBS 8580T | GCA_030568975.1 | 90.80 % | 90.70 % | 5040 | 12701715 | 40.48 | C. succicola clade | NCBI |
| Candida saopauloensis | NRRL Y-27815T | GCA_030582915.1 | 92.00 % | 91.90 % | 5099 | 12020785 | 44.26 | C. succicola clade | NCBI |
| Candida succicola | CBS 11726T | GCA_030563905.1 | 91.20 % | 91.10 % | 5077 | 12130308 | 43.04 | C. succicola clade | NCBI |
| Candida touchengensis | CBS 10585T | GCA_030566735.1 | 91.30 % | 91.30 % | 5147 | 12016383 | 45.07 | C. succicola clade | NCBI |
| Metschnikowia saccharicola | CBS 12575T | GCA_030569455.1 | 91.80 % | 91.70 % | 5151 | 12154026 | 40.63 | C. succicola clade | NCBI |
| Candida savonica | NRRL Y-17077T | GCA_030570115.1 | 95.20 % | 95.00 % | 5537 | 12643935 | 50.52 | C. tanticharoeniae clade | NCBI |
| Candida tanticharoeniae | CBS 11574T | GCA_030558325.1 | 93.90 % | 93.70 % | 5366 | 12225802 | 51.84 | C. tanticharoeniae clade | NCBI |
| Candida mogii | NRRL Y-17032T | GCA_030573315.1 | 87.20 % | 87.10 % | 4970 | 11074207 | 46.2 | C. tolerans clade | NCBI |
| Candida tolerans | NRRL Y-48705 | GCA_030582955.1 | 90.30 % | 90.20 % | 5291 | 12803081 | 42 | C. tolerans clade | NCBI |
| Candida aechmeae | NRRL Y-48456 | GCA_030583085.1 | 90.30 % | 90.20 % | 5287 | 11247228 | 49.06 | C. ubatubensis clade | NCBI |
| Candida ubatubensis | NRRL Y-27812 | GCA_030567085.1 | 88.60 % | 88.50 % | 5266 | 11259751 | 49.93 | C. ubatubensis clade | NCBI |
| Candida linzhiensis | AS 2.3073T | NMDC60137104 | 95.40 % | 95.00 % | 5881 | 14916200 | 30.02 | C. sequanensis clade | this study |
| Candida sequanensis | NRRL Y-17682T | GCA_030557975.1 | 94.50 % | 94.30 % | 5902 | 15045357 | 31.4 | C. sequanensis clade | NCBI |
| Candida sp. | XZY238F3 | NMDC60146131 | 94.70 % | 94.50 % | 5758 | 14038224 | 33.86 | C. sequanensis clade | this study |
| Candida auris | B11221 | GCA_002775015.1 | 94.40 % | 94.00 % | 5521 | 12741178 | 45.32 | CAH clade | NCBI |
| Candida chanthaburiensis | NBRC 102176T | NMDC60046445 | 90.20 % | 90.00 % | 5338 | 13025691 | 46.68 | CAH clade | this study |
| Candida duobushaemuli | B09383 | GCA_002926085.1 | 88.80 % | 88.50 % | 5173 | 12580400 | 46.84 | CAH clade | NCBI |
| Candida haemuli | SLLAear13-1 | NMDC60046450 | 91.60 % | 91.40 % | 5515 | 13278047 | 45.2 | CAH clade | this study |
| Candida haemuli var. haemuli | B11899 | GCA_002926055.1 | 90.40 % | 90.30 % | 5249 | 13314323 | 45.19 | CAH clade | NCBI |
| Candida haemuli var. vulneris | K1 | GCA_012184645.1 | 93.50 % | 93.40 % | 5502 | 13207566 | 45.21 | CAH clade | NCBI |
| Candida heveicola | AS 2.3483T | GCA_003708405.1 | 89.90 % | 89.50 % | 5274 | 13065558 | 47.2 | CAH clade | NCBI |
| Candida heveicola | SLLAear14-10 | NMDC60046449 | 90.60 % | 90.20 % | 5323 | 13075462 | 47.2 | CAH clade | this study |
| Candida khanbhai | AFear10 | NMDC60046448 | 90.30 % | 90.10 % | 5201 | 12119318 | 47.52 | CAH clade | this study |
| Candida khanbhai | CBS 16213T | NMDC60137105 | 89.80 % | 89.60 % | 5223 | 12474519 | 47.44 | CAH clade | this study |
| Candida khanbhai | CBS 16555 | NMDC60137106 | 90.10 % | 89.90 % | 5250 | 12332639 | 47.56 | CAH clade | this study |
| Candida konsanensis | NBRC 109082T | NMDC60046446 | 90.20 % | 89.90 % | 5344 | 13069910 | 46.71 | CAH clade | this study |
| Candida pseudohaemuli | UZ153 17 | GCA_002933435.1 | 90.10 % | 89.90 % | 5412 | 12690930 | 47.14 | CAH clade | NCBI |
| Candida ruelliae | CBS 10815T | NMDC60046447 | 88.40 % | 88.30 % | 5359 | 13541693 | 46.84 | CAH clade | this study |
| Candida vulturna | CBS 14366T | GCA_030585165.1 | 90.80 % | 90.60 % | 5423 | 12642490 | 46.97 | CAH clade | NCBI |
| Candida asparagi | NRRL Y-48714T | GCA_030573135.1 | 89.40 % | 89.30 % | 5047 | 11455834 | 48.68 | Clavispora s.str. clade | NCBI |
| Candida carvajalis | NRRL Y-48694T | GCA_030581635.1 | 89.50 % | 89.30 % | 4949 | 11240520 | 48.27 | Clavispora s.str. clade | NCBI |
| Candida phyllophila | CBS 12671T | SRR16989024 | 90.40 % | 90.00 % | 5263 | 12079776 | 48.02 | Clavispora s.str. clade | NCBI |
| Candida vitiphila | CBS 12672T | GCA_030557995.1 | 91.80 % | 91.60 % | 5038 | 11158879 | 48.2 | Clavispora s.str. clade | NCBI |
| Clavispora fructus | NRRL Y-17072T | GCA_003707795.1 | 88.00 % | 87.20 % | 4931 | 11424019 | 49.03 | Clavispora s.str. clade | NCBI |
| Clavispora lusitaniae | CBS 6936T | GCA_001673695.2 | 95.10 % | 95.00 % | 5537 | 11992787 | 44.53 | Clavispora s.str. clade | NCBI |
| Clavispora opuntiae | NRRL Y-11820T | GCA_030574075.1 | 92.40 % | 92.30 % | 5181 | 11556692 | 42.54 | Clavispora s.str. clade | NCBI |
| Clavispora paralusitaniae | NYNU 161120T | GCA_022058765.1 | 87.10 % | 86.70 % | 5259 | 12616965 | 44.51 | Clavispora s.str. clade | NCBI |
| Clavispora santaluciae | A1.5 | GCA_022577645.1 | 91.00 % | 90.50 % | 4978 | 11018616 | 49.7 | Clavispora s.str. clade | NCBI |
| Danielozyma ontarioensis | NRRL YB-1246T | GCA_003706395.1 | 95.40 % | 95.10 % | 5544 | 10694567 | 46.57 | Danielozyma | NCBI |
| Danielozyma pruni | NYUN 218101T | NMDC60146132 | 92.60 % | 92.50 % | 5403 | 11944991 | 58.53 | Danielozyma | this study |
| Debaryomyces hansenii | CBS 767T | GCA_000006445.2 | 98.70 % | 98.30 % | 6272 | 12152486 | 36.35 | Debaryomycetaceae | NCBI |
| Hyphopichia gotoi | NRRL Y-27225T | GCA_003708205.1 | 96.10 % | 95.70 % | 5823 | 13294060 | 40.54 | H. heimiiclade | NCBI |
| Hyphopichia heimii | NRRL Y-7502T | GCA_003706925.2 | 95.60 % | 95.40 % | 5863 | 12888973 | 40.37 | H. heimiiclade | NCBI |
| Hyphopichia pseudorhagii | NRRL YB-2076T | GCA_030449045.1 | 95.70 % | 95.50 % | 5759 | 12489910 | 40.75 | H. heimiiclade | NCBI |
| Hyphopichia rhagii | NRRL Y-2594T | GCA_003708185.2 | 95.60 % | 95.40 % | 5646 | 12379694 | 40.79 | H. heimiiclade | NCBI |
| Hyphopichia burtonii | NRRL Y-1933T | GCA_001661395.1 | 94.60 % | 94.20 % | 5996 | 12403110 | 34.99 | Hyphopichia s.str. clade | NCBI |
| Hyphopichia buzzinii | CBS 14300T | GCA_030556945.1 | 95.80 % | 95.40 % | 5829 | 12668805 | 41.65 | Hyphopichia s.str. clade | NCBI |
| Hyphopichia fennica | NRRL Y-7505T | GCA_030444945.1 | 96.10 % | 95.80 % | 5838 | 13952948 | 32.58 | Hyphopichia s.str. clade | NCBI |
| Hyphopichia homilentoma | JCM 1507T | GCA_001599095.1 | 94.40 % | 94.00 % | 5536 | 12176763 | 49.52 | Hyphopichia s.str. clade | NCBI |
| Hyphopichia khmerensis | CBS 9784T | GCA_030569195.1 | 95.50 % | 95.30 % | 5712 | 13329260 | 32.4 | Hyphopichia s.str. clade | NCBI |
| Hyphopichia pseudoburtonii | makgeolli | GCA_003856775.1 | 95.00 % | 94.60 % | 5831 | 15547333 | 36 | Hyphopichia s.str. clade | NCBI |
| Hyphopichia wangnamkhiaoensis | CBS 11695T | GCA_030578475.1 | 95.90 % | 95.50 % | 5829 | 14591231 | 37.39 | Hyphopichia s.str. clade | NCBI |
| Candida wancherniae | NRRL Y-48709T | GCA_003708715.2 | 85.90 % | 85.80 % | 4721 | 10260097 | 53.15 | M. agaves clade | NCBI |
| Metschnikowia agaves | UWOPS 92-207.1T | GCA_008065245.1 | 92.10 % | 92.00 % | 5172 | 11121255 | 44.54 | M. agaves clade | NCBI |
| Metschnikowia sp. | yHMJ9 | GCA_030444895.1 | 90.50 % | 90.30 % | 5327 | 11908917 | 51.39 | M. agaves clade | NCBI |
| Metschnikowia aberdeeniae | SUB 05-213.1 | GCA_002370615.1 | 91.20 % | 91.00 % | 5098 | 10651211 | 48.54 | M. arizonensis clade | NCBI |
| Metschnikowia amazonensis | UFMG-CM-6309T | GCA_008065195.1 | 87.90 % | 87.70 % | 6691 | 19103064 | 40.43 | M. arizonensis clade | NCBI |
| Metschnikowia arizonensis | UWOPS 99-103.4 | GCA_002370875.1 | 92.20 % | 92.10 % | 5716 | 16199712 | 41.67 | M. arizonensis clade | NCBI |
| Metschnikowia borealis | UWOPS 96-101.1 | GCA_002370855.1 | 91.10 % | 91.00 % | 6591 | 20526619 | 43.08 | M. arizonensis clade | NCBI |
| Metschnikowia bowlesiae | UWOPS 12-619.1 | GCA_002370295.1 | 89.30 % | 89.20 % | 5773 | 17200370 | 48.58 | M. arizonensis clade | NCBI |
| Metschnikowia cerradonensis | UFMG 03-T67.1T | GCA_002370635.1 | 92.70 % | 92.50 % | 6559 | 20691529 | 42.89 | M. arizonensis clade | NCBI |
| Metschnikowia colocasiae | UWOPS 03-202.1 | GCA_002370175.1 | 91.10 % | 90.90 % | 5546 | 14917106 | 47.01 | M. arizonensis clade | NCBI |
| Metschnikowia continentalis | UWOPS 95-402.1T | GCA_002370835.1 | 92.50 % | 92.30 % | 6934 | 22098529 | 42.3 | M. arizonensis clade | NCBI |
| Metschnikowia cubensis | MUCL 45753T | GCA_002374405.1 | 91.20 % | 91.10 % | 6383 | 20567312 | 43.53 | M. arizonensis clade | NCBI |
| Metschnikowia dekortorum | UWOPS 01-142b3T | GCA_002374455.1 | 89.80 % | 89.60 % | 5524 | 16339066 | 48.73 | M. arizonensis clade | NCBI |
| Metschnikowia drakensbergensis | EBD-CdVSA10-2A | GCA_002370475.1 | 91.80 % | 91.70 % | 5255 | 11864716 | 48.2 | M. arizonensis clade | NCBI |
| Metschnikowia hamakuensis | UWOPS 04-199.1 | GCA_002370815.1 | 90.30 % | 90.20 % | 6157 | 18736887 | 43.9 | M. arizonensis clade | NCBI |
| Metschnikowia hawaiiana | NRRL Y-27473T | GCA_003708615.1 | 91.40 % | 91.30 % | 5019 | 11848248 | 48.16 | M. arizonensis clade | NCBI |
| Metschnikowia hawaiiensis | UWOPS 87-2203.2 | GCA_002370325.1 | 89.70 % | 89.60 % | 6019 | 18360083 | 44.5 | M. arizonensis clade | NCBI |
| Metschnikowia hibisci | UWOPS 95-797.2T | GCA_002374725.1 | 91.80 % | 91.70 % | 5187 | 11402227 | 42.13 | M. arizonensis clade | NCBI |
| Metschnikowia ipomoeae | NRRL Y-27455T | GCA_030566495.1 | 92.20 % | 92.10 % | 6257 | 19124835 | 43.62 | M. arizonensis clade | NCBI |
| Metschnikowia ipomoeae | UWOPS 10-104.1 | GCA_002374715.1 | 92.60 % | 92.40 % | 6211 | 19000479 | 43.55 | M. arizonensis clade | NCBI |
| Metschnikowia kamakouana | UWOPS 04-112.5T | GCA_002374535.1 | 92.00 % | 91.90 % | 5713 | 15746623 | 44.81 | M. arizonensis clade | NCBI |
| Metschnikowia kipukae | UWOPS 00-669.2T | GCA_002370135.1 | 92.20 % | 92.00 % | 5181 | 11229551 | 45.13 | M. arizonensis clade | NCBI |
| Metschnikowia lochheadii | UWOPS 03-167a3 | GCA_002374545.1 | 92.40 % | 92.30 % | 6647 | 20911142 | 41.89 | M. arizonensis clade | NCBI |
| Metschnikowia matae | UFMG-CM-Y395T | GCA_002374375.1 | 92.30 % | 92.20 % | 6851 | 21250408 | 40.91 | M. arizonensis clade | NCBI |
| Metschnikowia mauinuiana | UWOPS 04-110.4 | GCA_002370755.1 | 91.20 % | 91.00 % | 5961 | 17251389 | 43.88 | M. arizonensis clade | NCBI |
| Metschnikowia orientalis | UWOPS05-269.1 | GCA_002893685.1 | 88.30 % | 88.20 % | 4975 | 12453693 | 49.39 | M. arizonensis clade | NCBI |
| Metschnikowia proteae | EBD-T1Y1T | GCA_002370515.1 | 91.50 % | 91.40 % | 5263 | 12400777 | 48.51 | M. arizonensis clade | NCBI |
| Metschnikowia santaceciliae | UWOPS 01-517a1T | GCA_002374485.1 | 92.20 % | 92.00 % | 6644 | 20559367 | 43.32 | M. arizonensis clade | NCBI |
| Metschnikowia similis | UWOPS 03-133.4 | GCA_002370765.1 | 88.60 % | 88.50 % | 5637 | 17255585 | 48.72 | M. arizonensis clade | NCBI |
| Metschnikowia shivogae | UWOPS 04-310.1T | GCA_002374645.1 | 91.30 % | 91.10 % | 5108 | 10759625 | 49.88 | M. arizonensis clade | NCBI |
| Candida golubevii | NRRL Y-48707T | GCA_003708755.1 | 91.40 % | 91.40 % | 5532 | 14776713 | 45.12 | M. bicuspidata clade | NCBI |
| Metschnikowia andauensis | CBS 10809T | GCA_030568715.1 | 66.50 % | 65.90 % | 8207 | 17931512 | 45.44 | M. bicuspidata clade | NCBI |
| Metschnikowia anglica | CBS 15342T | GCA_030573055.1 | 93.00 % | 92.90 % | 5379 | 13582273 | 47.05 | M. bicuspidata clade | NCBI |
| Metschnikowia australis | UFMG-CM-Y6158 | GCA_002073855.1 | 87.60 % | 87.50 % | 4828 | 14350488 | 47.21 | M. bicuspidata clade | NCBI |
| Metschnikowia baotianmanensis | CBS 15869 | GCA_030565705.1 | 88.70 % | 73.70 % | 7503 | 19862458 | 45.9 | M. bicuspidata clade | NCBI |
| Metschnikowia bicuspidata | NRRL YB-4993T | GCA_001664035.1 | 92.70 % | 92.30 % | 5838 | 16055203 | 47.85 | M. bicuspidata clade | NCBI |
| Metschnikowia chrysomelidarum | NRRL Y-27749T | GCA_030582795.1 | 89.90 % | 89.40 % | 6328 | 16371206 | 44.48 | M. bicuspidata clade | NCBI |
| Metschnikowia chrysoperlae | NRRL Y-27615T | GCA_030674525.1 | 81.50 % | 81.20 % | 7409 | 17268004 | 46.24 | M. bicuspidata clade | NCBI |
| Metschnikowia corniflorae | NRRL Y-27750T | GCA_030581935.1 | 84.20 % | 82.40 % | 7901 | 28064570 | 44.83 | M. bicuspidata clade | NCBI |
| Metschnikowia fructicola | NRRL Y-27328T | GCA_030556695.1 | 83.00 % | 66.50 % | 8438 | 20112834 | 45.77 | M. bicuspidata clade | NCBI |
| Metschnikowia gelsemii | NRRL Y-48212T | GCA_030561745.1 | 92.90 % | 92.70 % | 6021 | 18115406 | 43.39 | M. bicuspidata clade | NCBI |
| Metschnikowia gruessii | NRRL Y-17809T | GCA_030563445.1 | 90.20 % | 90.10 % | 7143 | 22233920 | 42.45 | M. bicuspidata clade | NCBI |
| Metschnikowia henanensis | CBS 12677T | GCA_030674755.1 | 81.40 % | 64.90 % | 8030 | 20688651 | 46.77 | M. bicuspidata clade | NCBI |
| Metschnikowia kofuensis | NRRL Y-27226T | GCA_030564885.1 | 87.70 % | 78.70 % | 7168 | 18128511 | 45.35 | M. bicuspidata clade | NCBI |
| Metschnikowia koreensis | CBS 8854T | GCA_030569435.1 | 92.30 % | 92.10 % | 6024 | 14348919 | 41.75 | M. bicuspidata clade | NCBI |
| Metschnikowia krissii | NRRL Y-5389T | GCA_030561945.1 | 89.30 % | 89.20 % | 4865 | 13288520 | 45.31 | M. bicuspidata clade | NCBI |
| Metschnikowia kunwiensis | NRRL Y-48698T | GCA_030583255.1 | 84.70 % | 84.50 % | 5080 | 14222198 | 48.91 | M. bicuspidata clade | NCBI |
| Metschnikowia lachancei | NRRL Y-27242T | GCA_030572615.1 | 90.40 % | 86.70 % | 8305 | 23926579 | 42.49 | M. bicuspidata clade | NCBI |
| Metschnikowia lunata | NRRL Y-7131T | GCA_030583235.1 | 92.00 % | 91.90 % | 5953 | 16680955 | 44.15 | M. bicuspidata clade | NCBI |
| Metschnikowia noctiluminum | NRRL Y-27753T | GCA_030578735.1 | 90.30 % | 88.80 % | 6171 | 16712092 | 44.81 | M. bicuspidata clade | NCBI |
| Metschnikowia peoriensis | CBS 15345T | GCA_030573015.1 | 91.10 % | 81.50 % | 7691 | 17708309 | 41.8 | M. bicuspidata clade | NCBI |
| Metschnikowia persimmonesis | KIOM_G15050 | GCA_014905795.1 | 78.00 % | 73.80 % | 6939 | 16473584 | 45.81 | M. bicuspidata clade | NCBI |
| Metschnikowia picachoensis | NRRL Y-27607T | GCA_030556465.1 | 91.00 % | 87.20 % | 7540 | 21509105 | 44.7 | M. bicuspidata clade | NCBI |
| Metschnikowia pimensis | NRRL Y-27619T | GCA_030556455.1 | 91.00 % | 90.60 % | 6503 | 18519651 | 44.73 | M. bicuspidata clade | NCBI |
| Metschnikowia pulcherrima | NRRL Y-7111T | GCA_030583425.1 | 90.60 % | 90.40 % | 6048 | 15504344 | 45.81 | M. bicuspidata clade | NCBI |
| Metschnikowia aff.pulcherrima | APC 1.2 | GCA_004217705.1 | 89.40 % | 89.20 % | 5800 | 15801215 | 45.88 | M. bicuspidata clade | NCBI |
| Metschnikowia reukaufii | MR1 | GCA_003401635.1 | 89.90 % | 89.50 % | 5978 | 15552339 | 41.85 | M. bicuspidata clade | NCBI |
| Metschnikowia rubicola | CBS 15344T | GCA_030557065.1 | 87.80 % | 83.10 % | 7583 | 18345891 | 45.69 | M. bicuspidata clade | NCBI |
| Metschnikowia shanxiensis | NRRL Y-48710T | GCA_030578695.1 | 86.40 % | 76.10 % | 7440 | 17950602 | 45.74 | M. bicuspidata clade | NCBI |
| Metschnikowia sinensis | NRRL Y-48711T | GCA_030583125.1 | 89.70 % | 89.30 % | 6121 | 15503712 | 45.76 | M. bicuspidata clade | NCBI |
| Metschnikowia vanudenii | NRRL Y-17036T | GCA_030583145.1 | 93.50 % | 93.20 % | 6550 | 20514424 | 42.71 | M. bicuspidata clade | NCBI |
| Metschnikowia viticola | NRRL Y-48693T | GCA_030556725.1 | 82.00 % | 81.70 % | 6090 | 16028048 | 45.27 | M. bicuspidata clade | NCBI |
| Metschnikowia zobellii | gsMetZobe1.1 | GCA_939531405.1 | 88.40 % | 88.20 % | 4913 | 13653384 | 47.69 | M. bicuspidata clade | NCBI |
| Candida hainanensis | NRRL Y-48715T | GCA_030561765.1 | 89.90 % | 89.90 % | 5012 | 10423758 | 50.33 | M. caudata clade | NCBI |
| Metschnikowia caudata | EBD-CdVSA08-1T | GCA_008065185.1 | 84.50 % | 84.40 % | 4576 | 10906389 | 55.99 | M. caudata clade | NCBI |
| Metschnikowia lopburiensis | CBS 12574T | GCA_030563105.1 | 90.70 % | 90.70 % | 4975 | 10450139 | 50.35 | M. caudata clade | NCBI |
| Metschnikowia drosophilae | UWOPS83-1135.3T | GCA_002893735.1 | 90.70 % | 90.60 % | 4905 | 10543888 | 52.8 | M. drosophilae clade | NCBI |
| Metschnikowia laotica | CBS 12961T | GCA_030563125.1 | 82.00 % | 81.90 % | 5449 | 10521976 | 49.27 | M. drosophilae clade | NCBI |
| Metschnikowia torresii | CBS 5152T | GCA_002893725.1 | 90.40 % | 90.30 % | 4994 | 10913326 | 51.24 | M. drosophilae clade | NCBI |
| Metahyphopichia laotica | CBS 13022T | NMDC60146133 | 95.30 % | 95.00 % | 5622 | 11063518 | 42.17 | Metahyphopichia | this study |
| Metahyphopichia silvanorum | NRRL Y-7782T | GCA_030574095.1 | 95.10 % | 94.90 % | 5700 | 11457544 | 38.51 | Metahyphopichia | NCBI |
| Metschnikowia sp. | yHQL527 | GCA_030578455.1 | 91.50 % | 91.10 % | 5516 | 11974810 | 35.23 | Metahyphopichia | NCBI |
| Metschnikowia sp. | yHKB443 | GCA_030444905.1 | 77.20 % | 77.00 % | 4528 | 11390856 | 53.78 | Metschnikowia sp. yHKB443 | NCBI |
CompareM v. 0.1.2 (https://github.com/dparks1134/CompareM) was used to assess the AAI values ( Liu et al. 2024) among Metschnikowiaceae with default parameters. The method for calculating the percentage of conserved protein (POCP) was done according to Qin et al. (2014). The proteomes of each combination of two strains were compared using Blastp ( Tatusova & Madden 1999). The conserved proteins were identified based on aligned length (50 %), identity (> 40 %) and e-value (< 1 × 10−5). POCP was calculated as the ratio of conserved proteins to the total number of two proteomes as published on GitHub (https://github.com/hoelzer/pocp ) and was used to verify the results.
Presence-absence patterns of orthologs (PAPO) were made according to the method described by Takashima et al. (2019). Ortho Finder v. 2.5.4 ( Emms & Kelly 2019) was used to identify the orthologous genes (OGs) that were indicated as 0 (zero) in case of ‘absence’ OGs and 1 (one) as ‘presence’ OGs. We identified the number of unique and shared proteins in the CAH clade, and the other 23 clades in the Metschnikowiaceae (Table 2), to clarify the boundaries between the clades. The core genome was considered as the conserved OGs within a clade and the pan genome was considered as the OGs found in at least one strain in a clade and the unique genome was the OGs found in all strains of a clade but not in any other clades. The software eggNOG-mapper v. 2.0 ( Cantalapiedra et al. 2021) was used to annotate the unique groups of orthologous genes (OGs) to obtain the function of genes in eggNOG, KEGG, Gene Ontology (GO) and Pfam domain. We selected OG with the same annotation function or OG with more than 30 % identity as the genus- specific OGs (Table 3). The identity of OG is calculated using EMBOSS water alignment tool ( Madeira et al. 2019).
Table 2.
List of the AAI, POCP and PAPO values of genera and clades in Metschnikowiaceae.
| Genera and clades | AAI | POCP | PAPO (unique genes) |
|---|---|---|---|
| C. blattae clade+Candida citri | 69.25–98.86 % | 83.47–99.07 % | 0 |
| C. blattae clade | 70.19–98.86 % | 83.47–99.07 % | 2 |
| C. entomophila clade | 67.93–67.93 % | 88.63–88.63 % | 4 |
| C. melibiosica clade | 72.05–97.51 % | 89.13–98.18 % | 15 |
| C. oregonensis clade | 72.8–72.8 % | 89.11–89.11 % | 4 |
| C. succicola clade | 70.79–96.43 % | 89.21–98.64 % | 11 |
| C. tanticharoeniae clade | 83.56–83.56 % | 96.84–96.84 % | 55 |
| C. tolerans clade | 71.98–71.98 % | 89.12–89.12 % | 22 |
| C. ubatubensis clade | 91.22–91.22 % | 97.48–97.48 % | 52 |
| CAH clade | 74.67–100.0 % | 90.87–99.65 % | 24 |
| CAH clade+C. tolerans clade | 64.14–100.0 % | 81.38–99.65 % | 3 |
| Clavispora | 65.0–94.98 % | 77.05–95.37 % | 0 |
| Clavispora s.str. clade | 67.26–94.98 % | 88.35–96.19 % | 6 |
| Hyphopichia | 63.8–95.63 % | 82.11–98.59 % | 0 |
| Hyphopichia s.str. clade | 65.53–81.68 % | 83.07–94.87 % | 4 |
| H. heimii clade+C. sequanensis clade | 65.74–95.63 % | 83.00–98.59 % | 0 |
| C. sequanensis clade | 71.95–81.98 % | 89.07–92.47 % | 13 |
| H. heimii clade | 80.42–95.63 % | 95.13–98.59 % | 41 |
| Metahyphopichia | 68.78–73.3 % | 90.16–92.21 % | 11 |
| Metschnikowia | 57.46–99.37 % | 55.16–98.91 % | 0 |
| M. arizonensis clade | 69.02–99.78 % | 71.01–98.29 % | 4 |
| M. arizonensis clade+M. caudata clade | 63.23–99.78 % | 63.38–98.75 % | 0 |
| M. caudata clade | 78.73–99.02 % | 92.82–98.75 % | 12 |
| M. drosophilae clade+Metschnikowia sp. yHKB443 | 62.86–91.0 % | 74.64–97.79 % | 0 |
| M. drosophilae clade | 84.44–91.0 % | 94.66–97.79 % | 43 |
| M. bicuspidata clade+M. agaves clade+Candida danieliae | 64.5–99.37 % | 71.33–98.91 % | 0 |
| M. bicuspidata clade+M. agaves clade | 64.5–99.37 % | 71.33–98.91 % | 1 |
| M. agaves clade | 73.43–74.58 % | 88.37–90.80 % | 5 |
| M. bicuspidata clade | 67.98–99.37 % | 72.26–98.91 % | 2 |
| Danielozyma | 74.43–74.43 % | 92.98–92.98 % | 16 |
Table 3.
List of the genus-specific OGs (unique genes) to use as diagnostic characters for the newly proposed genera.
| Clade | OGs to use in describing genus as diagnostic characters |
|---|---|
| C. sequanensis clade | OG0009095 |
| C. blattae clade | OG0005896 |
| C. entomophila clade | OG0010431; OG0010436 |
| C. eppingiae clade | OG0008853; OG0014363; OG0014397; OG0007521 |
| CAH clade | OG0005701; OG0005971; OG0005961 |
| C. kutaoensis clade | OG0010973; OG0011060; OG0011082; OG0011085; OG0011093 |
| Clavispora s.str. clade | OG0006565; OG0006567 |
| C. melibiosica clade | OG0007152 |
| C. oregonensis clade | OG0011188 |
| C. succicola clade | OG0006718; OG0006721 |
| C. tanticharoeniae clade | OG0011372; OG0011374; OG0011341 |
| C. tolerans clade | OG0009123 |
| C. ubatubensis clade | OG0006898 |
| H. heimii clade | OG0007683; OG0008295; OG0007296 |
RESULTS AND DISCUSSION
Genome assemblies and annotation
The newly sequenced genomes of C. chanthaburiensis NBRC 102176T, C. eppingiae JCM 17241T, C. haemuli SLLAear13-1, C. heveicola SLLAear14-10, C. khanbhai AFear10, CBS 16213T, CBS 16555, C. konsanensis NBRC 109082T, C. linzhiensis AS 2.3073T, C. melibiosica JCM 9558T and C. ruelliae CBS 10815T, Candida sp. XZY238F3, Danielozyma pruni NYUN 218101T and Metahyphopichia laotica CBS 13022T were assembled and ranged in size from 11.02 Mb to 14.92 Mb, and the number of predicted genes varied between 5 049 and 5 881. For detailed information on these genomes see Table 1.
Phylogenomic analysis
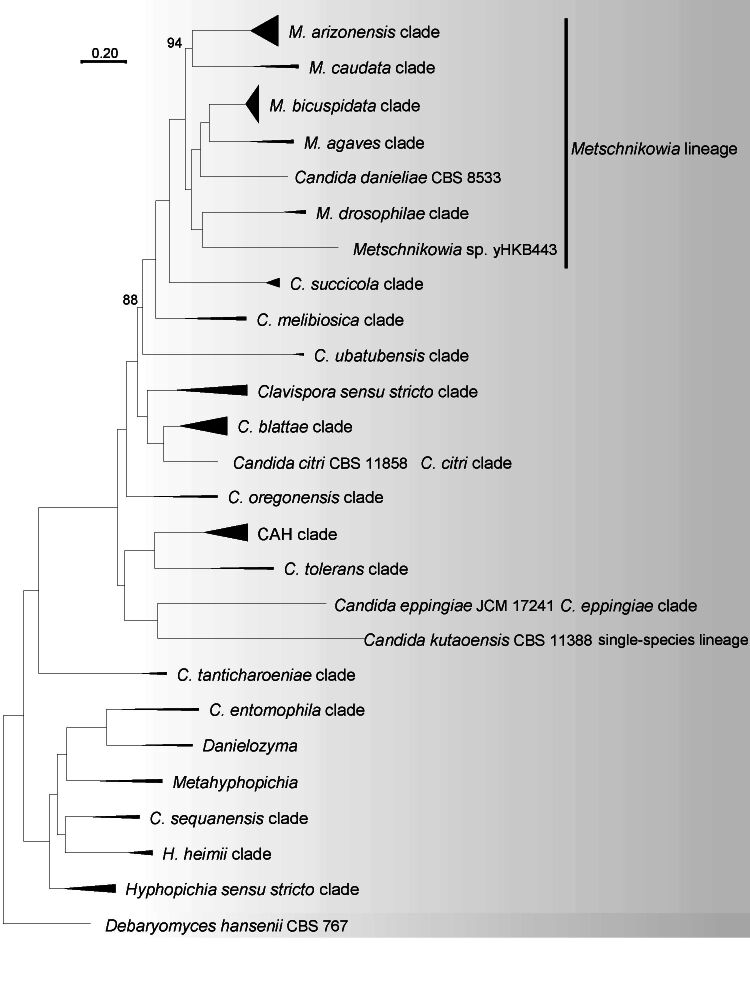
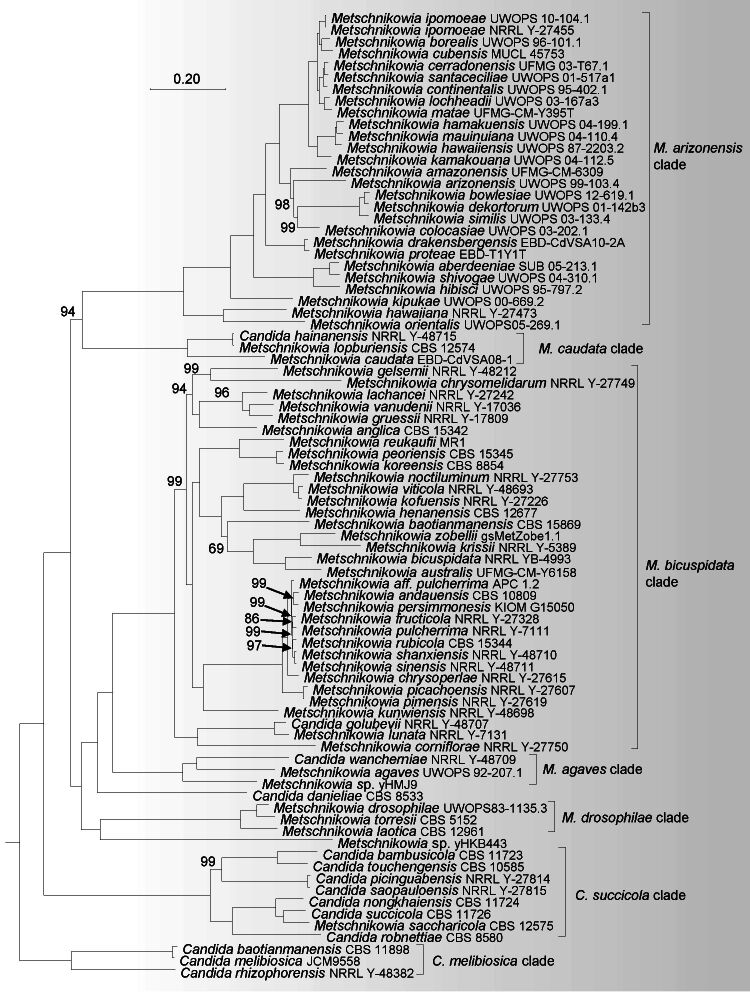
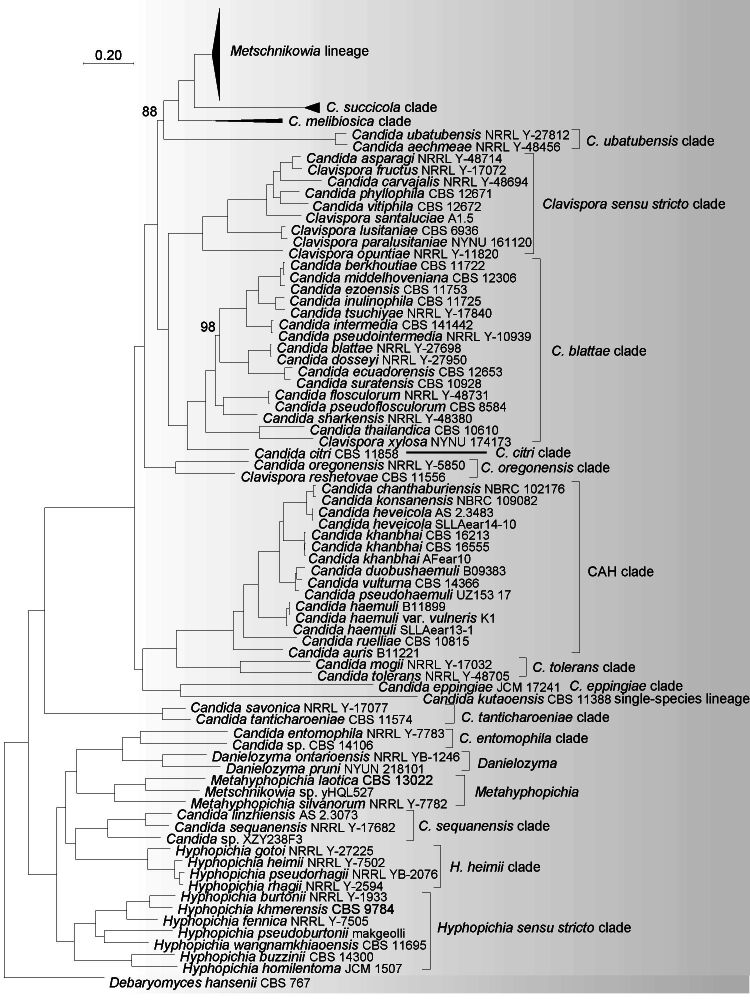
A total of 304 single-copy orthologue sequences were obtained from single-copy BUSCO proteins collected from 155 strains in the Metschnikowiaceae and Debaryomycetaceae (Table 1), which were used to construct the ML genome-based tree (Fig. 1). The BUSCO completeness of 155 genomes used in the analysis exceeded 60 %. Six Candida species and six Metschnikowia species, namely Candida akabanensis, C. bromeliacearum, C. metrosideri, C. ohialehuae, C. xinjiangensis, C. xylosifermentans, Metschnikowia colchici, M. maroccana, M. miensis, M. persici, M. rancensis and M. taurica were not included in the phylogenomic analysis as no genome data were available. Phylogenetic trees including those species for which a genome is lacking were constructed based on ITS, D1/D2 and ITS+D1/D2 rDNA sequences (Fig. S1–S3). In phylogenomic analyses, the bootstrap values obtained for the branches in the phylogenetic trees are expected to be high because phylogenomic-based analysis minimizes sampling error ( Salichos & Rokas 2013, Lachance 2022), hence lower bootstrap values indicate potential incongruences between gene loci in phylogenomic analyses. For this reason, only bootstraps < 100 % are shown on the nodes of the phylogenomic tree. The genera Danielozyma and Hyphopichia were assigned to the family Debaryomycetaceae ( Groenewald et al. 2023), but our phylogenomic analysis showed that these two genera formed a well-supported lineage with other lineages in Metschnikowiaceae (Fig. 1). Hence, we conclude that these two genera belong to Metschnikowiaceae, which is in agreement with the earlier results by Kurtzman (2011a), Shen et al. (2018) and Opulente et al. (2023).
Fig. 1.
Phylogenomic tree inferred using the 304 single copy orthologue proteins showed the phylogenetic relationship between the genera and clades in the Metschnikowiaceae. Bootstrap percentages of maximum likelihood analysis below 100 % from 1 000 bootstrap replicates are shown on the major branches. Bar = 0.2 substitutions per nucleotide position. a. An outline of the phylogeny of Metschnikowiaceae showing the phylogenetic relationship of the genera and clades; b. a subtree of Metschnikowia lineage, C. melibiosica clade and C. succicola clade; c. a tree of CAH clade, C. citri clade, C. blattae clade, C. entomophila clade, C. eppingiae clade, C. oregonensis clade, C. sequanensis clade, C. tanticharoeniae clade, C. tolerans clade, C. ubatubensis clade, Candida kutaoensis single-species lineage, Clavispora s.str. clade, Danielozyma, Metahyphopichia, Hyphopichia s.str. clade and H. heimii clade.
Our phylogenomic analysis showed that genera Clavispora, Hyphopichia, Metschnikowia and Candida in the Metschnikowiaceae are heterogenous and to some extent polyphyletic (Fig. 1a–c). They occurred in 21 clades or single species lineage, namely the C. blattae clade, the C. citri clade, the C. ento mophila clade, the C. eppingiae clade, the C. melibiosica clade, the C. oregonensis clade, the C. sequanensis clade, the C. succicola clade, the C. tanticharoeniae clade, the C. tolerans clade, the C. ubatubensis clade, the CAH clade, the Clavispora s.str. clade, the Hyphopichia s.str. clade, the H. heimii clade, the M. agaves clade, the M. arizonensis clade, the M. bicuspidata clade, the M. caudata clade, the M. drosophilae clade and the C. kutaoensis single-species lineage (Fig. 1a–c).
The genus Clavispora contains seven species at present ( Drumonde-Neves et al. 2020, Chai et al. 2022). Several Candida species were placed close to Clavispora species in previous analyses, but not transferred to the genus as it was preferred by those authors to await further phylogenetic analyses using housekeeping genes to circumscribe both genera reliably ( Daniel et al. 2014). Based on a phylogenetic analysis of concatenated ACT1, EF2, Mcm7 and RPB2 sequences, Guzmán et al. (2013) showed that the genus Clavispora is not monophyletic. The LSU rDNA analysis placed Clavispora reshetovae ( Yurkov et al. 2009) in a distinct lineage from the clade containing the nomenclatural type of the genus, Clavispora lusitaniae, and the species Clavispora opuntiae ( Guzmán et al. 2013). The phylogenomic analysis in this study supported the polyphyletic nature of Clavispora (Fig. 1a, c). Four Clavispora species, viz., Cl. fructus, Cl. lusitaniae, Cl. opuntiae and Cl. paralusitaniae, and four Candida species, viz., C. asparagi, C. carvajalis, C. phyllophila and C. vitiphila, formed a well-supported clade (Fig. 1c). The Clavispora s.str. clade clustered with the C. citri clade and the C. blattae clade consisting of 15 more Candida species, namely C. berkhoutiae, C. blattae, C. dosseyi, C. ecuadorensis, C. ezoensis, C. flosculorum, C. inulinophila, C. intermedia, C. middelhoveniana, C. pseudointermedia, C. pseudoflosculorum, C. sharkensis, C. suratensis, C. thailandica, C. tsuchiyae and Clavispora xylosa. Clavispora reshetovae and Candida oregonensis formed a well-supported clade, namely the C. oregonensis clade that is positioned in a separate branch in the phylogenomic tree, namely at position basal to Metschnikowia and Clavispora (Fig. 1a, c). It is important to note that the C. blattae clade included the recently described asexual species Cl. xylosa ( Chai et al. 2022). Based on the results of our phylogenomic analysis, it is possible either to merge the C. blattae clade with the genus Clavispora or to classify them as two different genera and transfer Cl. xylosa to a newly erected genus for the C. blattae clade. Members of the Clavispora s.str. clade contain CoQ8 as the major component of ubiquinone system ( Lachance 2011a, Lachance et al. 2011, Limtong & Kaewwichian 2013), whereas species in the C. blattae clade, i.e., C. berkhoutiae, C. ezoensis, C. intermedia, C. inulinophila, C. pseudointermedia, C. thailandica and C. tsuchiyae have CoQ9 ( Jindamorakot et al. 2007, Lachance et al. 2011, Nakase et al. 2011). In the past the CoQ composition has been used as an additional criterion to classify yeasts at the generic level ( Yamada & Kondo 1972, Yamada et al. 1976, Billon-Grand 1985, 1989, Suzuki & Nakase 1986), and this biochemical characteristic has also been used to support the circumscription of several genera of basidiomycetous yeasts ( Wang et al. 2015a, b, Takashima et al. 2019).
A few discrepancies between the phylogenomic and the combined ribosomal ITS and LSU rDNA-based tree (Fig. S1) have been observed. While the entire clade comprising Clavispora s.str. clade, the C. blattae clade, the C. citri clade and the C. oregonensis clade received good support in both analyses (100 % in the phylogenomic tree and 91 % in the rDNA analysis, respectively), the two analyses showed different phylogenetic relationships within this large clade (Fig. 1, S1). Specifically, C. citri and C. xylosifermentans occupied a basal position in the ITS+D1/D2 rDNA tree and other species were distributed between two moderately supported clades, namely the C. blattae clade (77 % support) and a second cluster of clades (76 % support) comprising the C. melibiosica clade, the C. oregonensis clade, the C. ubatubensis clade and the Clavi spora s.str. clade (Fig. S1). The position of the C. citri clade was remarkably different between our phylogenomic analysis (Fig. 1) and the ITS+D1/D2 rDNA tree (Fig. S1).
Based on the phylogenomic analysis alone, the discrepancy between the two phylogenetic analyses brought some uncertainty to the question whether it is reasonable to merge C. citri with the C. blattae clade. Therefore, we evaluated other genome-based statistics. No clade-specific OGs were found for the C. blattae clade + C. citri in the PAPO analysis described below, which suggests that it is better to separate C. citri from the C. blattae clade as it indicates that no genomic synapomorphic genes occur in the C. blattae clade + C. citri. However, considering the unavailability of a genome of C. xylosifermentans, the second known member of the clade, we presently keep the taxonomic position of the C. citri clade, and suggest to resolve the position of this clade in the future.
All Metschnikowia species that produce needle-shaped ascospores and four related Candida species, namely C. danieliae, C. golubevii, C. hainanensis and C. wancherniae, formed a well-supported lineage, namely the Metschnikowia lineage, in our phylogenomic analysis (Fig. 1a, b). The genus Metschnikowia includes two groups that produce large spores and small spores, respectively ( Lachance 2011b, Guzmán et al. 2013, Lachance et al. 2016, Lee et al. 2018). The large-spored group included the M. arizonensis clade and the M. caudata clade, while the small-spored group contained C. danieliae, the M. agaves clade, the M. bicuspidata clade, the M. drosophilae clade and Metschnikowia sp. yHKB443 (Fig. 1b). The M. caudata clade, the M. drosophilae clade and Metschnikowia sp. yHKB443 clustered together in the previous analysis of Opulente et al. (2023) that used a phylogenomic analysis indicated that the taxonomic relationships of those clades need to be addressed further by exploring more taxa. Therefore, the reclassification of the Metschnikowia lineage was not further considered in this study.
Kaewwichian et al. (2012) described Metschnikowia saccharicola, a species with no known sexual morph, based on a combined analysis of the ITS and D1/D2 rDNA sequences. Our rDNA sequence and phylogenomic analyses showed that this species is part of a highly supported (100 % support in both analyses) clade (Fig. 1b, S1), namely the C. succicola clade, with Candida bambusicola, C. nongkhaiensis, C. picinguabensis, C. robnettiae, C. saopauloensis, C. succicola, C. tocantinsensis and C. touchengensis. However, the clade was located outside the core Metschnikowia lineage, suggesting that the placement of M. saccharicola in the genus Metschnikowia needs to be reconsidered. Notably, all members of the C. succicola clade are asexually reproducing species as far as presently known. In our phylogenomic analysis, this clade occurred as the first branching lineage to the core Metschnikowia lineage (Fig. 1a, b).
Within the Metschnikowiaceae the Candida species occur dispersed. Besides the Candida species in the above-discussed C. blattae clade, the C. citri clade, the C. oregonensis clade, the C. succicola clade, the Clavispora s.str. clade and the Metschnikowia lineage, more than 20 Candida species occurred in eight well-supported clades and one single-species lineage (Fig. 1b, c). Among them, C. baotianmanensis, C.melibiosica and C. rhizophorensis formed a separate lineage, namely the C. melibiosica clade, which occurred as the first branching lineage in the Metschnikowia lineage and the C. succicola clade (Fig. 1b). Daniel et al. (2014) concluded that the C. melibiosica clade and the C. succicola clade formed two lineages that were separated by long distances from the core Metschnikowia clade and suggested that they might be considered as two new genera. Our data support this notion (Fig. 1, S1). Candida aechmeae and C. ubatubensis formed a small clade on a long branch that was placed outside the Metschnikowia lineage, the C. melibiosica clade and the C. succicola clade in the Metschnikowiaceae (Fig. 1). A search among public sequences revealed at least five undescribed potential new species in the C. ubatubensis clade represented by strains Candida sp. UFMGCMY6390, Candida sp. MCB1C2(5), Candida sp. UFMGF12, Candida aff. ubatubensis IMUFRJ 51945 and ‘Clavispora’ sp. UFMGCMY3120 (Fig. S2), which indicated that new species in the C. ubatubensis clade might be described in the future.
The phylogenomic analysis showed that the CAH clade received good support and was phylogenetically positioned remotely from the genera Clavispora, Danielozyma, Hyphopichia, Metschnikowia (Fig. 1a, c). As far as is known at present, members of the CAH clade reproduce asexually, unlike the phylogenetic closely related genera Clavispora and Metschnikowia that reproduce sexually with asci and ascospores. Two species, C. mogii and C. tolerans, formed a well-supported clade closely related to the CAH clade (Fig. 1c). Until now, the phylogenetic position of these species remained uncertain. Sugita & Nakase (1999) placed C. mogii in a basal position with affinities to the genus Clavispora based on the analysis of the small subunit (SSU) rDNA sequence, whereas Kurtzman & Robnett (1998) suggested a weak connection to C. haemuli (CAH clade) based on a phylogenetic analysis of sequences of the D1/D2 domains of LSU rDNA. Our ITS+D1/D2 rDNA sequence analyses showed that C. mogii and C. tolerans formed a well-supported (100 % support) clade distinct from the CAH clade together with sequences of four unpublished and potentially new species labelled as Candida cf. tolerans UWO(PS)99-704.2, Candida sp. 1A1, ‘Clavispora’ sp. 111180 and ‘Clavispora’ sp. 111221 (Fig. S1).
A previous phylogenetic analysis of D1/D2 LSU rDNA sequences indicated that C. savonica and C. tanticharoeniae were closely related to the genus Kodamaea in Debaryomycetaceae ( Nakase et al. 2010), but our phylogenomic analysis showed that they occupied a basal position in the Metschnikowiaceae with high statistical support (Fig. 1c). Phylogenetic analysis of combined ITS and D1/D2 sequence data showed that three other and potentially new species occurred in the C. tanticharoeniae clade (Fig S2, S3). Both C. eppingiae and C. kutaoensis formed a single-species lineage on a long branch adjacent to the CAH and C. tolerans clades (Fig. 1c), but this topology received low bootstrap support in analyses based on rDNA sequences ( Groenewald et al. 2011, Yuan et al. 2012; Fig. S1–S3). Although C. bromeliacearum was not included in our phylogenomic analysis due to the lack of genome data, this species and C. eppingiae clustered together with high (100 % BP) support in the combined ITS+D1/D2 rDNA-based tree constructed by Groenewald et al. (2011), suggesting that C. bromeliacearum and C. eppingiae belong together to this clade. An unpublished strain Candida sp. UWO(PS)00-137.1 clustered with the C. eppingiae clade in our D1/D2 LSU rDNA-based phylogenetic analysis (Fig. S2) and likely represents another member of the C. eppingiae clade. Opulente et al. (2023) previously observed that C. kutaoensis occurred on a long branch next to the C. ubatubensis clade. Based on the D1/D2 LSU rDNA sequence analysis (Fig. S2) Candida aff. kutaonensis UCDFST: 62–304 likely represents a different species than C. kutaoensis because its D1/D2 sequence differs from that of the type strain of C. kutaoensis by more than 4 % of the nucleotides.
Hyphopichia species were placed in two clades in this and a previous phylogenomic analysis ( Opulente et al. 2023; Fig. 1c). Specifically, the nomenclatural type H. burtonii, H. buzzinii, H. fennica, H. homilentoma, H. khmerensis, H. lachancei, H. pseudoburtonii and H. wangnamkhiaoensis formed the Hyphopichia s.str. clade (Fig. 1c, S1, S2). The second clade comprised H. heimii, H. gotoi, H. rhagii, H. paragotoi and H. pseudorhagii, and is called the H. heimii clade (Fig. 1c, S1, S2). Candida linzhiensis and C. sequanensis grouped together and formed the C. sequanensis clade with high support and positioned in a basal position to the H. heimii clade (Fig. 1c). Together the H. heimii and C. sequanensis clades built a larger well-supported clade with Danielozyma, the C. entomophila clade and Metahyphopichia (Fig. 1c), which agrees with the result of Opulente et al. (2023). A controversial position of Danielozyma ontarioensis was found in the phylogenetic analyses by Shen et al. (2018) and Li et al. (2021) based on genome data as the species occurred nested in the genus Hyphopichia. In our phylogenomic analysis, Danielozyma was found to be closely related to C. entomophila and Candida sp. CBS 14106 that clustered together with high support. Our analyses also revealed two potentially new species in this clade represented by strains labelled as Danielozyma aff. ontarioensis UCDFST:681027.2 and Danielozyma sp. DMKUSK8 (Fig. S1). The combined ITS+D1/D2 sequence analysis showed that Candida xinjiangensis described by Zhu et al. (2017) and C. entomophila formed a well-supported clade closely related to the genus Danielozyma but lacking statistical support (Fig. S1). The D1/D2 rDNA sequence analysis indicated that four strains, namely Candida sp. UFMG-CM-Y7109, Candida sp. UFMG-CM-Y6230, Candida sp. UFMG-CM-Y605 and Candida sp. GE17L14 may represent three new species in the C. entomophila clade (Fig. S2). Daniel et al. (2014) concluded that C. entomophila did not cluster confidently with any existing genera in the phylogenetic analysis of D1/D2 rDNA sequences. The data presented above and the results of our phylogenomic analysis suggested that the C. entomophila clade may represent a new genus comprising at least five species. Sipiczki & Tap (2016) described Metahyphopichia, a genus closely related to Danielozyma and Hyphopichia. Recently, Candida silvanorum was transferred to Metahyphopichia ( Khunnamwong et al. 2022). Our phylogenomic analysis showed that Metahyphopichia laotica, Metahyphopichia silvanorum and ‘Metschnikowia’ sp. yHQL527 clustered together closely related to the genus Danielozyma and the C. entomophila clade (Fig. 1c).
Delineation of genera based on genomic-based metrics
Although the taxonomic relationship of most clades in the Metschnikowiaceae was well resolved based on the above phylogenomic analysis, the results revealed several small clades that were found to be closely related to existing genera or clades occurring on rather long branches, i.e., i) the CAH clade and the C. tolerans clade; and ii) the H. heimii clade and the C. sequanensis clade. Recently, several genomic metrics have been explored to test phylogenetic hypotheses ( Takashima et al. 2019, Liu et al. 2024). Here we explored the use of these genomics-based statistical analyses, namely the AAI, POCP and PAPO approaches, which have been recently used to test the boundaries of generally well-accepted genera in the Saccharomycetaceae ( Liu et al. 2024), to address the taxonomic relationship between the various genera and clades in the Metschnikowiaceae in more detail.
In the AAI analysis, species in the genera Clavispora, Hyphopichia and Metschnikowia had 65.0–94.98 %, 63.8–95.63 % and 57.46–99.37 % AAI values, respectively (Table 2, S2), which all are lower than the observed values (about 70 %) for well-accepted genera in the Saccharomycetaceae ( Liu et al. 2024). The AAI values of the C. blattae clade, the C. entomophila clade, the C. melibiosica clade, the C. oregonensis clade, the C. sequanensis clade, the C. succicola clade, the C. tanticharoeniae clade, the C. tolerans clade, the C. ubatubensis clade, the CAH clade, the Clavispora s.str. clade, the Hyphopichia s.str. clade, the H. heimii clade, the M. agaves clade, the M. caudata clade, the M. arizonensis clade, the M. bicuspidata clade, the M. drosophilae clade, the Danielozyma clade and the Metahyphopichia clade were all within in the range suggested by Liu et al. (2024) (Table 2, S2).
Protein sequence similarity was analyzed among 150 yeast species belonging to the 23 clades or genera using the POCP method (Table 2, S3). The results showed that the POCP values in the C. blattae clade, the C. entomophila clade, the C. melibiosica clade, the C. oregonensis clade, the C. sequanensis clade, the C. succicola clade, the C. tanticharoeniae clade, the C. tolerans clade, the C. ubatubensis clade, the CAH clade, the Clavispora s.str. clade, the Hyphopichia s.str. clade, the H. heimii clade, the M. agaves clade, the M. caudata clade, the M. drosophilae clade, the genus Danielozyma and Metahyphopichia were > 80 % (Table 2). However, the M. arizonensis clade and the M. bicuspidata clade had POCP values of 71.01–98.29 % and 72.26–98.91 %, respectively, which are lower than values observed for well-accepted genera of Saccharomycetaceae with POCP > 80 % ( Liu et al. 2024). The range of the POCP values for all species presently classified in the genus Metschnikowia was larger, namely 55.16 % to 98.91 % (Table 2). The analysis indicated that the genus Metschnikowia is genetically far more heterogeneous than any of its closely related genera or clades, possibly due to the presence of lineages that are phylogenetically very different, i.e., those containing large-spored and small-spored species, respectively ( Guzmán et al. 2013, Lachance et al. 2016, Lee et al. 2018). Our data suggest that this genus needs to be reclassified in the future.
The PAPO analysis has been used by Takashima et al. (2019) and Liu et al. (2024) to delimit yeast genera in Trichosporonales and Saccharomycetaceae, respectively. With this method, the number of unique genes (also known as unique orthologs and genus-specific genes) present in clades and genera is examined. Here, we performed this analysis using 154 strains of species belonging to Metschnikowiaceae. We identified unique, core, and pan-genomics genes based on OrthoFinder OGs results (Table S3). More than two unique genes were found in the C. blattae clade, the C. entomophila clade, the C. melibiosica clade, the C. oregonensis clade, the C. sequanensis clade, the C. succicola clade, the C. tanticharoeniae clade, the C. tolerans clade, the C. ubatubensis clade, the CAH clade, the Clavispora s.str. clade, the Hyphopichia s.str. clade, the H. heimii clade, the M. agaves clade, the M. caudata clade, the M. arizonensis clade, the M. bicuspidata clade, the M. drosophilae clade, the Danielozyma clade and the Metahyphopichia clade. No unique genes were found in the genera Clavispora, Hyphopichia and Metschnikowia, indicating that these genera are genetically more diverse than any other such group, suggesting that these genera are in need of reclassification.
The above results indicate that the three genera Clavispora, Hyphopichia and Metschnikowia display a notable degree of genomic heterogeneity. Previous research showed that the relative evolutionary divergence (RED) in genera Clavispora (RED = 0.903), Hyphopichia (RED = 0.859) and Metschnikowia (RED = 0.914) is closer to that of family-level ranks in Fungi, median RED = 0.889 ( Li et al. 2021), which is an indication that these genera are underclassified. Further investigation of the AAI and POCP values of the genera Clavispora and Metschnikowia showed that these were lower than those obtained for any other closely related clade or genus (Table 2). The phylogenomic analysis indicated that Clavispora was polyphyletic, and Cl. reshetovae and Cl. xylosa belonged to the C. oregonensis clade and the C. blattae clade, respectively. The high heterogeneity of the genomic indices suggests that the genus Clavispora should be restricted to the Clavispora s.str. clade containing the type species Cl. lusitaniae. The situation with the genus Metschnikowia is less trivial. The genus is monophyletic, although it contains two large clades comprising species having large spores and small spores, respectively. The divergence of the genus at the level of rDNA sequences has been acknowledged before, as well as the similarity of growth responses and ecology ( Lachance 2011b). The phylo genomic analysis performed in our study resolved the two major clades but they seem to be still heterogeneous (Fig. 1b). The M. caudata clade was found to be separated from most other species of the large-spored clade. Similarly, the small-spored clade contained three well-supported clades, namely the M. bicuspidata clade, the M. agaves clade, and the M. drosophilae clade. When we analyzed these five clades within the genus Metschnikowia separately, the number of unique genes increased to two and more, the AAI values were close to 70 %, and the POCP values were higher than 80 %, except for the two large M. arizonensis and M. bicuspidata clades (Table 2), the cores of the large- and small-spored clades, respectively. These results showed that the genus Metschnikowia as it is presently recognized is characterized by significant genetic and phylogenetic divergence that may have various causes. For instance, the rates of sequence divergence may differ between older and newer phylogenetic lineages, as well as those undergoing hybridization and speciation (e.g., Shen et al. 2018). In particular, a strong effect is observed in lineages with short generation times, such as microorganisms including yeasts (e.g., Shen et al. 2018, Steenwyk & Rokas 2023). However, evolutionary rates among protein-coding genes of different yeast classes are rather universal, with only a minor shift toward higher rates in ‘younger’ gene classes ( Wolf et al. 2009). A recent RED analysis ( Groenewald et al. 2023) showed that the RED values obtained for families Metschnikowiaceae, Saccharomycodaceae (incl. Hanseniaspora) and Saccharomycetaceae are in the same range as those of other major fungal lineages. While relatively similar evolutionary divergence levels can be consistent across large lineages (e.g., taxonomic ranks of family and order), specific genetic features involved in genome recombination may further contribute to unique divergence patterns in single genera. Recently, it has been demonstrated that the ascomycetous yeast genus Hanseniaspora exhibits high molecular evolutionary rates and is characterized by extensive loss of cell-cycle and DNA repair genes ( Steenwyk et al. 2019). Among the two lineages in the genus, the fast-evolving one lost more genes associated with the cell cycle and genome integrity. The phenomenon may not be restricted to Hanseniaspora, but also be present in other lineages of ascomycetous yeasts. Whether or not other lineages characterized by a high genetic divergence, like the aforementioned Metschnikowia, underwent an accelerated evolution due to reduced repair mechanisms requires further investigation.
The CAH and C. tolerans clades, and the H. heimii and C. sequanensis clades formed two well-supported lineages, respectively (Fig. 1). However, no unique genes were observed for the H. heimii + C. sequanensis clade. The CAH clade + C. tolerans clade had three unique genes, but their respective AAI values were rather low, 64.14–100.0 % (Table 2). Considering phylogenetic distance, low similarity in genetic composition and overall sequence similarity, we conclude that H. heimii and C. sequanensis should preferably be accommodated in different genera. The phenotypic comparison (see Taxonomy section) revealed that the CAH clade and the C. tolerans clade can be distinguished by the assimilation of melezitose, which is positive for the CAH clade and negative for the C. tolerans clade.
Due to the lack of a sufficient number of genome sequences for species belonging to the C. eppingiae clade and C. kutaoensis single-species lineage, results of AAI, POCP and PAPO values were not available. However, the phylogenomic and the rDNA sequence analyses (Fig. 1, S1–S3) both suggest that the two clades represent two distinct lineages in the Metschnikowiaceae.
Morphological, biochemical, and physiological characteristics have traditionally served as primary criteria for circumscribing yeast genera. With a growing number of species, features associated with sexual reproduction and other diagnostic characteristics, including ascospore morphology and rare physiological properties do not apply universally to all species anymore (e.g., Giménez-Jurado et al. 2003, Garcia-Acero et al. 2024). While most species may still exhibit the major traits in common (plesiomorphies), this tendency potentially increases the heterogeneity within genera and complicates the demarcation of generic boundaries. The family Metschnikowiaceae contains three teleomorphic genera, Clavispora, Hyphopichia and Metschnikowia. The morphological characteristics of these yeasts, particularly those pertaining to sexual reproduction, exhibit greater diversity compared to a few distinctive physiological and biochemical characteristics, such as glucose fermentation, assimilation of nitrate and major respiratory ubiquinone system ( Kurtzman 2011b, Lachance 2011a, b). In the absence of sexual morphology, identifying physiological attributes for clades consisting solely of asexual species (such as the former Candida) poses a significant challenge, as larger clades tend to exhibit fewer shared traits in common. A few morphological and physiological features alone may not always suffice for the accurate circumscription of asexual genera (as seen in examples from basidiomycetous genera like Bullera, Cryptococcus, Dioszegia, Rhodotorula and Sporobolomyces). In such cases, additional methods such as molecular techniques may be necessary for the definitive classification of these yeasts. For the resolved in the phylogenomic analysis Candida clades of Metschnikowiaceae, molecular metrics remain the most reliable tool for identification. Maintaining Candida species outside the family Debaryomycetaceae is not sustainable due to the increasing heterogeneity of the genus and the growing uncertainty in identification at the genus level with the discovery of new species. Accommodating asexual species within sexual genera is feasible to a certain extent. However, clades that cannot be assigned to any previously described genus would necessitate the creation of a new name, like the previously described Danielozyma and Metahyphopichia (Kurtzman & Robnett 2014, Sipiczki et al. 2016).
Taking together the results of our phylogenomic analysis, the statistical evaluation of the variability of genomic metrics, and phenotypic characters, we propose 13 new genera in the Metschnikowiaceae to improve the taxonomy of these ascomycetous yeasts (see Taxonomy section below). The representative unique genes (genus-specific protein families, OGs) of each clade (Table 3) were used to diagnose the new genera in the taxonomy section. Detailed information on those OGs is given in Table S4.
Benefits of renaming yeast taxa
Changing the name of fungi may be confusing for the end-users in the applied field, be it clinical or industrial, and this certainly may be true in a short time frame. A concern was raised about the disconnect between newly introduced names and the practical needs of end-users (e.g., De Hoog et al. 2023). However, the long-term negative effects of newly introduced names were not supported by several recent surveys ( Chang et al. 2021, Chen et al. 2021, Kidd et al. 2022, 2023, Carroll et al. 2023). Appropriate and accurate name changes that reflect the proper evolutionary relationships among organisms may provide benefits for the broader user community ( Lücking et al. 2021). Such changes support the fundamental disciplines of taxonomy and nomenclature, ensuring the communication of accurate information to the end-users ( Carroll et al. 2023).
Fungal taxonomy, including that of yeasts, has experienced many name changes in the recent past. Application of new techniques and tools led to changing generic concepts by adapting broader or more narrow circumscriptions of genera like Saccharomyces, Kluyveromyces and Pichia to name a few ( Kurtzman 2003, Kurtzman et al. 2008). The concern about renaming, splitting, and lumping genera is understandable. However, the history of the genus Candida is different from that of many other yeast genera. The broad definition of the genus that is based on a few phenotypic characteristics, namely the absence of a sexual state, together with the past concept of ‘dual nomenclature’ ( Hawksworth et al. 2011, Lücking et al. 2021) has made this genus large and phylogenetically heterogeneous ( Daniel et al. 2014). The polyphyletic nature of the genus Candida has been recognized over the last decades ( Lachance et al. 2011, Daniel et al. 2014) and attempts to reclassify this genus have been made ( Takashima & Sugita 2022). The CAH clade is phylogenetically related to the genera Clavispora and Metschnikowia, and distantly positioned from the C. albicans clade (or Lodderomyces clade) representing the genus Candida in the strict sense as C. vulgaris, a current synonym under C. tropicalis and the type of the genus, belongs to the C. albicans/Lodderomyces clade. Because of the significant phylogenetic distance between the CAH clade and the C. albicans/Lodderomyces clade, the separation of the CAH clade from the genus Candida into its own genus is warranted. A wide range of knowledge suggests that apart from their morphological appearance on some culture media, members of the CAH and the C. albicans/Lodderomyces clade do not share common characteristics, including many physiological properties and resistance to antimycotics (see below). The proposal presented here is based on solid data and widely accepted taxonomic practice in mycology and zymology. Below we propose a new genus Candidozyma to accommodate the species in the CAH clade.
Species in a phylogenetically defined genus usually share genetic properties and evolutionary traits, including pheno typic ones. In other words, species within different genera have gained different genetic and phenotypic characteristics from distinct and not necessarily closely related recent ancestors during evolution. As such, genera can be seen as centers of speciation in which the species are genetically more closely related than species from other such centers. Thus, a generic name is used to communicate traits that are common for a species or strains in the genus, i.e., synapomorphies. This is very much true for the genus Candida which is most referred to as representing yeasts of clinical importance. However, among single-celled fungi, i.e., the yeasts, such phenotypic expressions may not always be clear. Here it may help to consider non-standard datasets, such as antifungal susceptibility data. For example, the Candida species that belong to different families, such as C. albicans (Debaryomycetaceae) and C. auris (Metschnikowiaceae) have different antifungal susceptibility patterns ( Schmalreck et al. 2014, Stavrou et al. 2019, Kidd et al. 2023). Therefore, separating distantly related species, like those of the CAH clade from those of the C. albicans/Lodderomyces clade, into a new genus confirms that these yeasts possess diverse properties and require distinct treatment options ( Schmalreck et al. 2014, Stavrou et al. 2019, Lücking et al. 2021). Furthermore, organisms bearing the same generic name are expected to share similar properties, including those useful for biotechnological and agricultural applications, fermentations, and biological safety concerns. In the field of fungal biotechnology, appropriate fungal name changes can help to predict and search for novel production organisms and their applications ( Kurtzman et al. 2015) and may assist researchers to identify and track the different species of yeast with their intrinsic properties, which can help in identifying novel production organisms that may have been previously overlooked or misidentified ( Houbraken et al. 2014). The case of renaming so-called Candida species that do not belong to the core C. albicans/Lodderomyces clade will also be beneficial for the application of such organisms in biotechnology, for example, due to a separation from pathogenic clades containing important opportunists. Presently, the name Candida is strongly associated with candidiasis, an infection caused by several human opportunistic yeasts. Renaming the bulk of species that presently are classified in the genus Candida may boost their biotechnological applications, facilitate general acceptance and ease the authorization process for use in production processes.
TAXONOMY
Validated taxa
Among species considered in the present study, three species names are presently indicated in the nomenclatural repositories Index Fungorum and MycoBank as invalid according to 40.7 of the ICNafp Shenzhen Code (Turland et al. 2018). The interpretation of the wording of the ICNafp applied to descriptions of yeast fungi has been a matter of recent debate. A group of yeast taxonomists argued that they disagree with some strict and literal interpretations of the ICNafp requirements and wordings, which made a few hundred names of yeast species (and some genera) invalid, despite that they have been documented, safely preserved and with accessible authentic type material in their descriptions ( Yurkov et al. 2021). By co-incidence, the names of several species revised in the present study are controversially declared invalid. In addition, it turned out that the orthography of the epithet ‘haemulonii’, for which also other orthographic variants were in use, e.g., ‘haemuloni’ and ‘haemulonis’, needs a correction. The epithet refers to the fish genus Haemulon, a word derived from the Greek neuter noun haema (= blood). Like other Greek-derived genera (e.g., Rhododendron, Agropyron) it is latinized as 2nd declension neuter noun with the correct genitive case ‘haemuli’. Epithets derived from this name will be corrected as C. haemuli, C. pseudohaemuli and C. pseuduobushaemuli.
Australozyma Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 852145
Etymology. The genus is named based on the yeasts in this lineage having been isolated from the southern Hemisphere.
Type species. Australozyma succicola (Nakase et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the C. succicola clade, which is in a separate lineage from the C. melibiosica clade and the Metschnikowia lineage (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2), and the presence of genus-specific protein families OG0006718 and OG0006721 (Table 3, S4).
Sexual reproduction not known. Colonies white to grayish white, butyrous, smooth. Multilateral budding cells present. Hyphae not produced, but pseudohyphae present or not. Growth in the presence of 50 % glucose (osmotolerance) and 15 % NaCl (halotolerance). The major ubiquinon coenzyme Q-9.
Notes — The genus Australozyma differs from its closely related genus Helenozyma (i.e., the C. melibiosica clade) by lack of assimilation of N-acetyl-D-glucosamine, whereas the genus Helenozyma can use this compound (Table S5).
Australozyma bambusicola (Nakase et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852146
Basionym. Candida bambusicola Nakase et al., J. Gen. Appl. Microbiol. 57: 234. 2011.
Australozyma nongkhaiensis (Nakase et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852147
Basionym. Candida nongkhaiensis Nakase et al., J. Gen. Appl. Microbiol. 57: 237. 2011.
Australozyma picinguabensis (Ruivo et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852148
Basionym. Candida picinguabensis Ruivo et al., Int. J. Syst. Evol. Microbiol. 56: 1149. 2006.
Australozyma robnettiae (M. Groenew. et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852149
Basionym. Candida robnettiae M. Groenew. et al., Int. J. Syst. Evol. Microbiol. 61: 2020. 2011.
Australozyma saccharicola Kaewwich. ex Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, sp. nov. — MycoBank MB 852211
Holotype. NBRC 108904 (preserved in a metabolically inactive state), National Institute of Technology and Evaluation (NITE), Kisarazu, Chiba, Japan.
Synonym. Metschnikowia saccharicola Kaewwich., Antonie van Leeuwenhoek 102: 746. 2012. Nom. inval., Art. 40.7 (Melbourne).
For a description see Antonie van Leeuwenhoek 102: 746. 2012.
Australozyma saopauloensis (Ruivo et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852160
Basionym. Candida saopauloensis Ruivo et al. (as ‘saopaulonensis’), Int. J. Syst. Evol. Microbiol. 56: 1150. 2006.
Australozyma succicola (Nakase et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852161
Basionym. Candida succicola Nakase et al., J. Gen. Appl. Microbiol. 57: 238. 2011.
Australozyma touchengensis (Nakase et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852162
Basionym. Candida touchengensis Nakase et al., J. Gen. Appl. Microbiol. 57: 240. 2011.
Candidozyma Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 848168.
Etymology. The genus is named for the asexual morphology like that found in the genus Candida.
Type species. Candidozyma auris (Satoh & Makimura) Q.M. Wang, Yurkov, Boekhout, & F.Y. Bai.
This genus is proposed to accommodate members of the CAH clade, which is closely related to the C. tolerans clade (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2) and the presence of genus-specific protein families OG0005701, OG0005971 and OG0005961 (Table 3, S4).
Sexual reproduction not known, but most mating and meiosis genes are conserved and MTLa and MTLα mating loci are present in different populations ( Muñoz et al. 2018). Therefore, mating might be expected, and a sexual state might be inducible under appropriate conditions. Colonies cream to yellowish cream, white, butyrous. Budding multilateral. Pseudohyphae and hyphae are usually not produced, but occur in special conditions, such as when growing aerobically.
Notes — The genus Candidozyma differs from its closely related genus Osmozyma (i.e., the C. tolerans clade) by assimilation of melezitose, whereas the genus Osmozyma does not assimilate this compound (Table S5). Most species in the Candidozyma are clinically important and are resistant to multiple antifungal drugs, which seems to be a unique feature compared to other genera in the Metschnikowiaceae.
Candidozyma auris (Satoh & Makimura) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848169
Basionym. Candida auris Satoh & Makimura, Microbiol. Immunol. 53: 43. 2009.
Synonym. Candida auris Satoh & Makimura ex F. Hagen, Med. Mycol. 61: myad009, 7. 2023. Nom. illegit., Art 53.
Candidozyma chanthaburiensis (Limtong et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848170
Basionym. Candida chanthaburiensis Limtong et al., Med. Mycol. 61: myad009, 7. 2023.
Synonym. Candida chanthaburiensis Limtong & Yongman., Antonie van Leeuwenhoek 98: 383. 2010. Nom. inval., Art. 40.7 (Shenzhen).
Candidozyma duobushaemuli (Cend.-Bueno et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848171
Basionym. Candida duobushaemuli Cend.-Bueno et al. (as ‘duobushaemulonii’), J. Clin. Microbiol. 50: 3646. 2012.
Candidozyma haemuli (Uden & Kolip.) Q.M. Wang, Yurkov, Boekhout, F.Y. Bai, comb. nov. — MycoBank MB 848173
Basionym. Torulopsis haemuli Uden & Kolip. (as ‘haemulonii’), Antonie van Leeuwenhoek 28: 78. 1962.
Synonym. Candida haemuli (Uden & Kolip.) S.A. Mey. & Yarrow (as ‘haemulonii’), Int. J. Syst. Bacteriol. 28: 612. 1978.
Candidozyma haemuli var. vulneris (Cend.-Bueno et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848174
Basionym. Candida haemuli var. vulneris Cend.-Bueno et al., J. Clin. Microbiol. 50: 3648. 2012.
Candidozyma heveicola (F.Y. Bai & S.A. Wang) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848175
Basionym. Candida heveicola F.Y. Bai & S.A. Wang, Antonie van Leeuwenhoek 94: 263. 2008.
Candidozyma khanbhai (A.W. de Jong & F. Hagen) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848176
Basionym. Candida khanbhai A.W. de Jong & F. Hagen, Med. Mycol. 61: myad009, 5. 2023.
Candidozyma konsanensis (Sarawan et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848177
Basionym. Candida konsanensis Sarawan et al., Med. Mycol. 61: myad009, 7. 2023.
Synonym. Candida konsanensis Sarawan et al., World J. Microbiol. Biotechnol. 29: 1483. 2013. Nom. inval., Arts 40.7, F.5.1 (Shenzhen).
Candidozyma metrosideri (J. Klaps et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848178
Basionym. Candida metrosideri J. Klaps et al., Med. Mycol. 61: myad009, 7. 2023.
Synonym. Candida metrosideri J. Klaps et al., PLoS ONE 15: e0240093, 11. 2020. Nom. inval., Art. 36.1(a) (Shenzhen).
Candidozyma ohialehuae (J. Klaps et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848179
Basionym. Candida ohialehuae Klaps et al., Med. Mycol. 61: myad009, 7. 2023.
Synonym. Candida ohialehuae J. Klaps et al., PLoS ONE 15: e0240093, 11. 2020. Nom. inval., Art. 36.1(a) (Shenzhen).
Candidozyma pseudohaemuli (Sugita, M. Takash. et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848180
Basionym. Candida pseudohaemuli Sugita, M. Takash. et al. (as ‘pseudohaemulonii’), Microbiol. Immunol. 50: 472. 2006.
Candidozyma ruelliae (Saluja & G.S. Prasad) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848181
Basionym. Candida ruelliae Saluja & G.S. Prasad, FEMS Yeast Res. 8: 664. 2008.
Candidozyma vulturna (Sipiczki et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848182
Basionym. Candida vulturna Sipiczki et al., Med. Mycol. 61: myad009, 7. 2023.
Synonym. Candida vulturna Sipiczki & Tap, Int. J. Syst. Evol. Microbiol. 66: 4014. 2016. Nom. inval., Arts 36.1(b), 40.7 (Shenzhen).
Clavispora Rodr. Mir., Antonie van Leeuwenhoek 45: 480. 1979, emend. Q.M. Wang, Yurkov, Boekhout & F.Y. Bai
Type species. Clavispora lusitaniae Rodr. Mir.
This genus is emended to accommodate the Clavispora s.str. clade including sexual and asexual members (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). Genus-specific protein families OG0006565, OG0006567 (Table 3).
Sexual reproduction is observed in some species. For sexual taxa, conjugation of haploid cells of opposite mating types usually precedes ascus formation. Bud-parent conjugation is also possible. Ascospores usually clavate, rarely ovoid to spherical. One or two (rarely three or four) ascospores per ascus ( Lachance 2011a). The spore wall may have small warts, which are visible by electron microscopy. Colonies white to cream, butyrous. Budding multilateral. Pseudohyphae may be formed but hyphae are not formed. The major ubiquinone is coenzyme Q-8.
Clavispora asparagi (F.Y. Bai & H.Z. Lu) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852163
Basionym. Candida asparagi F.Y. Bai & H.Z. Lu, Int. J. Syst. Evol. Microbiol. 54: 1413. 2004.
Clavispora carvajalis (S.A. James et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852164
Basionym. Candida carvajalis S.A. James et al., FEMS Yeast Res. 9: 786. 2009.
Clavispora phyllophila Limtong & Kaewwich. ex Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, sp. nov. — MycoBank MB 852212
Holotype. CBS 12671 (preserved in a metabolically inactive state), Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands.
Synonym. Candida phyllophila Limtong & Kaewwich., Curr. Microbiol. 59: 194. 2013. Nom. inval., Art. 40.7 (Melbourne).
For a description see Curr. Microbiol. 59: 194. 2013.
Clavispora vitiphila (Limtong & Kaewwich.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852165
Basionym. Candida vitiphila Limtong & Kaewwich., Curr. Microbiol. 59: 195. 2013.
Gaillardinia Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 852166
Etymology. The genus is named in honor of Claude Gaillardin for his contribution to yeast genomics.
Type species. Gaillardinia entomophila (D.B. Scott et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the C. entomophila clade, which is in a separate lineage closely related to Danielozyma and Metahyphopichia (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by the phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2), and the presence of genus-specific protein families OG0010431, OG0010436 (Table 3).
Sexual reproduction not known. Colonies white, butyrous, smooth. Multilateral budding cells and blastoconidia are present. Pseudohyphae and hyphae are present. Spherical to ellipsoidal asexual endospores are formed in hyphal strands. The major ubiquinone is coenzyme Q-8.
Notes — The genus Gaillardinia differs from Danielozyma and Metahyphopichia by assimilation of L-rhamnose, lactose and soluble starch (Table S5). All members of Gaillardinia do not use soluble starch, but the species of Danielozyma and Metahyphopichia assimilate this compound.
Gaillardinia entomophila (D.B. Scott et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852167
Basionym. Candida entomophila D.B. Scott et al., Antonie van Leeuwenhoek 37: 456. 1971.
Gaillardinia xinjiangensis (X.F. Zhu et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852168
Basionym. Candida xinjiangensis X.F. Zhu et al., Arch Microbiol. 199: 379. 2017.
Danielia Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB848188
Etymology. The genus is named in honor of Heide-Marie Daniel for her contribution to yeast taxonomy.
Type species. Danielia oregonensis (Phaff & Carmo Souza) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the C. oregonensis clade, which was resolved as a separate lineage in Metschnikowiaceae (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by the phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2), and the presence of genus-specific protein family OG0011188 (Table 3, S4).
Ascus formation may be preceded by conjugation between independent cells or between a parent cell and a bud. Asci with two ovoid ascospores with a small ring, and after maturation, ascospores are liberated from the ascus and tend to agglutinate ( Yurkov et al. 2009). Colonies white to cream, butyrous. Multilateral budding cells present. Hyphae not formed. Pseudo-hyphae present or not.
Danielia oregonensis (Phaff & Carmo Souza) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848189
Basionym. Candida oregonensis Phaff & Carmo Souza, Antonie van Leeuwenhoek 28: 206. 1962.
Danielia reshetovae (Yurkov et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852214
Basionym. Clavispora reshetovae Yurkov et al., Persoonia 23: 183. 2009.
Helenozyma Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 852177
Etymology. The genus is named in honor of Helen R. Buckley for her contribution to yeast taxonomy.
Type species. Helenozyma melibiosica (H.R. Buckley & Uden) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the C. melibiosica clade, which is in a separate lineage near C. succicola clade and Metschnikowia lineage (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2), and the presence of genus-specific protein family OG0007152 (Table 3, S4).
Asci present with one or two ellipsoidal to elongate ascospores or absent. Colonies white, butyrous, smooth. Multilateral budding cells present. Hyphae not produced, pseudohyphae present. The major ubiquinon coenzyme Q-9.
Notes — The genus Helenozyma differs from its closely related genus Australozyma (i.e., the C. succicola clade) by assimilation of N-acetyl-D-glucosamine. The former assimilates it, but the latter does not (Table S5).
Helenozyma baotianmanensis F.L. Hui & T. Ke ex Q.M. Wang, Yurkov, Boekhoutt & F.Y. Bai, sp. nov. — MycoBank MB 852234
Holotype. CGMCC 2.4378 (preserved in a metabolically inactive state), China General Microbiological Culture Collection Center, Beijing, China.
Synonym. Candida baotianmanensis F.L. Hui & T. Ke, J. Gen. Appl. Microbiol. 58: 61. 2012. Nom. inval., Art. 40.7 (Melbourne).
For a description see J. Gen. Appl. Microbiol. 58: 61. 2012.
Helenozyma melibiosica (H.R. Buckley & Uden) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852178
Basionym. Candida melibiosica H.R. Buckley & Uden, Mycopathol. Mycol. Appl. 36: 264. 1968.
Helenozyma rhizophorensis (Fell et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852179
Basionym. Candida rhizophorensis Fell et al. (as ‘rhizophoriensis’), Antonie van Leeuwenhoek 99: 545. 2011.
Hermanozyma Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB852180
Etymology. The genus is named in honor of Herman J. Phaff for his contribution to yeast taxonomy.
Type species. Hermanozyma ubatubensis (Ruivo et al., Pagnocca, Lachance & C.A. Rosa) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the C. ubatubensis clade, which is in a separate lineage affinity with C. melibiosica clade, C. succicola clade and Metschnikowia lineage (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by the phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2), and the presence of genus-specific protein families OG0006898 (Table 3, S4).
Sexual reproduction not known. Colonies white to cream, butyrous, smooth. Multilateral budding cells present. Hyphae and pseudohyphae present or not.
Notes — The genus Hermanozyma differs from its closely related genera Australozyma (i.e., the C. succicola clade) and Helenozyma (i.e., C. melibiosica clade) by assimilation of erythritol and L-rhamnose, whereas the latter two genera cannot use those carbon sources (Table S5).
Hermanozyma aechmeae (Landell & P. Valente) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852181
Basionym. Candida aechmeae Landell & P. Valente, Int. J. Syst. Evol. Microbiol. 60: 246. 2010.
Hermanozyma ubatubensis (Ruivo et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852182
Basionym. Candida ubatubensis Ruivo et al., Int. J. Syst. Evol. Microbiol. 55: 2216. 2005.
Isabelozyma Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 852183
Etymology. The genus is named in honor of Isabel Spencer-Martins for her contribution to yeast taxonomy.
Type species. Isabelozyma heimii (Pignal) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the H. heimii clade, which is in a separate lineage related to the C. sequanensis clade (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by the phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2), and the presence of genus-specific protein family OG0007683, OG0008295 and OG0007296 (Table 3, S4).
If sexual reproduction is present, asci are formed with one to four hat-shaped ascospores. Colonies white to tannish white, butyrous, smooth to somewhat convoluted. Multilateral budding cells and blastoconidia present. Hyphae not formed, pseudo-hyphae present.
Notes — The genus Isabelozyma differs from its relative Soucietia (i.e., the C. sequanensis clade) by positive assimilation of sucrose, whereas the genus Soucietia does not use sucrose (Table S5).
Isabelozyma gotoi (Nakase & M. Suzuki) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852184
Basionym. Candida gotoi Nakase & M. Suzuki, Microbiol. Culture Coll. 13: 110. 1997.
Synonym. Hyphopichia gotoi (Nakase & M. Suzuki) L.R. Ribeiro et al., Antonie van Leeuwenhoek 110: 992. 2017.
Isabelozyma heimii (Pignal) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852185
Basionym. Pichia heimii Pignal, Antonie van Leeuwenhoek 36: 525. 1970.
Synonym. Hyphopichia heimii (Pignal) Kurtzman, Antonie van Leeuwenhoek 88: 123. 2005.
Isabelozyma paragotoi F.L. Hui et al. ex Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, sp. nov. — MycoBank MB 852235
Holotype. CICC 33048 (preserved in a metabolically inactive state), China Centre of Industrial Culture Collection, Beijing, China.
Synonym. Hyphopichia paragotoi F.L. Hui et al., Int. J. Syst. Evol. Microbiol. 65: 2879. 2015. Nom. inval., Art. 40.7 (Melbourne).
For a description see Int. J. Syst. Evol. Microbiol. 65: 2879. 2015.
Isabelozyma pseudorhagii (Kurtzman) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852186
Basionym. Candida pseudorhagii Kurtzman, Antonie van Leeuwenhoek 88: 123. 2005.
Synonym. Hyphopichia pseudorhagii (Kurtzman) L.R. Ribeiro et al., Antonie van Leeuwenhoek 110: 992. 2017.
Isabelozyma rhagii (Diddens & Lodder) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB852188
Basionym. Candida tropicalis var. rhagii Diddens & Lodder, Die Hefasammlung des ‘Centraalbureau voor Schimmelcultures’: Beitrage zu einer Monographie der Hefearten. II. Teil. Die anaskosporogenen Hefen. Zweite Halfte: 488. 1942.
Synonym. Hyphopichia rhagii (Diddens & Lodder) L.R. Ribeiro et al., Antonie van Leeuwenhoek 110: 992. 2017.
Osmozyma Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 852189
Etymology. The genus is named because of the osmotolerant character of the species in this lineage.
Type species. Osmozyma mogii (Vidal-Leir.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the C. tolerans clade, which are in a separate lineage closely related to the CAH clade (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by the phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2), and the presence of genus-specific protein family OG0009123 (Table 3, S4).
Sexual reproduction not known. Colonies white, butyrous, smooth. Multilateral budding cells and blastoconidia are present. Hyphae not produced, pseudohyphae are present. Growth in the presence of 50 % glucose (osmotolerance) and 15 % NaCl (halotolerance). The major ubiquinone is coenzyme Q-9.
Notes — The related genus Candidozyma (the CAH clade) differs from Osmozyma by assimilation of melezitose, which is not utilized by the latter (Table S5).
Osmozyma mogii (Vidal-Leir.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852190
Basionym. Candida mogii Vidal-Leir., Antonie van Leeuwenhoek 33: 342. 1967.
Osmozyma tolerans Lachance et al. ex Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, sp. nov. — MycoBank MB 852236
Holotype. CBS 8613 (preserved in a metabolically inactive state), Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands.
Synonym. Candida tolerans Lachance et al., Canad. J. Microbiol. 45: 173. 1999. Nom. inval., Art. 40.3 (Melbourne).
For a description see Canad. J. Microbiol. 45: 173. 1999.
Soucietia Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 852191
Etymology. The genus is named in honor of Jean-Luc Souciet for his contribution to yeast genomics.
Type species. Soucietia sequanensis (Saëz & Rodr. Mir.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the C. sequanensis clade, which is in a separate lineage closely related to the H. heimii clade (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by the phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2), and the presence of genus-specific protein family OG0009095 (Table 3, S4).
Sexual reproduction not known. Colonies white, butyrous, smooth. Multilateral budding cells and blastoconidia are present. Hyphae and pseudohyphae are present.
Notes — The genus Soucietia differs from its closely related genus Isabelozyma (i.e., the H. heimii clade) by lack of assimilation of sucrose, but that can be used by the latter (Table S5).
Soucietia linzhiensis (F.Y. Bai & Z.W. Wu) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852192
Basionym. Candida linzhiensis F.Y. Bai & Z.W. Wu, Int. J. Syst. Evol. Microbiol. 56: 1155. 2006.
Soucietia sequanensis (Saëz & Rodr. Mir.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852193
Basionym. Candida sequanensis Saëz & Rodr. Mir., Antonie van Leeuwenhoek 50: 379. 1984.
Sungouiella Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 848198
Etymology. The genus is named in honor of Sung-Oui Suh for his contribution to yeast taxonomy.
Type species. Sungouiella intermedia (Cif. & Ashford) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the C. blattae clade, which is in a separate lineage closely related to Clavispora s.str. clade (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by the phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2), and the presence of genus-specific protein family OG0005896 (Table 3).
Sexual reproduction not known. Colonies white, cream, yellowish gray, butyrous. Multilateral budding cells present. Hyphae not formed, pseudohyphae present. The major ubiquinone coenzyme Q-9.
Notes — The genus Sungouiella has CoQ 9 as major ubiquinone, which differs from the presence of CoQ 8 formed by its relative Clavispora s.str. (Table S5).
Sungouiella akabanensis (Nakase et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848199
Basionym. Candida akabanensis Nakase et al., Microbiol. Cult. Collect. 10: 36. 1994.
Sungouiella berkhoutiae (Nakase et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852425
Basionym. Candida berkhoutiae Nakase et al., J. Gen. Appl. Microbiol. 57: 76. 2011.
Sungouiella blattae (N.H. Nguyen et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848201
Basionym. Candida blattae N.H. Nguyen et al., Mycologia 99: 853. 2008.
Sungouiella dosseyi (N.H. Nguyen et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852196
Basionym. Candida dosseyi N.H. Nguyen et al., Mycologia 99: 853. 2008.
Sungouiella ecuadorensis (S.A. James) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852197
Basionym. Candida ecuadorensis S.A. James (as ‘ecuadoriensis’), Int. J. Syst. Evol. Microbiol. 63: 396. 2013.
Sungouiella ezoensis (Nakase et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852198
Basionym. Candida ezoensis Nakase et al., J. Gen. Appl. Microbiol. 57: 78. 2011.
Sungouiella flosculorum (C.A. Rosa et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852199
Basionym. Candida flosculorum C.A. Rosa et al., Int. J. Syst. Evol. Microbiol. 57: 2972. 2007.
Sungouiella intermedia (Cif. & Ashford) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848202
Basionym. Blastodendrion intermedium Cif. & Ashford (as ‘intermedius’), Porto Rico J. Publ. Health Trop. Med. 5: 103. 1929.
Synonym. Candida intermedia (Cif. & Ashford) Langeron & Guerra, Ann. Parasitol. Humaine Comp. 16: 461. 1938.
Sungouiella inulinophila (Nakase et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852200
Basionym. Candida inulinophila Nakase et al., J. Gen. Appl. Microbiol. 57: 79. 2011.
Sungouiella middelhoveniana (J.R.A. Ribeiro et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852201
Basionym. Candida middelhoveniana J.R.A. Ribeiro et al., Antonie van Leeuwenhoek 100: 343. 2011.
Sungouiella pseudoflosculorum (M. Groenew. et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852202
Basionym. Candida pseudoflosculorum M. Groenew. et al., Int. J. Syst. Evol. Microbiol. 61: 2020. 2011.
Sungouiella pseudointermedia (Nakase et al.) Q.M. Wang, Yurkov, Boekhout, F.Y. Bai, comb. nov. — MycoBank MB 848203
Basionym. Candida pseudointermedia Nakase et al., J. Gen. Appl. Microbiol. 22: 178. 1976.
Sungouiella sharkensis (Fell et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852203
Basionym. Candida sharkensis Fell et al. (as ‘sharkiensis’), Antonie van Leeuwenhoek 99: 542. 2011.
Sungouiella suratensis Limtong & Yongman. ex Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, sp. nov. — MycoBank MB 852237
Holotype. CBS 10928 (preserved in a metabolically inactive state), Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands.
Synonym. Candida suratensis Limtong & Yongman., Antonie van Leeuwenhoek 98: 386. 2010. Nom. inval., Art. 40.7 (Melbourne).
For a description see Antonie van Leeuwenhoek 98: 386. 2010.
Sungouiella thailandica Jindam. et al. ex Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, sp. nov. — MycoBank MB 848219
Holotype. NBRC 102562 (preserved in a metabolically inactive state), National Institute of Technology and Evaluation (NITE), Kisarazu, Chiba, Japan.
Synonym. Candida thailandica Jindam. et al., FEMS Yeast Res. 7: 1411. 2007. Nom. inval., Art. 40.7 (Shenzhen).
For a description see Jindamorak et al., FEMS Yeast Res. 7: 1411. 2007.
Sungouiella tsuchiyae (Nakase & M. Suzuki) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852204
Basionym. Candida tsuchiyae Nakase & M. Suzuki, J. Gen. Appl. Microbiol. 31: 508. 1985.
Sungouiella xylosa (F.L. Hui & C.Y. Chai) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 848204
Basionym. Clavispora xylosa F.L. Hui & C.Y. Chai, Frontiers Microbiol. 13(no. 1019599): 7. 2022.
Tanozyma Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 852206
Etymology. The genus is named in honor of Chen Shuhui Tan for her contributions to The Yeasts, A taxonomic Study, and her contributions to the cryopreservation of microbes.
Type species. Tanozyma kutaoensis S.A. Wang & F.L. Li ex Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the C. kutaoensis single-species lineage, which is in a separate lineage positioned near the C. eppingiae clade (Fig. 1a, c). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by the phylogenomic analysis and the presence of genus-specific protein families OG0010973, OG0011060, OG0011082, OG0011085, OG0011093 (Table 3, S4).
Sexual reproduction not known. Colonies white, butyrous, smooth. Multilateral budding cells present. Hyphae not produced, pseudohyphae are present.
Tanozyma kutaoensis S.A. Wang & F.L. Li ex Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, sp. nov. — MycoBank MB 852216
Holotype. AS 2.4027 (preserved in a metabolically inactive state), China General Microbiological Culture Collection Center, Beijing, China.
Synonym. Candida kutaoensis S.A. Wang & F.L. Li (as ‘kutaonensis’), Appl. Microbiol. Biotechn. 96: 1522. 2012. Nom. inval., Art. 40.7 (Melbourne).
For a description see Appl. Microbiol. Biotechn. 96: 1522. 2012.
Gabaldonia Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 852207
Etymology. The genus is named in honor of Toni Gabaldón for his contribution to yeast genomics and biology, especially of hybrids.
Type species. Gabaldonia eppingiae (M. Groenew. et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the C. eppingiae clade, which is in a separate branch closely to the C. kutaoensis single-species lineage (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by the phylogenomic analysis and the presence of genus-specific protein families OG0008853, OG0014363, OG0014397 and OG0007521 (Table 3, S4).
Sexual reproduction not known. Colonies white, butyrous, smooth. Multilateral budding cells and blastoconidia present. Hyphae not present, pseudohyphae present.
Gabaldonia eppingiae (M. Groenew. et al.) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB852208
Basionym. Candida eppingiae M. Groenew. et al., Int. J. Syst. Evol. Microbiol. 61: 2021. 2011.
Wilhelminamyces Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, gen. nov. — MycoBank MB 852209
Etymology. The genus is named in honor of Wilhelmina Ch. Slooff for her contribution to yeast taxonomy.
Type species. Wilhelminamyces savonicus (Sonck) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai.
This genus is proposed for the species in the C. tanticharoeniae clade, which is in a separate lineage near the C. eppingiae clade and the C. kutaoensis single-species lineage (Fig. 1a, b). Member of the Metschnikowiaceae (Serinales, Pichiomycetes). The genus is mainly circumscribed by the phylogenomic analysis, the genome-based metrics AAI, POCP and PAPO (Table 2), and the presence of genus-specific protein families OG0011372, OG0011374 and OG0011341 (Table 3, S4).
Sexual reproduction not known. Colonies white to brownish grey, butyrous, smooth. Multilateral budding cells and blastoconidia present. Hyphae not present, pseudohyphae present. The major ubiquinone coenzyme Q-9.
Wilhelminamyces savonicus (Sonck) Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, comb. nov. — MycoBank MB 852210
Basionym. Candida savonica Sonck, Antonie van Leeuwenhoek 40: 543. 1974.
Wilhelminamyces tanticharoeniae Nakase et al. ex Q.M. Wang, Yurkov, Boekhout & F.Y. Bai, sp. nov. — MycoBank MB 852215
Holotype. BCC 11806 (preserved in a metabolically inactive state), BIOTEC Culture Collection, Pathumthani, Thailand.
Synonym. Candida tanticharoeniae Nakase et al., J. Gen. Appl. Microbiol. 56: 90. 2010. Nom. inval., Art. 40.7 (Melbourne).
For a description see J. Gen. Appl. Microbiol. 56: 90. 2010.
Contributions
Q.-M.W. conceived and designed the project. W.-N.Z. and Z.-X.F. and F.-L.H. performed yeast isolation and phenotypic comparison. F.L., Z.-D.H. and X.-M.Z. performed genomic metrics analysis. K.B. worked on nomenclatural matters. Q.-M.W., A.Y., T.B., S.A. and F.-Y.B. wrote the paper. Q.-M.W., A.Y. and T.B. revised the paper.
Acknowledgments
We thank Dr. Marizeth Groenewald, the yeast curator of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands for supplying us with the strains to include in the phylogenomic analyses of the revised version and we thank Dr. Masako Takashima for her help in checking the original page of the description of Candida akabanensis and Candida pseudoglaebosa. We gratefully acknowledge the help of Shaun Pennycook and Uwe Braun with checking the correct orthography of the epithet ‘haemuli ’. This study was supported by grants No. 31770018, No. 31961133020 and No. 32370015 from the National Natural Science Foundation of China (NSFC), No. 2021FY100900 from the Ministry of Science and Technology of China, No. 521000981388 from Advanced Talents Incubation Program of the Hebei University. The authors are solely responsible for the content of this work. TB and SA thank the Distinguished Scientists Fellowship Program (DSFP) of King Saud University for financial support.
Declaration on conflict of interest
The authors declare that there is no conflict of interest.
Supplementary material
Fig. S1–S3, Table S1–S5 and all OGs used in the description of genera genus as diagnostic characters are deposited in the Figshare repository: https://doi.org/10.6084/m9.figshare.25132418. The genome sequences of 14 strain have been deposited in National Microbiology Data Center (NMDC) with project number NMDC10018537, NMDC10018368 and NMDC10018700 (https://nmdc.cn/resource/en/genomics/project/detail/NMDC10018537, https://nmdc.cn/resource/en/genomics/project/detail/NMDC10018368, https://nmdc.cn/resource/en/genomics/project/detail/NMDC10018700).
List of yeast species and Genbank numbers used in the ITS and D1/D2 analyses.
The matrix of AAI and POCP values in the Metschnikowiaceae. The upper right corner represents the AAI values and the lower left corner represents the POCP values.
The PAPO analysis of the clades in the Metschnikowiaceae.
The annotation results of clade-specific OGs of the 17 clades/genera or single species lineages in the Metschnikowiaceae.
The phenotypic characteristics of different genera and clades in Metschnikowiaceae.
Phylogenetic tree inferred using the ITS and D1/D2 domain of 26S rDNA gene showed the positions of the genera and clades in the Metschnikowiaceae. Bootstrap percentages of maximum likelihood analysis over 50 % from 1000 bootstrap replicates are shown on the major branches. Bar = 0.05 substitutions per nucleotide position.
Phylogenetic tree inferred using the D1/D2 domain of 26S rDNA gene showed the positions of the genera and clades in the Metschnikowiaceae. Bootstrap percentages of maximum likelihood analysis over 50 % from 1000 bootstrap replicates are shown on the major branches. Bar = 0.05 substitutions per nucleotide position.
Phylogenetic tree inferred using the ITS gene showed the positions of the genera and clades in the Metschnikowiaceae. Bootstrap percentages of maximum likelihood analysis over 50 % from 1000 bootstrap replicates are shown on the major branches. Bar = 0.05 substitutions per nucleotide position.
REFERENCES
- Arora P, Singh P, Wang Y, et al. 2021. Environmental isolation of Candida auris from the Coastal Wetlands of Andaman Islands, India. mBio 12: e03181–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, et al. 2012. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco RA, Garrity GM, Scott JJ, et al. 2020. A genus definition for bacteria and archaea based on a standard genome relatedness index. mBio 11: e02475–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon-Grand G. 1985. Coenzyme Q de quelques espèces du genre Pichia: Détermination qualitative et quantitative. Mycopathologia 90: 101–106. [DOI] [PubMed] [Google Scholar]
- Billon-Grand G. 1989. A new ascosporogenous yeast genus: Yamadazyma gen. nov. Mycotaxon 35: 201–204. [Google Scholar]
- Cantalapiedra CP, Hernandez-Plaza A, Letunic I, et al. 2021. eggNOGmapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Molecular Biology and Evolution 38: 5825–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KC, Munson E, Butler-Wu SM, et al. 2023. Point-Counterpoint: What’s in a name? Clinical Microbiology Laboratories should use nomenclature based on current taxonomy. Journal of Clinical Microbiology 61: e0173222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, et al. 2012. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: Three multiresistant human pathogenic yeasts. Journal of Clinical Microbiology 50: 3641–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai CY, Li Y, Yan ZL, et al. 2022. Phylogenetic and genomic analyses of two new species of Clavispora (Metschnikowiaceae, Saccharomycetales) from Central China. Frontiers in Microbiology 13: 1019599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Blyth CC, Chen SC, et al. 2021. Introduction to the updated Australasian consensus guidelines for the management of invasive fungal disease and use of antifungal agents in the haematology/oncology setting. Internal Medicine Journal 51: 3–17. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, et al. 2018. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, Perfect J, Colombo AL, et al. 2021. Global guideline for the diagnosis and management of rare yeast infections: An initiative of the ECMM in cooperation with ISHAM and ASM. The Lancet Infectious Diseases 21: e375–e386. [DOI] [PubMed] [Google Scholar]
- Clancy CJ, Nguyen MH. 2017. Emergence of Candida auris: An international call to arms. Clinical Infectious Diseases 64: 141–143. [DOI] [PubMed] [Google Scholar]
- Daniel HM, Lachance MA, Kurtzman CP. 2014. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie van Leeuwenhoek 106: 67–84. [DOI] [PubMed] [Google Scholar]
- De Hoog S, Walsh TJ, Ahmed SA, et al. 2023. A conceptual framework for nomenclatural stability and validity of medically important fungi: a proposed global consensus guideline for fungal name changes supported by ABP, ASM, CLSI, ECMM, ESCMID-EFISG, EUCAST-AFST, FDLC, IDSA, ISHAM, MMSA, and MSGERC. Journal of Clinical Microbiology 61: e0087323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong AW, Al-Obaid K, Mohd Tap R, et al. 2023. Candida khanbhai sp. nov., a new clinically relevant yeast within the Candida haemulonii species complex. Medical Mycology 61: myad009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drumonde-Neves J, Čadež N, Reyes-Domínguez Y, et al. 2020. Clavispora santaluciae f.a., sp. nov., a novel ascomycetous yeast species isolated from grapes. International Journal of Systematic and Evolutionary Microbiology 70: 6307–6312. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biology 20: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Acero AM, Morais CG, Souza GFL, et al. 2024. Ogataea nonmethanolica f.a, sp. nov., a novel yeast species isolated from rotting wood in Brazil and Colombia. International Journal of Systematic and Evolutionary Microbiology 74: 006273. [DOI] [PubMed] [Google Scholar]
- Giménez-Jurado G, Kurtzman CP, Starmer WT, et al. 2003. Metschnikowia vanudenii sp. nov. and Metschnikowia lachancei sp. nov., from flowers and associated insects in North America. International Journal of Systematic and Evolutionary Microbiology 53: 1665–1670. [DOI] [PubMed] [Google Scholar]
- Groenewald M, Hittinger CT, Bensch K, et al. 2023. A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina. Studies in Mycology 105: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewald M, Robert V, Smith MT. 2011. Five novel Wickerhamomyces- and Metschnikowia-related yeast species, Wickerhamomyces chaumierensis sp. nov., Candida pseudoflosculorum sp. nov., Candida danieliae sp. nov., Candida robnettiae sp. nov. and Candida eppingiae sp. nov., isolated from plants. International Journal of Systematic and Evolutionary Microbiology 61: 2015–2022. [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, et al. 2013. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán B, Lachance MA, Herrera CM. 2013. Phylogenetic analysis of the angiosperm-floricolous insect-yeast association: Have yeast and angiosperm lineages co-diversified? Molecular Phylogenetics and Evolution 68: 161–175. [DOI] [PubMed] [Google Scholar]
- Hawksworth DL, Crous PW, Redhead SA, et al. 2011. The Amsterdam declaration on fungal nomenclature. IMA fungus 2: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Sant’Anna F, Bach E, Porto RZ, et al. 2019. Genomic metrics made easy: What to do and where to go in the new era of bacterial taxonomy. Critical Reviews in Microbiology 45: 182–200. [DOI] [PubMed] [Google Scholar]
- Houbraken J, De Vries RP, Samson RA. 2014. Modern taxonomy of bio-technologically important Aspergillus and Penicillium species. Advances in Applied Microbiology 86: 199–249. [DOI] [PubMed] [Google Scholar]
- Jindamorakot S, Limtong S, Yongmanitchai W, et al. 2007. Two new anamorphic yeasts, Candida thailandica sp. nov. and Candida lignicola sp. nov., isolated from insect frass in Thailand. FEMS Yeast Research 7: 1409–1414. [DOI] [PubMed] [Google Scholar]
- Kaewwichian R, Yongmanitchai W, Kawasaki H, et al. 2012. Metschnikowia saccharicola sp. nov. and Metschnikowia lopburiensis sp. nov., two novel yeast species isolated from phylloplane in Thailand. Antonie van Leeuwenhoek 102: 743–751. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khunnamwong P, Savarajara A, Jindamorakot S, et al. 2022. Metahyphopichia suwanaadthiae sp. nov., an anamorphic yeast species in the order Saccharomycetales and reassignment of Candida silvanorum to the genus Metahyphopichia. International Journal of Systematic and Evolutionary Microbiology 72: 005183. [DOI] [PubMed] [Google Scholar]
- Kidd SE, Abdolrasouli A, Hagen F. 2023. Fungal nomenclature: Managing change is the name of the game. Open Forum Infectious Diseases 10: ofac559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd SE, Halliday CL, Haremza E, et al. 2022. Attitudes of Australasian clinicians and laboratory staff to changing fungal nomenclature: Has myco-logical correctness really gone mad? Microbiology Spectrum 10: e0237721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaps J, De Vega C, Herrera CM, et al. 2020. Candida metrosideri pro tempore sp. nov. and Candida ohialehuae pro tempore sp. nov., two anti-fungal-resistant yeasts associated with Metrosideros polymorpha flowers in Hawaii. PLoS ONE 15: e0240093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP. 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Research 4: 233–245. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. 2011a. Discussion of teleomorphic and anamorphic ascomycetous yeasts and yeast-like taxa. In: Kurtzman CP, Fell JW, Boekhout T. (eds), The yeasts, A taxonomic study, 5th edn: 293–307. Elsevier, The Netherlands. [Google Scholar]
- Kurtzman CP. 2011b. Hyphopichia von Arx & van der Walt (1976). In: Kurtzman CP, Fell JW, Boekhout T. (eds), The yeasts, A taxonomic study, 5th edn: 4355–4438. Elsevier, The Netherlands. [Google Scholar]
- Kurtzman CP, Mateo RQ, Kolecka A, et al. 2015. Advances in yeast systematics and phylogeny and their use as predictors of biotechnologically important metabolic pathways. FEMS Yeast Research 15: fov050. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73: 331–371. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. 2014. Three new anascosporic genera of the Saccharomycotina: Danielozyma gen. nov., Deakozyma gen. nov. and Middelhovenomyces gen. nov. Antonie van Leeuwenhoek 105: 933–942. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ, Basehoar-Powers E. 2008. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Research 8: 939–954. [DOI] [PubMed] [Google Scholar]
- Lachance MA. 2011a. Clavispora Rodrigues de Miranda (1979). In: Kurtzman CP, Fell JW, Boekhout T. (eds), The yeasts, A taxonomic study, 5th edn: 349–353. Elsevier, The Netherlands. [Google Scholar]
- Lachance MA. 2011b. Metschnikowia Kamienski (1899). In: Kurtzman CP, Fell JW, Boekhout T. (eds), The yeasts, A taxonomic study, 5th edn: 575–620. Elsevier, The Netherlands. [Google Scholar]
- Lachance MA. 2022. Phylogenies in yeast species descriptions: In defense of neighbor-joining. Yeast 39: 513–520. [DOI] [PubMed] [Google Scholar]
- Lachance MA, Boekhout T, Scorzetti G, et al. 2011. Candida Berkhout. In: Kurtzman CP, Fell JW, Boekhout T. (eds), The yeasts, A taxonomic study, 5th edn: 987–1278. Elsevier, The Netherlands. [Google Scholar]
- Lachance MA, Hurtado E, Hsiang T. 2016. A stable phylogeny of the large-spored Metschnikowia clade. Yeast 33: 261–275. [DOI] [PubMed] [Google Scholar]
- Lee DK, Hsiang T, Lachance MA. 2018. Metschnikowia mating genomics. Antonie van Leeuwenhoek 111: 1935–1953. [DOI] [PubMed] [Google Scholar]
- Li Y, Steenwyk JL, Chang Y, et al. 2021. A genome-scale phylogeny of the kingdom Fungi. Current Biology 31: 1653–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limtong S, Kaewwichian R. 2013. Candida phyllophila sp. nov. and Candida vitiphila sp. nov., two novel yeast species from grape phylloplane in Thailand. Journal of General and Applied Microbiology 59: 191–197. [DOI] [PubMed] [Google Scholar]
- Limtong S, Yongmanitchai W. 2010. Candida chanthaburiensis sp. nov., Candida kungkrabaensis sp. nov. and Candida suratensis sp. nov., three novel yeast species from decaying plant materials submerged in water of mangrove forests. Antonie van Leeuwenhoek 98: 379–388. [DOI] [PubMed] [Google Scholar]
- Liu F, Hu ZD, Yurkov A, et al. 2024. Saccharomycetaceae: delineation of fungal genera based on phylogenomic analyses, genomic relatedness indices and genomics-based synapomorphies. Persoonia 52: 1–21. [Google Scholar]
- Lockhart SR, Etienne KA, Vallabhaneni S, et al. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clinical Infectious Diseases 64: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücking R, Aime MC, Robbertse B, et al. 2021. Fungal taxonomy and sequence-based nomenclature. Nature Microbiology 6: 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Rodriguez-R LM, Konstantinidis KT. 2014. MyTaxa: An advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Research 42: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, et al. 2019. The EMBL–EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research 268: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, et al. 2021. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Molecular Biology and Evolution 38: 4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill J, Barrie FR, Buck WR, et al. 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). Koelz Scientific Books, Koenigstein, Germany. [Google Scholar]
- Meier-Kolthoff JP, Göker M. 2019. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nature Communications 10: 2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, et al. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz JF, Gade L, Chow NA, et al. 2018. Genomic insights into multidrug- resistance, mating and virulence in Candida auris and related emerging species. Nature Communications 9: 5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy A, Rao M, Chakrabarti A, et al. 2022. Case report: Catheter related blood stream infection caused by Candida vulturna. Medical Mycology Case Reports 36: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase T, Jindamorakot S, Am-In S, et al. 2010. Candida tanticharoeniae sp. nov., a novel anamorphic yeast species found in Thailand. The Journal of General and Applied Microbiology 56: 89–92. [DOI] [PubMed] [Google Scholar]
- Nakase T, Jindamorakot S, Am-In S, et al. 2011. Three novel species of the anamorphic yeast genus Candida in the Candida intermedia clade found in Japan, Thailand and Taiwan. Journal of General and Applied Microbiology 57: 73–81. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Sangal V. 2022. Advanced prokaryotic systematics: The modern face of an ancient science. New Microbes and New Infections 49–50: 101036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opulente DA, LaBella AL, Harrison MC, et al. 2023. Genomic and ecological factors shaping specialism and generalism across an entire subphylum. BioRxiv. Preprint. [Google Scholar]
- Parks DH, Chuvochina M, Rinke C, et al. 2022. GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Research 50: D785–D794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Waite DW, et al. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nature Biotechnology 36: 996–1004. [DOI] [PubMed] [Google Scholar]
- Qin QL, Xie BB, Zhang XY, et al. 2014. A proposed genus boundary for the prokaryotes based on genomic insights. Journal of Bacteriology 196: 2210–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan AA, Eljaaly K, Alfouzan WA, et al. 2023. Psychogenetic, genetic and epigenetic mechanisms in Candida auris: Role in drug resistance. Journal of Infection and Public Health 16: 257–263. [DOI] [PubMed] [Google Scholar]
- Salichos L, Rokas A. 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497: 327–331. [DOI] [PubMed] [Google Scholar]
- Saluja P, Prasad GS. 2008. Candida ruelliae sp. nov., a novel yeast species isolated from flowers of Ruellia sp. FEMS Yeast Research 8: 660–666. [DOI] [PubMed] [Google Scholar]
- Sarawan S, Mahakhan P, Jindamorakot S, et al. 2013. Candida konsanensis sp. nov., a new yeast species isolated from Jasminum adenophyllum in Thailand with potentially carboxymethyl cellulase-producing capability. World Journal of Microbiology & Biotechnology 29: 1481–1486. [DOI] [PubMed] [Google Scholar]
- Satoh K, Makimura K, Hasumi Y, et al. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiology and Immunology 53: 41–44. [DOI] [PubMed] [Google Scholar]
- Schmalreck AF, Lackner M, Becker K, et al. 2014. Phylogenetic relationships matter: Antifungal susceptibility among clinically relevant yeasts. Antimicrobial Agents and Chemotherapy 58: 1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XX, Opulente DA, Kominek J, et al. 2018. Tempo and mode of genome evolution in the budding yeast subphylum. Cell 175: 1533–1545.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M, Pfliegler WP, Safar SVB, et al. 2016. Metahyphopichia laotica gen. nov., sp. nov., a polymorphic yeast related to Hyphopichia. International Journal of Systematic and Evolutionary Microbiology 66: 2550–2557. [DOI] [PubMed] [Google Scholar]
- Sipiczki M, Tap RM. 2016. Candida vulturna pro tempore sp. nov., a dimorphic yeast species related to the Candida haemulonis species complex isolated from flowers and clinical sample. International Journal of Systematic and Evolutionary Microbiology 66: 4009–4015. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrou AA, Lackner M, Lass-Flörl C, et al. 2019. The changing spectrum of Saccharomycotina yeasts causing candidemia: Phylogeny mirrors anti-fungal susceptibility patterns for azole drugs and amphothericin B. FEMS Yeast Research 19: foz037. [DOI] [PubMed] [Google Scholar]
- Steenwyk JL, Opulente DA, Kominek J, et al. 2019. Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts. PLoS Biology 17: e3000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Rokas A. 2023. The dawn of relaxed phylogenetics. PLoS Biology 21: e3001998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita T, Nakase T. 1999. Non-universal usage of the leucine CUG codon and the molecular phylogeny of the genus Candida. Systematic and Applied Microbiology 22: 79–86. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Nakase T. 1986. Heterogeneity of ubiquinone systems in the genus Sporothrix. Journal of General and Applied Microbiology 32: 165–168. [Google Scholar]
- Takashima M, Manabe RI, Nishimura Y, et al. 2019. Recognition and delineation of yeast genera based on genomic data: Lessons from Trichosporonales. Fungal Genetics and Biology 130: 31–42. [DOI] [PubMed] [Google Scholar]
- Takashima M, Sugita T. 2022. Taxonomy of pathogenic yeasts Candida, Cryptococcus, Malassezia, and Trichosporon. Medical Mycology Journal 63: 119–132. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiology Letters 174: 247–250. [DOI] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, et al. 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Research 18: 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turland N, Wiersema J, Barrie FR, et al. 2018. International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nine- teenth International Botanical Congress Shenzhen, China, July 2017. Koeltz Botanical Books, Glashütten. [Google Scholar]
- Van Uden, Kolipinski MC. 1962. Torulopsis haemulonii nov. spec., a yeast from the Atlantic Ocean. Antonie van Leeuwenhoek 28: 78–80. [DOI] [PubMed] [Google Scholar]
- Varghese NJ, Mukherjee S, Ivanova N, et al. 2015. Microbial species delineation using whole genome sequences. Nucleic Acids Research 43: 6761–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Bai FY. 2008. Molecular phylogeny of basidiomycetous yeasts in the Cryptococcus luteolus lineage (Tremellales) based on nuclear rRNA and mitochondrial cytochrome b gene sequence analyses: Proposal of Derxomyces gen. nov. and Hannaella gen. nov., and description of eight novel Derxomyces species. FEMS Yeast Research 8: 799–814. [DOI] [PubMed] [Google Scholar]
- Wang QM, Begerow D, Groenewald M, et al. 2015a. Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Studies in Mycology 81: 55–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Yurkov AM, Göker M, et al. 2015b. Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Studies in Mycology 81: 149–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SA, Jia JH, Bai FY. 2008. Candida alocasiicola sp. nov., Candida hainanensis sp. nov., Candida heveicola sp. nov. and Candida musiphila sp. nov., novel anamorphic, ascomycetous yeast species isolated from plants. Antonie van Leeuwenhoek 94: 257–265. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Novichkov PS, Karev GP, et al. 2009. The universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages. Proceedings of the National Academy of Sciences of the United States of America 106: 7273–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A, Jain K, Wang Y, et al. 2022. Candida auris on apples: Diversity and clinical significance. mBio 13: e0051822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Arimoto M, Kondo K. 1976. Coenzyme Q system in the classification of apiculate yeasts in the genera Nadsonia, Saccharomycodes, Hanseniaspora, Kloeckera and Wickerhamia. Journal of General & Applied Microbiology 22: 293–299. [Google Scholar]
- Yamada Y, Kondo K. 1972. Taxonomic significance of the coenzyme Q system in yeasts and yeast-like fungi (2). In: Proceedings of The IVth International Fermentation Symposium, Osaka, Japan: 781–784. [Google Scholar]
- Yuan B, Hu N, Sun J, et al. 2012. Purification and characterization of a novel extracellular inulinase from a new yeast species Candida kutaonensis sp. nov. KRF1T. Applied Microbiology and Biotechnology 96: 1517–1526. [DOI] [PubMed] [Google Scholar]
- Yurkov A, Alves A, Bai FY, et al. 2021. Nomenclatural issues concerning cultured yeasts and other fungi: why it is important to avoid unneeded name changes. IMA Fungus 12: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkov A, Schäfer AM, Begerow D. 2009. Clavispora reshetovae A. Yurkov, A.M. Schäfer & Begerow, sp. nov. Fungal Planet 35. Persoonia 23: 182–183. [Google Scholar]
- Zhu XF, Zhang DP, Yang S, et al. 2017. Candida xinjiangensis sp. nov., a new anamorphic yeast species isolated from Scolytus scheryrewi Semenov in China. Archives of Microbiology 199: 377–383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of yeast species and Genbank numbers used in the ITS and D1/D2 analyses.
The matrix of AAI and POCP values in the Metschnikowiaceae. The upper right corner represents the AAI values and the lower left corner represents the POCP values.
The PAPO analysis of the clades in the Metschnikowiaceae.
The annotation results of clade-specific OGs of the 17 clades/genera or single species lineages in the Metschnikowiaceae.
The phenotypic characteristics of different genera and clades in Metschnikowiaceae.
Phylogenetic tree inferred using the ITS and D1/D2 domain of 26S rDNA gene showed the positions of the genera and clades in the Metschnikowiaceae. Bootstrap percentages of maximum likelihood analysis over 50 % from 1000 bootstrap replicates are shown on the major branches. Bar = 0.05 substitutions per nucleotide position.
Phylogenetic tree inferred using the D1/D2 domain of 26S rDNA gene showed the positions of the genera and clades in the Metschnikowiaceae. Bootstrap percentages of maximum likelihood analysis over 50 % from 1000 bootstrap replicates are shown on the major branches. Bar = 0.05 substitutions per nucleotide position.
Phylogenetic tree inferred using the ITS gene showed the positions of the genera and clades in the Metschnikowiaceae. Bootstrap percentages of maximum likelihood analysis over 50 % from 1000 bootstrap replicates are shown on the major branches. Bar = 0.05 substitutions per nucleotide position.