Abstract
A correct classification of fungi, including yeasts, is of prime importance to understand fungal biodiversity and to communicate about this diversity. Fungal genera are mainly defined based on phenotypic characteristics and the results of single or multigene-based phylogenetic analyses. However, because yeasts often have less phenotypic characters, their classification experienced a strong move towards DNA-based data, from short ribosomal sequences to multigene phylogenies and more recently to phylogenomics. Here, we explore the usefulness of various genomics-based parameters to circumscribe fungal genera more correctly taking the yeast domain as an example. Therefore, we compared the results of a phylogenomic analysis, average amino acid identity (AAI) values, the presence of conserved signature indels (CSIs), the percentage of conserved proteins (POCP) and the presence-absence patterns of orthologs (PAPO). These genome-based metrics were used to investigate their usefulness in demarcating 13 hitherto relatively well accepted genera in Saccharomycetaceae, namely Eremothecium, Grigorovia, Kazachstania, Kluyveromyces, Lachancea, Nakaseomyces, Naumovozyma, Saccharomyces, Tetrapisispora, Torulaspora, Vanderwaltozyma, Zygosaccharomyces and Zygotorulaspora. As a result, most of these genera are supported by the genomics-based metrics, but the genera Kazachstania, Nakaseomyces and Tetrapisispora were shown to be genetically highly diverse based on the above listed analyses. Considering the results obtained for the presently recognized genera, a range of 80–92 % POCP values and a range of 60–70 % AAI values might be valuable thresholds to discriminate genera in Saccharomycetaceae. Furthermore, the genus-specific genes identified in the PAPO analysis and the CSIs were found to be useful as synapomorphies to characterize and define genera in Saccharomycetaceae. Our results indicate that the combined monophyly-based phylogenomic analysis together with genomic relatedness indices and synapomorphies provide promising approaches to delineating yeast genera and likely those of filamentous fungi as well. The genera Kazachstania, Nakaseomyces and Tetrapisispora are revised and we propose eight new genera and 41 new combinations.
Citation: Liu F, Hu Z-D, Yurkov A, et al. 2024. Saccharomycetaceae: delinaeation of fungal genera based on phylogenomic analyses, genomic relatedness indices and genomics-based synapomorphies. Persoonia 52: 1–21. https://doi.org/10.3767/persoonia.2024.52.01.
Keywords: AAI, barcode, CSI, new taxa, PAPO, phenotypic comparisons, phylogenomics, POCP, taxonomy, yeasts
Introduction
Generic demarcation is fundamental in the taxonomy and phylogeny of fungi, including yeasts. Historically, the assignment of yeasts to genera was based on the use of morphological, physiological and biochemical characteristics ( Boekhout et al. 2021). Unique phenotypic properties, including nutritional growth patterns, morphology (including sexual reproduction), genetic properties (e.g., mating compatibility, karyotyping), but also biochemical features, e.g., the number of isoprenologues of the coenzyme Q system, have been used to delimit yeast species and circumscribe genera (e.g., Kurtzman et al. 2011). During the last two decades the importance of DNA-based features in the classification of yeasts became more important ( Kurtzman 2011, Boekhout et al. 2021). The taxonomy and approaches for the delimitation of yeast genera showed a strong shift towards DNA-based methods ( Boekhout et al. 2021) starting with GC-content estimations introduced in the 1970s and DNA-DNA hybridization results in the 1980s, to ribosomal DNA sequences and single-gene phylogenies in the 1990s and the early 2000s. Recently multigene and whole-genome-based phylogenies gained importance in the last decade ( Kurtzman 2011, Kurtzman et al. 2011, Groenewald et al. 2023). With such molecular data in hands, it has been convincingly demonstrated that many important yeast genera, for example, the ascomycetous genera Candida, Pichia and Saccharomyces, and the basidiomycetous genera Cryptococcus and Rhodotorula, were (and some still are) largely polyphyletic (e.g., Kurtzman & Robnett 2003, Lachance et al. 2011, Daniel et al. 2014, Liu et al. 2015b, Wang et al. 2015a, b, d, Shen et al. 2018). As a result, dozens of new yeast genera have been erected to recognize smaller monophyletic groups to reduce the taxonomic heterogeneity of large, polyphyletic yeast genera. The application of the ‘One fungus, one name’ principle affected fungi with yeast morphs and facilitated such reclassifications leading either to the merging of sexual and asexual species or to the reinstatement of previous generic synonyms to genera that were apparently wrongly synonymized. These taxonomic proposals heavily relied on the availability of authentic reference material, such as type strains, nucleotide sequence data and reliable phylogenetic analyses, and, accordingly, new genera were attributed to well-supported monophyletic clades ( Kurtzman et al. 2008, Liu et al. 2015b, Wang et al. 2015b, d, Boekhout et al. 2021).
The application of molecular tools in the field of prokaryotic taxonomy is developing faster than that in the domain of eukaryotes, such as fungi. Indeed, fungal taxonomists repeatedly adapted methods, which were previously successfully used for prokaryotes, for example, GC-content, cell-wall composition, DNA-DNA hybridization, and ribosomal DNA gene sequences. Fast progress in the whole-genome sequencing of prokaryotes ( Wu et al. 2009, Wu & Ma 2019) facilitated the development of computational tools to discriminate species, genera and higher taxa in that domain (e.g., Meier-Kolthoff & Göker 2019, Parks et al. 2022). The following genomics-based indices have been employed to delimit new genera of prokaryotes based on the analysis of whole-genome data ( Luo et al. 2014, Varghese et al. 2015, Parks et al. 2018, Hayashi Sant’Anna et al. 2019, Barco et al. 2020, Nouioui & Sangal 2022): the average amino acid identity (AAI) values, the percentage of conserved proteins (POCP) and conserved signature indels (CSIs). With these new genomic indices and distance measurements, several thresholds have been introduced. Luo et al. (2014) and Rodriguez-R & Konstantinidis (2014) proposed to apply an AAI threshold range of 60–80 % to distinguish between prokaryote genera, but this cut-off value did not become a universal threshold for all bacteria ( Skennerton et al. 2015, Orata et al. 2018, Wirth & Whitman 2018, Xu et al. 2019). Nevertheless, AAI values and other related parameters have since been used as a useful approach to delimit genera for bacteria in taxonomic lineages for which this measurement is applicable ( Kuzmanovic et al. 2022, Montecillo 2023). Qin et al. (2014) used the POCP value for prokaryotic generic delineation to estimate their evolutionary and phenotypic distances and proposed a POCP value of 50 % as a boundary to distinguish between bacterial genera. The CSIs are unique insertions or deletions present in gene/protein sequences as derived molecular markers (i.e., synapomorphies) shared among organisms of common evolutionary descent ( Gupta 2016). Many studies showed that CSIs are robust markers useful to circumscribe genera or higher taxonomic ranks of bacteria ( Naushad et al. 2015, Alnajar & Gupta 2017, Patel & Gupta 2018) and animals ( Gupta & Suggett 2022). For basidiomycetous yeasts, Takashima et al. (2019) proposed the presence-absence patterns of orthologs (PAPO) to select genus-specific genes to be used as synapomorphies in a taxonomic analysis to delineate genera in the Trichosporonales.
Several other studies indicated that the use of genomics-based metrics can be a robust approach to delimit the boundary of genera for yeasts and other fungi (Matute & Sepúlveda 2019, Passer et al. 2019, Takashima et al. 2019, Lachance et al. 2020, Libkind et al. 2020, Xu 2020, Boekhout et al. 2021, Wibberg et al. 2021, De Albuquerque & Haag 2022, Stengel et al. 2022), but this approach is still hardly used and the utility of the above-mentioned genomic indices has not been sufficiently tested in Fungi.
Here, we present results from a comparative genomics-based taxonomy study in which we tested the circumscription of several generally well-accepted genera of Saccharomycetaceae. This family includes 18 genera, namely Cyniclomyces, Eremothecium, Grigorovia, Hagleromyces, Kazachstania, Kluyveromyces, Lachancea, Nakaseomyces, Naumovozyma, Saccharomyces, Savitreea, Stenotrophomyces, Tetrapisispora, Torulaspora, Vanderwaltozyma, Yueomyces, Zygosaccharomyces and Zygotorulaspora ( Kurtzman 2003, Kurtzman et al. 2011, Groenewald et al. 2023, Heidler von Heilborn et al. 2023).
Genera in Saccharomycetaceae have been traditionally recognized based on their morphology (including sexual morphs) and physiological traits. Classification of these yeasts went through several periods of splitting and lumping of genera applying either broad or narrow generic concepts for genera such as Kluyveromyces, Saccharomyces and Zygosaccharomyces. Using a multigene-based phylogeny, Kurtzman (2003) revised the genera in the Saccharomycetaceae and proposed five new genera, viz., Lachancea, Nakaseomyces, Naumovozyma (= Naumovia nom. inval.), Vanderwaltozyma and Zygotorulaspora, that accommodated species that before were classified in the genera Kluyveromyces, Saccharomyces and Zygosaccharomyces ( Kurtzman & Robnett 2003). Later Gouliamova & Dimitrov (2020) transferred four Kazachstania species into a newly described genus, Grigorovia, based on a combined phylogenetic analysis of the internal transcribed spacer region, including the 5.8S rDNA (ITS) and the D1/D2 domains of the large subunit rDNA, and physiological profiles. Recently four monotypic genera, i.e., Hagleromyces, Savitreea, Stenotrophomyces and Yueomyces, were proposed by Sousa et al. (2014), Sakpuntoon et al. (2020), Heidler von Heilborn et al. (2023) and Wang et al. (2015c), respectively, based on multigene-based phylogenetic analyses.
The genomes of most species in the above genera, except for the monotypic Cyniclomyces, Savitreea and Stenotrophomyces, are available at present ( Shen et al. 2018, Li et al. 2021, Opulente et al. 2023, Yu et al. 2023, https://www.ncbi.nlm.nih.gov/datasets/genome/). In order to address the potential application of the phylogenomics and genomics-based metrics to delimitate yeast genera, we explored the approaches of using the AAI and POCP statistics and CSIs synapomorphies that have been used for the demarcation of genera among prokaryotes ( Luo et al. 2014, Qin et al. 2014, Naushad et al. 2015, Alnajar & Gupta 2017, Patel & Gupta 2018, Kuzmanovic et al. 2022, Montecillo 2023), and the PAPO value that has been applied to delineate the genera in the Trichosporonales ( Takashima et al. 2019). For this, we used genome data of 13 widely accepted genera in the Saccharomycetaceae, namely Eremothecium, Grigorovia, Kazachstania, Kluyveromyces, Lachancea, Nakaseomyces, Naumovozyma, Saccharomyces, Tetrapisispora, Torulaspora, Vanderwaltozyma, Zygosaccharomyces and Zygotorulaspora, and we compared the results with DNA-barcode data and results of a polyphasic approach using phenotypic data, such as morphology and sexual reproduction.
MATERIALS AND METHODS
Ribosomal DNA (rDNA) and multi-gene phylogenetic analysis
The sequences of the ITS (including 5.8S), D1/D2 domains of large subunit (LSU) and the small subunit (SSU) rDNA, the largest subunits of DNA polymerase II (RPB1), the second largest subunits of DNA polymerase II (RPB2) and the translation elongation factor 1-α (TEF1) (Table S1) were aligned using the MAFFT program G-INS-i ( Katoh & Standley 2013). RAxML v. 8.2.12 ( Stamatakis 2014) was used to construct a Maximun Likelihood (ML) tree with the GRT+I+G model. The confidence levels of these phylogenetic branches were estimated through 1 000 repeated bootstrap analyses ( Felsenstein 1985).
Genome assembly and annotation
Nuclear DNA was extracted using the method described previously by Wang & Bai (2008). Genomic libraries (150 bp paired-end) were constructed following the manufacturer’s protocols of TruSeq Nano DNA library prep kit (Illumina) and sequenced on an Illumina HiSeq 2000 platform using TruSeq SBS Kit (Illumina). Fastp v. 0.20.1 was used to remove low-quality and adapter sequences with default parameters ( Chen et al. 2018). The genome of the yeast species Naumovozyma baii was assembled using the SPAdes v. 3.15.0 ( Bankevich et al. 2012) with the following parameters: ‘--memory 800 -k 21,33,55,77,99 --careful --cov-cutoff auto’. GeneMark-ES ( Ter-Hovhannisyan et al. 2008) was used for gene prediction.
Phylogenomic analysis and comparative genomics
To evaluate the phylogenetic relationship of members of Saccharomycetaceae, we identified single copy orthologs in 137 genomes (Table 1). BUSCO v. 5.3.2 ( Manni et al. 2021) was applied to evaluate the completeness and obtain single copy BUSCO sequences. Single copy orthologues were aligned using the MAFFT v. 7.475 program G-INS-i ( Katoh & Stand-ley 2013), concatenated with Perl scripts (https://github.com/Liufei0823/Single_Copy_Orthologue/), and an ML gene tree was constructed using RAxML v. 8.2.12 ( Stamatakis 2014) with model PROTGAMMALGX with a total of 100 bootstrap replicates. The alignment and the phylogenomics-based tree were deposited in TreeBASE (www.treebase.org, No. 30680).
Table 1.
List of yeast strains and genomes used in this study.
| Species | Strain | Assembly | Complete BUSCOs | Duplicated BUSCOs | Protein nums | Contig nums | Total length | GC (%) | N50 |
|---|---|---|---|---|---|---|---|---|---|
| Eremothecium aceris | ATCC 10895 | GCA_000412225.2 | 91.70 % | 0.10 % | 4487 | 8 | 8894523 | 51.19 | 1493473 |
| Eremothecium coryli | CBS 5749 | GCA_000710315.1 | 92.40 % | 0.00 % | 4485 | 19 | 9094934 | 41.58 | 1035239 |
| Eremothecium cymbalariae | DBVPG 7215 | GCA_000235365.1 | 90.50 % | 0.10 % | 4434 | 8 | 9669424 | 40.32 | 1193613 |
| Eremothecium gossypii | ATCC 10895 | GCA_000091025.4 | 95.70 % | 0.10 % | 4776 | 8 | 9119312 | 51.7 | 1519140 |
| Eremothecium sinecaudum | ATCC 58844 | GCA_001548555.1 | 95.00 % | 0.10 % | 4536 | 8 | 8948761 | 40.15 | 1398029 |
| Grigorovia humatica | NRRL Y-48839 | GCA_030462875.1 | 78.20 % | 1.90 % | 4489 | 574 | 12469330 | 33.06 | 111969 |
| Grigorovia jiainica | NRRL Y-48843 | GCA_030571715.1 | 84.30 % | 3.20 % | 6075 | 948 | 14480493 | 27.56 | 62478 |
| Grigorovia transvaalensis | NRRL Y-17245 | GCA_003708445.2 | 80.70 % | 1.90 % | 4776 | 518 | 12472218 | 33.31 | 136221 |
| Grigorovia yakushimaensis | NRRL Y-48837 | GCA_003709265.1 | 85.10 % | 1.70 % | 5423 | 530 | 12823882 | 30.46 | 249423 |
| Hagleromyces aurorensis | yHDO579 | SRR16974332 | 94.00 % | 0.00 % | 4988 | 297 | 11792941 | 43.03 | 112499 |
| Kazachstania aerobia | NRRL Y-27976 | GCA_003708495.1 | 88.80 % | 2.50 % | 5102 | 454 | 12056888 | 33.87 | 115019 |
| Kazachstania africana | CBS 2517 | GCA_000304475.1 | 95.70 % | 3.20 % | 5378 | 12 | 11130140 | 36.29 | 1026673 |
| Kazachstania aquatica | NRRL Y-27993 | GCA_030571515.1 | 86.90 % | 3.20 % | 5555 | 618 | 12940683 | 34.5 | 84279 |
| Kazachstania barnettii | CLIB 1767 | GCA_903064755.1 | 94.80 % | 3.70 % | 5269 | 14 | 12616033 | 33.63 | 1404614 |
| Kazachstania bovina | CBS 16326 | GCA_023309525.1 | 90.20 % | 3.20 % | 5528 | 210 | 15883906 | 31.72 | 203893 |
| Kazachstania bromeliacearum | NRRL Y-48836 | GCA_003708535.2 | 91.70 % | 3.50 % | 5222 | 412 | 11280051 | 34.63 | 173093 |
| Kazachstania bulderi | CLIB 596 | GCA_933962305.1 | 93.10 % | 5.80 % | 5611 | 30 | 14529293 | 33.13 | 911116 |
| Kazachstania exigua | NRRL Y-12640 | GCA_030580595.1 | 90.10 % | 72.30 % | 10428 | 1185 | 25075894 | 32.33 | 53120 |
| Kazachstania gamospora | NRRL Y-48841 | GCA_030462905.1 | 87.90 % | 2.80 % | 4947 | 654 | 11378243 | 27.32 | 44099 |
| Kazachstania hellenica | NRRL Y-48844 | GCA_030571615.1 | 89.10 % | 4.20 % | 4947 | 454 | 11951485 | 28.95 | 84326 |
| Kazachstania heterogenica | NRRL Y-27499 | SRR16974533 | 89.30 % | 1.60 % | 5117 | 816 | 13294299 | 30.33 | 28932 |
| Kazachstania humilis | CLIB 1323 | GCA_933934105.1 | 85.90 % | 6.80 % | 5329 | 16 | 13969787 | 49.15 | 1009204 |
| Kazachstania ichnusensis | CBS 11859 | GCA_030580495.1 | 90.50 % | 2.80 % | 4934 | 278 | 10116563 | 37.38 | 527079 |
| Kazachstania intestinalis | NRRL Y-48847 | GCA_003708845.2 | 91.20 % | 2.60 % | 4928 | 218 | 9892576 | 41.56 | 136819 |
| Kazachstania jinghongensis | CBS 15232 | GCA_030572895.1 | 93.40 % | 4.20 % | 5360 | 562 | 11642923 | 35.51 | 55136 |
| Kazachstania kunashirensis | NRRL Y-27209 | GCA_003708465.1 | 91.50 % | 4.30 % | 5173 | 281 | 10958485 | 32.32 | 237337 |
| Kazachstania lodderae | NRRL Y-8280 | GCA_030571655.1 | 93.60 % | 4.70 % | 5401 | 251 | 12092871 | 33.65 | 177372 |
| Kazachstania martiniae | NRRL Y-409 | GCA_003708925.2 | 92.80 % | 4.90 % | 5663 | 408 | 11743474 | 33.69 | 205775 |
| Kazachstania naganishii | CBS 8797 | GCA_000348985.1 | 94.00 % | 3.40 % | 5321 | 13 | 10845821 | 45.89 | 856010 |
| Kazachstania piceae | NRRL Y-17977 | SRR16974347 | 94.50 % | 3.90 % | 5367 | 411 | 12655860 | 32.81 | 68608 |
| Kazachstania pintolopesii | NCYC 4417 | GCA_950065675.1 | 90.10 % | 1.60 % | 5099 | 28 | 13998629 | 30.59 | 948874 |
| Kazachstania pseudohumilis | CBS 11404 | GCA_030579215.1 | 91.20 % | 5.30 % | 5723 | 816 | 13806380 | 44.87 | 59499 |
| Kazachstania psychrophila | CBS 12689 | GCA_030579255.1 | 93.70 % | 4.20 % | 5114 | 99 | 10504644 | 33.29 | 324962 |
| Kazachstania rosinii | NRRL Y-17919 | GCA_003708425.2 | 92.70 % | 3.60 % | 5434 | 341 | 12228026 | 40.46 | 195966 |
| Kazachstania saulgeensis | CLIB 1764 | GCA_900180425.1 | 95.20 % | 3.90 % | 5329 | 17 | 12935755 | 32.51 | 1371409 |
| Kazachstania serrabonitensis | UFMG-CM-Y273 | GCA_030571355.1 | 94.40 % | 3.60 % | 5462 | 308 | 13240085 | 31.98 | 127329 |
| Kazachstania servazzii | PF 9 W20 | GCA_028408395.1 | 90.60 % | 2.30 % | 5166 | 22 | 12334243 | 34.35 | 981509 |
| Kazachstania siamensis | NRRL Y-48842 | GCA_003708905.2 | 88.80 % | 2.30 % | 5057 | 483 | 11808792 | 32.99 | 107908 |
| Kazachstania sinensis | NRRL Y-27222 | SRR16974264 | 91.80 % | 2.90 % | 5392 | 315 | 11520106 | 46.15 | 95921 |
| Kazachstania slooffiae | NRRL Y-4349 | GCA_030580615.1 | 81.90 % | 19.10 % | 9633 | 5317 | 23922660 | 30.04 | 10822 |
| Kazachstania solicola | NRRL Y-27207 | GCA_003708835.2 | 90.10 % | 2.80 % | 5803 | 890 | 13004848 | 35.68 | 117325 |
| Kazachstania spencerorum | NRRL Y-17920 | GCA_003708825.2 | 92.90 % | 5.20 % | 6431 | 1151 | 12991956 | 33.39 | 111153 |
| Kazachstania taianensis | NRRL Y-48846 | GCA_003708865.1 | 88.90 % | 2.30 % | 5352 | 263 | 13582649 | 43.21 | 190094 |
| Kazachstania telluris | UCD400 | GCA_009394695.1 | 90.20 % | 1.60 % | 5226 | 730 | 13895863 | 31.8 | 48784 |
| Kazachstania turicensis | NRRL Y-48834 | GCA_003708545.1 | 91.70 % | 3.70 % | 5734 | 564 | 14081953 | 33.21 | 150948 |
| Kazachstania unispora | NRRL Y-1556 | GCA_003708525.2 | 90.40 % | 2.50 % | 5220 | 382 | 12259717 | 32.25 | 159570 |
| Kazachstania viticola | NRRL Y-27206 | GCA_003708455.1 | 90.10 % | 5.90 % | 5648 | 680 | 12089988 | 32.66 | 91405 |
| Kazachstania yasuniensis | CBS 13946 | GCA_030558655.1 | 90.10 % | 2.50 % | 5298 | 364 | 12450102 | 31.11 | 195526 |
| Kluyveromyces aestuarii | NRRL YB-4510 | GCA_003707555.1 | 93.80 % | 0.10 % | 4768 | 93 | 10039207 | 38.32 | 516559 |
| Kluyveromyces dobzhanskii | CBS 2104 | GCA_000820885.1 | 95.10 % | 1.10 % | 4957 | 86 | 10741898 | 41.25 | 493016 |
| Kluyveromyces lactis | NRRL Y-1140 | GCA_000002515.1 | 96.20 % | 0.20 % | 5076 | 6 | 10689156 | 38.76 | 1753957 |
| Kluyveromyces marxianus | NRRL Y-6860 | GCA_002356615.1 | 94.00 % | 0.20 % | 4881 | 8 | 10837618 | 40.2 | 1406771 |
| Kluyveromyces nonfermentans | NRRL Y-27343 | GCA_003670155.1 | 94.50 % | 0.10 % | 4654 | 104 | 9522276 | 35.92 | 432147 |
| Kluyveromyces siamensis | CBS 10860 | GCA_030579315.1 | 92.90 % | 0.10 % | 4957 | 360 | 10079274 | 38.38 | 231898 |
| Kluyveromyces starmeri | UFMG-CM-Y3682 | GCA_008973615.1 | 93.30 % | 0.10 % | 4725 | 47 | 9518874 | 43.94 | 729094 |
| Kluyveromyces wickerhamii | UCD 54-210 | GCA_000179415.1 | 91.10 % | 0.00 % | 4987 | 510 | 9807744 | 40.87 | 36691 |
| Lachancea cidri | NCYC 2875 | GCA_947297695.1 | 89.80 % | 5.90 % | 6746 | 2195 | 12003368 | 41.21 | 9179 |
| Lachancea dasiensis | CBS 10888 | GCA_900074725.1 | 98.30 % | 0.10 % | 5096 | 8 | 10701617 | 45.16 | 1410526 |
| Lachancea fermentati | CBS 6772 | GCA_900074765.1 | 98.80 % | 0.00 % | 5233 | 8 | 10264457 | 42.57 | 1346284 |
| Lachancea kluyveri | NRRL Y-12651 | GCA_000149225.2 | 97.20 % | 0.50 % | 5261 | 34 | 11538858 | 41.59 | 1295560 |
| Lachancea lanzarotensis | CBS 12615 | GCA_000938715.1 | 98.20 % | 0.10 % | 5058 | 24 | 11092131 | 44.28 | 910667 |
| Lachancea meyersii | CBS 8951 | GCA_900074715.1 | 98.80 % | 0.10 % | 4998 | 8 | 11261819 | 45.33 | 2013154 |
| Lachancea mirantina | CBS 11717 | GCA_900074745.1 | 98.10 % | 0.00 % | 5056 | 8 | 10117267 | 45.1 | 1414338 |
| Lachancea nothofagi | CBS 11611 | GCA_900074755.1 | 98.60 % | 0.00 % | 5154 | 8 | 11313798 | 43.72 | 1763880 |
| Lachancea quebecensis | CBS 14088 | GCA_002900925.1 | 98.70 % | 0.10 % | 5074 | 51 | 10229370 | 46.71 | 533706 |
| Lachancea sp. | CBS 6924 | GCA_900074735.1 | 98.10 % | 0.00 % | 5059 | 7 | 11336659 | 44.49 | 2184418 |
| Lachancea sp. | yHQL494 | GCA_030562185.1 | 95.00 % | 0.10 % | 4900 | 203 | 11060012 | 43.11 | 286026 |
| Lachancea thermotolerans | CBS 6340 | GCA_000142805.1 | 98.80 % | 0.10 % | 5092 | 8 | 10392862 | 47.3 | 1513537 |
| Lachancea waltii | NCYC 2644 | GCA_000167115.1 | 92.20 % | 0.60 % | 5296 | 713 | 10912112 | 44.29 | 62747 |
| Nakaseomyces bacillisporus | CBS 7720 | GCA_001046975.1 | 90.50 % | 2.20 % | 4796 | 182 | 10838378 | 36.59 | 145552 |
| Nakaseomyces bracarensis | CBS 10154 | GCA_001077315.1 | 93.90 % | 2.70 % | 5157 | 250 | 12229116 | 36.13 | 109957 |
| Nakaseomyces castellii | CBS 4332 | GCA_001046935.1 | 86.90 % | 1.20 % | 4570 | 101 | 10201440 | 40.86 | 351735 |
| Nakaseomyces delphensis | CBS 2170 | GCA_001039675.1 | 92.90 % | 2.50 % | 4949 | 177 | 10867124 | 38.87 | 129090 |
| Nakaseomyces glabratus | CBS 138 | GCA_000002545.2 | 95.60 % | 2.40 % | 5202 | 13 | 12318245 | 38.65 | 1100349 |
| Nakaseomyces kungkrabaensis | CBS 10927 | GCA_030556385.1 | 93.30 % | 2.70 % | 5135 | 200 | 11724688 | 37.03 | 243157 |
| Nakaseomyces nivariensis | CBS 9983 | GCA_017309295.1 | 94.10 % | 2.90 % | 5104 | 16 | 11832599 | 37.11 | 885783 |
| Nakaseomyces sp. | UFMG-CM-Y6046 | GCA_030571395.1 | 94.40 % | 2.80 % | 5368 | 376 | 12947746 | 37.93 | 88113 |
| Nakaseomyces uthaithaninus | CBS 10932 | GCA_030564085.1 | 92.80 % | 2.30 % | 5100 | 314 | 12455243 | 42.15 | 88913 |
| Naumovozyma baii | AS 2.4520 | NMDC20081875 | 94.60 % | 4.90 % | 5322 | 203 | 11260751 | 34.79 | 161193 |
| Naumovozyma castellii | CBS 4309 | GCA_000237345.1 | 96.20 % | 5.20 % | 5592 | 10 | 11219539 | 36.76 | 1245273 |
| Naumovozyma dairenensis | CBS 421 | GCA_000227115.2 | 95.80 % | 4.20 % | 5548 | 11 | 13527580 | 34.15 | 1230053 |
| Saccharomyces arboricola | yHDPN432 | GCA_918268255.1 | 95.40 % | 4.20 % | 5251 | 17 | 11461381 | 38.77 | 894440 |
| Saccharomyces cerevisiae | S288C | GCA_016858165.1 | 97.20 % | 5.00 % | 5850 | 53 | 12862231 | 38.41 | 929257 |
| Saccharomyces eubayanus | FM 1318 | GCA_001298625.1 | 94.60 % | 4.00 % | 5379 | 24 | 11734173 | 39.86 | 896107 |
| Saccharomyces jurei | CBS 14759 | GCA_900290405.1 | 94.80 % | 4.50 % | 5370 | 18 | 11938758 | 37.9 | 738741 |
| Saccharomyces kudriavzevii | CBS 8840 | GCA_918252685.1 | 95.30 % | 4.60 % | 5253 | 23 | 11698107 | 39.6 | 875623 |
| Saccharomyces mikatae | IFO1815 | GCA_918250775.1 | 94.80 % | 4.60 % | 5275 | 27 | 11963209 | 37.72 | 827134 |
| Saccharomyces paradoxus | CBS 432 | GCA_002079055.1 | 97.00 % | 4.50 % | 5528 | 17 | 12092810 | 38.54 | 903028 |
| Saccharomyces uvarum | CBS 7001 | GCA_027557585.1 | 97.40 % | 5.10 % | 5580 | 17 | 12081644 | 40.06 | 917875 |
| Tetrapisispora arboricola | NRRL Y-27308 | GCA_030557565.1 | 93.00 % | 3.70 % | 5127 | 231 | 12739091 | 31.55 | 259939 |
| Tetrapisispora blattae | CBS 6284 | GCA_000315915.1 | 92.10 % | 2.90 % | 5389 | 10 | 14048593 | 31.74 | 1449145 |
| Tetrapisispora fleetii | NRRL Y-27350 | GCA_003707605.1 | 89.70 % | 3.00 % | 4982 | 324 | 12055180 | 32.76 | 482824 |
| Tetrapisispora iriomotensis | NRRL Y-27309 | GCA_003705975.1 | 93.50 % | 7.70 % | 5357 | 144 | 11946050 | 32.39 | 451623 |
| Tetrapisispora namnaoensis | NRRL Y-27982 | GCA_003705985.1 | 91.90 % | 3.30 % | 5193 | 291 | 12471591 | 32.36 | 466427 |
| Tetrapisispora nanseiensis | NRRL Y-27310 | GCA_030568035.1 | 93.20 % | 3.50 % | 5385 | 482 | 13482527 | 30.89 | 121629 |
| Tetrapisispora phaffii | CBS 4417 | GCA_000236905.1 | 95.40 % | 3.80 % | 5253 | 17 | 12115070 | 33.56 | 815984 |
| Tetrapisispora pingtungensis | CBS 12780 | GCA_030573885.1 | 93.90 % | 3.50 % | 5173 | 271 | 12565781 | 29.14 | 168631 |
| Tetrapisispora taiwanensis | CBS 10586 | GCA_030573835.1 | 93.30 % | 3.70 % | 5263 | 320 | 12640076 | 27.38 | 172981 |
| Torulaspora delbrueckii | CBS 1146 | GCA_000243375.1 | 98.00 % | 0.10 % | 4972 | 8 | 9220678 | 42.02 | 1218070 |
| Torulaspora franciscae | CBS 2926 | GCA_013387355.1 | 95.00 % | 0.20 % | 4735 | 81 | 9205904 | 45.05 | 481156 |
| Torulaspora globosa | CBS 764 | GCA_014133895.1 | 96.70 % | 0.10 % | 4931 | 8 | 9281121 | 46.01 | 1122226 |
| Torulaspora indica | CBS 12408 | GCA_931305995.1 | 95.10 % | 0.10 % | 4688 | 58 | 9110689 | 45.76 | 593772 |
| Torulaspora maleeae | CBS 10694 | GCA_003708055.2 | 94.20 % | 0.10 % | 4721 | 54 | 9217477 | 45.78 | 764704 |
| Torulaspora microellipsoides | NRRL Y-1549 | GCA_003707085.1 | 96.10 % | 4.50 % | 5289 | 120 | 10927271 | 38.7 | 506894 |
| Torulaspora pretoriensis | CBS 2187 | GCA_012851205.1 | 95.40 % | 0.10 % | 4800 | 20 | 9367368 | 44.93 | 1253998 |
| Torulaspora quercuum | UCD657 | GCA_946403475.1 | 96.00 % | 0.10 % | 4903 | 9 | 10364244 | 41.38 | 1208319 |
| Torulaspora sp. | CBS 2947 | GCA_013694445.1 | 97.00 % | 0.10 % | 4938 | 8 | 9264691 | 42.42 | 1146439 |
| Torulaspora sp. | yHMJ407 | GCA_030580195.1 | 95.30 % | 0.20 % | 4766 | 183 | 9065090 | 44.45 | 923141 |
| Vanderwaltozyma polyspora | DSM 70294 | GCA_000150035.1 | 91.80 % | 4.60 % | 5367 | 281 | 14674591 | 33.02 | 126622 |
| Vanderwaltozyma tropicalis | NRRL Y-63776 | GCA_030555675.1 | 94.60 % | 4.80 % | 5275 | 330 | 11169621 | 30.69 | 188659 |
| Vanderwaltozyma verrucispora | NRRL Y-63795 | GCA_030565105.1 | 93.40 % | 4.50 % | 5229 | 467 | 11912226 | 29.39 | 81885 |
| Vanderwaltozyma yarrowii | NRRL Y-17763 | GCA_030568135.1 | 94.20 % | 5.60 % | 5439 | 577 | 12611320 | 30.55 | 68148 |
| Yueomyces silvicola | MN-29 | GCA_030179955.1 | 82.90 % | 2.00 % | 4484 | 176 | 11594790 | 36.63 | 642877 |
| Yueomyces sinensis | NRRL Y-17406 | GCA_003707995.1 | 83.90 % | 2.30 % | 5086 | 510 | 12915648 | 29.6 | 177725 |
| Zygosaccharomyces bailii | CBS 680 | GCA_000442885.1 | 93.90 % | 0.20 % | 4723 | 27 | 10268813 | 42.48 | 932251 |
| Zygosaccharomyces bisporus | NRRL Y-12626 | GCA_003707595.1 | 95.60 % | 0.10 % | 4981 | 185 | 10539560 | 43.94 | 157422 |
| Zygosaccharomyces gambellarensis | CBS 2191 | GCA_030571545.1 | 94.80 % | 0.20 % | 4863 | 125 | 9918755 | 38.99 | 468129 |
| Zygosaccharomyces kombuchaensis | NRRL YB-4811 | GCA_003705955.1 | 94.70 % | 0.10 % | 4908 | 252 | 10225954 | 44.56 | 108046 |
| Zygosaccharomyces lentus | NRRL Y-27276 | GCA_030568175.1 | 94.30 % | 0.20 % | 4918 | 180 | 10214768 | 45.22 | 170809 |
| Zygosaccharomyces mellis | CBS 736 | GCA_020521395.1 | 95.90 % | 0.10 % | 4734 | 79 | 9559548 | 38.78 | 413958 |
| Zygosaccharomyces parabailii | ATCC 60483 | GCA_001984395.2 | 98.20 % | 92.00 % | 10086 | 18 | 20864403 | 42.48 | 1283838 |
| Zygosaccharomyces parabailii | ZPA 3699 DN | GCA_949129065.1 | 97.10 % | 82.20 % | 9519 | 21 | 20977846 | 42.29 | 1359109 |
| Zygosaccharomyces pseudobailii | PF2202 | GCA_023629055.1 | 96.10 % | 87.70 % | 9509 | 322 | 20001422 | 42.35 | 141095 |
| Zygosaccharomyces pseudobailii | ZPS 3697 DN | GCA_949129085.1 | 96.10 % | 86.00 % | 9522 | 19 | 21347288 | 42.24 | 1405639 |
| Zygosaccharomyces pseudobailii | Zpse1 | GCA_900408955.1 | 96.30 % | 89.70 % | 9526 | 95 | 20217079 | 42.38 | 684448 |
| Zygosaccharomyces pseudorouxii | NRRL Y-63794 | GCA_030572675.1 | 95.50 % | 0.50 % | 4967 | 220 | 10017212 | 39.87 | 314261 |
| Zygosaccharomyces rouxii | NRRL Y-64007 | GCA_021535285.1 | 97.10 % | 0.20 % | 5001 | 8 | 9952157 | 39.12 | 1530681 |
| Zygosaccharomyces sapae | ABT301 | GCA_900465325.1 | 96.90 % | 83.70 % | 11904 | 52 | 24741993 | 39.57 | 1409619 |
| Zygosaccharomyces sapae | CBS 12607 | GCA_020521375.1 | 97.00 % | 94.80 % | 13915 | 356 | 27714775 | 39.48 | 309874 |
| Zygosaccharomyces siamensis | MinabeTanabe | GCA_013423405.1 | 95.10 % | 0.10 % | 4752 | 110 | 9666950 | 38.83 | 342483 |
| Zygotorulaspora chibaensis | CBS 15364 | GCA_030565665.1 | 95.10 % | 0.20 % | 4937 | 142 | 10874240 | 41.58 | 655227 |
| Zygotorulaspora danielsina | CBS 15365 | GCA_030572985.1 | 94.80 % | 0.10 % | 4869 | 203 | 10517112 | 40.38 | 858518 |
| Zygotorulaspora florentina | NRRL Y-1560 | GCA_003671575.2 | 95.40 % | 0.10 % | 5030 | 199 | 11024643 | 40.97 | 562868 |
| Zygotorulaspora mrakii | NRRL Y-6702 | GCA_013402915.1 | 97.20 % | 0.10 % | 5041 | 9 | 10450160 | 39.96 | 1312970 |
| Zygotorulaspora sp. | UFMG-CM-Y6047 | GCA_030571275.1 | 95.40 % | 0.10 % | 4908 | 130 | 10629834 | 37.1 | 379747 |
| Hanseniaspora osmophila | NRRL Y-1613 | GCA_003707715.1 | 82.50 % | 2.20 % | 4654 | 390 | 11743089 | 37.12 | 139256 |
| Saccharomycodes ludwigii | NBRC 1722 | GCA_020623625.1 | 87.90 % | 0.10 % | 5031 | 8 | 12500424 | 30.85 | 1848403 |
To assess the amino acid identity (AAI) of the 13 genera in Saccharomycetaceae, namely Eremothecium, Grigorovia, Kazachstania, Kluyveromyces, Lachancea, Nakaseomyces, Naumovozyma, Saccharomyces, Tetrapisispora, Torulaspora, Vanderwaltozyma, Zygosaccharomyces and Zygotorulaspora, we used CompareM v. 0.1.2 (https://github.com/dparks1134/CompareM) with defaulted parameters.
To predict Orthologous Groups (OGs) all proteins were clustered using OrthoFinder v. 2.5.4 ( Emms & Kelly 2019). Presence-absence patterns of orthologs (PAPO) were constructed using the method described by Takashima et al. (2019). According to the OGs results of OrthoFinder, ‘absence’ OGs were denoted as 0 (zero) and ‘Presence’ OGs denoted as 1 (one). To examine the OGs relationship of the emerging clade, we identified the number of unique and shared proteins of the 13 genera of Saccharomycetaceae. The OGs that were fully conserved within a clade were considered core proteins, whereas the OGs found in at least one strain in a clade were considered pan proteins, and the OGs found in all strains of a clade but not in another clade were considered as unique proteins for that clade.
The percentage of conserved proteins (POCP) was calculated following Qin et al. (2014). The proteins of the two strains were compared with each other using BLASTp ( Tatusova & Madden 1999). The conserved proteins were identified based on identity (> 40 %), aligned length (50 %) and e-value (< 1 × 10−5). POCP was calculated by the ratio of the total number of conserved proteins in the two proteomes and verified by the POCP calculation method (https://github.com/hoelzer/pocp).
The identification of CSIs was carried out according to the method described by Gupta (2014). The creation of multiple sequence alignments (MSA) for amino acid sequences of each of the 115 OGs using MAFFT v. 7.475 ( Katoh & Standley 2013) with default options is the first step to finding the conserved Indels (CSIs). Next, the genus-specific CSIs were identified from MSA carried out by visual inspection using MEGA v. 7 ( Kumar et al. 2016). In general, the sequences of 20–30 bp around CSIs are relatively conservative and marked by a short line (-). The indel length is generally 1bp to very large indels (> 20 aa). The ‘genus-specific signature nucleotides’ (GSNs) of the rDNA (LSU and SSU) were detected in the same way as CSI.
D1/D2 LSU and ITS sequence similarity analysis
We compared the sequence similarity and nucleotide variations in the ITS and D1/D2 LSU among the 13 genera in Saccharomycetaceae (Table S1) using the EMBOSS water alignment tool ( Madeira et al. 2019, Li et al. 2020) to run the local alignment for the calculation of the sequence similarities and nucleotide variation including substitutions and deletions.
Phenotypic characteristics comparison
The morphological and physiological data used in the phenotypic characteristics analysis were collected from The yeasts, a taxonomic study ( Kurtzman et al. 2011) and the Yeasts Trust Database (http://theyeasts.org/).
RESULTS AND DISCUSSION
Genome assembly and annotation
The genome of Naumovozyma baii AS 2.4520 was newly sequenced with the Illumina HiSeq 2000 platform. The other genomes were downloaded from the NCBI genome database (https://www.ncbi.nlm.nih.gov/datasets/genome/). All 137 genomes belonged to two families (15 genera belong to Saccharomycetaceae and 2 genera belonged to Saccharomycodaceae for outgroups) and ranged in size from 8.89 Mb to 27.71 Mb. The number of predicted proteins of the studied species ranged from 4 434 to 13 915 (Table 1). The G+C content of all genomes ranges from 27.32 to 51.7 %. To search for single copy orthologs and remove hybrid genomes, we retained only those genomes that contained ≤ 20 % duplicated BUSCOs. The genomes of Kazachstania exigua, Zygosaccharomyces parabailii, Zygosacchromyces pseudobailii and Zygosaccharomyces sapae were discarded because their duplicated BUSCOs content ranged from 72.3 % to 94.8 %.
rDNA, multigene and phylogenomic analyses
Kazachstania exigua, Z. parabailii, Z. pseudobailii and Z. sapae were not included in the phylogenomic analysis because they are diploid or hybrids containing two copies of orthologous genes. The concatenation-based phylogenomic analysis was based on 115 single-copy orthologous genes present in 129 strains belonging to 15 genera of Saccharomycetaceae and two genera of Saccharomycodaceae (Table 1). For comparison, two datasets were used for phylogenetic analyses: i) the ITS+D1/D2 LSU rDNA-based tree; and ii) a multigene-based dataset comprising three fragments of the rDNA repeat, namely the SSU, ITS, D1/D2 LSU, and partial sequences of the RPB1, RPB2 and TEF1 genes. The taxon sampling in the latter two trees was larger than that used in the phylogenomic analysis.
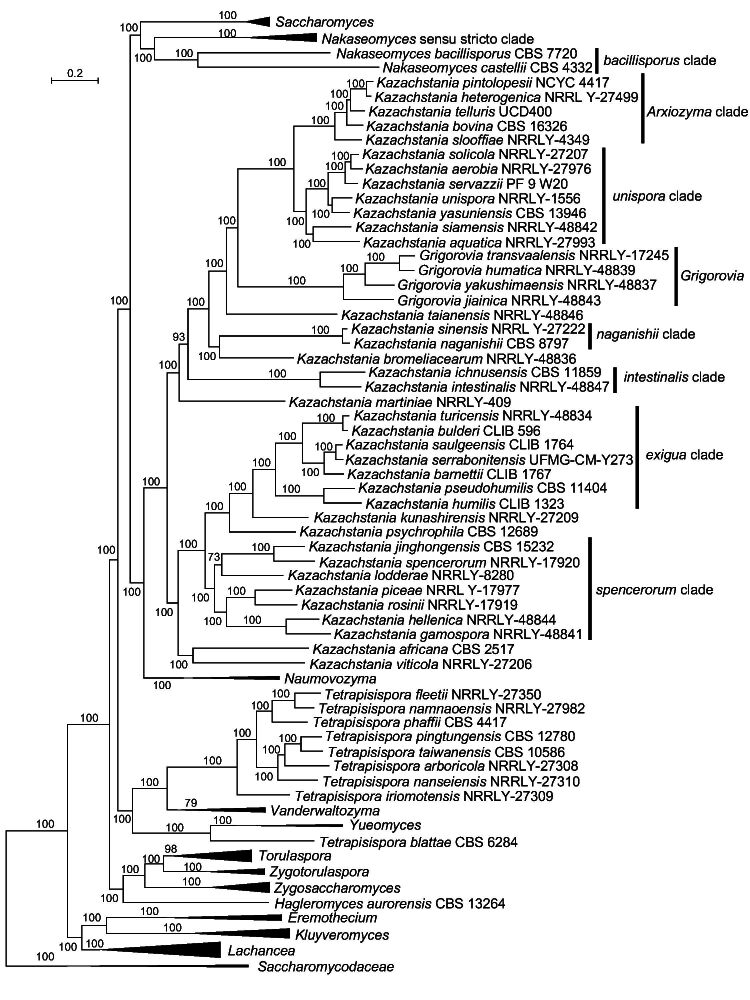
The phylogenomic analysis showed that most traditionally recognized genera of Saccharomycetaceae received high supported values (i.e., 98 to 100 % bootstrap), but the genus Vanderwaltozyma had moderate support (79 %), and Kazachstania and Tetrapisispora were found to be heterogeneous and polyphyletic (Fig. 1). The phylogenetic relationships among the genera in the Saccharomycetaceae were found to be incongruent as demonstrated by multigene phylogenetic analyses ( Kurtzman & Robnett 2003, 2013, Kurtzman 2011). Although most of the established clades were found to be robust, the phylogenetic network analysis by Wu et al. (2008) revealed a conflict between mitochondrial- and nuclear-encoded genes, and complex patterns due to hybridization and introgression in the family Saccharomycetaceae, i.e., Nakaseomyces and Tetrapisispora. The genera Kazachstania, Nakaseomyces, Naumovozyma and Saccharomyces formed a poorly supported clade in the study of Kurtzman & Robnett (2003), but in another study Kurtzman & Robnett (2013) showed that the genus Nakaseomyces was phylogenetically remotely related to Kazachstania, Naumovozyma and Saccharomyces, but clustered with Cyniclomyces. The genus Zygosaccharomyces was found to be phylogenetically distinct from the genera Torulaspora and Zygotorulaspora when using LSU, SSU, ITS, TEF1, RPB2, and mitochondrial-encoded small-subunit rDNA (Sm rDNA) and cytochrome oxidase II (COX II) sequences ( Kurtzman & Robnett 2003). However, the results from Kurtzman & Robnett (2013), using LSU, SSU, RPB1, RPB2 and TEF1 sequences, clustered these three genera together. Eremothecium, Kluyveromyces and Lachancea formed three distinct branches using LSU, SSU, ITS, TEF1, RPB2, Sm rDNA and COX II sequences ( Kurtzman 2003), but they clustered together as sister genera when using LSU, SSU, RPB1, RPB2 and TEF1 sequences ( Kurtzman & Robnett 2013). Using SSU and D1/D2 LSU sequences, the genus Hagleromyces was added to the family Saccharomycetaceae and placed in a clade with Cyniclomyces guttulatus, though the position of this clade in the family remained unclear ( Sousa et al. 2014). Our phylogenomic analysis supported sister relationships of the genera Eremothecium, Kluyveromyces and Lachancea, and for the genera Torulaspora, Zygosaccharomyces and Zygotorulaspora (Fig. 1). Hagleromyces aurorensis was found to be located on a basal branch more closely related to Torulaspora, Zygosaccharomyces and Zygotorulaspora. The genus Nakaseomyces was found to be phylogenetically closely related to Saccharomyces (Fig. 1). The genera Kazachstania and Naumovozyma formed a strongly supported lineage (Fig. 1). The phylogenomic analysis showed that the genera Yueomyces, Tetrapisispora and Vanderwaltozyma clustered together (Fig. 1). The results of the above phylogenomic analyses are in agreement with the results from Shen et al. (2018) and Opulente et al. (2023).
Fig. 1.
Phylogenomics tree inferred using 115 single copy orthologue proteins showing the phylogenetic relationship between genera in Saccharomycetaceae. Bootstrap percentages of maximum likelihood analysis over 50 % from 1 000 bootstrap replicates are shown on the major branches. Bar = 0.2 substitutions per nucleotide position.
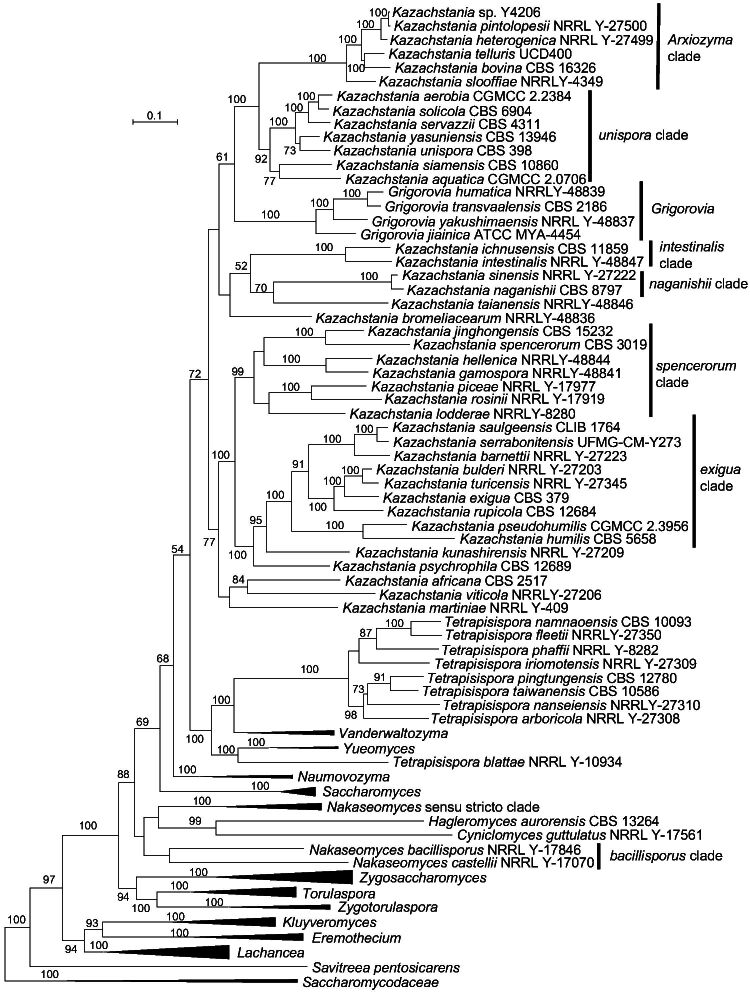
Tetrapisispora blattae formed a separate and long branch closely related to Yueomyces and less to the other Tetrapisispora species, a result that agrees with data from Shen et al. (2018) and Opulente et al. (2023). Tetrapisispora blattae was previously named Kluyveromyces blattae ( Henninger & Windisch 1976) and was transferred to Tetrapisispora based on the outcome of a multigene analysis ( Kurtzman 2003, Kurtzman & Robnett 2003), but in those studies it also formed a basal and long branch when compared to the rest of the Tetrapisispora species. It must be mentioned that the circumscription of the genus Tetrapisispora was amended to accommodate this species that differed from other species in ascospore shape and ascus properties, and some assimilation tests. Our multigene phylogenetic analysis also showed that T. blattae was phylogenetically distinct from other Tetrapisispora species, and was found to be closely related to Yueomyces (Fig. 2).
Fig. 2.
Phylogenetic tree inferred using a combined dataset of SSU, ITS, D1/D2, RPB1, RPB2 and TEF1 nucleotide sequences, showing the phylogenetic relationship between genera in Saccharomycetaceae. Bootstrap percentages of maximum likelihood analysis over 50 % from 1 000 bootstrap replicates are shown on the major branches. Bar = 0.1 substitutions per nucleotide position.
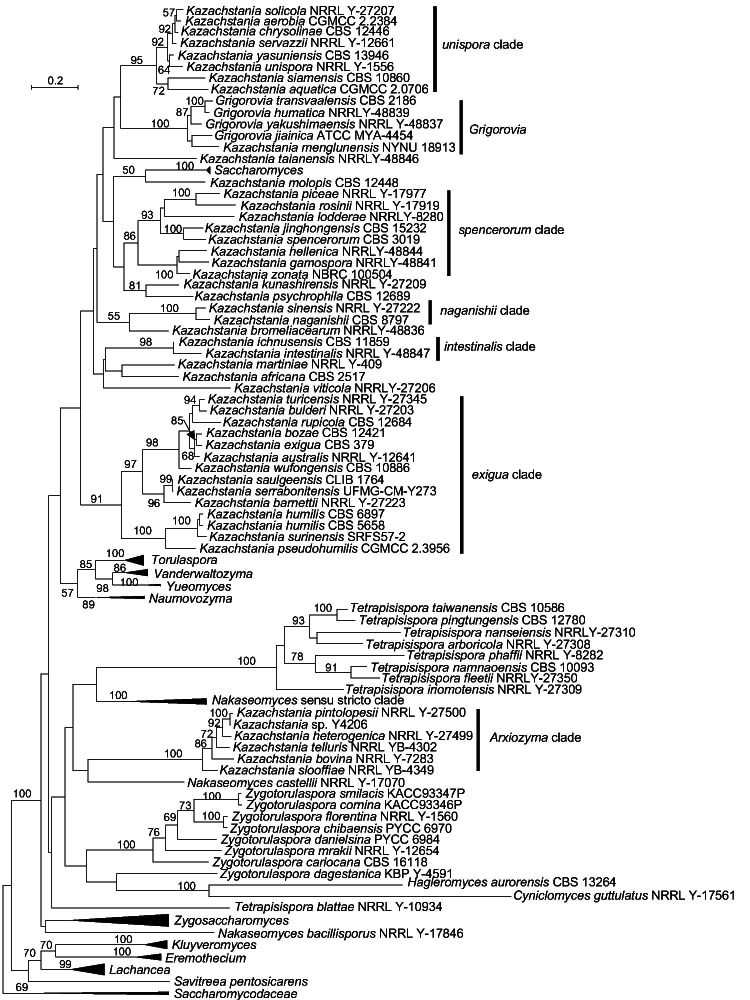
The genus Nakaseomyces included two clades, namely the bacillisporus and Nakaseomyces s.str. clades (Fig. 1). The bacillisporus clade contained Nakaseomyces bacillisporus and Nakaseomyces castellii, which formed a long branch distinct from the Nakaseomyces s.str. clade including the type species of Nakaseomyces, namely Nakaseomyces delphensis. Nakaseomyces bacillisporus and N. castellii clustered with the Nakaseomyces s.str. clade with moderate bootstrap support in the multigene analyses of Kurtzman (2003) and Kurtzman & Robnett (2003), but it is phylogenetically distinct from the Nakaseomyces s.str. clade in our multigene analysis (Fig. 2). This separation was also found in the single-gene analysis by Kurtzman (2003), Kurtzman & Robnett (2003) and Wu et al. (2008), and in the ITS and D1/D2 LSU-based analysis in this study (Fig. 3). These results suggest that the two species of the bacillisporus clade may represent one or two distinct genera.
Fig. 3.
Phylogenetic tree inferred using the concatenated ITS and D1/D2 sequences showing the phylogenetic relationship between genera in Saccharomycetaceae. Bootstrap percentages of maximum likelihood analysis over 50 % from 1 000 bootstrap replicates are shown on the major branches. Bar = 0.2 substitutions per nucleotide position.
The genus Kazachstania turned out to be heterogeneous in the phylogenomic analysis with the genus Grigorovia nested in this clade (Fig. 1; Shen et al. 2018, Opulente et al. 2023). Kurtzman (2003) revised Kazachstania to include members of Kazachstania and species that were previously classified in the genera Kluyveromyces and Saccharomyces, and it received moderate support in the multigene analysis. Although this clade was treated as a single genus by him, Kurtzman (2003) stated that the relationship of some species in this clade was phylogenetically unstable and that this clade might be resolved into three main lineages. In a consensus NJ network analysis using LSU, SSU, ITS, TEF1, RPB2, Sm rDNA and COX II sequences, the genus Kazachstania appeared as a single, but diverse lineage ( Wu et al. 2008). Using LSU sequences, Vaughan-Martini et al. (2011) showed that at least five separate clades occurred in the Kazachstania lineage, and she expected that new sister genera within this lineage could be recognized with the discovery of additional species. James et al. (2015) suggested that the Kazachstania unispora clade represents a separate genus in the Kazachstania lineage, and, in addition, a number of new sister genera would be created with further multigene analysis and additional species descriptions. Recently, Gouliamova & Dimitrov (2020) transferred Kazachstania humatica, Kazachstania jiainica, Kazachstania transvaalensis and Kazachstania yakushimaensis into Grigorovia, a newly created genus that was established based on an analysis of the combined ITS and LSU rDNA sequence similarities among Kazachstania species. The same authors also suggested the presence of four clades in Kazachstania, including Grigorovia. Our combined ITS and D1/D2 LSU rDNA sequence analysis supported Grigorovia as a distinct clade and, in addition, Kazachstania menglunensis clustered with Grigorovia (Fig. 3), which agrees with the results from Ke et al. (2019). The monotypic genus Arxiozyma accommodated the species Arxiozyma telluris ( Van der Walt & Yarrow 1984), but this species was transferred to Kazachstania as a new combination Kazachstania telluris ( Kurtzman 2003). However, it is phylogenetically positioned far away from the type species of Kazachstania (Kazachstania viticola) in the phylogenomic tree (Fig. 1) as well as in the multigene-based trees (Fig. 2; Kurtzman 2003, Kurtzman & Robnett 2003). Except Grigorovia and Arxiozyma, five clades, namely exigua, intestinalis, naganishii, spencerorum and unispora, received moderate to high support and seven well-separated lineages distinct from the one containing the generic type species K. viticola were observed in the ITS+D1/D2 LSU rDNA, the multigene, and the phylogenomics-based trees (Fig. 1–3). The Arxiozyma and unispora clades clustered together in the phylogenomic and multigene-based analyses, but they occurred distantly from each other in the ITS+D1/D2 LSU analysis (Fig. 3). The result agrees with the result of James et al. (2015) who suggested that the unispora clade represented a distinct genus in the Kazachstania lineage. Likely, and in agreement with all previous studies, the large and polymorphic genus Kazachstania needs to be revised.
Zygotorulaspora dagestanica clustered with C. guttulatus and H. aurorensis, and was found to be distinct from the genus Zygotorulaspora (with type Z. mrakii) in the ITS+D1/D2 LSU rDNA tree (Fig. 3). This species was placed as a basal long branch with other Zygotorulaspora species in the multigene analysis by Kachalkin et al. (2021), but the proper phylogenetic position of Z. dagestanica and Cyniclomyces can only be resolved when the genomes of these species become available.
Below we will explore some genome parameters that can be used to reclassify these and other yeast genera.
AAI analysis
The AAI values among species of the genera compared were as follows, viz., Eremothecium, 63.08–90.35 %; Grigorovia, 70.06–86.8 %; Kazachstania, 57.98–93.86 %; Kluyveromyces, 63.89–91.29 %; Lachancea, 61.75–89.7 %; Nakaseomyces, 57.34–96.91 %; Naumovozyma, 64.17–73.47 %; Saccharomyces, 82.41–92.71 %; Tetrapisispora, 57.1–83.97 %; Torulaspora, 68.08–91.18 %; Vanderwaltozyma, 64.16–71.58 %; Zygosaccharomyces, 71.68–95.57 % and Zygotorulaspora, 71.99–87.98 % (Table 2, S2). The estimated intergeneric AAI values between the above 13 genera were 54.49–69.16 % (Table S2).
Table 2.
List of the AAI, POCP, PAPO, CSIs and RED values of genera in Saccharomycetaceae.
| Genera or clades | AAI | POCP | PAPO (genus-specific genes) | CSIs | ITS similarity | D1/D2 similarity | REDa |
|---|---|---|---|---|---|---|---|
| Eremothecium | 63.08–90.35 % | 85.28–96.84 % | 44 | 4 | 81.1–97.5 % | 94–99.5 % | 0.90 |
| Grigorovia | 70.06–86.8 % | 80–93.51 % | 50 | 18 | 84.6–93.7 % | 96.5–99.5 % | \ |
| Kazachstania | 57.98–93.86 % | 60.38–97.64 % | 0 | 0 | 49–99.7 % | 86.2–99.8 % | 0.89 |
| Arxiozyma clade | 78.69–93.86 % | 86.35–97.41 % | 6 | 1 | 78.8–94.9 % | 93–97.3 % | \ |
| exigua clade | 68.35–93.46 % | 82.25–97.64 % | 4 | 0 | 66.8–99.7 % | 95.5–99.8 % | \ |
| intestinalis clade | 76.15 % | 96.19 % | 62 | 2 | 92.10 % | 98.60 % | \ |
| naganishii clade | 93.71 % | 95.33 % | 63 | 2 | 86.10 % | 99.10 % | \ |
| spencerorum clade | 63.77–76.12 % | 79.9–92.68 % | 2 | 0 | 69.5–87.7 % | 94.3–98.8 % | \ |
| unispora clade | 71.86–89.94 % | 87.79–95.2 % | 9 | 0 | 72.8–97.3 % | 95.7–99.8 % | \ |
| Kluyveromyces | 63.89–91.29 % | 86.63–97.26 % | 47 | 5 | 85.9–99.2 % | 95.8–100 % | 0.92 |
| Lachancea | 61.75–89.7 % | 80.91–97.71 % | 15 | 0 | 85–99.7 % | 95–99.6 % | 0.85 |
| Nakaseomyces | 57.34–96.91 % | 65.11–98.40 % | 0 | 0 | 58.4–97.1 % | 85.3–98.5 % | 0.89 |
| Nakaseomyces s.str. | 69.04–96.91 % | 89.19–98.40 % | 95 | 0 | 62.6–97.1 % | 91.9–98.5 % | \ |
| bacillisporus clade | 59.05 % | 72.68 % | 8 | 0 | 62 % | 90.70 % | \ |
| Naumovozyma | 64.17–73.47 % | 82.58–91.78 % | 21 | 3 | 73.6–77.6 % | 95.7–97.4 % | 0.92 |
| Saccharomyces | 82.41–92.71 % | 94.11–97.83 % | 57 | 7 | 96.8–99.9 % | 97.3–99.8 % | 0.97 |
| Tetrapisispora | 57.1–83.97 % | 62.77–96.25 % | 1 | 0 | 58.8–89.9 % | 80.5–99.1 % | \ |
| Tetrapisispora s.str. | 68.24–83.97 % | 90.74–96.25 % | 36 | 0 | 68.3–89.9 % | 90.3–99.1 % | 0.95 |
| Torulaspora | 68.08–91.18 % | 89.95–98.5 % | 2 | 0 | 83–99.6 % | 96.3–99.5 % | 0.93 |
| Vanderwaltozyma | 64.16–71.58 % | 83.34–92.44 % | 4 | 0 | 76.4–98.0 % | 95.6–99.6 % | \ |
| Zygosaccharomyces | 71.68–95.57 % | 91.5–98.59 % | 7 | 5 | 61.7–94.6 % | 86.4–99.8 % | 0.94 |
| Zygotorulaspora | \ | \ | \ | \ | 51.4–99.5 % | 88.7–98.6 % | \ |
| Zygotorulaspora s.str. | 71.99–87.98 % | 92.98–96.83 % | 6 | 1 | 71.6–99.5 % | 92.1–98.6 % | 0.95 |
adata from Li et al. (2021).
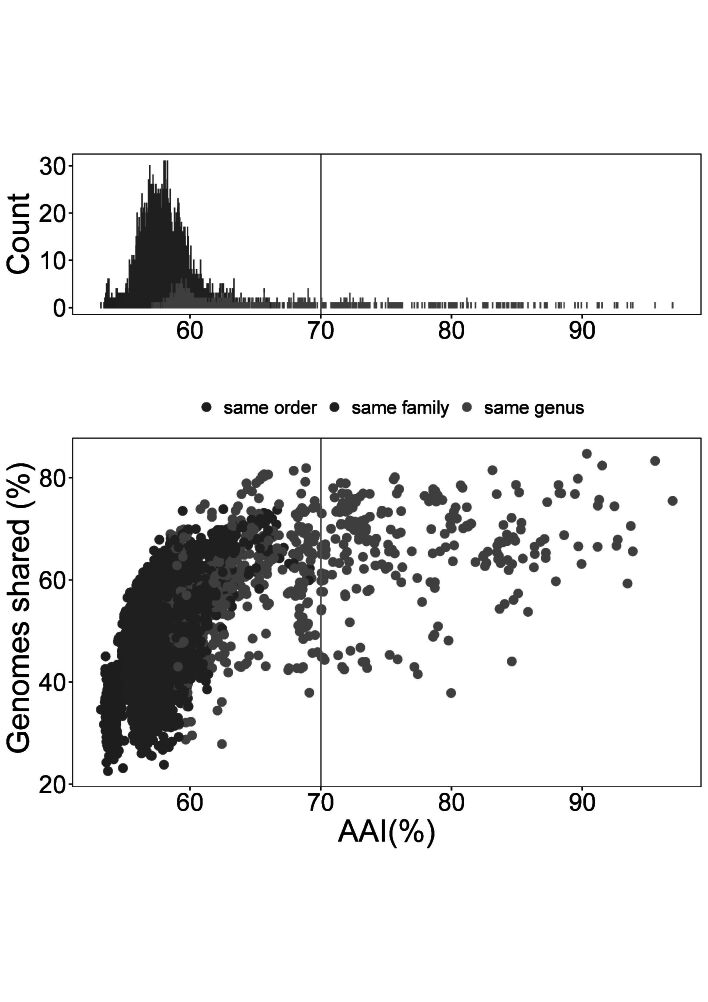
As indicated in the above described phylogenomic analyses Kazachstania, Nakaseomyces and Tetrapisispora were found to be heterogenic or polyphyletic, and they, thus, showed lower intrageneric AAI values than the other genera studied (Table 2, S2). When Nakaseomyces was divided into the bacillisporus and Nakaseomyces s.str. clades (Fig. 1), the AAI values of the bacillisporus clade were 59.05 %, which is still more divergent than observed in any of the other genera studied. On the contrary, the Nakaseomyces s.str. clade showed AAI values being in the range detected for the other genera, namely 69.04–96.91 % (Table 2, S2). Thus, the genomic heterogeneity of the Nakaseomyces is higher than in any other lineage of Saccharomycetaceae, and the bacillisporus clade might represent at least one new genus, or possibly two. The above indicated clades in the genus Kazachstania, i.e., Arxiozyma, exigua, intestinalis, naganishii, spencerorum and unispora showed AAI values of 78.69–93.86 %, 68.35–93.46 %, 76.15 %, 93.71 %, 63.77–76.12 % and 71.86–89.94 %, respectively (Table 2, S2). The Tetrapisispora s.str. clade, excluding T. blattae, displayed an AAI value range of 68.24–83.97 % (Table 2, S2). The analysis of the interrelationship between AAI and shared gene content (Fig. 4) showed that the inter-genus AAI values found were generally below 70 %, and the examples of lower AAI values were observed to occur among species from large heterogenic genera, like Kazachstania. A range of 60–70 % for the AAI values might be a good empirical value to distinguish between intrageneric and intergeneric relationships for genera in Saccharomycetaceae. The lower values as observed in the genera Kazachstania, Nakaseomyces and Tetrapisispora agree with earlier taxonomic views on these genera that already suggested that they need to be reclassified in the future.
Fig. 4.
Interrelationship between shared gene content and AAI values of 13 well-defined genera of Saccharomycetaceae. The X-axis displays the AAI similarity between strains. The Y-axis shows the rate of genome sharing between strains (the genome sharing rate = number of orthologous genes/the genes number in the minimum genome between two strains). Green dots indicate AAI values between strains of the same genus, but not of the same species. The blue dots indicate the AAI values between strains of the same family and different genera. The red dots indicate the AAI values of the same order and different family (Saccharomycetaceae and Saccharomycodaceae).
The applicability of AAI in Fungi is limited yet, but the available results are intriguing. Recently, Wibberg et al. (2021) revealed a 75 % AAI value as a threshold of intergeneric and interfamilial boundary in Hypoxylaceae (Ascomycota). For Ustilaginaceae (Basidiomycota), Ullmann et al. (2022) obtained a similar result as Wibberg et al. (2021), and an AAI value above 74.6 % was observed in the Ustilaginaceae when excluding Ustanciosporium gigantosporium and Ustilago xerochloae that are both species with a diploid genome. Although representing distant evolutionary lineages, a similar AAI threshold range of 60–80 % has been proposed to separate related but different genera of prokaryotes ( Luo et al. 2014, Rodriguez-R & Konstantinidis 2014). Specific AAI boundaries have been proposed for generic delineation in different bacterial families, for example, 71 % AAI in the family Methylococcaceae ( Orata et al. 2018), 70 % AAI in the family Methylothermaceae ( Skennerton et al. 2015), 80 % AAI in the family Rhodobacteraceae ( Wirth & Whitman 2018) and 64.6–77 % AAI in the family Geobacteraceae ( Xu et al. 2019). These examples may also implicate that specific AAI boundaries may occur in different groups of yeasts, and fungi in general as well.
POCP analysis
The following POCP results values were observed for the studied genera, viz., Eremothecium, 85.28 – 96.84 %; Grigorovia, 80–93.51 %; Kazachstania, 60.38–97.64 %; Kluyveromyces, 86.63–97.26 %; Lachancea, 80.91–97.71 %; Nakaseomyces, 65.11–98.40 %, Naumovozyma, 82.58–91.78 %; Saccharomyces, 94.11–97.83 %; Tetrapisispora, 62.77–96.25 %; Torulaspora, 89.95–98.5 %; Vanderwaltozyma, 83.34–92.44 %; Zygosaccharomyces, 91.5 – 98.59 % and Zygotorulaspora, 92.98–96.83 % (Table 2, S3). The intergeneric POCP values between those genera were 49.85–92.11 % (Table S3). The lower boundaries and broader ranges for POCP intrageneric values were observed for the genera Kazachstania, Nakaseomyces and Tetrapisispora, namely 60.38 %, 65.11 % and 62.77 %, respectively. As in the case of the above listed AAI values, lower POCP values than those observed for the remaining genera indicate once again excessive genetic heterogeneity in those three genera, thus suggesting that they might need to be restructured taxonomically.
The POCP values of Tetrapisispora s.str., excluding T. blattae, were in the range 90.74–96.25 %. The bacillisporus and Nakaseomyces s.str. clades of Nakaseomyces showed 72.68 % and 89.19–98.40 % POCP values, respectively. Like in the aforementioned AAI analysis, the bacillisporus clade had a rather low POCP value, which suggests that this clade is heterogeneous with respect to the POCP values, and needs to be revised. In the genus Kazachstania, the Arxiozyma, exigua, intestinalis, naganishii, spencerorum and unispora clades had POCP values of 80 % and higher, when analyzed separately, namely, 86.35–97.41 %, 82.25–97.64 %, 96.19 %, 95.33 %, 79.9–92.68 % and 87.79–95.2 %, respectively. The POCP analysis showed that there was some overlap between the observed intrageneric and intergeneric POCP values for species belonging to Kazachstania, Nakaseomyces and Tetrapisispora. While most genera in Saccharomycetaceae displayed POPC values within the range of 80–92 %, values of the same order of magnitude were observed for the various clades recognized among the three genera. If these genera will be taxonomically revised, the range in POCP values of 80–92 % may indicate the generic boundaries for these, and other genera belonging to Saccharomycetaceae.
To delimitate prokaryotic genera with POCP values, a 50 % boundary has been proposed as a genomic relatedness index ( Qin et al. 2014). However, for several prokaryotic families, e.g., Neisseriaceae and Rhodobacteraceae, it was found not to be an appropriate metric to delineate genera ( Aliyu et al. 2016, Li et al. 2017, Lopes-Santos et al. 2017, Orata et al. 2018, Wirth & Whitman 2018). The recent experience from filamentous fungi ( Wibberg et al. 2021, Ullmann et al. 2022) was not conclusive with regard to a common value or range of POCP values for delimitation of genera, but a 70 % POCP value was proposed to define families within Xylariales ( Wibberg et al. 2021).
PAPO analysis
Following the approach used by Takashima et al. (2019) to delimit genera in Trichosporonales, we tested the applicability of a PAPO analysis for the delimitation of genera in Saccharomycetaceae. The unique (genus-specific genes), core and pan proteins were determined based on the number of OrthoFinder OGs results (Table S4). Kazachstania, Nakaseomyces and Tetrapisispora contained zero, zero and one unique gene, respectively, whereas the other genera in the family had more than two genus-specific genes (Table 2). Taking into consideration the results of AAI and POPC analyses from above, we explored how the results of PAPO analyses will be impacted by changing the circumscription of these three heterogeneous genera. The exclusion of T. blattae from the genus Tetrapisispora increased the number of genus-specific genes from one to 36 in this genus. When evaluated separately, the bacillisporus and Nakaseomyces s.str. clades of Nakaseomyces contained 8 and 95 specific genes, respectively. The unique genes for the clades of the genus Kazachstania were as follows: Arxiozyma six, exigua four, intestinalis 62, naganishii 63, spencerorum two and unispora nine. These data additionally confirmed that Kazachstania, Nakaseomyces and Tetrapisispora in the present form contain species that are evolutionary too distantly related to be considered congeneric when compared to all other genera in Saccharomycetaceae.
Takashima et al. (2019) revised and characterized the genera in the Trichosporonales by using the PAPO analysis and comparing the phylogenomic analysis with the CoQ system present in these yeasts. The genus-specific genes analysis by Takashima et al. (2019) supported the delimitation of the genera Apiotrichum (with 24 specific genes) and Trichosporon (with 285 specific genes) that were recognized based on earlier multigene analyses ( Liu et al. 2015b), but they argued that the genus Cutaneotrichosporon (with only one specific gene) needed to be revised and they excluded C. guehoae from the genus. Consequently, two more monotypic genera, Pascua and Prillingera were described to accommodate those divergent line ages and to make the core of the genus Cutaneotrichosporon phylogenetically more homogeneous. Similar to the case of the genus Cutaneotrichosporon, our PAPO results indicated that the genera Kazachstania, Nakaseomyces and Tetrapisispora need to be revised to achieve a consistent delimitation of genera in Saccharomycetaceae.
CSIs analysis
A total of 43 conserved signature indels (CSIs) were identified from the 115 protein sequences in the seven genera listed below. Each CSIs had at least 4–5 conserved amino acids in the 40–50 amino acids adjacent to each other, either upstream or downstream. The CSIs characteristics corresponding to each clade are shown in Table 3. For Eremothecium four CSIs were found, for Grigorovia 18, for Kluyveromyces five, for Naumovozyma three, for Saccharomyces seven, for Zygosaccharomyces five and for Zygotorulaspora one (Table 2, 3). All sequence alignments of the clade-specific CSIs were provided in the supplemental data (Fig. S1). Kazachstania, Nakaseomyces and Tetrapisispora had zero CSIs, which agrees with the PAPO analysis that detected no genus-unique genes. The Arxiozyma, intestinalis and naganishii clades of the genus Kazachstania had one, two, and two CSIs, respectively (Table 2, 3) that are shown in the supplemental data (Fig. S1). In the genera Lachancea and Torulaspora, no CSIs were found, though all these genera were confidently supported by PAPO, POCP and AAI analyses.
Table 3.
Conserved signature indels specific for the genus Eremothecium, Grigorovia, Kluyveromyces, Naumovozyma, Saccharomyces, Zygosaccharomyces, Zygotorulaspora, Arxiozyma clade, intestinalis clade and naganishii clade.
| Serial number | Indel size | Indel position | Sequence alignments of CSIs |
|---|---|---|---|
| CSIs specific for the genus Eremothecium | |||
| 199929at4890 | 2 aa del | 705–760 | Fig. S1-1 |
| 30409at4890 | 1 aa ins | 1350–1399 | Fig. S1-2 |
| 41603at4890 | 1 aa ins | 1810–1850 | Fig. S1-3 |
| 36866at4890 | 3 aa ins | 3205–3245 | Fig. S1-4 |
| CSIs specific for the genus Grigorovia | |||
| 219278at4890 | 4 aa ins | 480–527 | Fig. S1-5 |
| 222386at4890 | 1 aa ins | 241–320 | Fig. S1-6 |
| 222386at4890 | 26 aa ins | 241–320 | Fig. S1-7 |
| 222386at4890 | 1 aa ins | 380–432 | Fig. S1-8 |
| 227580at4890 | 1 aa ins | 814–855 | Fig. S1-9 |
| 227580at4890 | 10 aa ins | 1160–1207 | Fig. S1-10 |
| 275358at4890 | 20 aa ins | 145–195 | Fig. S1-11 |
| 133107at4890 | 13 aa del | 633–679 | Fig. S1-12 |
| 139019at4890 | 3 aa ins | 1271–1310 | Fig. S1-13 |
| 148406at4890 | 3 aa del | 40–85 | Fig. S1-14 |
| 153452at4890 | 3 aa del | 70–113 | Fig. S1-15 |
| 306395at4890 | 2 aa del | 111–154 | Fig. S1-16 |
| 328156at4890 | 2 aa del | 195–240 | Fig. S1-17 |
| 60346at4890 | 12 aa ins | 3297–3375 | Fig. S1-18 |
| 60346at4890 | 3 aa ins | 3297–3375 | Fig. S1-18 |
| 17135at4890 | 6 aa ins | 820–870 | Fig. S1-19 |
| 39674at4890 | 1 aa ins | 2054–2106 | Fig. S1-20 |
| 41603at4890 | 8 aa ins | 1050–1090 | Fig. S1-21 |
| CSIs specific for the genus Kluyveromyces | |||
| 125930at4890 | 3 aa ins | 585–640 | Fig. S1-22 |
| 39674at4890 | 1 aa ins | 1946–1996 | Fig. S1-23 |
| 11957at4890 | 5 aa ins | 1303–1354 | Fig. S1-24 |
| 43781at4890 | 1 aa del | 2650–2700 | Fig. S1-25 |
| 41603at4890 | 3 aa del | 810–855 | Fig. S1-26 |
| CSIs specific for the genus Naumovozyma | |||
| 190878at4890 | 1 aa ins | 1320–1375 | Fig. S1-27 |
| 320265at4890 | 1 aa del | 410–470 | Fig. S1-28 |
| 320265at4890 | 1 aa ins | 460–500 | Fig. S1-29 |
| CSIs specific for the genus Saccharomyces | |||
| 230608at4890 | 1 aa ins | 1220–1780 | Fig. S1-30 |
| 235543at4890 | 1 aa ins | 945–1005 | Fig. S1-31 |
| 250301at4890 | 6 aa ins | 620–660 | Fig. S1-32 |
| 252424at4890 | 19 aa ins | 140–205 | Fig. S1-33 |
| 275223at4890 | 4 aa ins | 840–885 | Fig. S1-34 |
| 305650at4890 | 1 aa ins | 445–490 | Fig. S1-35 |
| 130793at4890 | 2 aa del | 280–330 | Fig. S1-36 |
| CSIs specific for the genus Zygosaccharomyces | |||
| 285587at4890 | 3 aa del | 180–240 | Fig. S1-37 |
| 344512at4890 | 1 aa del | 430–480 | Fig. S1-38 |
| 60152at4890 | 1 aa del | 714–751 | Fig. S1-39 |
| 85232at4890 | 1 aa del | 1600–1660 | Fig. S1-40 |
| 130323at4890 | 1 aa del | 1184–1231 | Fig. S1-41 |
| CSIs specific for the genus Zygotorulaspora | |||
| 25255at4890 | 1 aa ins | 426–481 | Fig. S1-42 |
| CSIs specific for the intestinalis clade | |||
| 199929at4890 | 1 aa ins | 663–720 | Fig. S1-43 |
| 230608at4890 | 1 aa ins | 1220–1278 | Fig. S1-44 |
| CSIs specific for the naganishii clade | |||
| 197945at4890 | 1 aa ins | 788–836 | Fig. S1-45 |
| 199929at4890 | 3 aa ins | 360–425 | Fig. S1-46 |
| CSIs specific for the Arxiozyma clade | |||
| 219278at4890 | 13 aa ins | 380–445 | Fig. S1-47 |
Genus-specific signature nucleotides of rDNA
Like CSIs, the ‘genus-specific signature nucleotides’ (GSNs) of the rDNA (LSU and SSU) were used as a molecular synapomorphy to distinguish different yeast genera ( Gueho et al. 1989, Kurtzman & Robnett 1991). We used their strategy to detect various GSNs of each genus in Saccharomycetaceae and found a region in the SSU rDNA (Fig. S2) that can distinguish those genera, except for the genera Kazachstania, Nakaseomyces, and the species T. blattae and Z. dagestanica. The GSNs in this region were as follows: ‘GA-T-T--TTCTTCGTGTACGGGA---------------------TC’ (Eremothecium), ‘A---T--CTTTCCGTGTACTGGTAT--------------------GCAACCGA’ (Grigorovia), ‘ATT-T--TATGTCGCGCACTGGTTT--------------------TCAACCGGAT’ (Kluyveromyces), ‘A-T-T—TTTTT(G) CGTGTACTGGA---------------------TC’ (Lachancea), ‘ATT--------------------CCAACCGGG’ (Naumovozyma), ‘ATT-------------------TCCAACGGGG’ (Saccharomyces), ‘CACGGAGGGCCGGTCC-GA---T--TATTTCGAGAACTGGA’ (Tetrapisispora s.str.), ‘A---T--TTTTTCGTGTACTGGTTT--------------------CC’ (Torulaspora), ‘CGGCCGGTCCGGA---T’ (Zygosaccharomyces) and ‘CCA(G)ACCGGGCCTT-TCCTTCTGGCTAACCTTGA(G)GTC-C-TTGT-GGCTCTT’ (Vanderwaltozyma), respectively. The Nakaseomyces s.str. clades of Nakaseomyces, and clades Arxiozyma, naganishii and unispora clades of Kazachstania had the following GSNs in this region: ‘AAT--------------------GCACCCGGGCCTT-TCCTTCTGGCTAACCCC-A’ (Nakaseomyces s.str. clade), ‘T--TTTTCCACGTACTGGGAT’ (Arxiozyma clade), ‘GCGTACTGGGAT’ (naganishii clade) and ‘CCACGTACTGGAAT--------------------GCAACCGGG’ (unispora clade). Our analysis did not reveal GSNs in SSU rDNA sequences for the other clades of Kazachstania, but they have at least one GSNs in the D1/D2 LSU rDNA (Table S5).
Barcode analysis for genus identification
The sequence analyses of the D1/D2 LSU and the ITS region of rDNA have been widely used for yeast identification and species delineation ( Kurtzman & Robnett 1998, Scorzetti et al. 2002). The pair-wise sequence similarities became a mainstream approach for identification of ascomycetous yeasts soon after Kurtzman & Robnett (1998) studied sequence variation in the D1/D2 LSU rDNA of c. 500 species of ascomycetous yeasts and compared the application of the Biological Species Concept with the amount of sequence divergence present in the D1/D2 LSU rDNA sequences (reviewed in Boekhout et al. 2021). It has been demonstrated that conspecific strains may differ by up to three nucleotide substitutions and distinct species by six or more substitutions of the approximate 600 nucleotides in LSU (i.e., roughly above 1 % divergence). This leaves a grey zone for the interpretation of four and five nucleotide differences. Despite some notable exceptions, the 1 % sequence divergence was often interpreted as the threshold for species delimitation in yeasts ( Boekhout et al. 2021, Lücking et al. 2021), but no cut-off value was ever suggested for generic borders. The analysis of more than 8 500 barcode sequences generated at the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands, demonstrated that species belonging to the same genus, as accepted at that time, can be correctly identified with the highest confidence at the genus level with a sequence similarity level of 93.7 % in ITS and 98.9 % in D1/D2 LSU rDNA sequences ( Boekhout et al. 2021). The observed quality of identification (confidence, F-value) of basidiomycetous yeasts was generally higher than that of ascomycetous yeasts. The confidence of identification for ascomycetous yeasts remained largely in a narrow range of 0.68 to approximately 0.5 (F-measure) for ITS with a flat distribution within a 90–98 % sequence similarity range, whereas the confidence of identification for D1/D2 LSU rDNA sequences showed a pronounced unimodal distribution (see Boekhout et al. 2021: f. 2). The results indicated the importance of the quality of taxonomic classifications for reliable genus identification using barcode sequences. The noted more reliable identification of basidiomycetous yeasts (i.e., higher confidence values) was explained by the recent re-classification of large, polyphyletic genera (Cryptococcus and Rhodotorula among others) that resulted in more homogeneous inter- and intrageneric distances. On the contrary, several clades of ascomycetous yeasts and the genus Candida in particular had heterogeneous sequences that substantially decreased the reliability of identification at the generic level ( Boekhout et al. 2021).
To address the reliability of those two barcodes for yeast genus identification in Saccharomycetaceae, a pairwise similarity comparison using the EMBOSS water alignment tool ( Madeira et al. 2019, Li et al. 2020) was performed. The sequence similarities in the LSU and the ITS region (ITS results in brackets) among the different genera studied were Eremothecium 94–99.5 % (81.1–97.5 %), Grigorovia 96.5–99.5 % (84.6–93.7 %), Kazachstania 86.2–99.8 % (49–99.7 %), Kluyveromyces 95.8– 100 % (85.9–99.2 %), Lachancea 95–99.6 % (85–99.7 %), Nakaseomyces 85.3–98.5 % (58.4–97.1 %), Naumovozyma 95.7–97.4 % (73.6–77.6 %), Saccharomyces 97.3–99.8 % (96.8–99.9 %), Tetrapisispora 80.5–99.1 % (58.8–89.9 %), Torulaspora 96.3–99.5 % (83–99.6 %), Vanderwaltozyma 95.6–99.6 % (76.4–98 %), Zygosaccharomyces 86.4–99.8 % (61.7–94.6 %) and Zygotorulaspora 88.7– 98.6 % (51.4 – 99.5 %) (Table 2, S6, S7). The sequences similarity of D1/D2 LSU and ITS rDNA sequences in the genus Saccharomyces, namely 97.3–99.8 % and 96.8–99.9 %, respectively, were in the optimal range of sequence similarity to identify ascomycetous yeasts at the genus level as predicted by Vu et al. (2016) and re-assessed in Boekhout et al. (2021). However, the sequence similarity values of those two barcodes were lower in the other genera when compared to the predicted most optimal values of 98.9 % and 93.7 % in the D1/D2 LSU and ITS regions, respectively. These results indicate a limited applicability of generalized thresholds or cut-off values for ascomycetous yeasts even in the family Saccharomycetaceae which contains well-circumscribed and generally accepted genera. The genera Kazachstania, Nakaseomyces, Tetrapisispora and Zygotorulaspora had values below 90 % and 60 % intrageneric sequence similarity in the D1/D2 LSU and ITS rDNA sequences, respectively. The Nakaseomyces s.str. clade, the Arxiozyma, exigua, intestinalis, naganishii, spencerorum and unispora clades of the genus Kazachstania, Tetrapisispora s.str. and Zygotorulaspora s.str. had up to 90 % intrageneric sequence similarity in the D1/D2 LSU rDNA, and most of them had up to 70 % sequence similarity in the ITS region of rDNA (Table 2). The analysis of the two DNA barcodes was consistent with results obtained with the other genomic tools studied as described above. Again, the DNA-barcode analysis indicated that those four genera are likely too heterogeneous to be considered one genus, and should be revised. The genus Zygosaccharomyces has a low intrageneric ITS sequence similarity, but the D1/D2 LSU sequence similarity was above 85 % and the other analyses described above supported that this is a well-defined genus.
Phenotypic characteristics analysis
Recognition of yeast genera using phenotypic properties is not an easy task due to the limited morphological characteristics as well as the restricted number of physiological characteristics that consistently differ among species belonging to Saccharomycetaceae ( Kurtzman 2003). New identification tools and the growing knowledge of yeast biology and genetics changed the views on the composition and circumscription of several old genera, like Kluyveromyces, Saccharomyces and Zygosaccharomyces ( Kurtzman 2003, Lachance 2011). The revision undertaken by Kurtzman (2003) resulted in the subdivision of the members of the Saccharomycetaceae into existing, reinstated and newly proposed genera creating the taxonomy that is still in use today. The complicated history of the circumscription and demarcation of genera in the family resulted from many variable features within the genera, be it morphological (ascus formation, ascospore number and shape), biochemical (coenzyme Q system, cell wall carbohydrates) and physiological characters. Here we compared the phenotypic data of some sister genera with the aim to search for morphological or physiological synapomorphies for their application in generic definitions in Saccharomycetaceae. The genus Eremothecium differs from other members of Saccharomycetaceae in its formation of fusiform or acicular (needle-shaped) ascospores ( Kurtzman & De Hoog 2011). All species of Eremothecium assimilate glycerol, malt-ose and sucrose (Table 4). The sister genera Kluyveromyces and Lachancea cannot be distinguished by morphology, but differ in the assimilation of nitrogen sources ethylamine and cadaverine (Table 4). Torulaspora, Zygosaccharomyces and Zygotorulaspora are sister genera and are difficult to recognize by morphology. However, they can be distinguished by some physiological tests. All species of Zygotorulaspora can ferment raffinose, whereas all members of Zygosaccharomyces cannot. Zygotorulaspora species grow well with 0.1 % cycloheximide, but Torulaspora species do not. Yueomyces, Tetrapisispora and Vanderwaltozyma formed a well-supported lineage in our study (Fig. 1). Yueomyces is characterized by bipolar budding, but is phylogenetically separated from Hanseniaspora, Nadsonia, Saccharomycodes and Wickerhamia that have a similar budding morphology as Yueomyces ( Wang et al. 2015c). The genus Yueomyces is also characterized by its inability to utilize ammonium which is usually a favorable nitrogen source for other yeast species (Yu et al. 2023). Tetrapisispora and Vanderwaltozyma have similar morphology, but all species of the former genus do not grow with 0.1 % cycloheximide and raffinose, whereas the members of the latter have variable utilization of those sources. Saccharomyces differs from its sister genus Nakaseomyces s.str. by fermentation of sucrose and assimilation of raffinose (Table 4) and forms globose to short ellipsoid ascospores. The genera Grigorovia, Kazachstania and Naumovozyma clustered together in the molecular phylogenies and have no distinct phenotypic characteristics.
Table 4.
The phenotypic characteristics of different genera and clades in Saccharomycetaceae.
| Taxon | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Eremothecium | v | v | v | + | v | v | v | + | + | v | v | n | n | n | n | n |
| Kluyveromyces | v | v | v | v | v | + | v | v | v | v | v | + | + | + | v | v |
| Lachancea | v | v | v | v | v | v | v | v | v | v | v | v | v | + | v | v |
| Saccharomyces | v | + | v | v | + | v | v | v | v | v | – | – | – | v | n | n |
| Nakaseomyces bacillisporus | – | v | – | n | v | – | – | – | + | – | – | – | – | – | – | – |
| Nakaseomyces s.str. | – | – | – | – | – | – | v | – | v | – | – | – | – | – | – | – |
| Naumovozyma | + | – | – | v | – | + | v | – | v | – | – | – | – | – | v | – |
| exigua clade | v | v | v | v | v | v | + | v | v | v | – | v | v | v | v | v |
| spencerorum clade | v | v | v | v | v | v | v | v | v | v | v | v | v | v | v | v |
| Arxiozyma clade | – | – | – | – | – | – | – | – | – | – | – | n | n | n | n | n |
| Grigorovia clade | + | – | – | – | – | + | v | – | – | – | – | v | v | v | v | v |
| unispora clade | + | – | – | v | v | v | v | – | v | v | – | v | v | v | v | v |
| naganishii clade | + | v | + | v | + | v | + | – | v | – | – | v | v | v | – | – |
| intestinalis clade | v | v | n | v | v | + | v | v | v | v | – | – | v | – | n | + |
| Torulaspora | v | v | v | v | v | v | v | v | v | v | v | – | n | n | v | – |
| Zygosaccharomyces | – | v | – | v | v | v | v | v | + | v | v | n | n | n | n | v |
| Zygotorulaspora | v | + | + | + | + | v | v | v | v | + | + | n | n | n | + | + |
| Vanderwaltozyma | v | v | v | v | v | + | v | – | v | – | – | – | – | – | v | v |
| Tetrapisispora | v | – | – | v | – | + | v | – | v | – | – | – | – | – | v | – |
| Yueomyces | + | – | – | – | – | + | – | – | – | – | – | – | – | n | – | – |
1: Fermentation of galactose; 2: fermentation of sucrose; 3: fermentation of raffinose; 4: sucrose; 5: raffinose; 6: galactose; 7: trehalose; 8: maltose; 9: glycerol; 10: D-mannitol; 11: D-glucitol; 12: cadaverine; 13: L-lysine; 14: ethylamine; 15: 0.01 % cycloheximide; 16: 0.1 % cycloheximide.
Abbreviations: + = positive; &x2013; = negative; v = variable; n = not available.
Monophyly, thresholds (cut-off values) and synapomorphies
A genus usually comprises genetically and phylogenetically closely related organisms that form a monophyletic clade and that are characterized by shared-derived characters (viz., synapomorphies) resulting from their shared evolutionary history ( Hennig 1966, Kitching et al. 1998, Wiley & Lieberman 2011). This is also true for fungi. Yeasts are predominantly single-cell fungi with limited morphological characteristics to distinguish different taxa morphologically. Taxa for which no sexual reproduction is documented show, by default, less morphological characters, and, hence, physiological tests largely replaced morphological characters for the classification and identification of asexually reproducing yeasts, certainly at the species level. However, many of the traditionally assigned yeast genera based on such phenotypic (viz., morphology and physiology) criteria appeared to be polyphyletic. Candida is a prime example of this. Although the recognition of smaller, monophyletic genera based on single-gene and multi-gene phylogenies reduced the taxonomic heterogeneity of those previous polyphyletic genera (discussed in Boekhout et al. 2021), in other cases the topology of the phylogenetic trees based on single or multi-gene phylogeny remained unstable ( Kurtzman 2003, 2011, Kurtzman & Robnett 2013) and the phylogenetic recognition of genera proved to be efficient only in well-resolved lineages. For example, several species rich genera like Kazachstania and Metschnikowia remained taxonomically unresolved because their internal phylogeny and delimitation clades, sub-clades and lineages were found to differ depending on the dataset used. For some pragmatic reason, distantly related single-species lineages were sometimes merged with already existing genera, e.g., Tetrapisispora, Torulaspora, Zygotorualspora, rather than accommodating them in new genera. Therefore, the analysis of more robust data sets, such as the phylogenetic analysis of high-quality whole genome data, can improve the taxonomy of several hitherto large and polyphyletic genera, such as Kazachstania, Nakaseomyces, Tetrapisispora and Candida, in order to enhance the accuracy of fungal identification, including yeasts. Our phylogenomic analyses indicated that three genera in the family Saccharomycetaceae, namely Kazachstania, Nakaseomyces and Tetrapisispora are genetically more heterogeneous than other genera of the family. Six clades, namely Arxiozyma, exigua, intestinalis, naganishii, spencerorum and unispora, may be separated from the core of the genus Kazachstania. The bacillisporus clade of the genus Nakaseomyces may represent one or two genus-level taxa. Similarly, T. blattae is a species phylogenetically located far away from other species of Tetrapisispora, including the generic type T. phaffii. It must be noted that the phylogenetic heterogeneity of these genera was well-documented in earlier studies that utilized rDNA sequences and multi-gene datasets ( Kurtzman & Robnett 1998, Kurtzman 2003). The phylogenomic tree obtained in the present study (Fig. 1) was different from the 6-gene-based tree (Fig. 2) regarding the position of Nakaseomyces and Hagleromyces. Although the cluster of Nakaseomyces, Hagleromyces and Cyniclomyces received no support in the 6-gene-based tree, the genus Hagleromyces was placed with strong statistical support as an early branching taxon in a clade with Torulaspora, Zygotorulaspora and Zygosaccharomyces. The phylogenetic position of Torulaspora close to Yueomyces and Vanderwaltozyma was only observed in the ITS-LSU rDNA-based tree (Fig. 3), but not in other trees (Fig. 1, 2). Our phylogenomic analyses provided a better placement and support for those genera, but several limitations of such phylogenomic analysis still occur with the main shortcoming to deciding as objectively as possible where to split the well-supported monophyletic clade from closely related clades at genus-level or higher ranks. In our view, the above-described genomics-based metrics may help to realize such a more objective classification of yeasts and other fungi.
The sequence identity thresholds (or cut-off values) for rDNA-barcode have been applied to identify or define the species, e.g., by applying the ‘1 % rule’ for ascomycetous yeasts. Delimitation of genera based on rDNA-barcode, i.e., sequence similarity and barcoding gap, was never applied to yeasts. In a pragmatic taxonomy, the Phylogenetic Genus Concept defined genera as monophyletic lineages, but irrespective of the size and genetic heterogeneity in these groups. Estimations of rDNA-barcode variability by Vu et al. (2016) and Boekhout et al. (2021) predicted that a random yeast sequence can be assigned to a genus with the best confidence at sequence similarity of 96.31 % (updated 93.7 %) for the ITS region and 97.11 % (updated 98.9 %) for the D1/D2 LSU rDNA sequences, respectively. Our case-by-case sequence similarity analyses (Table 2) showed that most genera have lower intrageneric ITS and LSU values than the above thresholds. It is important to mention that in the analysis by Boekhout et al. (2021), the confidence level of yeast identification with ITS sequences was very broad, suggesting a high level of ITS sequence heterogeneity in the currently recognized genera of ascomycetous yeasts. Next to this, ITS length polymorphism and intragenomic heterogeneity are well documented for Saccharomycotina (e.g., Boekhout et al. 2021, Lücking et al. 2021). Thus, finding one reliable cut-off for all yeasts, ascomycetous yeasts only, and even the single family Saccharomycetaceae, is unlikely.
Recently, indices based on genomic relatedness, such as the AAI and POCP approaches, have been used to delimit generic boundaries for Bacteria and Archaea ( Luo et al. 2014, Qin et al. 2014, Rodriguez-R & Konstantinidis 2014, Varghese et al. 2015, Parks et al. 2018, Hayashi Sant’Anna et al. 2019, Barco et al. 2020, Nouioui & Sangal 2022). We used genome data from 13 hitherto well-accepted genera of Saccharomycetaceae to test the applicability of these two approaches for fungi, taking yeasts as an example. Our result showed that the genera Eremothecium, Grigorovia, Naumovozyma, Saccharomyces, Torulaspora, Vanderwaltozyma, Zygosaccharomyces and Zygotorulaspora were well circumscribed, but that the genera Kazachstania, Nakaseomyces and Tetrapisispora were poorly delimited with lower AAI and POCP values than present in the other genera.
The relative evolutionary divergence (RED) approach, like the method using divergence times as a ranking criterion ( Tedersoo et al. 2018) and the phylogenetic rank boundary optimisation (PRBO) method to measures the divergence of each lineage ( Liu et al. 2015a), which normalize the inferred phylogenetic distances to reflect evolutionary divergence, has been used in the taxonomy of prokaryotes ( Parks et al. 2018, Rinke et al. 2021). Recently, the RED approach was applied to Fungi, including yeast taxa ( Li et al. 2021, Groenewald et al. 2023). Li et al. (2021) calculated the RED values of different ranks of Fungi based on a phylogenomic data matrix of 290 genes from the genomes of 247 genera distributed in 6 phyla, 14 classes, 41 orders and 90 families. The results showed that about 85 % of ranks were well-defined, and they fell within ± 0.1 of the median RED value for taxa at that rank, which indicated that those ranks had comparable levels of evolutionary divergence. Nearly 40 % (1 order, 5 families and 16 genera) of the under-classified ranks belonged to Saccharomycotina that included until recently only one class Saccharomycetes and one order Saccharomycetales ( Kurtzman et al. 2011). Groenewald et al. (2023) revised the taxonomy of Saccharomycetales and split them into seven classes and 12 orders mostly based on the RED approach. The following RED values for the genera calculated by Li et al. (2021) were considered in our study: Eremothecium 0.902, Kazachstania 0.89, Kluyveromyces 0.92, Lachancea 0.846, Nakaseomyces 0.886, Naumovozyma 0.921, Saccharomyces 0.97, Tetrapisispora s.str. 0.949, Torulaspora 0.934, Zygosaccharomyces 0.942 and Zygotorulaspora s.str. 0.948. According to these results, the genera Eremothecium, Kazachstania, Kluyveromyces, Lachancea, Nakaseomyces and Naumovozyma were under-classified corresponding rather to family level ( Li et al. 2021), but the PAPO, POCP and AAI analyses supported that Eremothecium, Kluyveromyces, Lachancea and Naumovozyma are well-defined genera.
A synapomorphy is a derived trait that has evolved within evolutionary-related organisms as shown by phylogenetics and that is not present in any other species outside that group ( Bern et al. 2006). ‘Signature sequences’ as unambiguous molecular synapomorphies have been used to assign species to genera. For example, Gueho et al. (1989) and Kurtzman & Robnett (1991) identified ‘genus-specific signature nucleotides’ (GSNs) of SSU and LSU rDNA to distinguish Sterigmatomyces and Fellomyces, and Saccharomyces and Debaryomyces, respectively. Our results showed that the well-accepted genera in the Saccharomycetaceae have one or more GSNs in the SSU and LSU rDNA regions (Table S5). With an increasing number of available fungal (including yeasts) genomes, comparative genomic analyses of protein signatures, such as CSIs, may reveal specific molecular markers belonging to different higher taxa (e.g., genus level and above), and this approach has been proven useful for bacterial evolutionary and systematic studies ( Gupta 2014, 2016, Gupta & Suggett 2022). Our study showed that this approach is potentially useful to demarcate genera among yeasts.
CONCLUSION
Our results showed that comparative phylogenomic analyses may be an improvement to more objectively address generic boundaries within and between genera of Saccharomycetaceae. The application of the tested tools and metrics may be an important step towards a pragmatic approach for the delimitation of yeast genera in yet unresolved lineages. The genomic metrics, including POCP, AAI, PAPO and CSIs, are robust approaches to delimit the yeast genera in Saccharomycetaceae and may detect genetic heterogeneity within a priori defined monophyletic group. Our results demonstrated that the range of 80–92 % POCP values and a range of 60–70 % of AAI are likely good criteria to define thresholds for a pragmatic genome-based discrimination of genera in Saccharomycetaceae. The ranges were observed in all genera of Saccharomycetaceae studied, except three. The genera Kazachstania, Nakaseomyces and Tetrapisispora were already known to be heterogeneous when compared to other genera of Saccharomycetaceae, and will be revised below to optimize the generic taxonomy analogous to the high-rank classification proposed by Groenewald et al. (2023). Based on our results, we propose that the combined monophyly-based phylogenomic analysis and genomic relatedness indices and synapomorphies should be tested in other groups of yeasts to explore the more general applicability of these parameters for a broader range of taxa in an attempt to move toward a more general taxogenomics approach ( Libkind et al. 2020).
For pragmatic reasons, the following six single-species lineages of Kazachstania, namely K. bromeliacearum, K. kunashirensis, K. martiniae, K. molopis, K. psychrophila and K. taianensis, will not yet be considered for reclassification, as discovery of more related species may provide a more convincing taxonomic case. Because the application of the tested metrics is ultimately dependent on the taxon sampling and is based on similarity, the discovery of new species may change the degree of relatedness as estimated by the genomic metrics. These species are listed as pro tempore in the genus Kazachstania, as proposed before for several basidiomycetous yeast species, e.g., some described within the genus Pseudozyma, but that likely represent other genera that need further confirmation by adding more species and markers ( Wang et al. 2015a).
TAXONOMY
Based on the phylogenomic and genome-based metrics analyses, the genus Arxiozyma was reinstated and eight new genera and 41 new combinations were proposed below, which were listed in alphabetical order.
Arxiozyma Van der Walt & Yarrow, S. African J. Bot. 3: 341. 1984 — MycoBank MB 25498
Type species. Arxiozyma telluris (Van der Walt) Van der Walt & Yarrow, S. African J. Sci. 3: 341. 1984.
Basionym. Saccharomyces telluris Van der Walt (as ‘tellustris’), Antonie van Leeuwenhoek 23: 27. 1957.
Synonym. Kazachstania telluris (Van der Walt) Kurtzman, FEMS Yeast Res. 4: 239. 2003.
Arxiozyma bovina (Kurtzman & Robnett) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851335
Basionym. Kazachstania bovina Kurtzman & Robnett, J. Clin. Microbiol. 43: 105. 2005.
Arxiozyma heterogenica (Kurtzman & Robnett) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851283
Basionym. Kazachstania heterogenica Kurtzman & Robnett, J. Clin. Microbiol. 43: 107. 2005.
Arxiozyma pintolopesii (Kurtzman et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851284
Basionym. Kazachstania pintolopesii Kurtzman et al., J. Clin. Microbiol. 43: 108. 2005.
Arxiozyma slooffiae (Kurtzman & Robnett) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851285
Basionym. Kazachstania slooffiae Kurtzman & Robnett, J. Clin. Microbiol. 43: 109. 2005.
Cylindricascospora Q.M. Wang, Yurkov & Boekhout, gen. nov. — MycoBank MB 851287
Etymology. The genus is named based on the shape of ascospores, cylindrical to bacilliform.
Type species. Cylindricascospora bacillispora (Lachance et al.) Q.M. Wang, Yurkov & Boekhout.
This genus is proposed for the species Nakaseomyces bacillisporus, which formed a separate branch closely related to Nakaseomyces castellii (Fig. 1). It is a member of the Saccharomycetaceae (Saccharomycetales). The genus is mainly circumscribed by phylogenomic and AAI analyses, and phenotypic characteristics (see below).
Multilateral budding on a narrow base. Asci arise directly from diploid cells. Four, occasionally six to eight, cylindrical to bacilliform ascospores are formed. The spores are liberated from the ascus and tend to agglutinate (Lachance 2011). Colonies are butyrous, glabrous, and white. Hyphae and pseudohyphae not formed. The major ubiquinone is Q-6.
Notes — Ascus and ascospore formation is not known in the related N. castellii (reclassified in Oligophagozyma, see below).
Cylindricascospora bacillispora (Lachance et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851288
Basionym. Kluyveromyces bacillisporus Lachance et al., Int. J. Syst. Bacteriol. 43: 116. 1993.
Synonym. Nakaseomyces bacillisporus (Lachance et al.) Kurtzman, FEMS Yeast Res. 4: 240. 2003.
Grigorovia menglunensis (T. Ke et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851286
Basionym. Kazachstania menglunensis T. Ke et al., Int. J. Syst. Evol. Microbiol. 69: 3625. 2019.
Henningerozyma Q.M. Wang, Yurkov & Boekhout, gen. nov. — MycoBank MB 851289
Etymology. The genus is named in honour of W. Henninger for his contribution to yeast taxonomy.
Type species. Henningerozyma blattae (Henninger & Windisch) Q.M. Wang, Yurkov & Boekhout.
This genus is proposed for the species Tetrapisispora blattae, which formed a separate branch closely related to Yueomyces (Fig. 1). It is a member of the Saccharomycetaceae (Saccharomycetales). The genus is mainly circumscribed by phylogenomic analysis and phenotypic characteristics (see below).
Asci arise directly from diploid cells. One to eight or more spherical to ellipsoid ascospores are formed. The spores are liberated from the ascus soon after formation and tend to agglutinate (Vaughan-Martini et al. 2011). Colonies white to cream, butyrous, glossy. Multilateral budding cells present. Hyphae and pseudohyphae not formed. The major ubiquinone is Q-6.
Notes — The related genus Yueomyces differs from Henningerozyma by bipolar budding.
Henningerozyma blattae (Henninger & Windisch) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851290
Basionym. Kluyveromyces blattae Henninger & Windisch, Arch. Mikrobiol. 109: 155. 1977.
Synonym. Tetrapisispora blattae (Henninger & Windisch) Kurtzman, FEMS Yeast Res. 4: 241. 2003.
Huiozyma Q.M. Wang, Yurkov & Boekhout, gen. nov. — MycoBank MB 851292
Etymology. The genus is named in honour of F.L. Hui for his contribution to yeast taxonomy.
Type species. Huiozyma naganishii (Mikata et al.) Q.M. Wang, Yurkov & Boekhout.
This genus is proposed for species of the naganishii clade, which formed a separate lineage in Kazachstania as previously defined (Fig. 1, 3). It is a member of the Saccharomycetaceae (Saccharomycetales). The genus is mainly circumscribed by phylogenomic and genome metrics-based analyses.
Budding cells transform directly into asci containing one to four globose, subglobose or cylindrical ascospores. Colonies cream to tan, butyrous. Multilateral budding cells present. Hyphae and pseudohyphae not formed. The major ubiquinone is Q-6.
Huiozyma naganishii (Mikata et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851293
Basionym. Saccharomyces naganishii Mikata et al., Int. J. Syst. Evol. Microbiol. 51: 2191. 2001.
Synonym. Kazachstania naganishii (Mikata et al.) Kurtzman, FEMS Yeast Res. 4: 238. 2003.
Huiozyma sinensis (M.X. Li et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851295
Basionym. Kluyveromyces sinensis M.X. Li et al., Acta Microbiol. Sin. 30: 96. 1990.
Synonym. Kazachstania sinensis (M.X. Li et al.) Kurtzman, FEMS Yeast Res. 4: 238. 2003.
Jamesozyma Q.M. Wang, Yurkov & Boekhout, gen. nov. — MycoBank MB 851297
Etymology. The genus is named in honour of S.A. James for his contribution to yeast taxonomy.
Type species. Jamesozyma piceae (G. Weber & Spaaij) Q.M. Wang, Yurkov & Boekhout.
This genus is proposed for the species in the spencerorum clade of Kazachstania as previously defined, which formed a separate lineage closely related to the exigua clade (Fig. 1, 3). It is a member of the Saccharomycetaceae (Saccharomycetales). The genus is mainly circumscribed by phylogenomic and genome metrics-based analyses.
Asci containing one or two, sometimes up to four, globose to ovoid ascospores (Vaughan-Martini et al. 2011, Jacques et al. 2016). Colonies white to cream, butyrous. Multilateral budding cells present. Hyphae not produced. Pseudohyphae present or not. The major ubiquinone is Q-6.
Jamesozyma gamospora (Imanishi et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851298
Basionym. Kazachstania gamospora Imanishi et al., FEMS Yeast Res. 7: 336. 2007.
Jamesozyma hellenica (Nisiotou & Nychas) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851299
Basionym. Kazachstania hellenica Nisiotou & Nychas, Int. J. Syst. Evol. Microbiol. 58: 1265. 2008.
Jamesozyma jinghongensis (F.L. Hui & L.N. Huang) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851300
Basionym. Kazachstania jinghongensis F.L. Hui & L.N. Huang, Int. J. Syst. Evol. Microbiol. 69: 3625. 2019.
Jamesozyma lodderae (Van der Walt & Tscheuschner) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851301
Basionym. Saccharomyces lodderae Van der Walt & Tscheuschner (as ‘lodderi’), Antonie van Leeuwenhoek 23: 188. 1957.
Synonym. Kazachstania lodderae (Van der Walt & Tscheuschner) Kurtzman, FEMS Yeast Res. 4: 238. 2003.
Jamesozyma piceae (G. Weber & Spaaij) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851302
Basionym. Kluyveromyces piceae G. Weber & Spaaij, Antonie van Leeuwenhoek 62: 240. 1992.
Synonym. Kazachstania piceae (G. Weber & Spaaij) Kurtzman, FEMS Yeast Res. 4: 238. 2003.
Jamesozyma rosinii Vaughan-Mart., Barcaccia & Pollacci ex Q.M. Wang, Yurkov & Boekhout, sp. nov. — MycoBank MB 851303
Holotype. CBS 7127, preserved in a metabolically inactive state at Westerdijk Institute.
For a detailed description see Vaughan-Martini et al., Int. J. Syst. Bacteriol. 46: 616. 1996.
Notes — Originally described as Saccharomyces rosinii Vaughan-Mart., Barcaccia & Pollacci, Int. J. Syst. Bacteriol. 46: 616. 1996, nom. inval., Art. 40.7 (Shenzhen) and Kazachstania rosinii Vaughan-Mart. ex Kurtzman, FEMS Yeast Res. 4: 238. 2003, nom. inval., Arts 40.1 (Melbourne).
Jamesozyma spencerorum Vaughan-Mart. ex Q.M. Wang, Yurkov & Boekhout, sp. nov. — MycoBank MB 851304
Holotype. CBS 3019, preserved in a metabolically inactive state at Westerdijk Institute.
For a detailed description see Vaughan-Martini, Antonie van Leeuwenhoek 68: 116. 1995.
Notes — Originally described as Saccharomyces spencerorum Vaughan-Mart., Antonie van Leeuwenhoek 68: 116. 1995, nom. inval., Art. 40.7 (Shenzhen) and Kazachstania spencerorum Vaughan-Mart. ex Kurtzman, FEMS Yeast Res. 4: 238. 2003, nom. inval., Art. 40.1 (Melbourne).
Jamesozyma zonata (Imanishi et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851305
Basionym. Kazachstania zonata Imanishi et al., FEMS Yeast Res. 7: 335. 2007.
Maudiozyma Q.M. Wang, Yurkov & Boekhout, gen. nov. — MycoBank MB 851306
Etymology. The genus is named in honour of Maudy Th. Smith for her contribution to yeast taxonomy.
Type species. Maudiozyma humilis (E.E. Nel & Van der Walt) Q.M. Wang, Yurkov & Boekhout.
This genus is proposed for the species in the exigua clade of Kazachstania as previously defined, which formed a well-supported lineage closely related to spencerorum clade, K. kunashirensis and K. psychrophila (Fig. 1, 3). It is a member of the Saccharomycetaceae (Saccharomycetales). The genus is mainly circumscribed by phylogenomic and genomic metrics-based analyses.
Asci contain one to four globose to ovoid or ellipsoidal ascospores. Multilateral budding cells present. Colonies cream to tan, butyrous. Hyphae and pseudohyphae not formed. The major ubiquinone is Q-6.
Maudiozyma australis N. Jacques et al. ex Q.M. Wang, Yurkov & Boekhout, sp. nov. — MycoBank MB 851307
Holotype. CLIB 162, preserved in a metabolically inactive state at INRA Montpellier, France.
For description see Jacques et al., Int. J. Syst. Evol. Microbiol. 66: 5198. 2016.
Notes — Originally described as Kazachstania australis N. Jacques et al., Int. J. Syst. Evol. Microbiol. 66: 5198. 2016, nom. inval., Art. 40.7 (Melbourne).
Maudiozyma barnettii Vaughan-Mart. ex Q.M. Wang, Yurkov & Boekhout, sp. nov. — MycoBank MB851308
Holotype. CBS 5648, preserved in a metabolically inactive state at Westerdijk Institute.
For a detailed description see Vaughan-Martini, Antonie van Leeuwenhoek 68: 116. 1995.
Notes — Originally described as Saccharomyces barnettii Vaughan-Mart. (as ‘barnetti’), Antonie van Leeuwenhoek 68: 116. 1995, nom. inval., Art. 40.7 (Shenzhen) and Kazachstania barnettii Vaughan-Mart. ex Kurtzman, FEMS Yeast Res. 4: 238. 2003, nom. inval., Art. 40.1 (Shenzhen).
Maudiozyma bozae (Gouliamova & Dimitrov) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851309
Basionym. Kazachstania bozae Gouliamova & Dimitrov, Index Fungorum 432: 1. 2020.
Maudiozyma bulderi (Middelhoven et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851310
Basionym. Saccharomyces bulderi Middelhoven et al., Antonie van Leeuwenhoek 77: 224. 2000.
Synonym. Kazachstania bulderi (Middelhoven et al.) Kurtzman, FEMS Yeast Res. 4: 238. 2003.
Maudiozyma exigua (Kurtzman) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851311
Basionym. Kazachstania exigua Kurtzman, FEMS Yeast Res. 4: 238. 2003.
Synonym. Saccharomyces exiguus Reess ex E.C. Hansen, Compt. Rend. Lab. Carlsberg, Physiol. 2: 146. 1888, nom. illegit., Art. 53.1.
Maudiozyma humilis (E.E. Nel & Van der Walt) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851312
Basionym. Torulopsis humilis E.E. Nel & Van der Walt, Mycopathol. Mycol. Appl. 36: 95. 1968.
Synonym. Kazachstania humilis (E.E. Nel & Van der Walt) N. Jacques, Sarilar & Casarég., Int. J. Syst. Evol. Microbiol. 66: 5199. 2016.
Maudiozyma pseudohumilis (F.Y. Bai et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851313
Basionym. Candida pseudohumilis F.Y. Bai et al., FEMS Yeast Res. 9: 1325. 2009.
Synonym. Kazachstania pseudohumilis (F.Y. Bai et al.) N. Jacques, Sarilar & Casarég., Int. J. Syst. Evol. Microbiol. 66: 5199. 2016.
Maudiozyma rupicola Safar et al. ex Q.M. Wang, Yurkov & Boekhout, sp. nov. — MycoBank MB 851314
Holotype. CBS 12684, preserved in a metabolically inactive state at Westerdijk Institute.
For description see Safar et al., Int. J. Syst. Evol. Microbiol. 63: 1167. 2013.
Notes — Originally described as Kazachstania rupicola Safar, F.C.O. Gomes, C.A.R. Rosa & Lachance, Int. J. Syst. Evol. Microbiol. 63: 1167. 2013, nom. inval., Art. 40.7 (Melbourne).
Maudiozyma saulgeensis N. Jacques et al. ex Q.M. Wang, Yurkov & Boekhout, sp. nov. — MycoBank MB 851315
Holotype. CLIB 1764, preserved in a metabolically inactive state at INRA Montpellier, France.
For description see Jacques et al., Int. J. Syst. Evol. Microbiol. 66: 5196. 2016.
Notes — Originally described as Kazachstania saulgeensis N. Jacques et al., Int. J. Syst. Evol. Microbiol. 66: 5196. 2016, nom. inval., Art. 40.7 (Melbourne).
Maudiozyma serrabonitensis M.R. Lopes et al. ex Q.M. Wang, Yurkov & Boekhout, sp. nov. — MycoBank MB 851316
Holotype. CLIB 1783, preserved in a metabolically inactive state at INRA Montpellier, France.
For description see Jacques et al., Int. J. Syst. Evol. Microbiol. 66: 5197. 2016.
Notes — Originally described as Kazachstania serrabonitensis M.R. Lopes et al., Int. J. Syst. Evol. Microbiol. 66: 5197. 2016, nom. inval., Art. 40.7 (Melbourne).
Maudiozyma surinensis (S. Punyauppa-path et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851317
Basionym. Kazachstania surinensis S. Punyauppa-path et al., Int. J. Syst. Evol. Microbiol. 72: 6. 2022.
Maudiozyma turicensis (Wyder et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851318
Basionym. Saccharomyces turicensis Wyder et al., Syst. Appl. Microbiol. 22: 423. 1999.
Synonym. Kazachstania turicensis (Wyder et al.) Kurtzman, FEMS Yeast Res. 4: 239. 2003.
Maudiozyma wufongensis C.F. Lee ex Q.M. Wang, Yurkov & Boekhout, sp. nov. — MycoBank MB 851319
Holotype. CBS 10886, preserved in a metabolically inactive state at Westerdijk Institute.
For description see Lee et al., Antonie van Leeuwenhoek 95: 338. 2009.
Notes — Originally described as Kazachstania wufongensis C.F. Lee, Antonie van Leeuwenhoek 95: 338. 2009, nom. inval., Art. 40.7 (Melbourne).
Monosporozyma Q.M. Wang, Yurkov & Boekhout, gen. nov. — MycoBank MB 851320
Etymology. The genus is named based on the feature of forming one ascospore for species in this lineage.
Type species. Monosporozyma unispora (Henninger & Windisch) Q.M. Wang, Yurkov & Boekhout.
This genus is proposed for the species in the unispora clade of Kazachstania as previously defined, which formed a well-supported lineage closely related to Arxiozyma clade (Fig. 1, 3). It is a member of the Saccharomycetaceae (Saccharomycetales). The genus is mainly circumscribed by phylogenomic and genome metric-based analyses.
Budding cells transform directly into persistent asci containing one globose to ellipsoidal ascospore, but occasionally up to four (Vaughan-Martini et al. 2011). Colonies cream to tan, butyrous, semi-glossy to glossy. Multilateral budding cells present. Hyphae and pseudohyphae not formed. The major ubiquinone is Q-6.
Monosporozyma aerobia (F.Y. Bai & Y.M. Cai) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851321
Basionym. Kazachstania aerobia F.Y. Bai & Y.M. Cai, Int. J. Syst. Evol. Microbiol. 54: 2434. 2004.
Monosporozyma aquatica (F.Y. Bai & Z.W. Wu) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851322
Basionym. Kazachstania aquatica F.Y. Bai & Z.W. Wu, Int. J. Syst. Evol. Microbiol. 55: 2221. 2005.
Monosporozyma chrysolinae (Gouliamova & Dimitrov) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851323
Basionym. Kazachstania chrysolinae Gouliamova & Dimitrov, Index Fungorum 432: 1. 2020.
Monosporozyma servazzii (Capr.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851324
Basionym. Saccharomyces servazzii Capr., Ann. Microbiol. Enzimol. 17: 79. 1967.
Synonym. Kazachstania servazzii (Capr.) Kurtzman, FEMS Yeast Res. 4: 238. 2003.
Monosporozyma siamensis Limtong et al. ex Q.M. Wang, Yurkov & Boekhout, sp. nov. — MycoBank MB 851325
Holotype. NBRC 101968, preserved in a metabolically inactive state at NBRC, Japan.
For description see Limtong et al., Int. J. Syst. Evol. Microbiol. 57: 421. 2007.
Notes — Originally described as Kazachstania siamensis Limtong et al., Int. J. Syst. Evol. Microbiol. 57: 421. 2007, nom. inval., Art. 40.7 (Melbourne).
Monosporozyma solicola (F.Y. Bai & Z.W. Wu) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851326
Basionym. Kazachstania solicola F.Y. Bai & Z.W. Wu, Int. J. Syst. Evol. Microbiol. 55: 2222. 2005.
Monosporozyma unispora (A. Jörg.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851327
Basionym. Saccharomyces unisporus A. Jörg., Mikrosk. Betriebsk. Gährung. (Berlin) (5te Aufl.): 371. 1909.
Synonym. Kazachstania unispora (A. Jörg.) Kurtzman, FEMS Yeast Res. 4: 239. 2003.
Monosporozyma yasuniensis (S.A. James) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851328
Basionym. Kazachstania yasuniensis S.A. James, Int. J. Syst. Evol. Microbiol. 65: 1308. 2014.
Oligophagozyma Q.M. Wang, Yurkov & Boekhout, gen. nov. — MycoBank MB 851329
Etymology. The genus is named for the species in this lineage as they only assimilate few carbon and nitrogen sources.
Type species. Oligophagozyma castellii (Capr.) Q.M. Wang, Yurkov & Boekhout.
This genus is proposed for the species Nakaseomyces castellii, which formed a separate branch closely related to Nakaseomyces bacillisporus (Fig. 1). It is a member of the Saccharomycetaceae (Saccharomycetales). The genus is mainly circumscribed by phylogenomic and AAI analyses.
Sexual reproduction not known. Colonies white to cream, soft, smooth. Budding cells present. Pseudohyphae and hyphae not formed. The major ubiquinone is Q-6.
Oligophagozyma castellii (Capr.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851331
Basionym. Torulopsis castellii Capr., J. Gen. Microbiol. 26: 42. 1961.
Synonym. Candida castellii (Capr.) S.A. Mey. & Yarrow, Int. J. Syst. Bacteriol. 28: 612. 1978.
= Nakaseomyces castellii (Capr.) Sugita & M. Takash., Med. Mycol. J. 63: 126. 2022.
Sungouiozyma Q.M. Wang, Yurkov & Boekhout, gen. nov. — MycoBank MB 851332
Etymology. The genus is named in honor of Sung-Oui Suh for his contribution to yeast taxonomy.
Type species. Sungouiozyma intestinalis (Henninger & Windisch) Q.M. Wang, Yurkov & Boekhout.
This genus is proposed for species of the intestinalis clade of Kazachstania as previously defined, which formed a well-supported separate lineage (Fig. 1, 3). It is a member of the Saccharomycetaceae (Saccharomycetales). The genus is mainly circumscribed by phylogenomic and genome metrics-based analyses.
Asci form directly from diploid yeast cells or after conjugating. Asci persistent, with up to four globose to subglobose ascospores ( Suh & Zhou 2011, Cardinali et al. 2012). Colonies white to cream, butyrous, smooth. Multilateral budding cells present. Hyphae and pseudohyphae not formed.
Sungouiozyma ichnusensis (Cardinali et al.) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851333
Basionym. Kazachstania ichnusensis Cardinali et al., Int. J. Syst. Evol. Microbiol. 62: 722. 2012.
Sungouiozyma intestinalis (S.O. Suh & J.J. Zhou) Q.M. Wang, Yurkov & Boekhout, comb. nov. — MycoBank MB 851334
Basionym. Kazachstania intestinalis S.O. Suh & J.J. Zhou, Index Fungorum 335: 1. 2017.
Acknowledgments
We gratefully acknowledge the help of Konstanze Bensch with checking the nomenclatural matters. This study was supported by grants No. 31961133020, No. 32370015 and No. 31770018 from the National Natural Science Foundation of China (NSFC), No. 2021FY100900 from the Ministry of Science and Technology of China, No. 521000981388 from Advanced Talents Incubation Program of the Hebei University. The authors are solely responsible for the content of this work. TB would like to thank the Distinguished Scientists Fellowship Program (DSFP) of King Saud University, Riyadh, Saudi Arabia for financial support.
Declaration on conflict of interest
The authors declare that there is no conflict of interest.
Supplementary material
All supplementary files are deposited at Figshare repository: https://doi.org/10.6084/m9.figshare.24955620.
List of yeast species and GenBank numbers used in the combined ITS and D1/D2 and multi-gene analyses.
Matrix of the AAI values between Saccharomycetaceae.
Matrix of the POCP values between Saccharomycetaceae.
The PAPO analysis of the genera in Saccharomycetaceae.
List of the genus-specific signature nucleotides (GSNs) of rDNA.
Number of nucleotide variation and sequence similarities in the D1/D2 domain among species in Saccharomycetaceae.
Number of nucleotide variation and sequence similarities in the ITS region among species in Saccharomycetaceae.
Conserved signature indels specific for the genus Eremothecium, Grigorovia, Kluyveromyces, Naumovozyma, Saccharomyces, Zygosaccharomyces, Zygotorulaspora, intestinalis clade, naganishii clade, and Arxiozyma clade.
The SSU rDNA region containing the genus-specific signature nucleotides (GSNs) of the genera in Saccharomycetaceae.
REFERENCES
- Aliyu H, Lebre P, Blom J, et al. 2016. Phylogenomic re-assessment of the thermophilic genus Geobacillus. Systematic and Applied Microbiology 39: 527–533. [DOI] [PubMed] [Google Scholar]
- Alnajar S, Gupta RS. 2017. Phylogenomics and comparative genomic studies delineate six main clades within the family Enterobacteriaceae and support the reclassification of several polyphyletic members of the family. Infection, Genetics and Evolution 54: 108–127. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco RA, Garrity GM, Scott JJ, et al. 2020. A genus definition for bacteria and archaea based on a standard genome relatedness index. mBio 11: e02475–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern M, Goldberg D, Lyashenko E. 2006. Data mining for proteins characteristic of clades. Nucleic Acids Research 34: 4342–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhout T, Aime MC, Begerow D, et al. 2021. The evolving species concepts used for yeasts: from phenotypes and genomes to speciation networks. Fungal Diversity 109: 27–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali G, Antonielli L, Corte L, et al. 2012. Kazachstania ichnusensis sp. nov., a diploid homothallic ascomycetous yeast from Sardinian lentisk rhizosphere. International Journal of Systematic and Evolutionary Microbiology 62: 722–727. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, et al. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel HM, Lachance MA, Kurtzman CP. 2014. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie van Leeuwenhoek 106: 67–84. [DOI] [PubMed] [Google Scholar]
- De Albuquerque NRM, Haag KL. 2022. Using average nucleotide identity (ANI) to evaluate microsporidia species boundaries based on their genetic relatedness. The Journal of Eukaryotic Microbiology 70: e12944. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology 20: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Gouliamova D, Dimitrov R. 2020. Kazachstania chrysolinae and Kazachstania bozae two new yeast species of the genus Kazachstania. transfer of four Kazachstania species to Grigorovia gen. nov. as new combinations. Comptes Rendus de l’Académie Bulgare des Sciences 73: 48–57. [Google Scholar]
- Groenewald M, Hittinger CT, Bensch K, et al. 2023. A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina. Studies in Mycology 105: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueho E, Kurtzman CP, Peterson SW. 1989. Evolutionary affinities of heterobasidiomycetous yeasts estimated from 18S and 25S ribosomal RNA sequence divergence. Systematic and Applied Microbiology 12: 230–236. [Google Scholar]
- Gupta RS. 2014. Identification of conserved indels that are useful for classification and evolutionary studies. Methods in Microbiology 153–182. [Google Scholar]
- Gupta RS. 2016. Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin’s views on classification. FEMS Microbiology Reviews 40: 520–553. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Suggett C. 2022. Conserved signatures in protein sequences reliably demarcate different clades of Rodents/Glires species and consolidate their evolutionary relationships. Genes 13: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Sant’Anna F, Bach E, Porto RZ, et al. 2019. Genomic metrics made easy: what to do and where to go in the new era of bacterial taxonomy. Critical Reviews in Microbiology 45: 182–200. [DOI] [PubMed] [Google Scholar]
- Heidler von Heilborn D, Reinmüller J, Yurkov A, et al. 2023. Fungi under modified atmosphere – the effects of Co2 stress on cell membranes and description of new yeast Stenotrophomyces fumitolerans gen. nov., sp. nov. Journal of Fungi 9: 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig W. 1966. Phylogenetic systematics. University of Illinois Press, Urbana. [Google Scholar]
- Henninger W, Windisch S. 1976. Kluyveromyces blattae sp. n., a new multi-spored yeast for Blatta orientalis (author’s transl). Archives of Microbiology 109: 153–156. [DOI] [PubMed] [Google Scholar]
- Jacques N, Sarilar V, Urien C, et al. 2016. Three novel ascomycetous yeast species of the Kazachstania clade, Kazachstania saulgeensis sp. nov., Kazachstania serrabonitensis sp. nov. and Kazachstania australis sp. nov. Reassignment of Candida humilis to Kazachstania humilis f.a. comb. nov. and Candida pseudohumilis to Kazachstania pseudohumilis f.a. comb. nov. International Journal of Systematic and Evolutionary Microbiology 66: 5192–5200. [DOI] [PubMed] [Google Scholar]
- James SA, Carvajal Barriga EJ, Portero Barahona P, et al. 2015. Kazachstania yasuniensis sp. nov., an ascomycetous yeast species found in mainland Ecuador and on the Galápagos. International Journal of Systematic and Evolutionary Microbiology 65: 1304–1309. [DOI] [PubMed] [Google Scholar]
- Kachalkin AV, Abdullabekova DA, Magomedova ES, et al. 2021. Zygotorulaspora dagestanica sp. nov., a novel ascomycetous yeast species associated with the Georgian honeysuckle (Lonicera iberica M. Bieb.). International Journal of Systematic and Evolutionary Microbiology 71: 004785. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke T, Zhai YC, Yan ZL, et al. 2019. Kazachstania jinghongensis sp. nov. and Kazachstania menglunensis f.a., sp. nov., two yeast species isolated from rotting wood. International Journal of Systematic and Evolutionary Microbiology 69: 3623–3628. [DOI] [PubMed] [Google Scholar]
- Kitching I, Williams DM, Kitching IJ, et al. 1998. Cladistics: the theory and practice of parsimony analysis. Quarterly Review of Biology 11: 69–86. [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP. 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Research 4: 233–245. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. 2011. Discussion of teleomorphic and anamorphic ascomycetous yeasts and yeast-like taxa. In: Kurtzman CP, Fell JW, Boekhout T. (eds), The yeasts, a taxonomic study, 5th edn: 293–307. Elsevier, The Netherlands. [Google Scholar]
- Kurtzman CP, De Hoog GS. 2011. Eremothecium Borziemend. Kurtzman In: Kurtzman CP, Fell JW, Boekhout T. (eds), The yeasts, a taxonomic study, 5th edn: 405–412. Elsevier, The Netherlands. [Google Scholar]
- Kurtzman CP, Fell JW, Boekhout T. 2011. The yeasts, a taxonomic study, 5th edn. Elsevier, The Netherlands. [Google Scholar]
- Kurtzman CP, Robnett CJ. 1991. Phylogenetic relationships among species of Saccharomyces, Schizosaccharomyces, Debaryomyces and Schwanniomyces determined from partial ribosomal RNA sequences. Yeast 7: 61–72. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73: 331–371. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. 2003. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Research 3: 417–432. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. 2013. Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Research 13: 23–33. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ, Basehoar-Powers E. 2008. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Research 8: 939–954. [DOI] [PubMed] [Google Scholar]
- Kuzmanovic N, Fagorzi C, Mengoni A, et al. 2022. Taxonomy of Rhizobiaceae revisited: proposal of a new framework for genus delimitation. International Journal of Systematic and Evolutionary Microbiology 72: 005243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance MA. 2011. Nakaseomyces Kurtzman (2003). In: Kurtzman CP, Fell JW, Boekhout T. (eds), The yeasts, a taxonomic study, 5th edn: 633–636. Elsevier, The Netherlands. [Google Scholar]
- Lachance MA, Boekhout T, Scorzetti G, et al. 2011. Candida Berkhout. In: Kurtzman CP, Fell JW, Boekhout T. (eds), The yeasts, a taxonomic study, 5th edn: 987–1278. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- Lachance MA, Lee DK, Hsiang T. 2020. Delineating yeast species with genome average nucleotide identity: a calibration of ANI with haplontic, heterothallic Metschnikowia species. Antonie van Leeuwenhoek 113: 2097–2106. [DOI] [PubMed] [Google Scholar]
- Li AH, Yuan FX, Groenewald M, et al. 2020. Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: Proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Studies in Mycology 96: 17–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Steenwyk JL, Chang Y, et al. 2021. A genome-scale phylogeny of the kingdom Fungi. Current Biology 31: 1653–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xue H, Sang SQ, et al. 2017. Phylogenetic analysis of family Neisseriaceae based on genome sequences and description of Populibacter corticis gen. nov., sp. nov., a member of the family Neisseriaceae, isolated from symptomatic bark of Populus × euramericana canker. PLoS ONE 12: e0174506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libkind D, Čadež N, Opulente DA, et al. 2020. Towards yeast taxogenomics: lessons from novel species descriptions based on complete genome sequences. FEMS Yeast Research 20: foaa042. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Wang QM, Göker M, et al. 2015a. Towards an integrated phylogenetic classification of the Tremellomycetes. Studies in Mycology 81: 85–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Wang QM, Theelen B, et al. 2015b. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Studies in Mycology 81: 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes-Santos L, Castro DBA, Ferreira-Tonin M, et al. 2017. Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Antonie van Leeuwenhoek 110: 727–736. [DOI] [PubMed] [Google Scholar]
- Lücking R, Aime MC, Robbertse B, et al. 2021. Fungal taxonomy and sequence-based nomenclature. Nature Microbiology 6: 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Rodriguez-R LM, Konstantinidis KT. 2014. MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Research 42: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, et al. 2019. The EMBL–EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research 268: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, et al. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Molecular Biology and Evolution 38: 4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR, Sepúlveda VE. 2019. Fungal species boundaries in the genomics era. Fungal Genetics and Biology 131: 103249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Göker M. 2019. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nature Communications 10: 2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecillo JAV. 2023. Phylogenomics and comparative genomic analyses support the creation of the novel family Ignatzschineriaceae fam. nov. comprising the genera Ignatzschineria and Wohlfahrtiimonas within the order Cardiobacteriales. Research in Microbiology 174: 103988. [DOI] [PubMed] [Google Scholar]
- Naushad S, Adeolu M, Wong S, et al. 2015. A phylogenomic and molecular marker based taxonomic framework for the order Xanthomonadales: proposal to transfer the families Algiphilaceae and Solimonadaceae to the order Nevskiales ord. nov. and to create a new family within the order Xanthomonadales, the family Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Antonie van Leeuwenhoek 107: 467–485. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Sangal V. 2022. Advanced prokaryotic systematics: the modern face of an ancient science. New Microbes and New Infections 49–50: 101036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opulente DA, LaBella AL, Harrison MC, et al. 2023. Genomic and ecological factors shaping specialism and generalism across an entire subphylum. bioRxiv. Preprint.
- Orata FD, Meier-Kolthoff JP, Sauvageau D, et al. 2018. Phylogenomic analysis of the gammaproteobacterial methanotrophs (Order Methylococcales) calls for the reclassification of members at the genus and species levels. Frontiers in Microbiology 9: 3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Rinke C, et al. 2022. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Research 50: D785–D794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Waite DW, et al. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nature Biotechnology 36: 996–1004. [DOI] [PubMed] [Google Scholar]
- Passer AR, Coelho MA, Billmyre RB, et al. 2019. Genetic and genomic analyses reveal boundaries between species closely related to Cryptococcus pathogens. mBio 10: e00764-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Gupta RS. 2018. Robust demarcation of fourteen different species groups within the genus Streptococcus based on genome-based phylogenies and molecular signatures. Infection, Genetics and Evolution 66: 130–151. [DOI] [PubMed] [Google Scholar]
- Qin QL, Xie BB, Zhang XY, et al. 2014. A proposed genus boundary for the prokaryotes based on genomic insights. Journal of Bacteriology 196: 2210–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Chuvochina M, Mussig AJ, et al. 2021. A standardized archaeal taxonomy for the genome taxonomy database. Nature Microbiology 6: 946–959. [DOI] [PubMed] [Google Scholar]
- Rodriguez-R LM, Konstantinidis KT. 2014. By passing cultivation to identify bacterial species. Microbe 9: 111–118. [Google Scholar]
- Sakpuntoon V, Angchuan J, Boonmak C, et al. 2020. Savitreea pentosicarens gen. nov., sp. nov., a yeast species in the family Saccharomycetaceae isolated from a grease trap. International Journal of Systematic and Evolutionary Microbiology 70: 5665–5670. [DOI] [PubMed] [Google Scholar]
- Scorzetti G, Fell JW, Fonseca A, et al. 2002. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Research 2: 495–517. [DOI] [PubMed] [Google Scholar]
- Shen XX, Opulente DA, Kominek J, et al. 2018. Tempo and mode of genome evolution in the budding yeast subphylum. Cell 175: 1533–1545.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skennerton CT, Ward LM, Michel A, et al. 2015. Genomic reconstruction of an uncultured hydrothermal vent gammaproteobacterial methanotroph (family Methylothermaceae) indicates multiple adaptations to oxygen limitation. Frontiers in Microbiology 6: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa FMP, Morais PB, Lachance MA, et al. 2014. Hagleromyces gen. nov., a yeast genus in the Saccharomycetaceae, and description of Hagleromyces aurorensis sp. nov., isolated from water tanks of bromeliads. International Journal of Systematic and Evolutionary Microbiology 64: 2915–2918. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel A, Stanke KM, Quattrone AC, et al. 2022. Improving taxonomic delimitation of fungal species in the age of genomics and phenomics. Frontiers in Microbiology 13: 847067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh SO, Zhou JJ. 2011. Kazachstania intestinalis sp. nov., an ascosporogenous yeast from the gut of passalid beetle Odontotaenius disjunctus. Antonie van Leeuwenhoek 100: 109–115. [DOI] [PubMed] [Google Scholar]
- Takashima M, Manabe RI, Nishimura Y, et al. 2019. Recognition and delineation of yeast genera based on genomic data: Lessons from Trichosporonales. Fungal Genetics and Biology 130: 31–42. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. 1999. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiology Letters 174: 247–250. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Sánchez-Ramírez S, Kõljalg U, et al. 2018. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Diversity 90: 135–159. [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, et al. 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Research 18: 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann L, Wibberg D, Busche T, et al. 2022. Seventeen Ustilaginaceae high-quality genome sequences allow phylogenomic analysis and provide insights into secondary metabolite synthesis. Journal of Fungi 8: 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Walt JP, Yarrow D. 1984. The genus Arxiozyma gen. nov. (Saccharomycetaceae). South African Journal of Botany 3: 340–342. [Google Scholar]
- Varghese NJ, Mukherjee S, Ivanova N, et al. 2015. Microbial species delineation using whole genome sequences. Nucleic Acids Research 43: 6761–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan-Martini A, Lachance MA, Kurtzman CP. 2011. Kazachstania Zubkova. In: Kurtzman CP, Fell JW, Boekhout T. (eds), The yeasts, a taxonomic study, 5th edn: 439–470. Elsevier, The Netherlands. [Google Scholar]
- Vu D, Groenewald M, Szöke S, et al. 2016. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Studies in Mycology 85: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Groenewald M, Wang QM. 2015c. Reclassification of Saccharomycodes sinensis, proposal of Yueomyces sinensis gen. nov., comb. nov. within Saccharomycetaceae (Saccharomycetales, Saccharomycotina). PLoS ONE 10: e0136987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Bai FY. 2008. Molecular phylogeny of basidiomycetous yeasts in the Cryptococcus luteolus lineage (Tremellales) based on nuclear rDNA and mitochondrial cytochrome b gene sequence analyses: proposal of Derxomyces gen. nov. and Hannaella gen. nov., and eight novel Derxomyces species. FEMS Yeast Research 8: 799–814. [DOI] [PubMed] [Google Scholar]
- Wang QM, Begerow D, Groenewald M, et al. 2015a. Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Studies in Mycology 81: 55–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Groenewald M, Takashima M, et al. 2015b. Phylogeny of yeasts and related filamentous fungi within Pucciniomycotina determined from multigene sequence analyses. Studies in Mycology 81: 27–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Yurkov AM, Göker M, et al. 2015d. Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Studies in Mycology 81: 149–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibberg D, Stadler M, Lambert C, et al. 2021. High quality genome sequences of thirteen Hypoxylaceae (Ascomycota) strengthen the phylogenetic fami ly backbone and enable the discovery of new taxa. Fungal Diversity 106: 7–28. [Google Scholar]
- Wiley EO, Lieberman BS. 2011. Phylogenetics: the theory and practice of phylogenetic systematics, 2nd edn. John Wiley & Sons Inc, Hoboken. [Google Scholar]
- Wirth JS, Whitman WB. 2018. Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. International Journal of Systematic and Evolutionary Microbiology 68: 2393–2411. [DOI] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, et al. 2009. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462: 1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Ma J. 2019. The Global Catalogue of Microorganisms (GCM) 10K type strain sequencing project: providing services to taxonomists for standard genome sequencing and annotation. International Journal of Systematic and Evolutionary Microbiology 69: 895–898. [DOI] [PubMed] [Google Scholar]
- Wu Q, James SA, Roberts IN, et al. 2008. Exploring contradictory phylo-genetic relationships in yeasts. FEMS Yeast Research 8: 641–650. [DOI] [PubMed] [Google Scholar]
- Xu J. 2020. Fungal species concepts in the genomics era. Genome 63: 459–468. [DOI] [PubMed] [Google Scholar]
- Xu Z, Masuda Y, Itoh H, et al. 2019. Geomonas oryzae gen. nov., sp. nov., Geomonas edaphica sp. nov., Geomonas ferrireducens sp. nov., Geomonas terrae sp. nov., four Ferric-Reducing bacteria isolated from paddy soil, and reclassification of three species of the genus geobacter as members of the genus Geomonas gen. nov. Frontiers in Microbiology 10: 2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HT, Shang YJ, Zhu HY, et al. 2023. Yueomyces silvicola sp. nov., a novel ascomycetous yeast species unable to utilize ammonium, glutamate, and glutamine as sole nitrogen sources. Yeast 40: 540–549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of yeast species and GenBank numbers used in the combined ITS and D1/D2 and multi-gene analyses.
Matrix of the AAI values between Saccharomycetaceae.
Matrix of the POCP values between Saccharomycetaceae.
The PAPO analysis of the genera in Saccharomycetaceae.
List of the genus-specific signature nucleotides (GSNs) of rDNA.
Number of nucleotide variation and sequence similarities in the D1/D2 domain among species in Saccharomycetaceae.
Number of nucleotide variation and sequence similarities in the ITS region among species in Saccharomycetaceae.
Conserved signature indels specific for the genus Eremothecium, Grigorovia, Kluyveromyces, Naumovozyma, Saccharomyces, Zygosaccharomyces, Zygotorulaspora, intestinalis clade, naganishii clade, and Arxiozyma clade.
The SSU rDNA region containing the genus-specific signature nucleotides (GSNs) of the genera in Saccharomycetaceae.