Abstract
Macaques which developed high-titer neutralizing antibodies (htNAb) after immunization with a virion-derived oligomeric envelope glycoprotein subunit vaccine were protected against a homologous simian immunodeficiency virus SIVmac challenge. Here we demonstrate that the htNAb could be overcome by V1-env region variants isolated ex vivo from an SIVmac-infected macaque. The results further suggest that the development of V1-env region neutralization escape mutants is also necessary for survival of the virus in infected macaques. The immunological capacity of a single variable region to induce neutralizing antibodies in vaccinated and infected macaques initiate new ideas for a successful vaccine strategy.
Infections of macaques with simian immunodeficiency virus (SIV) have been used extensively in the last decade as an animal model for vaccine development against human immunodeficiency virus (HIV) (12). Despite several vaccine trials performed in nonhuman primates, the immune mechanisms responsible for protective effects remain largely unknown. Recently we showed that a subunit vaccine consisting of virion-derived oligomeric gp130 (O-gp130) induced a sterilizing immunity against homologous challenge with the swarm virus SIVmac32H, whereas monomeric preparations did not (16, 22, 23). Vaccine protection could be strongly correlated to high-titer neutralizing antibodies (htNAb) but not to a proliferative T-cell response or to cytotoxic T lymphocytes. This was the first time that htNAb was described as the major component of a preventive vaccine which would induce sterilizing immunity against an immunodeficiency virus. The induction of such an htNAb response was highly dependent on a specific immunization schedule, and protection was observed mainly after a homologous virus challenge (16, 22). The protective capacity of htNAb in a homologous system was recently directly confirmed in passively immunized monkeys challenged with an HIV/SIV chimera (SHIV) (25).
We have now investigated whether the variability in critical neutralizing epitopes might be mainly responsible for the rather restricted breadth of protection observed in our vaccine trials. Which envelope glycoprotein epitopes may directly contribute to the vaccine failures observed in heterologous challenge systems remains unknown. Their identification and characterization are, however, important in order to understand the molecular mechanisms responsible for the presence of vaccine-resistant viruses. In a previous study we suggested that the first variable domain (V1 region) of the external glycoprotein of SIVmac is critical for the development of neutralization escape mutants (13). The V1 region is known to be highly variable (1, 6), and a substantial portion of the htNAb from the O-gp130-immunized macaques showing a sterilizing immunity was directed against this region (13). Therefore, we have now investigated whether mutations which naturally occur in the V1 region of SIVmac-infected macaques help the virus to escape from the htNAb. The experiments with sera from protected monkeys demonstrated that variations in the V1 region are sufficient for the virus to escape from htNAb. The same results were obtained with sera obtained from SIVmac-infected monkeys. Our results strongly indicate that the V1 region acts as an immunological shield for SIVmac. However, although the high genetic variability of the V1 region seems to be necessary for the virus to escape from the htNAb, we could additionally demonstrate that this epitope is essential for an efficient replication of SIVmac. Therefore, a V1 region multivalent O-gp130 preparation should offer greater protection than the vaccines tested so far.
MATERIALS AND METHODS
Monkey sera.
Monkey sera were obtained from SIVmac-infected rhesus macaques (Macaca mulatta) Mm1604 and Mm1708 or O-gp130-immunized animals Mm1698, Mm1701, and Mm1715 (13, 16, 22). In the cases of Mm1604 and Mm1078, the sera were obtained about 114 and 52 weeks postinfection (wpi), respectively. Sera from the immunized animals were collected on the day of challenge.
Cloning of the V1 region recombinant SIVmac239.
The wild-type V1 region from SIVmac239 (15) was replaced by corresponding regions isolated ex vivo from peripheral blood monocytes of an SIVmac-infected rhesus macaque, Mm1708 (13). The ex vivo V1 regions were obtained 1 year after infection when the animal had developed simian AIDS. Two different V1 regions from Mm1708 were used to construct the SIVmac239 recombinant viruses SIVmacV1-1708/2 and SIVmacV1-1708/4. Additionally, we prepared a chimera in which the wild-type V1 region was replaced by the corresponding region of SIVmac32H. The human T-cell line C8166 was infected with SIVmac32H. One week after infection, the V1 region was amplified, cloned, and sequenced from the SIVmac32H-infected cells as described elsewhere (23). The V1 region representing the major genotype found in C8166 cells was used for constructing the chimera. The different V1 regions were cloned in the wild-type clone SIVmac239. First, the V1 region 3′ and 5′ flanking regions of SIVmac239 were independently amplified using the 3′ clone p239SpE1 from SIVmac239 as the template DNA. The 3′ fragment was amplified with the primer pair P1 (located in the vector pBS+ bp 980 to 995; 5′-AACAGCTATGACCATG-3′) plus P2 (SIVmac239 bp 7115 to 7144; 5′-CTCAAAGAGTTGCCATACATCCTCTATTGC-3′); the 5′ fragment was amplified with the primer pair P5 (SIVmac239 bp 7350 to 7379; 5′-AAATGATAAGCTGTAAATTCAACATGACAG-3′) plus P6 (SIVmac239 bp 8408 to 8441; 5′-AGACCCCGAATTCATTTCCTGAGGTGCCACCAG-3′). The ex vivo-isolated V1 regions and that of SIVmac32H were amplified with the primer pair P3 (5′GCAATAGAGGATGTATGGCAACTCTTTGAGACCTCAATAAAGCCTTGTGTAAAATTATCC-3′) plus P4 (5′-CTGTCATGTTGAATTTACAGCTTATCATTTGCTCTTGTTCCAAGCC TGTGCAATTATTCT-3′), where the 5′ region of P3 overlaps with the 3′ region of P2 and the 3′ region of P4 overlaps with the 5′ region of P5. The recombinant env genes were constructed by hybridization PCR in which primers P1 and P6 were used for amplification. The resulting amplification product was digested with the restriction enzymes SphI and and ClaI and cloned into the proviral SIVmac239 clone. The V1 deletion mutant was also constructed by hybridization PCR. For this purpose, we performed two independent PCRs using the primer pairs P1-P7 (5′-CTGTCATGTTGAATTTACAGCTTATCATTTGCTCAAAGAGTTGCCATGCATCCTCTATTGC-3′) and P5-P6, where the 5′ region of P7 overlaps with the 3′ region of P5. In the case of all V1 region recombinant env genes, the cloning was confirmed by sequence analysis.
Production of virus stocks in COS-7 cells.
COS-7 cells were transfected by the DEAE-dextran method with the wild-type clone SIVmac239 and the V1 region recombinant proviral clones. In short, 105 cells were transfected with 5 μg of DNA and cultivated in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM glutamine, 50 U of penicillin/ml, 50 μg of streptomycin/ml, and 4.5 g of glucose/liter. Three days after transfection, cell culture supernatants were harvested and analyzed for virus release. Virus production was assessed by measuring cell-free p27, the viral core antigen of SIV, with a commercially available enzyme-linked immunosorbent assay (ELISA; Innogenetics, Zwijnaarde, Belgium). Virus-containing supernatants were stored at −80°C for infection experiments.
Infection of CD4+ T lymphocytes.
Replication capacities of the wild-type clone SIVmac239 and the chimeras were tested in the human T-cell line HUT-78 and the T-B hybrid cell line CEMx174, both maintained in RPMI 1640 medium. Infection of these cell lines with each virus was carried out in triplicate. For the infection experiments, the virus-containing COS-7 cell supernatants were normalized with respect to p27 concentration. Every 3 days, 0.2 ml of medium was removed and stored at −80°C for assaying virus production, and fresh medium was added to 5 ml. Viral replication was determined by measuring the p27 concentration in the cell culture supernatants.
Phenotype determination of the V1 region recombinant viruses in chemokine-expressing U87.CD4 cell lines.
To determine the phenotype of the V1 region recombinant viruses, we used CCR5- as well as CXCR4-expressing U87.CD4 cell lines (4). The wild-type and recombinant viruses used for the infection were expanded on CEMx174 cells and normalized with respect to p27 concentration. Infection was carried out with all viruses in each cell line in triplicate. The U87.CD4 cells were propagated in DMEM supplemented with 10% FCS, 500 μg of G418/ml, 1% penicillin, and 1% streptomycin.
Neutralization assay.
NAb against wild-type SIVmac239 and V1 region recombinant viruses were measured with the MT-4 cell assay. This assay is well established for in vitro evaluation of NAb against HIV and SIV (10, 13). Wild-type SIVmac239 and the V1 region recombinant viruses used in the MT-4 cell assay were propagated in CEMx174 cells. The assay was performed in flat-bottom 96-well microtiter plates. During cultivation, MT-4 cells were grown in RPMI 1640 medium supplemented with 25 mM HEPES buffer (pH 6.5) and 10% FCS. The macaque sera were heat inactivated (56°C, 30 min) and tested in triplicate serial twofold dilutions starting with 1:40. The sera were incubated with 100 50% tissue culture infectious doses of cell-free virus. After 1 h of incubation at 37°C, 3 × 104 MT-4 cells were added to each well. Three days after infection, fresh medium was added to each well; 4 days after infection, each well was supplemented with 0.1 μCi of [3H]thymidine (Amersham, Braunschweig, Germany). [3H]thymidine incorporation into cellular DNA was measured 20 h later, and neutralization was defined as 75% reduction of virus-induced cell death.
V1 region antibody recognition.
V1 region antibody recognition was investigated in the prokaryotic glutathione S-transferase (GST) expression system. The V1 regions of SIVmac239, SIVmac32H, and ex vivo isolates V1-1708/2 and V1-1708/4 were cloned into the expression vector pGEX-2T and expressed as GST fusion proteins as specified by the manufacturer (Pharmacia Biotech, Uppsala, Sweden). Escherichia coli was lysed, and the V1-GST fusion proteins were adsorbed to glutathione-Sepharose 4B and eluted with 50 mM Tris buffer (pH 8.0) containing 10 mM glutathione. The V1-GST fusion proteins were purified by affinity chromatography. The purified fusion proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose; for Western blot analysis, we used the same sera as for the neutralization assay.
RESULTS
Production of SIVmac V1 chimeras.
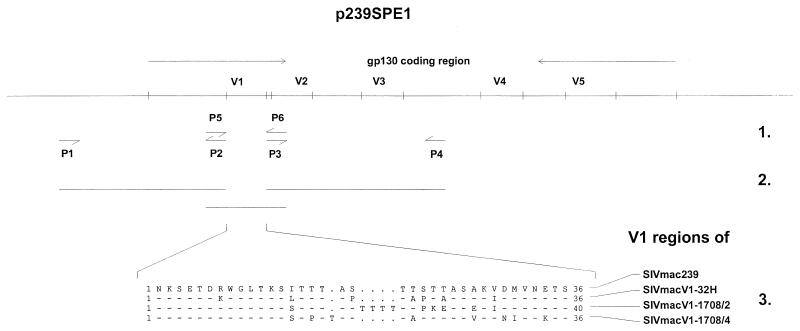
To investigate whether V1 variability is necessary for SIVmac replication in the presence of htNAb, we examined viruses which varied in this region only. All cloned viruses used herein were derived from SIVmac239 (15). For the chimera SIVmacV1-32H, the V1 of SIVmac239 was replaced by the corresponding region of SIVmac32H (Fig. 1). The V1 region of SIVmac32H used represented the major genotype present in about 80% of all proviral genomes of infected C8166 cells (data not shown). The sequence of this V1 region differed from the corresponding SIVmac239 sequence in seven positions, four of which were located in the serine- and threonine-rich region which might act as an O-linked glycosylation site. In contrast, no variations were present in the potential N-linked glycosylation sites located at the amino- and carboxy-terminal ends. The other two chimeras examined, SIVmacV1-1708/2 and SIVmacV1-1708/4, contained different ex vivo-isolated V1 regions from SIVmac-infected macaque Mm1708. The V1 regions from this animal were selected for the high genetic variability among the virus population present in this animal. As described previously (13), all clones analyzed 52 wpi when the animal had already developed simian AIDS showed unique V1 region sequences. In the case of SIVmacV1-1708/2, a sequence with the insertion of four threonines and preferentially serine/threonine mutations was used. Both insertions and mutations contributed to the establishment of a highly enriched potential O-linked glycosylation site. For the chimeric virus SIVmac1708/4, we selected a sequence with a fewer mutations in this site. To investigate whether the V1 region is essential for viral replication, we additionally constructed the V1 region deletion mutant SIVmacV1-del.
FIG. 1.
Construction of V1 chimeras. 1, each V1 region was amplified with the primer pair P5-P6; 5′ and 3′ flanking regions were amplified with primer pairs P1-P2 and P3-P4, respectively. 2, the V1 regions were introduced into the molecular clone SIVmac239 by recombinant PCR. 3, alignment of the V1 regions used for the chimeras, depicted in amino acid single-letter code.
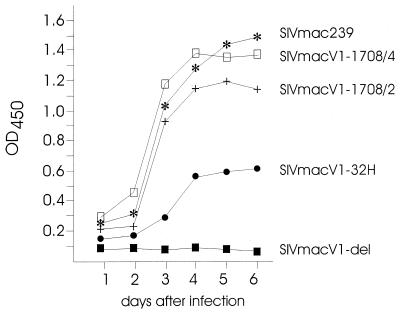
Infectious virus stocks were produced by transfecting COS-7 cells with each of the chimeras as well as the wild-type clone SIVmac239. After transfection, we detected release of the wild-type virus as well as the three recombinants, SIVmacV1-32H, SIVmacV1-1708/2, and SIVmacV1-1708/4 (Fig. 2). However, different concentrations of p27 viral core antigen in COS-7 cell supernatants indicated an influence of the V1 region on viral replication. The importance of the V1 region for SIVmac replication became more obvious when SIVmacV1-del-transfected COS-7 cells were examined for virus production. No p27 release was detected in the cell culture supernatants. Likewise, no virus production was observed when different CD4+ cell lines were transfected with SIVmacV1-del (data not shown).
FIG. 2.
Production of infectious virus stocks. COS-7 cells were transfected with the different viral clones. Supernatants were harvested over 6 days, and virus production was measured in triplicate by a commercially available p24 antigen ELISA. Here and in subsequent figures, OD450 denotes optical density at 450 nm.
Replication of V1 region chimeric viruses.
The virus-containing supernatants obtained after COS-7 cell transfection were normalized for p27 concentration and used to infect the human cell lines CEMx174 and HUT-78, which are both known to be susceptible to SIVmac infection. All four viruses were able to infect CEMx174 cells without obvious differences in the time course of infection, replication levels (Fig. 3), cell toxicity, and syncytium induction (data not shown). In HUT-78 cells, similar kinetics were measured for SIVmac239, SIVmacV1-32H, and SIVmacV1-1708/4, with the lowest replication level in the case of SIVmacV1-32H. In contrast, a delayed onset and decreased level of replication were observed for SIVmacV1-1708/2. Similar results were obtained with the permanent human T-cell lines MT-2 and MT-4 (data not shown).
FIG. 3.
Viral replication in permanent human T-cell lines. Virus-containing supernatants from COS-7 cells were normalized for p27 concentration and used to infect the CD4+ cell lines CEMx174 and HUT-78. Virus production was measured in triplicate over 31 days by a commercially available p24 antigen ELISA. Values represent the average and standard deviation from the mean of three experiments.
In further experiments, U87.CD4 cells expressing CCR5 or CXCR4 were infected to investigate coreceptor usage dependent on the V1 region. All four viruses replicated in CCR5-expressing U87.CD4 cells, whereas only marginal replication was detected in CXCR4-expressing cells (Fig. 4). In CCR5-expressing U87.CD4 cells, the four viruses replicated with similar kinetics but to different levels. We observed the highest activity for SIVmacV1-32H and the lowest for SIVmac239. Thus, the V1 region did not significantly change the cell tropism of SIVmac but rather influenced viral replication.
FIG. 4.
Coreceptor-dependent viral replication. Virus-containing supernatants from COS-7 cells were normalized for p27 concentration and used to infect U87.CD4 cells expressing CCR5 or CXCR4. Virus production was measured in triplicate over 17 days by a commercially available p24 antigen ELISA. Values represent the average and standard deviation from the mean of three experiments.
V1 region-mediated neutralization escape.
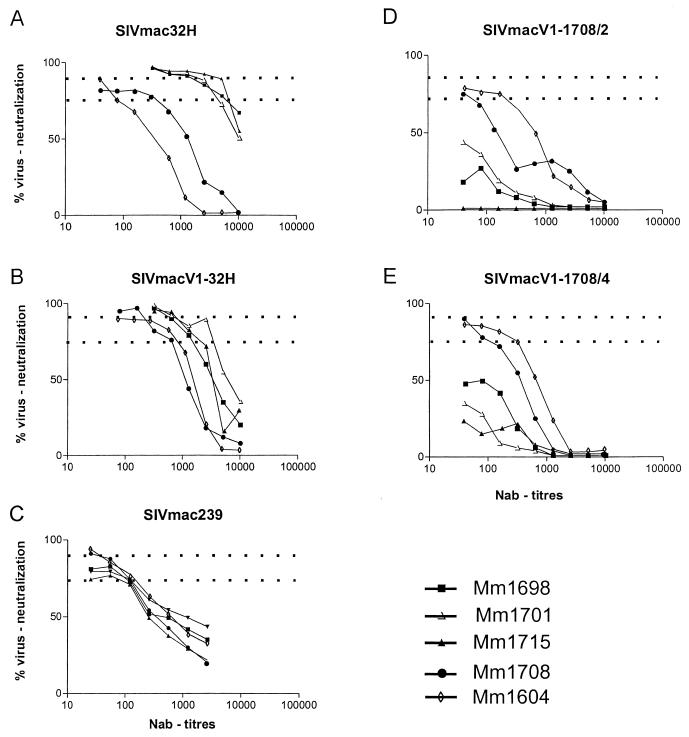
Recently we reported that sera from rhesus macaques Mm1698, Mm1701, and Mm1715 which were immunized with virus-derived O-gp130 contained htNAb directed against the V1 region of SIVmac32H (13). On the basis of these results, we suggested that variations in the V1 region influence the NAb susceptibility of SIVmac. The MT-4 cell assay performed with O-gp130 immune sera revealed remarkable differences in neutralization susceptibility among the five viruses tested. High ≥90% neutralization efficacies were detected against the swarm virus SIVmac32H, the SIVmacV1-32H chimera, and SIVmac239 (Fig. 5). This high neutralizing capacity was observed for the swarm virus SIVmac32H (Fig. 5A) and the chimera SIVmacV1-32H (Fig. 5B) up to titers of between 640 and 1,280. Although wild-type SIVmac239 was also neutralized with ≥90% efficacy, titers were significantly lower (between 40 and 160 [Fig. 5C]). Increased titers were found for these three viruses at a 75% neutralization cutoff (Table 1). The swarm virus SIVmac32H and the chimera SIVmacV1-32H were neutralized at high titers (1,280 to 5,120), whereas SIVmac239 was neutralized at lower titers (between 320 and 640). These results showed that neutralization susceptibility could be transferred by an exchange of the V1 region. In contrast, the chimeras SIVmacV1-1708/2 and SIVmacV1-1708/4 could not be neutralized with the O-gp130 sera, based on the 75% cutoff (Fig. 5D and F). Only at low titers (≤80) was a neutralization capacity of 50% obtained. It was further remarkable that the O-gp130 immune sera from animal Mm1715 failed to neutralize SIVmacV1-1708/2. Obviously, naturally occurring V1 region mutations strongly decreased neutralization susceptibility in the cases of SIVmacV1-1708/2 and SIVmacV1-1708/4.
FIG. 5.
Neutralization (MT-4 cell) assay. Neutralization was detected with different dilutions of sera from SIVmac-infected macaques Mm1708 and Mm1604 or O-gp130-immunized macaques Mm1698, Mm1701, and Mm1715. Dotted lines indicate the cutoff at 75 and 90% virus neutralization, respectively. Each NAb titer is the mean of three serum samples examined in parallel. In 82% of the assays, the mean deviation was 0; in the remaining assays, the results differed by one dilution step.
TABLE 1.
NAb titers of sera from O-gp130-immunized and SIVmac-infected rhesus macaquesa
| Virus | NAb titer
|
||||
|---|---|---|---|---|---|
| Mm1698 | Mm1701 | Mm1715 | Mm1708 | Mm1604 | |
| SIVmac239 | 5,120 | 2,560 | 2,560 | 320 | 320 |
| SIVmacV1-32H | 1,280 | 2,560 | 1,280 | 640 | 640 |
| SIVmacV1-1708/2 | <40 | <40 | <40 | 40 | 160 |
| SIVmacV1-1708/4 | <40 | <40 | <40 | 80 | 320 |
| SIVmac32H | 5,120 | 2,560 | 5,120 | 320 | 80 |
Serum samples were obtained from Mm1698, Mm1701, and Mm1715 after O-gp130 immunization; serum samples from Mm1708 and Mm1604 were obtained 52 and 114 wpi, respectively, after SIVmac infection. Neutralization was defined as 75% reduction of virus-induced cell death in the MT-4 cell assay.
In parallel, we tested sera obtained from the SIVmac-infected monkey Mm1708 at 52 wpi, when the V1-1708/2 and V1-1708/4 sequences were isolated from peripheral blood mononuclear cells from this animal. These sera exhibited only low to intermediate NAb titers (between 80 and 640), based on the 75% cutoff (Table 1). Although the NAb titers of serum from monkey Mm1708 were significantly lower than titers of the O-gp130 immune sera, the serum was able to neutralize the molecular clone SIVmac239, the swarm virus SIVmac32H, and all three chimeras. In the case of the molecular clone SIVmac239, the swarm virus SIVmac32H, and the chimera SIVmacV1-32H, neutralizing efficacies of ≥90% were measured at titers of between 320 and 640. These sera were also able to neutralize the chimeras SIVmacV1-1708/2 and SIVmacV1-1708/4 with an efficacy of ≥75% but at low titers (≤80). These results demonstrated an increased breadth of the V1-specific neutralizing humoral immune response in the SIVmac-infected monkey Mm1708 compared to that of the O-gp130 immune sera. Similar neutralization patterns were detected for sera obtained 114 wpi from the SIV-infected macaque Mm1604. The neutralization capacity in the presence of the chimeric viruses SIVmacV1-1708/2 and SIVmacV1-1708/4 was increased compared to the titers detected in the case of Mm1708. However, the titers of the SIVmac-infected monkey were marked lower than those obtained for the O-gp130-immunized animals. The data indicate that the V1 region variability enables the virus to circumvent the major neutralizing capacity of the immune serum from Mm1708, which was mainly directed against the original SIV isolate used for infection.
Antibody recognition of V1 region-encoded peptides.
In parallel, we tested whether the NAb susceptibility of the different viruses corresponded to a V1 region-specific antibody binding. The same sera which were investigated for the NAb titers were tested for their capacity to bind the different V1 region epitopes from SIVmac32H as well as those isolated ex vivo from animal Mm1708. The corresponding regions from SIVmacV1-32H, SIVmacV1-1708/2, and SIVmacV1-1708/4 were cloned as GST fusion proteins and tested for immune reactivity by Western blot analysis.
As expected, sera obtained from Mm1708 on the day of infection did not react with any of the V1 regions tested (Fig. 6C). During a 1-year infection period, the animal developed a strong immune reactivity directed against the V1-32H region that corresponds to the major sequence of the inoculum virus (Fig. 6C). Interestingly, at the time when the strong V1-32H-specific reactivity was observed, we did not detect the corresponding viral V1 sequence in this monkey (data not shown). These data indicate that the original immune response directed against the V1 region of the inoculum virus was stable over time despite the absence of the corresponding V1 sequence. In contrast, the V1 regions V1-1708/2 and V1-1708/4, present at 52 wpi in the peripheral blood of rhesus monkey Mm1708, did not react with sera obtained at the same time point. Binding to these two ex vivo-isolated V1 regions was observed, however, with serum from the Mm1604 obtained about 2 years after infection (Fig. 6B). Sequence analysis showed that genotypes at least similar to those of V1-1708/2 and V1-1708/4 had developed also in this animal (Fig. 7) and moreover suggested that the prolonged period of infection gave rise to the development of a broader immune response.
FIG. 6.
Antibody binding to GST-V1 fusion proteins. GST-V1 fusion proteins were expressed in E. coli, affinity purified, separated by SDS-PAGE, and transferred to a nitrocellulose membrane. Fusion proteins were stained with Coomassie blue (A) and detected with sera from SIVmac-infected Mm1604 (B) or SIVmac infected-Mm1708 (C) on the day of infection or 52 wpi, respectively, and with sera from the O-gp130-immunized animal Mm1698, Mm1701, or Mm1715. All sera were diluted 1:200 for Western blot analysis.
FIG. 7.
Alignment of predicted proviral V1 amino acid sequences (in single-letter code) from SIVmac-infected rhesus macaque Mm1604 and from SIVmacV1-1708/2, SIVmacV1-1708/4, and SIVmacV1-32H. Comparison was done with the program PILEUP of the Wisconsin Package (Genetics Computer Group, Madison, Wis.). Amino acid identities and deletions are denoted by dashes and periods, respectively.
Further Western blot analysis with the V1-GST fusion proteins showed that the sera obtained from the three O-gp130-immunized animals bound strongly to the V1-32H sequence (Fig. 6C). In contrast, no reactivity was observed with the ex vivo-isolated V1-1708/2 and V1-1708/4 regions (Fig. 6C). These results, in combination with those of the neutralization assays, showed that the vaccine produced on the basis of a virus isolate with a defined major viral genotype induced a restricted protective immune response that was circumvented by naturally occurring virus variants.
DISCUSSION
It has recently been proposed that htNAb are required to achieve protection against infection with HIV or SIV (19, 21). In the SIV-macaque model, we have found a correlation between htNAb exhibiting 90% efficacy and protection (16), showing for the first time that the induction of such htNAb was a consequence of a subunit vaccination. Moreover, the development of htNAb is dependent on the structure of the envelope glycoprotein gp130 present in the vaccine and on the immunization schedule. Whereas the protected macaques that were immunized with O-gp130 developed htNAb, the monomeric form induced very low, if any, NAb titers in the unprotected animals (22, 23). We further showed that the htNAb-correlated protective effect was high in the homologous system but rather low after challenge with a heterologous isolate (16). In more recent experiments, the protective activity of such htNAb has also been directly demonstrated by passive immunization experiments in the SHIV model with purified immunoglobulins (25). The immunoglobulins were obtained from an HIV-1-infected chimpanzee (5, 9) that had resisted successive virus challenges with different heterologous isolates. The chimpanzee immune serum also contained htNAb with an efficacy of 90%, which in general are not present in HIV-infected humans or SIV- or SHIV-infected macaques (25). However, since the breadth of protection was rather low, the presence of isolate-specific htNAb in the chimpanzee sera is probable.
Our previous studies had shown that a substantial part of the htNAb present in the O-gp130 immune sera of the protected animals were directed against the V1 region of the gp130 of the homologous challenge virus SIVmac32H (13). Therefore, the V1 region was an attractive candidate for such an isolate-specific strong neutralizing epitope. To explore this possibility, we constructed different V1 region chimeras based on the molecular clone SIVmac239. Three chimeras, SIVmacV1-32H, SIVmacV1-1708/2, and SIVmacV1-1708/4, were constructed using the V1 region of the wild-type SIVmac32H or isolated ex vivo 52 wpi from the SIVmac-infected macaque Mm1708. All chimeras were replication competent and exhibited a CCR5 tropism as reported for most SIV isolates (8, 17).
Although the variation in the V1 region had no marked influence on viral replication, a pronounced effect on neutralization susceptibility was observed. Whereas the chimera SIVmacV1-32H was neutralized with almost the same efficacy as the wild-type swarm virus SIVmac32H, the recombinant viruses SIVmacV1-1708/2 and SIVmacV1-1708/4 were highly resistant upon incubation with O-gp130 immune sera. The observation that the O-gp130 immune sera contained residual neutralizing activity was expected from a previous study in which the V1 region was described as a linear neutralizing epitope (13). Hence, the results of our previous and present studies suggest additional viral neutralizing epitopes outside the V1 region (2, 3, 11, 14, 18, 20) as targets for O-gp130 immune sera. The most pronounced neutralization escape was found with the chimera SIVmacV1-1708/2, containing the insertion of four threonine residues in the V1 region. These results support previous studies that the development of potential glycosylation sites in the V1 region are an important mechanism for SIV to circumvent the NAb in infected macaques (7, 24). However, the results obtained with SIVmacV1-1708/4, which showed a less pronounced variability in the V1 region and which was also strongly neutralization resistant, suggested an additional mechanism to overcome the activity of htNAb. In agreement with the results obtained in the neutralization assays, we observed, in parallel, that the V1 regions isolated ex vivo did not bind antibodies from the O-gp130 sera whereas the corresponding region from SIVmac32H reacted strongly with the same sera.
A similar pattern of neutralization and epitope binding was found for sera from the SIVmac-infected monkey Mm1708. Sera which obtained 52 wpi moderately neutralized the wild-type swarm virus SIVmac32H, the wild-type molecular clone SIVmac239, and the chimera SIVmacV1-32H. Whereas low neutralization was found when the same sera were tested against the chimeras SIVmacV1-1708/2 and SIVmacV1-1708/4, the ex vivo-isolated V1 regions failed to react with the sera obtained 52 wpi from Mm1708. The same sera showed, however, strong reactivity with the V1 region of the original swarm virus SIVmac32H. Interestingly, sera obtained from macaque Mm1604 at 114 wpi neutralized the chimeras SIVmacV1-1708/2 and SIVmacV1-1708/4 with increased efficacy. In Western blot analysis, these sera also reacted with the ex vivo-isolated regions V1-1708/2 and V1-1708/4. These results suggested that the increased antibody binding affinity and neutralization capacity developed within the second year of infection in the presence of V1-1708/2- and V1-1708/4-related V1 sequences in animal Mm1604. The results demonstrated furthermore that a V1-dependent neutralization escape plays a role not only after vaccination but also during infection.
Our results further demonstrated that the development of htNAb directed against a single V1 region was possible only in vaccine experiments including a minimum of six immunizations over a period of 6 months. Changes in the immunization schedule or the adjuvant resulted in a reduced development of NAb and the loss of sterilizing immunity (23). In infected animals, the V1 sequences present early in the infection obviously disappear with time and will be replaced by others. This observation suggests that the period given for each sequence is too short for the immune system to develop htNAb, which seems to be also necessary for efficient virus elimination.
Our vaccination experiments demonstrated that NAb induced by O-gp130 are mainly directed against an isolate-specific V1 region variant. Therefore, the efficacy of this vaccine is limited by the breadth of neutralization rather than by the NAb titer. This assumption, however, should be tested in an O-gp130 vaccine experiment where the immunized animals are challenged with the chimeras SIVmacV1-1708/2 and SIVmacV1-1708/4, to determine whether the in vitro experiments can be directly transferred to the in vivo situation of O-gp130-immunized animals. The limitation of the O-gp130 vaccine is further incentive to develop and test a multivalent V1 region-specific vaccine.
ACKNOWLEDGMENTS
SIVmac239 and SIVmac32H were obtained from Ronald Desrosiers (New England Regional Primate Research Center, Harvard Medical School, Southborough, Mass.) and Martin Cranage (Division of Pathology, PHLS Center for Applied Microbiological Research, Porton Down, Salisbury, United Kingdom). The U87.CD4 cell lines were kindly provided by Programme EVA Centralised Facility for AIDS Reagents. We thank Christiane Stahl-Hennig for supplying monkey sera, as well as Kerstin Wäse, Astrid Schäfer, and Karin Giller for excellent technical assistance.
This work was supported by a grant from the Bundesminister für Bildung, Wissenschaft Forschung und Technologie, Bonn/Jülich, Germany.
REFERENCES
- 1.Almond N, Jenkins A, Heath A B, Kitchin P. Sequence variation in the env gene of simian immunodeficiency virus recovered from immunized macaques is predominantly in the V1 region. J Gen Virol. 1993;74:865–871. doi: 10.1099/0022-1317-74-5-865. [DOI] [PubMed] [Google Scholar]
- 2.Babas T, Belhadj-Jrad B, Le Grand R, Dormont D, Montagnier L, Bahraoui E. Specificity and neutralizing capacity of the three monoclonal antibodies produced against the envelope glycoprotein of simian immunodeficiency virus isolate 251. Virology. 1995;211:339–344. doi: 10.1006/viro.1995.1414. [DOI] [PubMed] [Google Scholar]
- 3.Benichou S, Le Grand R, Nakagawa N, Faure T, Traincard F, Vogt G, Dormont D, Tiollais P, Kieny M-P, Madaule P. Identification of a neutralizing domain in the external envelope glycoprotein of simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1992;8:1165–1170. doi: 10.1089/aid.1992.8.1165. [DOI] [PubMed] [Google Scholar]
- 4.Bjorndal A, Deng H, Jansson M, Fiori J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruck C, Thiriart C, Fabry L, Francotte M, Pala P, VanOpstal O, Culp J, Rosenberg M, DeWilde M, Heidt P, Heeney J. HIV-1 envelope elicited neutralizing antibody titers correlate with protection and virus load in chimpanzees. Vaccine. 1994;12:1141–1148. doi: 10.1016/0264-410x(94)90185-6. [DOI] [PubMed] [Google Scholar]
- 6.Burns D P W, Desrosiers R C. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol. 1991;65:1843–1854. doi: 10.1128/jvi.65.4.1843-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chackerian B, Rudensey L M, Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J Virol. 1997;71:7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emini E A, Schleif W A, Nunberg J H, Conley A J, Eda Y, Tokiyoshi S, Putney S D, Matsushita S, Cobb K E, Jett C M, Eichberg J W, Murthy K K. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature. 1992;355:728–730. doi: 10.1038/355728a0. [DOI] [PubMed] [Google Scholar]
- 10.Harada S, Purtilo D T, Koyanagi Y, Sonnabend J, Yamamoto N. Sensitive assay for neutralizing antibodies against AIDS related viruses (HTLV-III/LAV) J Immunol Methods. 1986;92:177–181. doi: 10.1016/0022-1759(86)90163-8. [DOI] [PubMed] [Google Scholar]
- 11.Javaherian K, Langlois A J, Schmidt S, Kaufmann M, Cates N, Langedijk J P M, Meloen R H, Desrosiers R C, Burns D P W, Bolognesi D P, LaRosa G J, Putney S. The principle neutralization determinant of simian immunodeficiency virus differs from that of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:1418–1422. doi: 10.1073/pnas.89.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson R P. Macaque models for AIDS vaccine development. Curr Opin Immunol. 1996;8:554–560. doi: 10.1016/s0952-7915(96)80046-x. [DOI] [PubMed] [Google Scholar]
- 13.Jurkiewicz E, Hunsmann G, Schäffner J, Nißlein T, Lüke W, Petry H. Identification of the V1 region as a linear neutralizing epitope of the simian immunodeficiency virus SIVmac envelope glycoprotein. J Virol. 1997;71:9475–9481. doi: 10.1128/jvi.71.12.9475-9481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent K A, Rud E, Corcoran T, Powell C, Thiriart C, Collognon C, Stott E J. Identification of two neutralizing and 8 non-neutralizing epitopes on simian immunodeficiency virus envelope using monoclonal antibodies. AIDS Res Hum Retroviruses. 1992;8:1147–1151. doi: 10.1089/aid.1992.8.1147. [DOI] [PubMed] [Google Scholar]
- 15.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R C. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 16.Lüke W, Coulibaly C, Dittmer U, Voss G, Oesterle R, Makoschey B, Sauermann U, Jurkiewicz E, Stahl-Hennig C, Petry H, Hunsmann G. Simian immunodeficiency virus (SIV) gp130 oligomers protect rhesus macaques (Macaca mulatta) against the infection with SIVmac32H grown on T-cells or derived ex vivo. Virology. 1996;216:444–450. doi: 10.1006/viro.1996.0082. [DOI] [PubMed] [Google Scholar]
- 17.Marcon J, Choe H, Martin K A, Farzan M, Ponath P D, Wu L, Newman W, Gerard C, Sodroski J. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumi S, Matsushita S, Yoshimura K, Javaherian K, Takatsuki K. Neutralizing monoclonal antibody against an external envelope glycoprotein (gp110) of SIVmac251 of SIVmac251. AIDS Res Hum Retroviruses. 1995;11:501–508. doi: 10.1089/aid.1995.11.501. [DOI] [PubMed] [Google Scholar]
- 19.Moore J P, Burton D R. HIV-1 neutralizing antibodies: how full is the bottle? Nat Med. 1999;5:142–144. doi: 10.1038/5502. [DOI] [PubMed] [Google Scholar]
- 20.Palker T J, Muir A J, Spragion D E, Staats H F, Langlois A, Montefiori D C. The V3 domain of SIVmac251 gp120 contains a linear neutralizing epitope. Virology. 1996;224:415–426. doi: 10.1006/viro.1996.0548. [DOI] [PubMed] [Google Scholar]
- 21.Parren P W, Wang M, Trkola A, Binley J M, Katinger H, Moore J P, Burton D R. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72:10270–10274. doi: 10.1128/jvi.72.12.10270-10274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petry H, Stahl-Hennig C, Dittmer U, Jones D, Farrar G, Wachter H, Fuchs D, Nisslein T, Jurkiewicz E, Hunsmann G, Lüke W. A subunit vaccine consisting of gp130-oligomers but not of gp130-monomers protects rhesus macaques against the productive infection with SIVmac32H. AIDS. 1998;12:329–330. [PubMed] [Google Scholar]
- 23.Petry H, Dittmer U, Jones D, Farrar G, Wachter H, Fuchs D, Nißlein T, Jurkiewicz E, Hunsmann G, Stahl-Hennig C, Lüke W. Prechallenge high neutralizing antibodies and long-lasting immune reactivity to gp41 correlate with protection of rhesus monkeys against productive simian immunodeficiency virus infection or disease development. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:441–450. doi: 10.1097/00042560-199812150-00002. [DOI] [PubMed] [Google Scholar]
- 24.Reitter J N, Desrosiers R C. Identification of replication competent strains of simian immunodeficiency virus lacking multiple attachment sites for N-linked carbohydrates in variable regions 1 and 2 of the surface envelope protein. J Virol. 1998;72:5399–5407. doi: 10.1128/jvi.72.7.5399-5407.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibata R, Iagarshi T, Haighwood N, Buckler-White A, Ogert R, Ross W, Willey R, Cho M W, Martin M A. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;2:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]