Abstract
HIV-1 Tat binds human CyclinT1 and recruits the CDK9/P-TEFb complex to the viral TAR RNA in a step that links RNA polymerase II (RNAPII) C-terminal domain (CTD) Ser 2 phosphorylation with transcription elongation. Previous studies have suggested a connection between Tat and pre-mRNA splicing factors. Here we show that the splicing-associated c-Ski-interacting protein, SKIP, is required for Tat transactivation in vivo and stimulates HIV-1 transcription elongation, but not initiation, in vitro. SKIP associates with CycT1:CDK9/P-TEFb and Tat:P-TEFb complexes in nuclear extracts and interacts with recombinant Tat:P-TEFb:TAR RNA complexes in vitro, indicating that it may act through nascent RNA to overcome pausing by RNAPII. SKIP also associates with U5snRNP proteins and tri-snRNP110K in nuclear extracts, and facilitates recognition of an alternative Tat-specific splice site in vivo. The effects of SKIP on transcription elongation, binding to P-TEFb, and splicing are mediated through the SNW domain. HIV-1 Tat transactivation is accompanied by the recruitment of P-TEFb, SKIP, and tri-snRNP110K to the integrated HIV-1 promoter in vivo, whereas the U5snRNPs associate only with the transcribed coding region. These findings suggest that SKIP plays independent roles in transcription elongation and pre-mRNA splicing.
Keywords: HIV-1 Tat, CycT1:CDK9/P-TEFb, c-Ski-interacting protein, transcription elongation, alternative splicing, HIV-1 TAR RNA
The expression of mammalian RNA polymerase II (RNAPII) genes requires precise coordination between the different enzymatic complexes that mediate elongation, pre-mRNA processing (5′-end capping, splicing, and 3′-end polyadenylation/cleavage), RNA surveillance, and nuclear export (for reviews, see Sims et al. 2004; Zorio and Bentley 2004). An important question is how these steps are integrated at promoters that are regulated at the level of RNAPII elongation, including the HIV-1 and heat shock genes. The assembly of RNAPII elongation complexes is accompanied by specific modifications that target promoter-proximal nucleosomes and the C-terminal domain (CTD) of the large subunit of RNAPII, the functions of which are just beginning to be understood (for reviews, see Gerber and Shilatifard 2003; Sims et al. 2003).
The differential phosphorylation of RNAPII during promoter clearance and elongation establishes a checkpoint for packaging and processing of the nascent transcript. Ser 5 phosphorylation of the RNAPII CTD heptad repeat sequence (YSPTSP) by CDK7 signals 5′-end capping as well as H3K4 histone trimethylation, a hallmark of active genes (for reviews, see Gerber and Shilatifard 2003; Sims et al. 2003, 2004). Subsequent Ser 2 phosphorylation of the CTD by mammalian CycT1:CDK9/P-TEFb is required at many genes for productive transcription elongation (for review, see Shilatifard et al. 2003) and for pre-mRNA 3′-end processing (Ahn et al. 2004; Bird et al. 2004; Ni et al. 2004). The Ser 2-phosphorylated CTD recruits an H3-K36-specific histone methyltransferase as well as chromatin remodeling enzymes required for elongation and termination in yeast (Sims et al. 2004). P-TEFb can counteract the actions of the negative elongation factors, NELF and Spt5/DSIF, which otherwise arrest the Ser 5-phosphorylated RNAPII (for review, see Kim et al. 2001). A fraction of nuclear P-TEFb is found in an inactive form bound to 7SK RNA and can be released rapidly from these storage sites in response to various stress signals (Nguyen et al. 2001; Yang et al. 2001). P-TEFb is not universally required for transcription elongation, but rather plays a preferential role in 3′-end processing at some genes (Ahn et al. 2004; Ni et al. 2004) and also represses transcription of key regulators such as the nuclear receptor coactivator, PGC-1, in cardiac myocytes (Sano et al. 2004).
In contrast with 5′-end capping, less is known of the mechanisms that link transcription with pre-mRNA splicing in mammalian cells. A subset of mammalian serine- and arginine-rich (SR) proteins and pre-mRNA processing factors are targeted to active genes through direct binding to the Ser 2-phosphorylated CTD (for review, see Zorio and Bentley 2004). Yet another SR protein, U2AF65, cross-links directly to nascent transcripts in extracts and can reduce RNAPII pausing when bound to its specific RNA recognition site in vitro (Ujvari and Luse 2004). Recent studies have implicated a wide variety of splicing-associated or RNA-binding factors in promoter-specific gene regulation (Sune et al. 1997; Monsalve et al. 2000; Dellaire et al. 2002; Auboeuf et al. 2004; Kameoka et al. 2004), although in general little is known in detail about how these factors affect transcription.
The HIV-1 Tat-TAR regulatory system provides a useful model to study how transient binding of P-TEFb to nascent RNA may couple elongation with later RNA processing events. The high-affinity interaction between Tat and CycT1 directs transient, sequence-specific binding of the Tat:P-TEFb complex to the 5′-viral TAR RNA, stimulates RNAPII Ser 2 phosphorylation by CDK9 (for review, see Karn 1999), and alters the substrate specificity of CDK9 to include Ser 5 phosphorylation of the CTD (Garber et al. 2000; Zhou et al. 2000). As a consequence, Tat increases transcription elongation (Karn 1999), 5′-end capping (Chiu et al. 2002; Zhou et al. 2003), and histone methylation at the HIV-1 promoter (Zhou et al. 2004). Several studies have implicated a role for individual splicing factors in transcription elongation. For example, nuclear P-TEFb associates tightly with spliceosomal small nuclear ribonucleoproteins (snRNPs), and both cis-acting splice sites and snRNP-containing fractions enhance HIV-1 elongation in vitro (Fong and Zhou 2001). Two splicing-associated proteins, Tat-SF1 and CA150, associate with Tat:P-TEFb in extracts and stimulate HIV-1 transcription in vivo (Zhou and Sharp 1996; Sune et al. 1997; Zhou et al. 1998) and in vitro (Li and Green 1998). Tat-SF1 can interact with CycT1 (Zhou et al. 1998; Fong and Zhou 2000), and is present in large RNAPII elongation-splicing complexes that associate with 5′-splice sites (Kameoka et al. 2004).
One protein that has been strongly linked with transcription and splicing is the c-Ski-interacting protein, SKIP (Drosophila melanogaster BX42, Saccharomyces cerevisiae Prp45). SKIP is a coactivator of Notch (for review, see Hayward 2004) and vitamin D nuclear hormone receptor-dependent genes (VDR) (Barry et al. 2003; Zhang et al. 2003), but can also repress transcription of certain promoters (Figueroa and Hayman 2004; Leong et al. 2004). SKIP was identified as a constituent of activated spliceosomes and 35S-U5snRNP particles (Neubauer et al. 1998; Mintz et al. 1999; Makarov et al. 2002) and is required for splicing in S. cerevisiae (Albers et al. 2003) and in mammalian cells (Zhang et al. 2003; Nagai et al. 2004). Targeted elimination of SKIP by RNAi in Caenorhabditis elegans arrests gene expression early in development, indicating a widespread positive role in gene expression (Kostrouchova et al. 2002). SKIP binds to U5snRNPs through its conserved SNW domain (Zhang et al. 2003) and to the N-CoR and SMRT corepressors through an acidic N-terminal motif (Leong et al. 2004). SKIP can interact directly with Notch and the VDR (Barry et al. 2003; Hayward 2004) and is targeted to responsive promoters in vivo (Zhang et al. 2003; Fryer et al. 2004).
We previously reported that Notch activation of the human HES1 gene is accompanied by binding of SKIP as well as at least two transcription elongation factors, P-TEFb and FACT (Fryer et al. 2004), which raised the possibility that SKIP might belong to a subset of splicing factors that can modulate transcription elongation. We report here that SKIP stimulates basal and Tat-regulated HIV-1 transcription elongation in vivo and in vitro, without affecting RNA initiation. Interestingly, SKIP selectively associates, directly or indirectly, with P-TEFb and Tat:P-TEFb complexes in nuclear extracts. In addition, we find that recombinant SKIP interacts with Tat:P-TEFb:TAR RNA complexes in vitro. Nuclear SKIP also interacts with U5snRNPs and the tri-snRNP110K protein, but does not associate with a number of other splicing factors, including Tat-SF1 or CA150. Chromatin immunoprecipitation (ChIP) experiments reveal that Tat activation of the HIV-1 promoter in vivo is accompanied by binding of P-TEFb, SKIP, and tri-snRNP110K, whereas the U5snRNP proteins were detected only within the HIV-1/LacZ coding region. Taken together, these data suggest that SKIP is a dual-function protein that acts independently in P-TEFb-mediated transcription elongation and pre-mRNA splicing.
Results
SKIP is required for HIV-1 Tat transactivation in vivo
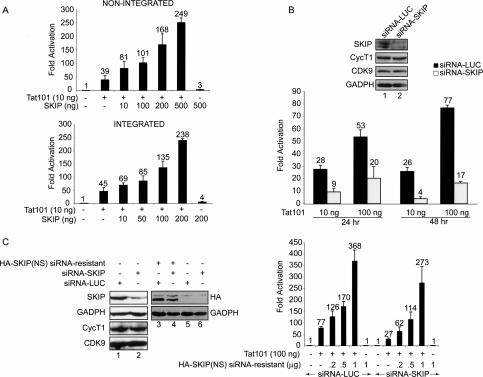
To investigate whether SKIP might regulate transcription elongation, we examined whether it is required for HIV-1 Tat activity in vivo. As shown in Figure 1A, ectopic expression of human SKIP enhanced Tat transactivation of both integrated and nonintegrated HIV-1 reporter genes in HeLa cells. Increasing amounts of the pSG5-HA-SKIP vector strongly enhanced HIV-1 LTR-LUC activity in the presence of Tat and also modestly stimulated basal HIV-1 transcription. A similar dose-dependent activation of basal and Tat-regulated transcription was observed for the integrated HIV-1 LTR:LacZ gene in HeLa P4 cells (Fig. 1A). To address whether endogenous SKIP is necessary for Tat transactivation in vivo, a SKIP-specific siRNA (siRNA-SKIP) was introduced into the HeLa P4 cells. As shown in Figure 1B (top panel), anti-SKIP siRNA decreased steady-state levels of endogenous SKIP (cf. lanes 1 and 2) without affecting the level of CycT1, CDK9, or GAPDH. Expression of the SKIP siRNA significantly impaired Tat transactivation at 24 and 48 h post-transfection, whereas Tat activity was unaffected by a control siRNA directed against luciferase (siRNA-LUC; Fig. 1B). In addition, Tat activity was selectively reduced in cells infected with a recombinant lentivirus expressing shSKIP (see Supplementary Fig. 1B, right panel).
Figure 1.
SKIP facilitates Tat transactivation of the HIV-1 promoter in vivo. (A) Transient expression of SKIP enhances Tat activation of a nonintegrated (top panel) or integrated (bottom panel) HIV-1 promoter. Hela cells were transfected with an HIV-1 LTR-luciferase reporter gene, alone or together with HIV-1 Tat101 and SKIP-expression vectors, as indicated. For normalization, cells were transfected with the control pRL-TK vector, and luciferase and renilla activities were analyzed 48 h after transfection. (Bottom panel) SKIP activity was analyzed in HeLa P4 cells, which contain a Lac-Z gene under control of an integrated HIV-1 LTR, β-galactosidase activity was measured in extracts derived from cells that transfected with Tat and SKIP expression vectors, as indicated. Fold activation refers to HIV-1 LTR activity relative to mock-transfected cells. (B) Native SKIP is required for Tat activity in vivo. HeLa P4 cells containing an integrated HIV-1 LTR-LacZ reporter were transfected with siRNAs specific for SKIP or luciferase, as indicated. (Lanes 1,2) Levels of SKIP, CycT1, CDK9, and GADPH proteins were determined by immunoblotting. The siRNA-treated HeLa P4 cells were mock-transfected or transfected with HIV-1 Tat101, as indicated, and β-galactosidase activity was measured 24 h and 48 h after transfection. Results are presented as fold activation relative to mock-transfected cells. (Right panel) The mean relative β-galactosidase activities were obtained from three independent experiments. (C) An siRNA-resistant SKIP(NS) can rescue Tat activity in siRNA-SKIP-treated cells. HeLa P4 cells were cotransfected with SKIP siRNA or control siRNA, together with the pHA-SKIP siRNA-resistant mutant or an empty vector, as indicated in the figure. (Left panel) At 48 h post-transfection, protein levels in cell lysates were analyzed by immunoblotting. Tat activity was analyzed 48 h post-transfection with 100 ng of Tat101, either alone or with the pHA-SKIP siRNA-resistant mutant. (Right panel) Fold activation was calculated relative to transfection in the absence of Tat101 expression plasmid.
The inhibitory effect of the anti-SKIP siRNA could be rescued by coexpression with an siRNA-resistant form of SKIP, designated HA-SKIP(NS), which is mutated in the siRNA recognition motif (Fig. 1C). The HA-SKIP(NS) protein also lacks the C terminus of SKIP and is more active than the full-length protein in vivo (see below). Immunoblot experiments confirmed that levels of HA-SKIP(NS) were not affected by the anti-SKIP siRNA (Fig. 1C, cf. lanes 3 and 4). Expression of the siRNA-resistant HA-SKIP(NS) protein increased Tat activity 10-fold in HeLa P4 cells (Fig. 1C).
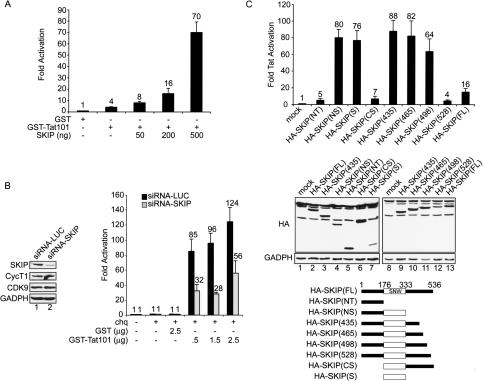
To exclude any possible unintended effects of SKIP on the expression of Tat in these cotransfection studies, the experiments were repeated using purified recombinant GST-Tat, which was introduced directly into cells via protein transduction (Becker-Hapak et al. 2001). As shown in Figure 2A, transient expression of SKIP enhanced GST-Tat101 activity as much as 17-fold in transduced HeLa cells. Moreover, anti-SKIP siRNA, but not a control anti-LUC siRNA, blocked Tat activity in Hela P4 cells (Fig. 2B). Although the anti-SKIP siRNA preferentially inhibited Tat-activated transcription, the low residual levels of SKIP may support basal transcription in these experiments.
Figure 2.
(A) SKIP enhances the activity of transduced HIV-1 Tat101 in vivo. HeLa cells were transfected with HIV-1 LTR-luciferase together with pHA-SKIP or an empty vector, as indicated. At 24 h post-transfection, HeLa cells were transduced with either 500 ng GST or GST-Tat101 in the presence of 100 μM of chloroquine, and luciferase activity was analyzed 48 h after transfection. Results are presented as fold activation relative to transduction with GST. (B) SKIP-specific siRNA blocks activation of the integrated HIV-1 LTR by transduced GST-Tat101. siRNA-treated HeLa P4 cells were transduced with the indicated amount of either GST or GST-Tat101 in the presence of 100 μM of chloroquine. Fold activation was calculated relative to transduction in the presence of chloroquine alone. (C) The SNW domain of SKIP enhances Tat transactivation in vivo. The HIV-1 LTR-Luc vector was transfected together with vectors expressing different HA-tagged SKIP mutants (as indicated schematically at the bottom of the panel), and HeLa cells were transduced with 500 ng GST-Tat101 at 24 h post-transfection. Luciferase activity was analyzed 48 h post-transfection, and results are presented as fold activation relative to transduction with GST-Tat101. Levels of expression of HA-tagged SKIP proteins were determined by immunoblot using an anti-HA antibody, and GADPH levels were analyzed as a loading control.
Analysis of mutant SKIP proteins in the Tat-transduced cells revealed that activation is strongly enhanced in vivo upon removal of a short (37-amino acid) motif at the very C terminus of the protein. Whereas Tat activity was enhanced fivefold by full-length SKIP, several SKIP mutants lacking the C terminus [HA-SKIP(NS), HA-SKIP(S), HA-SKIP(435), HA-SKIP(465), and HA-SKIP(498)] stimulated Tat activity as much as 80-fold in vivo (Fig. 2C). Proteins containing the SNW domain [HA-SKIP(S), and HA-SKIP(NS)] were much more active than constructs that included the C terminus [e.g., HA-SKIP(CS)]. Although the mechanism of inhibition is unclear, the C-terminal motif did not affect the expression of wild-type or mutant SKIP proteins as analyzed by Western blots from transfected cells (Fig. 2C, lanes 1–13). In addition, SKIP also activates transcription of the LEF1-reporter gene induced by β-catenin (Supplementary Fig. 1A,B). Collectively, these results indicate that SKIP functions as a positive coactivator of HIV-1 Tat and β-catenin in vivo.
SKIP enhances utilization of an alternative HIV-1 Tat splice site in vivo
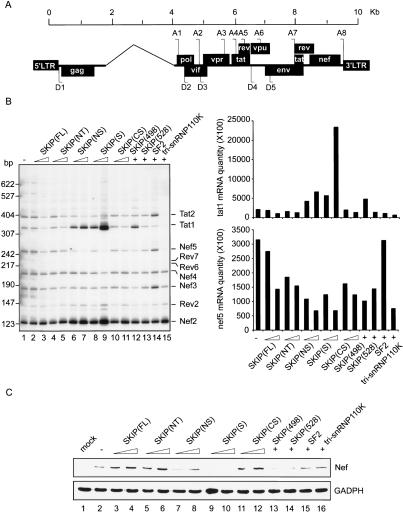
The pΔPSP plasmid, which includes eight HIV-1 splice acceptor sites and lacks the D1-A1 region (Fig. 3A), was used to examine the effect of SKIP on HIV-1 splicing. The different pΔPSP-derived RNAs were monitored by RT–PCR of total RNA using a reverse primer that encompasses the D4–A7 junction (Fig. 3B, lane 1). The pattern of HIV-1 transcripts derived from pΔPSP has been shown to be identical to that of authentic viral transcripts (Ropers et al. 2004). In these experiments we did not observe a reproducible effect of full-length SKIP on HIV-1 splice site choice; however, alternative splicing was strongly affected by the more active C-terminal truncated SKIP proteins. For example, the SKIP SNW domain [SKIP(S)] dramatically enhanced the use of a Tatspecific mRNA splice (tat2 mRNA; Fig. 3B, cf. lanes 8,9 and 1) that results from activation of the HIV-1 A3 splice acceptor. In addition, the SKIP SNW protein modestly reduced levels of other HIV-1 mRNAs, most notably the Nef5 mRNA (Fig. 3B, lane 14) and protein levels (Fig. 3C, cf. lanes 9,10 and 2).
Figure 3.
SKIP enhances alternative splicing of HIV-1 RNA in HeLa cells. (A) A schematic representation of the pΔPSP vector, indicating donor (D) and acceptor (A) sites. (B, left panel) HeLa cells were transfected with pΔPSP, either alone (lane 1), or together with plasmids encoding different SKIP truncation mutants (lanes 2–13), SF2 (lane 14), or tri-snRNP110K (lane 15), and equal amounts of total RNA were subjected to RT–PCR analysis using specific HIV-1 primers. Sizes of cDNA products are indicated to the left of the panel. The identity of the HIV-1 mRNAs corresponding to the fractionated cDNAs is indicated to the right of the panel. A histogram quantifying the levels of Tat1 and Nef5-specific transcripts is shown in the right panel. (C) Confirmation that Nef protein expression is down-regulated upon expression of the SKIP SNW domain. Total proteins were extracted 48 h post-transfection and separated by SDS-PAGE, and Nef was analyzed by immunoblot. GADPH levels were assessed as a loading control.
Comparison of several SKIP proteins indicates that the C-terminal motif interferes with splicing activity in vivo (Fig. 3B, cf. lanes 6–9 and 2,3, and lanes 12 and 10,11,13). The effect of the SKIP SNW domain on HIV-1 splicing resembles that described for two SR family members, SC35 and SRp40 (Ropers et al. 2004). A different SR protein, ASF/SF2, partially blocked the Tat1-splice, and enhanced the proportion of Tat2- and Nef5-spliced transcripts (Fig. 3B, lane 14), as described previously (Ropers et al. 2004). Taken together, these data indicate that the SKIP SNW domain promotes the use of the HIV-1 A3 acceptor site in vivo.
Recombinant SKIP stimulates basal and Tat-regulated HIV-1 transcription elongation in vitro, without affecting initiation
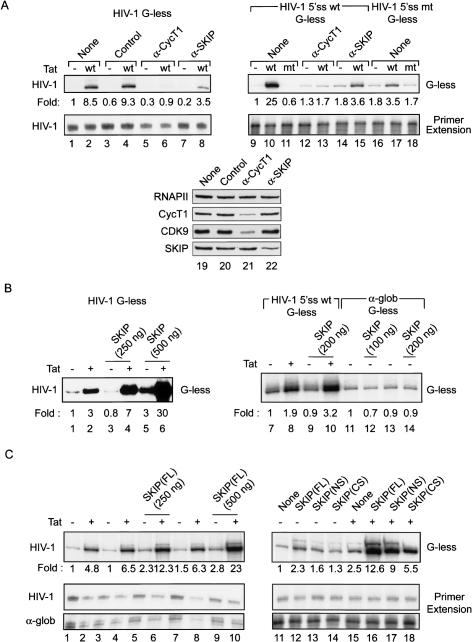
To address whether SKIP affects transcription elongation directly, we analyzed its activity in vitro. Immunodepletion of SKIP from HeLa nuclear extracts significantly reduced Tat-regulated transcription elongation from HIV-1 TAR-containing G-less templates that either lack (Fig. 4A, top left panel, cf. lanes 8 and 2,4) or contain (Fig. 4A, cf. lanes 15 and 10) the promoter-proximal HIV-1 5′-splice site. Consistent with a previous report (Fong and Zhou 2001), we find that mutation of the 5′-splice site reduces HIV-1 Tat activity in vitro (Fig. 4A, top right panel, cf. lanes 17 and 10). By comparison, Tat activity was eliminated in extracts lacking CycT1 (Fig. 4A, cf. lanes 6 and 2,4, and lanes 13 and 10). Immunodepletion of SKIP did not affect CycT1 levels, and vice versa (Fig. 4A, lanes 19–22) under these conditions. The reduced levels of SKIP and P-TEFb in these experiments were sufficient to support basal elongation rates (Fig. 4A, cf. lanes 9,12,14,16). In contrast, immunodepletion of either SKIP or P-TEFb had no effect on HIV-1 RNA initiation, as determined by primer extension analyses of RNA isolated from the same transcription reactions (Fig. 4A, bottom panels). Similarly, RNA initiation was unaffected by mutation of the 5′-splice site (Fig. 4A, bottom right panel, cf. lanes 17 and 10).
Figure 4.
SKIP facilitates basal and Tat-mediated HIV-1 transcription elongation in vitro. (A) Immunodepletion of SKIP reduces basal and Tat-mediated transcription in vitro. HeLa nuclear extracts were incubated with either 100 ng GST-Tat101 (wild type [wt]) or 100 ng GST (–), following depletion with either control (HA-specific) antibody (lanes 3,4), or with antisera specific to CycT1 (lanes 5,6) or SKIP (lanes 7,8), as indicated above each lane. (Bottom panel) HIV-1 RNA elongation was measured using “run-off” reactions containing the “G-less” reporter, whereas RNA initiation was analyzed by primer extension. (Lanes 19–22) Immunodepletion efficiency was assessed by immunoblots. (B) Recombinant SKIP activates basal and Tat-activated transcription elongation in vitro. HIV-1 transcription elongation was measured using the “G-less” reporter (lanes 1–6) in reactions containing 100 ng GST-Tat101 (wild type [wt]) or 100 ng GST (–), either alone or together with GST-SKIP, as indicated above each lane. (Lanes 7–14) The ability of recombinant GST-SKIP to activate α-globin RNA elongation was measured using “run-off” reactions containing the “G-less” reporter. (C, top panel) The ability of recombinant GST-SKIP to activate transcription was tested in lanes 1–10, where RNA elongation was measured using RNase T1-resistant “G-less” run-off transcripts and RNA initiation from the same reactions were analyzed by primer extension. RNA initiation from a human α-globin template added to the extract was included as a loading control. The ability of mutant GST-SKIP proteins to activate transcription elongation in vitro was measured in reactions containing 100 ng of either GST (lanes 11–14) or GST-Tat101 (lanes 15–18), together with 250 ng of either full-length GST-SKIP (lanes 12,16) or mutant GST-SKIP proteins (lanes 13,14,17,18), as indicated above each lane.
We next purified full-length and mutant GST-SKIP proteins and analyzed their transcriptional activity in vitro. As shown in Figure 4B, GST-SKIP stimulated basal and Tat-activated HIV-1 transcription by a factor of threefold and 10-fold, respectively, in experiments utilizing a “G-less” reporter template (cf. lanes 5,6 and 1,2). The effect of GST-SKIP on basal transcription was modest and observed only at relatively high levels of GST-SKIP, whereas the effect on HIV-1 Tat activity was robust and observed at levels of SKIP that did not affect HIV-1 basal promoter activity (Fig. 4B, cf. lanes 3,4 and 5,6) or that of the unrelated α-globin gene promoter (Fig. 4B, cf. lanes 7–10 and 11–14). Similar to its effect on HIV-1 basal promoter activity, GST-SKIP enhanced transcription elongation threefold in vitro from the adenovirus major late promoter (AdMLP), indicating that it can function on other genes (data not shown). In contrast with these effects on elongation, GST-SKIP did not affect RNA initiation from the HIV-1 promoter or control α-globin promoter (Fig. 4C, bottom left panel, cf. lanes 10 and 1) in these same reactions. Two different mutant GST-SKIP proteins (termed NS and CS) that contain the SNW domain had equivalent transcriptional activity (Fig. 4C, cf. lanes 15 and 16–18), indicating that the C terminus does not inhibit the function of the SNW domain in vitro. An N-terminal fragment of SKIP that lacks the SNW domain was inactive in vitro (data not shown). None of the mutant GST-SKIP proteins affected RNA initiation, as measured by primer extension (bottom panels). We conclude that GST-SKIP stimulates both basal and Tat-regulated transcription elongation in vitro.
SKIP associates with U5snRNPs and CycT1:CDK9/P-TEFb in nuclear extracts
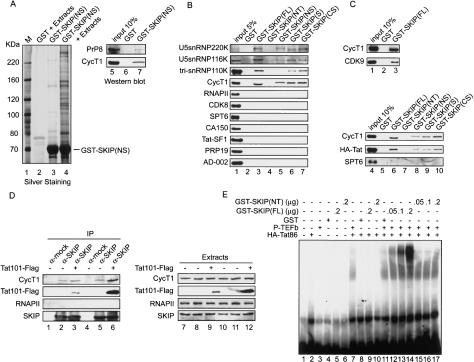
We next used a protein interaction assay to attempt to identify transcription factors that associate with GST-SKIP in extracts. Glutathione beads coated with purified GST-SKIP(NS) were incubated with the crude HeLa nuclear extract, and the interacting proteins that remained on the beads following stringent washes were eluted by boiling and analyzed by SDS-PAGE and silver staining (Fig. 5A). The most prominent SNW-associated factors were subjected to tryptic digestion and identified by MALDI-TOF mass-spectrometry. Among several proteins identified by this approach were the U5snRNP220K and hPrp8 proteins, which have been reported previously to bind the SKIP SNW domain (Zhang et al. 2003). Interestingly, the 86-kDa P-TEFb subunit, CycT1, was also present in these fractions. Both hPrp8 and CycT1 were confirmed by Western blots to associate with GST-SKIP(NS), and not with GST alone (Fig. 5A, lanes 5–7). We further observed that hPrp8 and CycT1 associate with full-length GST-SKIP and GST-SKIP(S), but not with GST-SKIP(NT) or GST alone (Fig. 5B).
Figure 5.
SKIP interacts with U5snRNP proteins and P-TEFb. (A) GST-SKIP(NS) interacts with U5-snRNP proteins and P-TEFb in nuclear extracts. Proteins bound to GST-SKIP(NS)-coupled glutathione beads were isolated and separated by SDS-PAGE prior to being analyzed by silver-staining (left panel) or by immunoblot with anti-Prp8 or anti-CycT1 antisera (right panel). (Right panel) The input HeLa extract is shown in lane 1. (Top panel, lane 1) M designates protein size markers. (Left panel) HeLa nuclear proteins bound to GST (lane 2) or GST-SKIP(NS) (lane 3) were identified by silver staining of the SDS-PAGE and identified by MALDI-TOF microsequencing (lane 4). (B) The SKIP SNW domain is necessary and sufficient to interact with U5-snRNPs, tri-snRNP110K, and P-TEFb. HeLa nuclear extract was incubated with either GST (lane 2) or various GST-SKIP-coupled glutathione beads (lanes 3–7) as indicated. The input HeLa extract is shown in lane 1. SKIP-interacting proteins were analyzed by Western blots, as indicated at the left of the panel. (C) HIV-1 Tat associates with GST-SKIP in extracts. Proteins interacting with GST-SKIP in HeLa nuclear extract (lanes 1–3) or with a nuclear extract from HeLa Tat-expressing cells (lanes 4–10) were analyzed by Western blot for the proteins indicated in each panel. (D) Coimmunoprecipitation of endogenous SKIP and CycT1 proteins in extracts that contain or lack exogenous HIV-1 Tat. HeLa cells (lanes 1–3,7–9) and HeLa P4 cells (lanes 4–6,10–12) were transfected with either an empty vector (lanes 1,2,4,5,7,8,10,11) or with the Flag-Tat101-coding vector (lanes 3,6,9,12). Cellular extracts prepared 48 h post-transfection were subjected to immunoprecipitation (IP; lanes 1–6) with an anti-SKIP antibody and analyzed by immunoblotting with antisera specific for the proteins indicated at the left of the panel. (Lanes 7–12) The input cell extract was also analyzed by direct immunoblot using the indicated antibodies. (E) SKIP associates with Tat:P-TEFb complex to the TAR RNA. Binding of recombinant Tat, CycT1:CDK9/P-TEFb, and SKIP(FL) or SKIP(NT) to HIV-1 TAR RNA was analyzed with gel-mobility shift experiments. Binding reactions contained 100 ng of HA-Tat86, 200 ng of CycT1:CDK9/P-TEFb, and indicated amount of either GST-SKIP(FL) (lanes 11–13) or GST-SKIP(NT) (lanes 15–17).
Where available, we tested additional antisera to examine whether other splicing factors also associate with GST-SKIP in the pull-down fraction. Interestingly, we identified a U4/U5.U6tri-snRNP component, termed tri-snRNP110K, that also associates with GST-SKIP (Fig. 5B). In contrast, SKIP does not interact with Tat-SF1, CA150, Prp19, AD-002, and CDC5 (Fig. 5B; data not shown). GST-SKIP also failed to interact with a number of other transcription factors we tested, including RNAPII, CDK8, Spt6, or p300 (Fig. 5B; data not shown). The catalytic subunit of P-TEFb, CDK9, was also present in the GST-SKIP pull-down reactions (Fig. 5C, lane 3), and we also recovered HA-Tat along with P-TEFb in extracts from Tat-expressing cells (Fig. 5C, lanes 6–10). Importantly, none of these interactions were affected by treating the extracts with RNaseA or ethidium bromide (data not shown), indicating that these protein interactions are not mediated through RNA or nucleic acids. We conclude that the SKIP SNW domain associates, directly or indirectly, with U5snRNPs and tri-snRNP110K, but not with the entire 35S tri-snRNP particle.
We further examined the interaction between SKIP and P-TEFb, or Tat:P-TEFb complexes by coimmunoprecipitation experiments. Interestingly, endogenous CycT1 protein was detected by Western blot in anti-SKIP immunoprecipitates from extracts without or with exogenously expressed HIV-1 Tat101 protein in both HeLa (Fig. 5D, lanes 2,3, respectively) and HeLa P4 cell extracts (Fig. 5D, lanes 5,6). In contrast, SKIP did not interact with RNAPII. To conclude, SKIP associates with native P-TEFb and Tat:P-TEFb complexes in vivo.
Recombinant SKIP interacts with Tat:P-TEFb:TAR RNA complexes in vitro
To further characterize the interaction with P-TEFb, we asked whether GST-SKIP can recognize the recombinant Tat:P-TEFb complex when bound to TAR RNA in vitro. We showed previously that an extended region of the CycT1 cyclin box domain (amino acids 1–301) mediates binding to Tat and TAR in vitro (Wei et al. 1998), whereas Tat:P-TEFb complexes containing full-length recombinant CycT1 protein (amino acids 1–726) are unable to bind TAR RNA (Garber et al. 2000). Relatively weak binding to the RNA is observed when these Tat:P-TEFb complexes are incubated with ATP (Garber et al. 2000); however, CDK9 activity is not required for Tat:P-TEFb:TAR complexes to form in crude extracts (see Supplementary Fig. 2), indicating that other factors may be necessary for stable binding to TAR in vitro.
For these experiments, HA-Tat86 was incubated with TAR RNA in the presence of purified baculovirus-expressed full-length human CycT1 and CDK9 proteins and either full-length GST-SKIP, or the transcriptionally inactive GST-SKIP(NT) protein. ATP was included in the reactions to enable binding of Tat:P-TEFb to RNA, and specific RNA-binding complexes were monitored by EMSA (Fig. 5E). Neither SKIP nor P-TEFb bound TAR RNA in the absence of Tat (Fig. 5E, cf. lanes 3,5,6 and 1). Neither GST-SKIP(FL), GST-SKIP(NT), nor GST alone had any effect on the relatively low-affinity Tat:TAR complex (Fig. 5E, cf. lanes 2 and 8–10). The full-length recombinant Tat:P-TEFb bound weakly to TAR RNA under these conditions (Fig. 5E, lane 7); however, a new, high-affinity complex was observed in the presence of SKIP (Fig. 5E, cf. lanes 12–14 and 7). SKIP did not bind TAR RNA in the absence of Tat or P-TEFb (Fig. 5E, lanes 5,9), and the Tat:P-TEFb:TAR complex was unaffected by the transcriptionally inert GST-SKIP(NT) protein (Fig. 5E, lanes 15–17). To date, we have not detected any direct interaction between SKIP and CycT1 (data not shown), and it remains to be determined whether recombinant SKIP directly interacts with P-TEFb, or may instead bind RNA sequences that are exposed upon binding of Tat:P-TEFb to TAR. We conclude that SKIP interacts directly with the Tat:P-TEFb:TAR complex in a manner that depends upon the SNW domain and correlates with transcriptional activation in vitro.
Tat recruits P-TEFb and SKIP to the integrated HIV-1 promoter in vivo
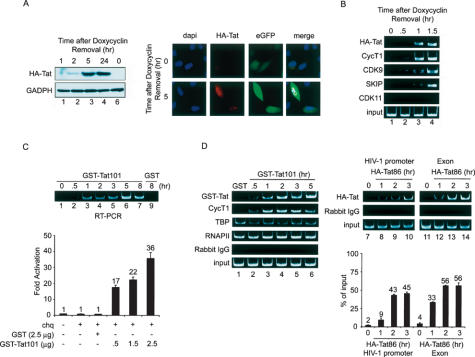
Taken together, these data indicate that SKIP enhances HIV-1 Tat-mediated transcription in conjunction with P-TEFb. To determine whether SKIP is recruited by Tat to the integrated HIV-1 LTR promoter in vivo, ChIP experiments were carried out using both a Tat-inducible HeLa cell line, as well as Tat-transduced HeLa P4 cells. To create an inducible Tet-off, Tat-on cell line, the HIV-1 HA-Tat86 protein was placed under control of the TRE promoter and individual subclones were selected in which Tat was induced strongly within 1–2 h following the removal of doxycycline (I. Turbachova and K.A. Jones, unpubl.). An HIV-1 LTR vector linked to the enhanced green fluorescence protein (eGFP) gene was then introduced into these cells, and transfectants were selected with blasticidin and sorted by fluorescence-activated cell sorting (FACS) for low eGFP activity in the presence of doxycycline. Induction of HA-Tat86 in the selected cells was confirmed by immunoblotting and immunohistochemical staining using anti-HA antiserum (Fig. 6A). Expression of HA-Tat was accompanied by increased eGFP expression from the HIV-1 LTR, as visualized by immunofluorescence (Fig. 6A). Formaldehyde cross-linked chromatin was isolated from these cells in the presence or absence of doxycycline treatment, immunoprecipitated with HA-tag-specific antibodies, and subjected to PCR using a primer pair specific for the HIV-1 LTR promoter (–149 to +69).
Figure 6.
(A) Induction of HA-Tat following removal of doxycycline in a stable cell line containing an integrated HIV-1 LTR-eGFP reporter gene. (Lanes 1–6) Levels of HA-Tat and control GAPDH proteins at different intervals following doxycycline removal were examined by Western blot, and the induced Tat and eGFP proteins were analyzed by immunofluorescence and deconvolution microscopy. (B, lanes 1–4) ChIP analysis of binding of factors to the integrated HIV-1 LTR promoter at different time points upon induction of Tat. (C, top panel) Purified recombinant HIV-1 HA-Tat86 induces HIV-1-LacZ mRNA, as determined by RT–PCR of total RNA isolated from HeLa P4 cells transduced with GST (lane 9) or GST-Tat101 (lanes 1–7) for the different times indicated above each lane. Recombinant Tat activates HIV-1 transcription in HeLa P4 cells transduced with GST (lane 1) or GST-Tat101 (lanes 2–4; the various time intervals following removal of doxycycline are indicated above each lane). Hela P4 cell lysates were analyzed for β-galactosidase activity at 48 h post-transfection. (Lower panel) Fold activation was calculated relative to β-galactosidase levels in nontransduced cells. (D) ChIP analysis of factors bound to the integrated HIV-1 LTR in HeLa P4 cells transduced with either GST-Tat101 (left panel) or HA-Tat86 (top right panel). Time intervals refer to total elapsed time following transduction of GST (lane 1), GST-Tat101 (lanes 2–6), or HA-Tat86 (lanes 8–10,12–14). Real-time PCR (lower right panel) was performed on the same DNA recovered from the HA-specific immunoprecipitation reactions (lanes 7–14, top right panel).
The ChIP experiments indicated that HA-Tat was present at the HIV-1 LTR within an hour of doxycycline removal (Fig. 6B, cf. lanes 3 and 1). Tat strongly increased the levels of CycT1 and CDK9 associated with the integrated HIV-1 promoter in vivo, and endogenous SKIP was also recruited to the promoter at this time (Fig. 6B). In contrast, we did not detect the splicing factor-associated kinase, CDK11, at the HIV-1 promoter, and no proteins were detected using control rabbit IgG antiserum (Fig. 6B; data not shown). Thus SKIP and P-TEFb are recruited with similar kinetics to the HIV-1 promoter upon induction of Tat in vivo.
To eliminate any possible effects of low levels of Tat on the basal conditions in these experiments, ChIPs were repeated in HeLa P4 cells transduced with recombinant Tat protein. Purified GST-Tat101, but not GST, strongly enhanced transcription from the integrated HIV-1 reporter within 0.5 h after transduction of the HeLa P4 cells, as determined by RT–PCR (Fig. 6C, cf. lanes 2–7 and 1,9) and β-galactosidase activity measurements (Fig. 6C, lower panel). Binding of GST-Tat101 to the HIV-1 LTR by ChIP was accompanied by a strong increase in the levels of CycT1 (Fig. 6D). In contrast, RNAPII and the TATA-binding protein, TBP, were constitutively present at HIV-1 LTR promoter in these experiments (Fig. 6D), consistent with the model that RNAPII complexes arrest at the HIV-1 promoter in the absence of Tat. The recruitment of HIV-1 HA-Tat86 to the HIV-1 promoter and exon regions observed by ethidium staining in these experiments (Fig. 6D, lanes 7–10,11–14, respectively) were also confirmed by real-time PCR measurements (Fig. 6D, bottom panel), which further established the validity of the ChIP conditions used in these experiments.
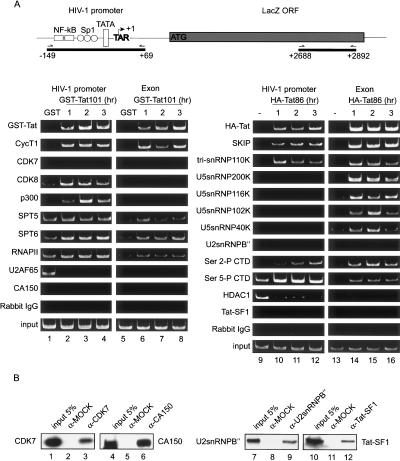
To evaluate the composition of basal and Tat-induced elongation complexes, a primer set specific for sequences near the end of the intronless LacZ gene was used in the ChIP experiments (Fig. 7, schematic diagram). Surprisingly, the Ser 5 CTD kinase, CDK7, was not detected at the HIV-1 promoter in either the presence or absence of Tat (Fig. 7, lanes 1–4), despite the presence of high levels of RNAPII (Fig. 7, lanes 1–4) that is Ser 5-phosphorylated (Fig. 7, lanes 9–12). The transcription elongation factors Spt5, Spt6, and NELF were also present at the promoter and within the gene, and levels of these proteins did not change upon Tat induction, consistent with their proposed role in RNAPII pausing (for review, see Kim et al. 2001). The levels of CDK8 and p300 at the promoter were higher in the presence of Tat (Fig. 7, lanes 1–4). As expected, none of these factors were found in elongation complexes within the body of the LacZ gene (Fig. 7, lanes 5–8). Interestingly, the U2AF65 splicing factor was readily detected at the HIV-1 promoter in the absence of Tat, but disappeared from the promoter upon Tat transactivation (Fig. 7, lanes 1–4).
Figure 7.
ChIP analysis of the recruitment of transcription and splicing factors to the HIV-1 promoter in the absence and presence of HIV-1 Tat. (A) A schematic diagram of the HIV-1 promoter LacZ gene indicating the primer sets used for ChIP (–149 to +69) or end of the LacZ ORF (+2688 to +2892) is shown above the panels. Purified GST (lanes 1,5) or GST-Tat101 (lanes 2–4,6–8) proteins were transduced into HeLa P4 cells for the different amounts of time indicated above each lane, and factors bound to the HIV-1 promoter or LacZ gene were analyzed by ChIP. The different proteins tested for binding to the HIV-1 promoter (lanes 1–4,9–12) or LacZ exon (lanes 5–8,13–16) are indicated to the left of each panel. (B) Control reactions testing the ability of the CDK7, CA150, U2snRNPB′′, and Tat-SF1 antisera to immunoprecipitate the appropriate protein, as determined by Western blot.
To determine whether the U5snRNP complex also accompanies SKIP to the HIV-1 promoter, ChIP experiments were carried out with HeLa P4 cells transduced with purified HA-Tat86 protein. As shown in Figure 7, endogenous SKIP appeared with Tat at the HIV-1 promoter and downstream region of the LacZ gene (lanes 13–16). Interestingly, the tri-snRNP110K protein appeared with Tat and SKIP at the HIV-1 promoter. In contrast, the U5-snRNP proteins were detected only within the body of the LacZ gene following Tat transactivation (Fig. 7, lanes 13–16). SKIP, as well as CycT1, was recruited to the activated c-Myc promoter, whereas the U5snRNP116K was detected only within the exon of the gene (Supplementary Fig. 1C). A gradual increase in Ser 2 CTD phosphorylation was observed in the presence of Tat, whereas levels of Ser 5 CTD phosphorylation were relatively unchanged (Fig. 7, lanes 9–12). HDAC1 was associated with the repressed promoter in the absence of Tat, and disappeared upon Tat induction, whereas Tat-SF1 and U2snRNPB′ were not detected at the HIV-1 promoter or within the LacZ gene in either the presence or absence of Tat. No signal was detected in reactions that contained rabbit IgG antiserum. As shown in Figure 7B, the antisera to CDK7, CA150, U2snRNPB”, and Tat-SF1 used in these experiments efficiently immunoprecipitate the appropriate proteins. These studies indicate that SKIP plays an important role in Tat-mediated transcription elongation, as presented in the model shown in Figure 8, and discussed below.
Figure 8.
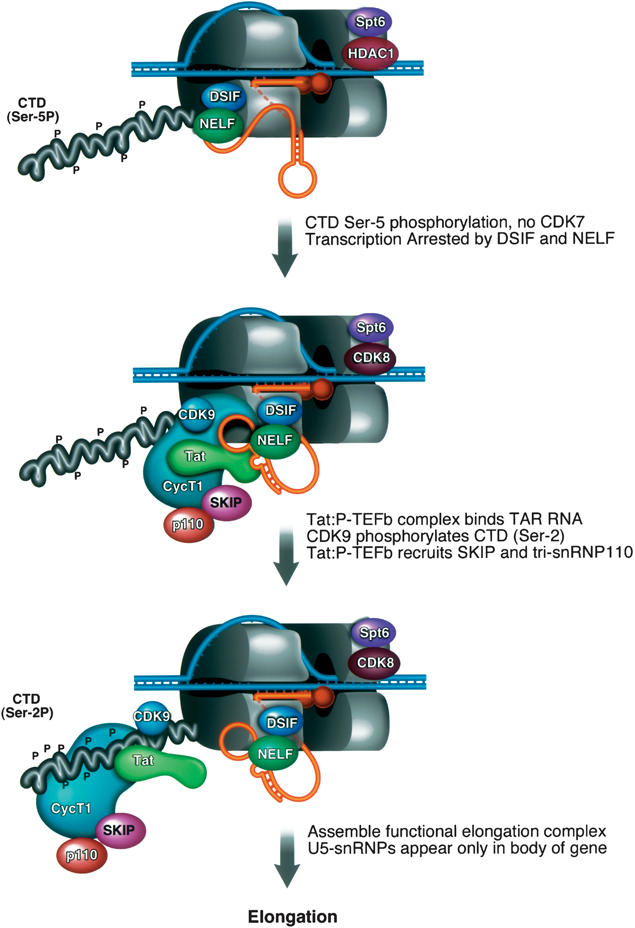
A model for the role of SKIP in HIV-1 Tat transactivation. In the absence of Tat, TBP and RNAPII are present at the promoter but transcription elongation is blocked. The RNAPII CTD is phosphorylated at the Ser 5 position, despite the absence of CDK7. In this state, RNAPII is proposed to be arrested prematurely following the actions of negative elongation factors, including DSIF and NELF. Tat interacts with CycT1 and binds cooperatively with P-TEFb to nascent TAR RNA in a step that also brings SKIP and tri-snRNP110K splicing proteins to the promoter. SKIP associates, directly or indirectly, with P-TEFb in cell extracts, and enhances transcription elongation in vitro. Recombinant SKIP can also interact with Tat:P-TEFb:TAR complexes in vitro, indicating that it may function through the nascent RNA. Although SKIP is present with P-TEFb at the viral promoter, the U5snRNPs are found only within the transcribed region of the gene, suggesting that SKIP acts at different times, and with different partners, to promote transcription elongation and splicing.
Discussion
The specific interaction between HIV-1 Tat and human CycT1 facilitates binding of Tat:P-TEFb to TAR RNA; however, little is known of the other factors that function at this step to promote elongation and subsequent processing of viral pre-mRNAs. Our observation that SKIP, a U5snRNP-associated splicing factor, physically associates with P-TEFb and enhances HIV-1 Tat-regulated elongation provides some insights into the role of transcription elongation and splicing factors at the HIV-1 promoter.
The process of Tat transactivation was examined by ChIP analysis of a Tet-off, Tat-on inducible HeLa cell line and in Hela P4 cells transduced with recombinant Tat, both of which carry an integrated HIV-1 LTR reporter gene. Collectively, these data suggest a model for Tat-regulated elongation that includes specific pre-mRNA splicing factors (Fig. 8). Consistent with a predominant role for Tat and P-TEFb in RNAPII elongation, Ser 5-phosphorylated RNAPII, TBP, and Spt5 were readily detected at the HIV-1 promoter in the absence of Tat. The CTD Ser 5 kinase, CDK7, was not detected in these experiments, which is consistent with an earlier report that CDK7 is not required for Tat activity in vitro (Chen and Zhou 1999). Consequently, the identity of the Ser 5 kinase at the HIV-1 promoter remains to be established. Our findings contrast with a recent report that Tat recruits RNAPII and TBP to the HIV-1 promoter in vivo (Raha et al. 2005). One possible reason for this discrepancy is that transfected HIV-1 promoter templates may not function identically to the integrated HIV-1 promoter that we examined.
Tat strongly increased the level of CycT1:CDK9/P-TEFb at the integrated HIV-1 promoter, accompanied by the appearance of at least two splicing factors, SKIP and tri-snRNP110K. We show here that SKIP enhances Tat-regulated elongation, and associates with P-TEFb and Tat:P-TEFb complexes in extracts. SKIP may engage Tat:P-TEFb on the nascent TAR RNA or could transfer with the complex to the RNAPII CTD, which serves as a scaffold for assembly of spliceosomal complexes (Zorio and Bentley 2004). Our findings also confirm earlier reports that Tat recruits histone acetyltransferases (p300, P/CAF) to the HIV-1 LTR (Lusic et al. 2003). In addition, PTEFb kinase activity was recently shown to be required for H3-K4 and H3-K36 methylation at the integrated HIV-1 promoter (Zhou et al. 2004). Thus, it will be important to establish how these various chromatin modifying enzymes function coordinately with P-TEFb to integrate CTD phosphorylation with changes in histone modification states.
A role for SKIP in RNAPII transcription elongation
SKIP has been characterized as a coactivator of Notch, VDR, and SMAD proteins and a corepressor for other genes (for review, see Folk et al. 2004); however, its role in transcription is not well defined mechanistically. We show here that SKIP activates HIV-1 and c-Myc transcription in vivo and that recombinant SKIP enhances basal- and Tat-regulated elongation at the HIV-1 promoter in vitro, without affecting RNA initiation. As was reported previously for Tat (Fong and Zhou 2000), we find that SKIP activity in vitro is optimal on HIV-1 templates that include the 5′-splice site (Fig. 4); however, both SKIP and Tat can activate HIV-1 templates that lack splice sites. Previous observations that SKIP appears at a relatively late stage in the VDR transcription cycle (Zhang et al. 2003), and binds to the Notch-regulated HES1 promoter simultaneously with P-TEFb (Fryer et al. 2004), are also consistent with a post-initiation role in transcription.
To better understand how SKIP functions in transcription, mass spectrometry and immunoblotting were used to identify proteins that interact, directly or indirectly, with GST-SKIP in pull-down experiments from nuclear extracts. SKIP has been identified as a stable component of 35S U5snRNP particles and 45S spliceosomes (Makarov et al. 2002), and associates with U5snRNP proteins, including hPrp8 and U5snRNP200K, in extracts (Zhang et al. 2003). Our data confirm the association with U5snRNP subunits, and show that SKIP also associates with CycT1:CDK9/P-TEFb and the 25S [U4/U6.U5] tri-snRNP protein, tri-snRNP110K. In contrast, GST-SKIP did not interact with several other splicing factors we tested, including hPrp19, AD-002, CDC5, Tat-SF1, and CA150 (Fig. 5; data not shown). Thus the SKIP SNW domain may independently engage P-TEFb and U5snRNPs at different stages during transcription and splicing. The ability of GST-SKIP to interact with Tat in nuclear extracts from HIV-1 Tat-expressing cells appears to be indirect and mediated through P-TEFb, and further suggests that binding of Tat to CycT1 does not affect the ability of SKIP to engage P-TEFb.
Whereas SKIP and tri-snRNP110K are recruited simultaneously with Tat and P-TEFb to the HIV-1 promoter in vivo, the U5snRNP proteins were detected only in the transcribed coding region (summarized in Fig. 8). These findings strongly suggest that SKIP associates independently with splicing and transcription complexes, consistent with the fact that P-TEFb was not identified proteomically as a spliceosomal component (Makarov et al. 2002). The association of P-TEFb with SKIP may contribute to its colocalization with splicing factor-rich speckles in the nucleus (Herrmann and Mancini 2001). The observation that U5snRNPs reside only within transcribed regions of the gene is also consistent with an earlier report that yeast U1snRNPs associate primarily with intronic and exonic regions of genes and are not efficiently recruited to promoters (Kotovic et al. 2003). We have not determined whether the interaction of U5snRNPs with the transcribed intronless HIV-1 reporter gene is mediated through DNA (i.e., the RNAPII elongation complex) or the pre-mRNA. To rule out the possibility that SKIP is recruited to the HIV-1 promoter though binding to the Ser 2-phosphorylated CTD, ChIP experiments were carried out in cells treated with the CDK9 inhibitor, flavopiridol (Chao and Price 2001). In the absence of CTD Ser 2 phosphorylation, SKIP and P-TEFb were readily detected at the HIV-1 promoter even in the absence of Tat, potentially as components of arrested basal transcription complexes (see Supplementary Fig. 2). We conclude that SKIP is not recruited to the HIV-1 promoter as an indirect consequence of Tat:P-TEFb-stimulated RNAPII phosphorylation. Most importantly, these data indicate that SKIP activates HIV-1 transcription independently of the U5snRNPs, and may engage transcription and splicing complexes through its SNW domain at different stages in the elongation process.
Interestingly, we find that the SKIP-associated tri-snRNP110K protein is recruited to the HIV-1 promoter, whereas other proteins implicated in splicing, including Tat-SF1, CA150, and the tri-snRNP CDC5 protein, were not (Fig. 7; other data not shown). The failure to detect Tat-SF1 and CA150 was surprising and is not readily reconciled with published reports that Tat-SF1 functions through binding to Tat and P-TEFb (Zhou et al. 1998; Fong and Zhou 2000). Although we cannot rule out the possibility that these proteins are present but failed to cross-link to the viral chromatin, the results are consistent with the fact that these splicing factors do not associate with SKIP (Fig. 5). Further work is required to establish whether tri-snRNP110K associates independently with SKIP and to identify any other proteins that may be present in SKIP transcription complexes. Tri-snRNP110K is an essential SR-like protein that recruits the tri-snRNP complex to prespliceosomes (Makarova et al. 2001). Our preliminary experiments indicate that tri-snRNP110K does not appear to alter Tat activity in vivo or in vitro (V. Brès and K.A. Jones, unpubl.), and it remains to be determined why it is recruited to the viral promoter. Taken together, these studies indicate that only a subset of splicing factors are recruited to promoters, and these proteins may play a direct role in transcription that is distinct from their subsequent role in splicing.
SKIP interacts with Tat:P-TEFb on TAR RNA in vitro
Interestingly, the ability of SKIP to activate transcription and to enhance binding of Tat:P-TEFb to TAR RNA in vitro are both mediated through the SNW domain. These findings indicate that binding of SKIP to Tat:P-TEFb on nascent RNA may underlie its strong enhancement of Tat-regulated transcription in vitro. The observation that SKIP can alter the use of alternative HIV-1 splice sites further suggests that it may function through RNA. To date we have not detected any SKIP-dependent change in RNAPII CTD phosphorylation in vivo (V. Brès and K.A. Jones, unpubl.). Thus, the Tat-associated protein complex on TAR may serve a dual function in recruiting P-TEFb and other elongation factors to the HIV-1 promoter and promoting the escape of the paused RNAPII complex.
A subset of splicing factors acts directly in transcription
Although the U5snRNPs do not interact with the HIV-1 promoter, other individual splicing-related proteins may play direct roles in transcription. For example, the U2snRNP spliceosomal protein, Sf3b1, was recently shown to be required for Polycomb-mediated repression of Hox genes and is recruited to repressed Hox gene promoters in vivo (Isono et al. 2005). Similarly, we detected high levels of U2AF65 at the HIV-1 promoter in the absence, but not presence, of Tat (Fig. 7), indicating that U2AF65 might repress or attenuate HIV-1 transcription in vivo. Interestingly, Sf3b1 has been shown to interact directly with U2AF65 (Gozani et al. 1998), which raises the possibility that the repressed HIV-1 promoter may be regulated by Polycomb protein complexes. The observation that U2AF65 is present together with the paused RNAPII complex is also interesting in light of a recent report that it can be cross-linked to emerging nascent transcripts in vitro (Ujvari and Luse 2004). By contrast, we failed to detect the U2snRNPB” spliceosomal protein at either the HIV-1 promoter or exonic region, indicating that U2snRNP recruitment may be intron dependent. The interaction between SKIP and P-TEFb may also influence other post-transcriptional events that require P-TEFb, including RNA 3′-end processing (Ahn et al. 2004; Bird et al. 2004; Ni et al. 2004). In this respect, it is interesting to note that a yeast ortholog of CDK9, Ctk1, copurifies with two TREX-associated factors that are required for elongation as well as RNA nuclear export (Hurt et al. 2004).
In summary, our data strongly suggest a role for SKIP at an early step in Tat-regulated elongation at the HIV-1 promoter. The ability of recombinant SKIP to enhance elongation correlates with its binding to the Tat:P-TEFb:TAR RNA complex. Because the integrated Tat-responsive HIV-1 reporter gene used here lacks the promoter-proximal viral 5′-splice site, it will be interesting to learn whether HIV-1 splice sites might function in vivo to recruit additional splicing factors, such as Tat-SF1 and CA150, which can stimulate transcription synergistically with Tat (Zhou and Sharp 1996; Sune et al. 1997; Zhou et al. 1998). It will also be important to compare the results obtained here with the more complex transcription factor interactions that occur on the native HIV-1 genome in the activated T cells, where both initiation and elongation are further up-regulated by enhancer factors such as NF-κB and NF-AT. Because our experiments were designed to analyze the events most directly involved in Tat transactivation, these results should provide a useful comparative framework for similar studies of the induced viral genome in activated T cells and macrophages.
Materials and methods
Plasmids and recombinant proteins
pLTR-luc, pTat101, pHA-SKIP, pΔPSP, pGST-Tat101, and pGST-HA-Tat86 were described previously (Wei et al. 1998; Fryer et al. 2004; Ropers et al. 2004). pGST-SKIP was generated by subcloning SKIP cDNA into EcoRI and XhoI sites of pGEX-KG (Pharmacia). Truncation mutants of pGST-SKIP and pHA-SKIP mutants were made using the QuickChange kit (Stratagene). The HIV-1 G-less plasmid was obtained by cloning a G-less cassette into EcoRV and ClaI sites of pLTR-Luc. HIV-1 5′ss wild-type G-less plasmid was generated by introducing a G-less cassette into pNL4-3. HIV-1 5′ss mutant G-less plasmid containing three mutations at the 5′ splice site was generated by using the wild-type vector as template. The α-globin G-less plasmid was created by inserting a G-less cassette 2.3 kb downstream of the promoter into the EcoRI site.
Transfection, transduction, and reporter assays
HeLa P4 cells, which contain the LacZ gene under control of the integrated HIV-1 LTR (Clavel and Charneau 1994), were propagated in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and transfected according to the manufacturer's protocol using Effectene (Qiagen). Transductions were carried out in presence of 100 μM chloroquine (Sigma) as previously described (Becker-Hapak et al. 2001). To assay luciferase activity, HeLa cells were transfected, lysed, and assayed for luciferase activity 48 h after transfection, according to the manufacturer's protocol (Promega). Luciferase activity was normalized to pRL-TK (Promega), which encodes the Renilla luciferase from TK promoter, as internal control. β-Galactosidase activity was measured in extracts of HeLa P4 cells 48 h after transfection, according to the manufacturer's protocol (Roche). For the experiments shown in Figures 1 and 2, mean relative luciferase or β-galactosidase activities were derived from at least three independent experiments.
Small-interfering RNAs
RNA oligonucleotides for luciferase (forward, 5′-CGUACGCG GAAUACUUCGATT-3′; reverse, 5′-UCGAAGUAUUCCGCG UACGTT-3′) and SKIP (forward, 5′-AUGUCGAAUGCGCUG GCCATT-3′; reverse, 5′-UGGCCAGCGCAUUCGACAUTT-3′) were synthesized (Proligo). The sense and antisense oligonucleotides were mixed in annealing buffer (30 mM HEPES at pH 7.4, 100 mM potassium acetate, and 2 mM magnesium acetate) before use. HeLa P4 cells were doubly transfected using Oligofectamine (Invitrogen) with 70 nM siRNAs, as indicated. At 24 h post-transfection, cells were split into 24-well plates and either transfected with expression plasmids using Effectene (Qiagen) or transduced with recombinant Tat, as indicated in Figure 2. Expression levels of SKIP, CDK9, CycT1, and GADPH proteins were monitored by immunoblot, and β-galactosidase activities were analyzed in the extracts 48 h after either transfection of pTat 101 plasmid or following transduction of GST-Tat101.
Coimmunoprecipitation experiments
Either HeLa cells or HeLa P4 cells, as indicated in Figure 5D, were transfected with 2 μg of wild-type Flag-HIV-1 Tat plasmid, or a control vector, using Effectene (Qiagen), and analyzed 48 h post-transfection. The cells were washed and suspended in EBC/120 buffer (50 mM Tris-HCl at pH 8, 120 mM NaCl, and 0.5% NP-40) containing 2 mM DTT, 0.1 mM PMSF, and protease inhibitors, frozen in liquid nitrogen, and thawed for 30 sec at 37°C. Lysates were precleared for 1 h at 4°C with proteinA/G (Santa Cruz) and incubated with anti-SKIP antibody for 2 h at 4°C, followed by the addition of protein A/G for1hat4°C. The beads were washed three times with EBC buffer containing 150 mM NaCl prior to elution of the proteins and analysis by SDS-PAGE and immunoblotting. Proteins were visualized by chemoluminescence.
In vitro transcription reactions
Two hundred nanograms of HIV-1 G-less plasmid was preincubated in a 25-μL reaction with HeLa transcription extract, recombinant proteins, and buffer containing 15 mM Tris-HCl at pH 7.9, 25 mM KCl, 6.25 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 10% glycerol, 3 mM ATP, 10 mM phospho-creatine, 5 μM CTP, and 600 μM ATP, GTP, and UTP. Transcriptional initiation was assayed by primer extension on the nascent transcripts using [γ32P]-labeled primer annealing at +75 nucleotides (nt) of the template. Elongated transcription was assayed by including 10 μCi [α32P]-CTP to the reaction. The mixture was incubated 30 min at 30°C to allow the transcription and digested with RNaseT1 (Roche). Both primer extension and RNaseT1-resistant G-less products were purified by phenol chloroform extraction and ethanol precipitation and resolved on 8 M urea sequencing gel followed by quantification using a Molecular dynamic phosphoimager.
Protein-interaction GST-pull-down experiments
HeLa nuclear extracts diluted 1:1 in binding buffer (50 mM Tris-HCl at pH 7.9, 120 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, 2 mM DTT, 0.1 mM PMSF, and 1× protease inhibitors) were incubated for 3 h at 4°C with 15 μL of Glutathione Sepharose 4B bead slurry saturated with bacterially expressed full-length or truncated GST-SKIP. Subsequently the beads were washed extensively (three times, 10 min at 4°C) in 400 μL wash buffer (20 mM Hepes at pH 8.0, 10% glycerol, 0.3 M KCl, 0.1% NP-40, 0.2 mM EDTA, 2 mM DTT, 0.1 mM PMSF, and 1× protease inhibitors) and for 5 min at 4°C in 400 μL wash buffer containing 0.1 M KCl. The pull-down samples were subjected to conventional SDS-PAGE and then silver stained or transferred to nitrocellulose for Western blotting analysis.
RNA-binding experiments
Gel mobility shift reactions were carried out as described previously (Garber et al. 2000). Bacterially expressed GST-HA-Tat86 was treated with thrombin prior to be used. Proteins were assembled in binding buffer (30 mM Tris at pH 8, 12% glycerol, 70 mM KCl, 0.03% NP-40, and 2 mM DTT) and incubated for 30 min at 30°C with TAR RNA probes labeled in vitro using a linearized DNA template and T7 RNA polymerase. RNA-bound complexes were separated on a prerun 4% Tris-Glycine gel.
Ex vivo splicing experiments
An HIV-1 plasmid deleted between nucleotides 1511 and 4551 referred to as pΔPSP (Ropers et al. 2004) was transfected into HeLa cells either alone or together with plasmids encoding SKIP proteins. Total cellular RNA was isolated from the transfected HeLa cells 48 h after transfection with the TRIzol reagent (Invitrogen) and was treated with DNase. Two micrograms of RNA were reverse transcribed by the MMLV reverse transcriptase according to the manufactured's protocol (Invitrogen), using oligonucleotide that spans the D4 and A7 splice sites as a primer. The Platinum PCR Super mix (Invitrogen) and the following primers, allowing the selective amplification of the 1.8-kb HIV size class mRNAs, were used to PCR ∼10% of each RT product: forward, 5′-GGCTTGCTGAAGCGCGCACGGCAAGAGG-3′; reverse, 5′-TTGGGAGGTGGGTTGCTTTGATAGAG-3′. Amplification products were radiolabeled by performing a single round of PCR with the addition of 10 μCi [α32P]-dCTP, and the products were analyzed by electrophoresis on 8 M urea sequencing gel.
ChIP
Tat reporter cells were derived by transfecting HeLa tat cells with plasmid LTR-eGFP, which contains the HIV-1 LTR linked to the eGFP gene. Transfectants were selected in medium (DMEM plus 10% FBS) containing 5 μg of blasticidin per milliliter. The resulting drug-resistant population was sorted by FACS for cells expressing low but detectable green fluorescence protein in the presence of 1 mg/mL doxycycline.
For the ChIP experiments, 5 × 107 HeLa P4 cells were cultured with 2 μg/mL of recombinant Tat protein for 0–3 h. Cells were fixed in 1% formaldehyde at room temperature for 15 min. The cross-linking was stopped by the addition of glycine to a final concentration of 0.125 M. The cells were lysed and sonicated as described (Fryer et al. 2004). Soluble chromatin was collected as supernatant after a 10-min centrifugation at 12,000 rpm. One milligram of total proteins was precleared and incubated with the appropriate antibodies overnight. Immunocomplexes were washed twice with RIPA buffer, four times with IP wash buffer, and twice more with RIPA buffer. Cross-links were reversed and DNA was purified by phenol chloroform extraction and ethanol precipitation and dissolved in 10 mM Tris-HCl (pH 8.5). The Platinum PCR Super mix (Invitrogen) and primers spanning either the promoter region (forward, 5′-CATCCGG AGTACTTCAAGAACTGC-3′; reverse, 5′-GGCTTAAGCAG TGGGTTCCCTAG-3′) or the end of the LacZ gene (forward, 5′-GATTGATGGTAGTGGTCAAATGGC-3′; reverse, 5′-CAT GTCTGACAATGGCAGATCCC-3′) were used to PCR the immunoprecipitated DNA. Thirty-five PCR cycles were performed, each cycle consisting of 1 min 94°C denaturation, 1 min annealing at 55°C, and 30 sec elongation at 72°C. PCR products were resolved using 2% polyacrylamide/TBE gels, stained with ethidium bromide. PCR with DNA prior to immunoprecipitation (input DNA) was used as a control for PCR amplification.
Real-time PCR was performed by combining a one-tenth aliquot of the ChIP reaction with 12.5 μL of SYBR Green PCR Master Mix (Applied Biosystems), 1.25 μL of 5 μM forward and reverse primers, and 5 μL H2O. A five-point serial dilution of each extract was run concurrently with each extract for every gene region tested. The primers used for the promoter are forward, 5′-CCGGAGTACTTCAAGAACTGCTG-3′; reverse, 5′-AGTCCCTTGTAGCAAGCTCGGATT-3′, or for the end of the LacZ gene forward, 5′-CGGGTAAACTGGCTCGGATT-3′; reverse, 5′-GGCGGTCGGGATAGTTTTCT-3′). The real-time PCR was performed in 96-well plates with the ABPrism 7500 Sequence Detection System, using SDS Software (v2.0; Applied Biosystems). The data are expressed as a point in the linear range of amplification for each sample (Ct value). The percentage input was determined by comparing the Ct value of each sample with a standard curve generated from a five-point serial dilution of the genomic input.
Acknowledgments
We are especially grateful to Evgeny Makarov and Reinhard Lührmann (Max-Plank Institute of Biophysical Chemistry, Gottingen, Germany) for sharing antisera to the U5snRNP and tri-snRNP proteins, and to R.L. for critical comments on the manuscript. We also thank Ivana Turbachova for the Tet-off Tat-on cell line, which was used to create the derivative HIV-1 eGFP LTR line characterized in the ChIP experiments; Wolfgang Fischer (Peptide Biology Laboratory, The Salk Institute) for microsequencing and mass-spectrometry; Jose Sierra for sharing protocol and primers used in the ChIP experiments on c-Myc gene; Matthias Kaeser for helping with the real-time PCR; Oded Singer and Inder Verma (The Salk Institute) for sharing plasmids and methods used to create the shRNA-SKIP lentivirus; Paul N. MacDonald (Case Western Reserve University) for SKIP reagents; and Jose-Ramon Suarez (Aventis) for flavopiridol. This work was funded by grants to K.A.J. from the NIH and the Universitywide Task Force on AIDS, and fellowship support to V.B. from the European Molecular Biology Organization.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1291705.
References
- Ahn S.H., Kim, M., and Buratowski, S. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13: 67–76. [DOI] [PubMed] [Google Scholar]
- Albers M., Diment, A., Muraru, M., Russell, C.S., and Beggs, J.D. 2003. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA 9: 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D., Dowhan, D.H., Kang, Y.K., Lee, J.W., Berget, S.M., and O'Malley, B.W. 2004. Differential recruitment of nuclear receptor co-activators may determine alternative RNA splice site choice in target genes. Proc. Natl. Acad. Sci. 101: 2270–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J., Leong, G.M., Church, W., Issa, L.L., Eisman, J.A., and Gardiner, E.M. 2003. Interactions of SKIP/NCoA-62, TFIIB, and retinoid X receptor with vitamin D receptor helix H10 residues. J. Biol. Chem. 278: 8224–8228. [DOI] [PubMed] [Google Scholar]
- Becker-Hapak M., McAllister, S., and Dowdy, S. 2001. TAT-mediated protein transduction into mammalian cells. Methods 24: 247–256. [DOI] [PubMed] [Google Scholar]
- Bird G., Zorio, D.A., and Bentley, D.L. 2004. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 24: 8963–8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao S. and Price. D. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276: 31793–31799. [DOI] [PubMed] [Google Scholar]
- Chen D. and Zhou, Q. 1999. Tat activates human immunodeficiency virus type 1 transcriptional elongation independent of TFIIH kinase. Mol. Cell. Biol. 19: 2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y., Ho, C., Saha, N., Schwer, B., Shuman, S., and Rana, T.M. 2002. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell 10: 585–597. [DOI] [PubMed] [Google Scholar]
- Clavel F. and Charneau, P. 1994. Fusion from without directed by human immunodeficiency virus particles. J. Virol. 68: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire G., Makarov, E.M., Cowger, J.J., Longman, D., Sutherland, H.G., Luhrmann, R., Torchia, J., and Bickmore, W.A. 2002. Mammalian PRP4 kinase co-purifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol. Cell. Biol. 22: 5141–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa J.D. and Hayman, M.J. 2004. Differential effects of the Ski-interacting protein (SKIP) on differentiation induced by transforming growth factor-β1 and bone morphogenetic protein-2 in C2C12 cells. Exp. Cell Res. 296: 163–172. [DOI] [PubMed] [Google Scholar]
- Folk P., Puta, F., and Skruzny, M. 2004. Transcriptional coregulator SNW/SKIP: The concealed tie of dissimilar pathways. Cell. Mol. Life Sci. 61: 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y. and Zhou, Q. 2000. Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20: 5897–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929–933. [DOI] [PubMed] [Google Scholar]
- Fryer C.J., White, J.B., and Jones, K.A. 2004. Mastermind recruits CycC:CDK8 to the HES1 promoter to co-ordinate Notch activation with turnover. Mol. Cell 16: 509–520. [DOI] [PubMed] [Google Scholar]
- Garber M.E., Mayall, T.P., Suess, E., Meisenhelder, J., Thompson, N.E., and Jones, K.A. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat–P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20: 6958–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber M. and Shilatifard, A. 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J Biol. Chem. 278: 26303–26306. [DOI] [PubMed] [Google Scholar]
- Gozani O., Potashkin, J., and Reed, R. 1998. A potential role for U2AF–SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 18: 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S.D. 2004. Viral interactions with the Notch pathway. Semin. Cancer Biol. 14: 387–396. [DOI] [PubMed] [Google Scholar]
- Herrmann C. and Mancini, M. 2001. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci. 114: 1491–1503. [DOI] [PubMed] [Google Scholar]
- Hurt E., Luo, M., Rother, S., Reed, R., and Strasser, K. 2004. Cotranscriptional recruitment of the serine–arginine (SR)-like proteins Gbp2 and Hrb1 to the nascent mRNA via the TREX complex. Proc. Natl. Acad. Sci. 101: 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K., Mizutani-Koseki, Y., Komori, T., Schmidt-Zachmann, M.S., and Koseki, H. 2005. Mammalian Polycomb-mediated repression of Hox genes requires the essential spliceosome protein Sf3b1. Genes & Dev. 19: 536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka S., Duque, P., and Konarska, M.M. 2004. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23: 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J. 1999. Tackling Tat. J. Mol. Biol. 293: 235–254. [DOI] [PubMed] [Google Scholar]
- Kim D.-K., Amaguchi, Y., Wada, T., and Handa. H. 2001. The regulation of elongation by eukaryotic RNA polymerase II: A recent view. Mol. Cell 11: 267–274. [PubMed] [Google Scholar]
- Kostrouchova M., Housa, D., Kostrouch, Z., Saudek, V., and Rall, J. 2002. SKIP is an indispensable factor for Caenorhabditis elegans development. Proc. Natl. Acad. Sci. 99: 9254–9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotovic K., Lockshon, D., Boric, L., and Neugebauer, K. 2003. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell. Biol. 23: 5768–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong G.M., Subramaniam, N., Issa, L.L., Barry, J.B., Kino, T., Driggers, P.H., Hayman, M.J., Eisman, J.A., and Gardiner, E.M. 2004. Ski-interacting protein, a bifunctional nuclear receptor coregulator that interacts with N-CoR/SMRT and p300. Biochem. Biophys. Res. Commun. 315: 1070–1076. [DOI] [PubMed] [Google Scholar]
- Li X.Y. and Green, M.R. 1998. The HIV-1 Tat cellular coactivator Tat-SF is a general transcription elongation factor. Genes & Dev. 12: 2992–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusic M., Marcello, A., Cereseto, A., and Giacca, M. 2003. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 22: 6550–6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., and Luhrmann, R. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Makarova O.V., Makarov, E.M., and Luhrmann, R. 2001. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J. 20: 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz P.J., Patterson, S.D., Neuwald, A.F., Spahr, C.S., and Spector, D.L. 1999. Purification and biochemical characterization of inter-chromatin granule clusters. EMBO J. 18: 4308–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve M., Wu, Z., Adelmant, G., Puigserver, P., Fan, M., and Spiegelman, B.M. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6: 307–316. [DOI] [PubMed] [Google Scholar]
- Nagai K., Yamaguchi, T., Takami, T., Kawasumi, A., Aizawa, M., Masuda, N., Shimizu, M., Tominaga, S., Ito, T., Tsukamoto, T., et al. 2004. SKIP modifies gene expression by affecting both transcription and splicing. Biochem. Biophys. Res. Commun. 316: 512–517. [DOI] [PubMed] [Google Scholar]
- Neubauer G., King, A., Rappsilber, J. Calvio, C., Watson, M., Ajuh, P., Sleeman, J., Lamond, A., and Mann, M. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20: 46–50. [DOI] [PubMed] [Google Scholar]
- Nguyen V., Kiss, T., Michels, A., and Bensaude, O. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414: 322–325. [DOI] [PubMed] [Google Scholar]
- Ni Z., Schwartz, B.E., Werner, J., Suarez, J.R., and Lis, J.T. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13: 55–65. [DOI] [PubMed] [Google Scholar]
- Raha T., Cheng, S.C., and Green, M.R. 2005. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 3: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers D., Ayadi, L., Gattoni, R., Jacquenet, S., Damier, L., Branlant, C., and Stevenin, J. 2004. Differential effects of the SR proteins 9G8, SC35, ASF/SF2, and SRp40 on the utilization of the A1 to A5 splicing sites of HIV-1 RNA. J. Biol. Chem. 279: 29963–29973. [DOI] [PubMed] [Google Scholar]
- Sano M., Wang, S.C., Shirai, M., Scaglia, F., Xie, M., Sakai, S., Tanaka, T., Kulkarni, P.A., Barger, P.M., Youker, K.A., et al. 2004. Activation of cardiac Cdk9 represses PGC-1 and confers a predisposition to heart failure. EMBO J. 23: 3559–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A., Conaway, R.C., and Conaway, J.W. 2003. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 72: 693–715. [DOI] [PubMed] [Google Scholar]
- Sims III R.J., Nishioka, K., and Reinberg, D. 2003. Histone lysine methylation: A signature for chromatin function. Trends Genet. 19: 629–639. [DOI] [PubMed] [Google Scholar]
- Sims III R.J., Belotserkovskaya, R., and Reinberg, D. 2004. Elongation by RNA polymerase II: The short and long of it. Genes & Dev. 18: 2437–2468. [DOI] [PubMed] [Google Scholar]
- Sune C., Hayashi, T., Liu, Y., Lane, W.S., Young, R.A., and Garcia-Blanco, M.A. 1997. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 17: 6029–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvari A. and Luse, D. 2004. Newly initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J. Biol. Chem. 279: 49773–49779. [DOI] [PubMed] [Google Scholar]
- Wei P., Garber, M.E., Fang, S., Fischer, W.H., and Jones, K.A. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92: 451–462. [DOI] [PubMed] [Google Scholar]
- Yang Z., Zhu, Q., Luo, K., and Zhou, Q. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414: 317–322. [DOI] [PubMed] [Google Scholar]
- Zhang C., Dowd, D.R., Staal, A., Gu, C., Lian, J.B., van Wijnen, A.J., Stein, G.S., and MacDonald, P.N. 2003. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 278: 35325–35336. [DOI] [PubMed] [Google Scholar]
- Zhou Q. and Sharp, P.A. 1996. Tat-SF1: Cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science 274: 605–610. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Chen, D., Pierstorff, E., and Luo, K. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17: 3681–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Halanski, M.A., Radonovich, M.F., Kashanchi, F., Peng, J., Price, D.H., and Brady, J.N. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20: 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Deng, L., Kashanchi, F., Brady, J.N., Shatkin, A.J., and Kumar, A. 2003. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc. Natl. Acad. Sci. 100: 12666–12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Deng, L., Lacoste, V., Park, H., Pumfery, A., Kashanchi, F., Brady, J., and Kumar, A. 2004. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J. Virol. 78: 13522–13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio D.A. and Bentley, D.L. 2004. The link between mRNA processing and transcription: Communication works both ways. Exp. Cell Res. 296: 91–97. [DOI] [PubMed] [Google Scholar]