Abstract
The E.3.3 mutation was generated in a Flp/FRT EMS screen for conditional mutations that cause growth and developmental defects in a genetic background that blocks apoptosis. The mutations were conditional, based on the Dark 82 allele being present on the starting chromosome, and blocking canonical apoptosis in a homozygous state. The E.3.3 mosaic eyes exhibit defects in eye development including patches of rough eye and irregular surface structure. Whole Genome Sequencing and complementation mapping revealed E.3.3 as an allele of prod . Prod is a DNA-binding protein that binds satellite repeats and is involved in chromocenter formation during mitosis. Here we present a novel allele of prod , prod E.3.3 , that disrupts the functional region of the Prod protein resulting in disruption of typical eye structure, likely due to disruption of chromatid separation during development.
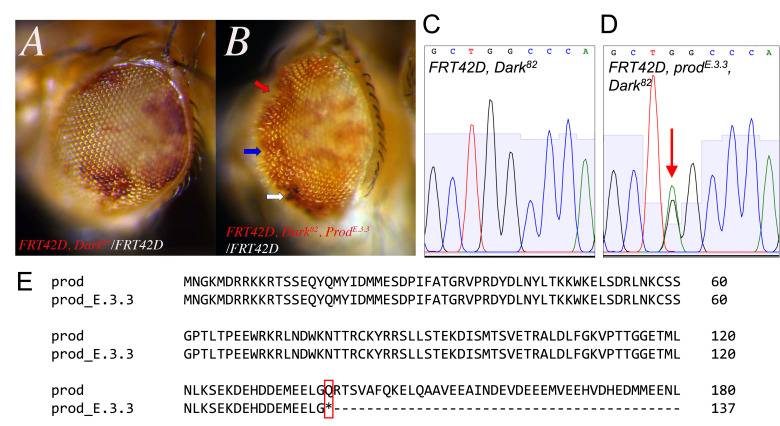
Figure 1. Characterization of the prod E.3.3 allele in mosaic eyes and genetic sequencing.
FRT42D, Dark 82 control mosaic eyes ( A ) have similar red (mutant cells): white (wild-type cells) ratio to FRT42D, prod E.3.3 , Dark 82 mosaic eyes ( B ). The FRT42D, prod E.3.3 , Dark 82 mosaic eyes are mis patterned (blue arrow), have potentially necrotic areas (white arrow), and have concave regions on the surface of the eye (red arrow). Sequencing of prod PCR products amplified from FRT42D, Dark 82 /CyO and FRT42D, prod E.3.3 , Dark 82 /CyO heterozygous flies identified a heterozygous peak ( D , G→A, red arrow) present in the mutant flies that was not present in the control ( C ). This nucleotide change at position 2R:18970530 creates a premature stop codon in the protein at amino acid 138 ( E , Q→STOP, red box).
Description
Undergraduate students in the Fly-CURE project (Merkle et al., 2023) characterize and map mutations identified in a screen for mutations that alter cell growth within the context of an undergraduate course. The mutant lines were generated in an EMS screen designed to identify genes that regulate cellular growth and development (Kagey et al., 2012) . The right arm of Chromosome 2 was screened using the Flp/FRT system to generate mitotic clones in a Dark 82 background that suppresses apoptosis (Kagey et al., 2012) . The Dark 82 loss-of-function mutation is the result of a transposon insertion that disrupts the Dark coding sequence, but also inserts the mini-white + gene allowing the identification of homozygous mutant tissue in the eye based on color. The growth defects that result from the E.3.3 mutation were examined by generating mitotic clones as a result of crossing w - , ey>FLP; FRT42D flies that express the FLP recombinase in the eye with flies that carry both the E.3.3 allele and Dark 82 allele on the FRT42D chromosome ( w - ; FRT42D, Dark 82 , E.3.3/CyO ). Tissue that was homozygous mutant for E.3.3 and Dark 82 was identified based on red color. These mosaic eyes were compared to the mosaic eyes from a control cross that did not include the E.3.3 mutation ( w - , ey>FLP; FRT42D x w - ; FRT42D, Dark 82 /CyO ). The E.3.3 mosaic eyes have a rough eye phenotype (blue arrow) with concave regions on the eye surface (red arrow) and some regions that appear glossy compared to the control mosaic eye (see Fig. 1A, B ). While the overall ratio of red to white tissue does not have a dramatic difference between the control and E.3.3 , the overall patterning and organization of the eye has been disrupted and the overall eye size does appear a bit smaller, suggesting E.3.3 has a role in eye development. Additionally, some mosaic E.3.3 eyes also have black spots suggesting potential necrotic tissue in the mutant eye (white arrow).
In parallel, female w - ; FRT42D, Dark 82 , E.3.3/CyO flies were crossed with males from the Bloomington Drosophila Stock Center 2R Deficiency Kit in order to map the E.3.3 mutation (Cook et al., 2012) . The Fly-CURE utilizes independent mapping experiments at each institution, so each cross was set up independently four times. For a cross to be designated a failure to complement, at least 50 curly winged flies have to be observed without any straight winged F1 progeny. These results were confirmed by independent replicates at each institution. Two of the 2R Deficiency Kit stocks failed to complement the FRT42D, Dark 82 , E.3.3 chromosome: Df(2R)ED2747 and Df(2R)BSC331 . Both of these deficiencies include the Dark locus (Gramates et al., 2022) , so it is likely that the failure to complement is due to the lethal Dark 82 allele. All of the remaining 2R Deficiency Kit stocks complemented the E.3.3 mutation, indicating that either the E.3.3 mutation lies in the same region of the chromosome as Dark , the E.3.3 mutation is not lethal (and, thus, could not be identified through complementation testing) or the 2R Deficiency Kit used did not cover the region of the chromosome that contained the E.3.3 . mutation. At this stage we took a similar approach to that of mutant B.2.16 (Vrailas-Mortimer et al. 2021) . Whole Genome Sequence analysis of the w - ; FRT42D, Dark 82 , E.3.3/CyO genome identified a point mutation (G→A) in the proliferation disrupter ( prod ) gene consistent with a premature stop codon (Vrailas-Mortimer et al. 2021) . The prod gene maps to nucleotides 18,969,404 to 18,971,133 on chromosome 2R ( D. melanogaster r6.56), which is located between two non-overlapping deficiencies in the Deficiency Kit, Df(2R)ED3610 and Df(2R)Exel6069 (Gramates et al., 2022) . The presence of this mutation in the prod gene and the location of prod in a region that is not covered by the Deficiency Kit used suggests that E.3.3 was an allele of prod , prod E.3.3 .
To confirm that prod E.3.3 is a lethal allele of prod , w - ; FRT42D, Dark 82 , E.3.3/CyO virgins were crossed with prod E (hypomorphic) and prod k08810 (null)males that carry lethal alleles of the prod gene (Török et al., 1997; Gramates et al., 2022) . The prod alleles did not complement prod E.3.3 , supporting the hypothesis that the lethal hit in E.3.3 is a null allele, prod E.3.3 . Based on newly available data on Flybase, there are only three of the ten prod alleles that have been characterized based on the nature of the allele. prod E.3.3 is the fourth characterized prod allele (Öztürk-Çolak et al. (2024)).
To confirm the molecular nature of the prod E.3.3 mutation, we sequenced PCR products from prod in w - ; FRT42D, Dark 82 , E.3.3/CyO flies and w - ; FRT42D, Dark 82 /CyO flies. Three different sets of primers were designed to ensure that the products overlapped and covered the entire coding sequence of the prod gene. PCR products of the expected size were visualized by gel electrophoresis and purified products were sequenced (Eurofins Genomics). The sequence chromatograms for both the mutant and control strains were examined for overlapping peaks that indicated heterozygosity. Only one heterozygous site was present in the E.3.3 sequences that was not present in the control sequences (see Fig. 1C, D ), and this site aligned with the mutation identified in the Whole Genome Sequence analysis. The sequenced mutation introduces a premature stop codon in the prod transcript ( Fig. 1E ) that interrupts the predicted coiled-coil domain of the protein, as well as terminates the protein sequence in a region that has been determined to be essential for DNA binding (Török et al., 2000). Thus, it is likely that the prod E.3.3 allele does not produce a functional Prod protein.
Based on both the genetic and molecular results, we conclude that prod E.3.3 is a newly identified allele of prod . The prod gene was initially identified as l(2)88/10 in a P-element screen that identified lethal P insertions with overgrowth phenotypes (Török et al., 1993) . It was later demonstrated to play a role in chromatid separation during mitosis, with homozygous mutants exhibiting a decrease in mitotic index and cell death in highly proliferative cell types (Török et al., 1997) . The previously identified functions of prod are consistent with the identification of an allele in a mosaic screen for growth defects.
Reagents
|
Bloomington Stock Center 2R Deficiency Kit | |||
|
Deficiency Stocks |
BDSC Stock # |
Chromosomal Deletions |
Complementation Results |
|
Df(2R)ED2747 |
9278 |
2R:16829073..17097303 |
Failed to complement (overlaps Dark 82 ) |
|
Df(2R)BSC331 |
24356 |
2R:16869330..17139923 |
Failed to complement (overlaps Dark 82 ) |
|
Mutant alleles of individual genes | |||
|
Gene |
BDSC Stock # |
Allele |
Complementation Results |
|
p rod |
42688 |
prod E |
Failed to complement |
|
prod |
10814 |
prod k08810 |
Failed to complement |
w - ; FRT42D, Dark 82 /CyO (Akdemir et al. 2006)
w - ; FRT42D, Dark 82 , E.3.3/CyO (this study)
w - , ey>FLP; FRT42D (BDSC 5616)
Bloomington Drosophila Stock Center 2R Deficiency Kit (Cook et al. 2012)
y 1 w 67c23 ; P{w[+mC]=lacW} prod k08810 /CyO (BDSC 10814)
y 1 w 67c23 ; prod E /CyO, y + (BDSC 42688)
prod forward primer 1: 5' CAT CGA GCA CGC AGG C 3'
prod reverse primer 1: 5' CTC CAT CTC GTC GTC GTG C 3'
prod forward primer 2: 5' GAT GCT CAA TCT GAA GTC CG 3'
prod reverse primer 2: 5' AGC TTA TTG CCG GAG GAG G 3'
prod forward primer 3: 5' ACG CCG TCG AGT ACG TCA C 3'
prod reverse primer 3: 5' CGA CTG CTT AGA CCC ACT GAT C 3'
Acknowledgments
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
References
- Akdemir F, Farkas R, Chen P, Juhasz G, Medved'ová L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006 Mar 15;133(8):1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13(3):R21–R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Agapite J, Attrill H, Calvi BR, Crosby MA, Dos Santos G, Goodman JL, Goutte-Gattat D, Jenkins VK, Kaufman T, Larkin A, Matthews BB, Millburn G, Strelets VB, the FlyBase Consortium. Fly Base: a guided tour of highlighted features. Genetics. 2022 Apr 4;220(4) doi: 10.1093/genetics/iyac035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Cummings R, Yamashita YM. The modular mechanism of chromocenter formation in Drosophila. Elife. 2019 Feb 11;8 doi: 10.7554/eLife.43938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey JD, Brown JA, Moberg KH. Regulation of Yorkie activity in Drosophila imaginal discs by the Hedgehog receptor gene patched. Mech Dev. 2012 Jun 15;129(9-12):339–349. doi: 10.1016/j.mod.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle JA, Devergne O, Kelly SM, Croonquist PA, Evans CJ, Hwalek MA, Straub VL, Hamill DR, Peister A, Puthoff DP, Saville KJ, Siders JL, Villanueva Gonzalez ZJ, Wittke-Thompson JK, Bieser KL, Stamm J, Vrailas-Mortimer AD, Kagey JD. Fly-CURE, a multi-institutional CURE using Drosophila, increases students' confidence, sense of belonging, and persistence in research. J Microbiol Biol Educ. 2023 Sep 21;24(3) doi: 10.1128/jmbe.00245-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öztürk-Çolak A, Marygold SJ, Antonazzo G, Attrill H, Goutte-Gattat D, Jenkins VK, Matthews BB, Millburn G, Dos Santos G, Tabone CJ, FlyBase Consortium FlyBase: updates to the Drosophila genes and genomes database. Genetics. 2024 Feb 1; doi: 10.1093/genetics/iyad211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török T, Tick G, Alvarado M, Kiss I. P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: isolation of lethals with different overgrowth phenotypes. Genetics. 1993 Sep 1;135(1):71–80. doi: 10.1093/genetics/135.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török T, Harvie PD, Buratovich M, Bryant PJ. The product of proliferation disrupter is concentrated at centromeres and required for mitotic chromosome condensation and cell proliferation in Drosophila. Genes Dev. 1997 Jan 15;11(2):213–225. doi: 10.1101/gad.11.2.213. [DOI] [PubMed] [Google Scholar]
- Török T, Gorjánácz M, Bryant PJ, Kiss I. Prod is a novel DNA-binding protein that binds to the 1.686 g/cm(3) 10 bp satellite repeat of Drosophila melanogaster. Nucleic Acids Res. 2000 Sep 15;28(18):3551–3557. doi: 10.1093/nar/28.18.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrailas-Mortimer AD, Aggarwal N, Ahmed NN, Alberts IM, Alhawasli M, Aljerdi IA, Allen BM, Alnajar AM, Anderson MA, Armstong R, Avery CC, Avila EJ, Baker TN, Basardeh S, Bates NA, Beidas FN, Bosler AC, Brewer DM, Buenaventura RS, Burrell NJ, Cabrera-Lopez AP, Cervantes-Gonzalez AB, Cezar RP, Coronel J, Croslyn C, Damery KR, Diaz-Alavez L, Dixit NP, Duarte DL, Emke AR, English K, Eshun AA, Esterly SR, Estrada AJ, Feng M, Freund MM, Garcia N, Ghotra CS, Ghyasi H, Hale CS, Hulsman L, Jamerson L, Jones AK, Kuczynski M, Lacey-Kennedy TN, Lee MJ, Mahjoub T, Mersinger MC, Muckerheide AD, Myers DW, Nielsen K, Nosowicz PJ, Nunez JA, Ortiz AC, Patel TT, Perry NN, Poser WSA, Puga DM, Quam C, Quintana-Lopez P, Rennerfeldt P, Reyes NM, Rines IG, Roberts C, Robinson DB, Rossa KM, Ruhlmann GJ, Schmidt J, Sherwood JR, Shonoda DH, Soellner H, Torrez JC, Velide M, Weinzapfel Z, Ward AC, Bieser KL, Merkle JA, Stamm JC, Tillett RL, Kagey JD. B.2.16 is a non-lethal modifier of the Dark 82 mosaic eye phenotype in Drosophila melanogaster . . MicroPubl Biol. 2021 Jan 18;2021 doi: 10.17912/micropub.biology.000359. [DOI] [PMC free article] [PubMed] [Google Scholar]