Abstract
Objectives
The rise in Carbapenem-resistant Enterobacterales (CRE) is perturbing. To curb the menace of CRE, a comprehensive understanding of its prevalence and epidemiology is crucial. As varying reports abound, the true prevalence of CRE in Nigeria remains unknown. Here, we conducted a systematic review and meta-analysis following standard guidelines to assess the situation of CRE in Nigeria.
Methods
We searched electronic databases including Pubmed, ScienceDirect, Scopus, Web of Science, and Google Scholar for articles providing information on CRE in Nigeria. The data gathered were analyzed using OpenMeta Analyst and Comprehensive Meta-Analysis software. The random-effect model was employed to calculate pooled resistance to carbapenem antibiotics.
Results
From 321 retrieved records, 57 were finally included. The studies were predominantly from the South-West region (n = 19). Escherichia coli and Klebsiella pneumoniae were the most frequently tested Enterobacterales among the included studies. The pooled prevalence estimate for imipenem resistance among CRE was 11.2 % (95 % CI: 7.9–15.7). Meropenem resistance had an estimate of 13.5 % (95 % CI: 9.1–19.6), whereas ertapenem and doripenem were estimated at 17.0 % (95 % CI: 9.9–27.7) and 37.9 % (95 % CI: 15.0–67.8), respectively. High heterogeneity (I2>85 %, p < 0.001) was observed for the estimates. The highest resistance rate to imipenem (28.4 %), meropenem (37.2 %) and ertapenem (46.5 %) were observed for the South-South region. Based on specific CRE genera, Morganella sp. was the most resistant (37.0 %) while Escherichia sp. was the least (9.4 %). Our analyses also revealed a progressive increase in resistance to carbapenem antibiotics over the years.
Conclusion
This study highlights carbapenem resistance as a concern in Africa's most populous nation, underscoring the need for proactive measures to address and mitigate the threat of CRE.
Keywords: Carbapenem, Enterobacterales, Resistance, Nigeria
Highlights
-
•
Over time, there has been a continuous rise in resistance to carbapenem antibiotics.
-
•
There is a high resistance to ertapenem and doripenem among CRE in Nigeria.
-
•
The South-South region of Nigeria is the most affected by CRE.
-
•
Morganella sp. is the most resistant CRE (37.0 %).
-
•
The true prevalence of CRE in Nigeria is before now, unknown.
1. Introduction
The global healthcare systems face substantial challenges due to antibiotic resistance. Based on predictive statistical models [1], bacterial antimicrobial resistance was linked to approximately 4.95 million deaths in 2019, with 1.27 million directly attributed to bacterial antimicrobial resistance. The rates of resistance are more elevated in low-income nations compared to their high-income counterparts. In numerous low-income settings, substantial data gaps exist, potentially indicating a more severe resistance situation in these countries than suggested by the available data [2].
In 2017, the World Health Organization identified carbapenem-resistant Enterobacterales (CRE) as one of the three primary categories of drug-resistant bacteria worldwide requiring immediate attention for the development of new antibiotics [3]. CRE, as defined by the Centers for Disease Control and Prevention (CDC) of the United States, are Enterobacterales exhibiting in vitro resistance to at least one carbapenem [4]. CRE resistance mechanisms encompass antibiotic degradation, hindrance of antibiotic penetration into bacterial cells, alteration of antibiotic binding targets, mutation or deletion of pore proteins, heightened activation of efflux pumps, and alteration of penicillin-binding proteins and biofilm composition [5]. Antibiotic degradation can result from the organism's production of carbapenemases, enzymes that degrade carbapenem antibiotics and render them ineffective. Notable examples include Klebsiella pneumoniae carbapenemase (KPC), New Delhi metallo-beta-lactamase (NDM), and oxacillinase-48 (OXA-48), produced by various Enterobacterales [5]. Porin proteins, which are channels in the bacterial cell wall that permits the entry of antibiotics may be mutated. These mutations can reduce carbapenem uptake, diminishing their effectiveness [5]. Additionally, bacteria can overexpress efflux pumps, increasing the expulsion of antibiotics from the cell before they can reach their targets [6]. This mechanism is significant in β-lactam resistance for pathogens like Pseudomonas aeruginosa, Escherichia coli, and Neisseria gonorrhoeae [6]. Furthermore, bacteria often combine these mechanisms, resulting in high-level resistance to carbapenems.
Carbapenems represent a highly effective category of broad-spectrum antibiotics [7]. They are regarded as one of the last-line antibiotics in many hospitals. However, the emergence of resistance to carbapenems poses a considerable restriction on the available antibiotic options for treating challenging infections. The prevalence of CRE has markedly increased over the years, and these resistant bacteria have been reported in virtually all parts of the world, including North America, Europe, the Mediterranean, South Asia, and Africa [[8], [9], [10], [11], [12], [13]]. This is particularly worrisome as CRE infections are linked to higher morbidity and mortality rates compared to other pathogens, especially among patients with severe underlying health conditions or those admitted to the intensive care unit [14,15].
The rise of CRE poses an escalating global threat to human health. In dealing with CRE infections, extended therapy durations become necessary, resulting in higher treatment costs and the need for approaches with elevated toxicities compared to strains susceptible to carbapenems. Given the rising challenges of carbapenem resistance and the limited therapeutic alternatives available, it is essential to completely understand the epidemiology of CRE infections. This information holds significance in enhancing infection prevention and control, implementing effective antimicrobial stewardship, and other strategic initiatives aimed at mitigating carbapenem resistance. Because varying prevalence rates have been reported in different parts of Nigeria, the true prevalence of CRE in Nigeria, and the different regions of the country is unknown. This knowledge is especially important because Nigeria, being the most populous country in Africa, sees a significant flow of people both into and out of the country for various reasons. Thus, in this study, we aimed to provide a comprehensive overview of the situation in the country. To our knowledge, this study represents the first meta-analysis to be conducted on this subject in Nigeria.
2. Methods
This systematic review and meta-analysis followed the guidelines established in the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) [16]. A study protocol was also developed and registered with PROSPERO (registration number: CRD42023418296) to avoid any redundancy with already existing or ongoing studies.
2.1. Literature search/sources of data
We employed a set of keywords, including “Carbapenem,” “Ertapenem,” “Imipenem,” “Meropenem,” “CRE,” “Enterobacterales,” “Enterobacteriaceae,” “Escherichia coli,” “E. coli,” “Enterobacter,” “Klebsiella,” “Shigella,” “Citrobacter,” and “Nigeria,” to conduct a thorough search across four electronic databases (Scopus, Google Scholar, Pubmed, Web of Science, and Science Direct) for studies that reported the prevalence of CRE in Nigeria. The detailed search strategy utilized for accessing all databases is available in the supplementary document (File S1). To ensure a comprehensive search, no filters based on study design, language, or publication year were applied. The initial search took place on February 25, 2023, and a final updated search, yielding a total of 321 records, was performed on April 14, 2023. The retrieved records from the databases were exported to EndNote X8 software for duplicate removal and initial screening.
2.2. Criteria for study eligibility
This study encompassed research focusing on CRE in the human population of Nigeria. We excluded (1) case reports, reviews, editorials, letters, book chapters, and opinions; (2) studies reporting CRE data from countries outside Nigeria; (3) studies evaluating CRE from sources other than humans; (4) articles with unavailable full texts; (5) investigations involving known cases of CRE that do not reflect prevalence among a sampled population.
All authors established the criteria for screening, selecting, and evaluating articles. Working independently, at least two authors conducted initial screening based on the title and abstract, followed by a thorough assessment of the full texts of the screened articles. In cases of disagreements during the screening and assessment process, the involvement of other authors was sought to reach a consensus.
2.3. Extraction of data and quality evaluation
Data extraction involved the utilization of a pre-established Excel spreadsheet. Independently, three authors extracted the following details from the incorporated studies: study ID, duration of the study, study location, study design, Enterobacterales members examined, method employed for resistance determination, sample type, count of resistant organisms, and the overall number of Enterobacterales evaluated.
The methodological quality of the incorporated studies was evaluated using the Joanna Briggs Institute (JBI) critical appraisal checklist designed for prevalence data [17] (File S2). Two authors conducted independent assessments, assigning a total quality score ranging from 0 to 9. Adequate quality was defined as a score of 7 or higher [18].
2.4. Quantitative analysis and synthesis of data
The collected data underwent scrutiny to identify and address potential duplications before being analyzed. OpenMeta Analyst and Comprehensive Meta-Analysis 3.0 (CMA 3.0) software were employed for analyses. Summary estimates regarding resistance to each of the carbapenems (imipenem, meropenem, ertapenem, and doripenem) were generated using the DerSimonian-Laird method of meta-analysis and the random-effect model. To assess potential publication bias, a funnel plot was generated. Egger's regression test was conducted to evaluate the plot's asymmetry, and a Trim and Fill sensitivity test was performed to assess changes in prevalence estimates. Cochran's Q test was utilized to examine heterogeneity in study-level estimates, with I2 statistics used for quantification; I2 values of 25 %, 50 %, and 75 % indicated low, moderate, and high heterogeneity, respectively [19,20]. Subgroup meta-analysis was performed to explore sources of heterogeneity and to determine the prevalence of CRE in various subgroups. In all tests, statistical significance was defined by a p-value of <0.05.
3. Results
3.1. Selection of studies
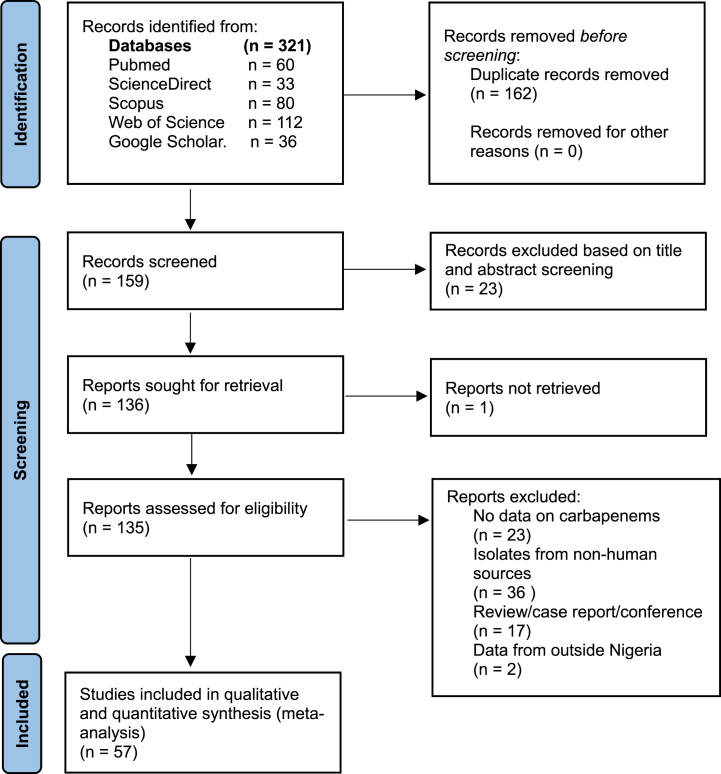
The process of article identification and selection in this study is outlined in Fig. 1. Our search across five electronic databases yielded 321 records. Following the removal of duplicates and exclusion of studies not meeting the predetermined inclusion criteria, we assessed the full text of 135 studies for eligibility. Fifty-seven studies met the eligibility criteria and were consequently included in both the qualitative and quantitative synthesis.
Fig. 1.
Overview of the process for searching and selecting articles.
3.2. Characteristics of the included studies
This study encompassed research conducted at various locations across Nigeria, with all regions of the country represented. While the majority of the reports originated after 2010, diverse sample types, such as urine, blood, sputum, stool, cerebrospinal fluid, wound swab, nasal swab, and rectal swab, among others, were utilized for assessing Enterobacterales. Carbapenem resistance evaluation encompassed a range of Enterobacterales, with E. coli and K. pneumoniae being the most frequently tested. Various testing methods, including disk diffusion techniques, broth microdilution method, Epsilometer test, and assessment using automated systems were employed. A comprehensive overview of the features of the included studies is provided in Table 1.
Table 1.
Major characteristics of the included studies.
| Study ID | Study period | Location | Study design | Organism tested | Method | Sample type | Total Enterobacterales | Reference |
|---|---|---|---|---|---|---|---|---|
| Adegoke 2022 | NR | Akwa Ibom | Cross-sectional | E. coli | DDM | Urine | 39 | [21] |
| Adekanmbi 2022 | 2019–2020 | Oyo | NR | K. pneumoniae, K. oxytoca, P. mirabilis, E. cloacae | DDM | Urine | 20 | [22] |
| Adekanmbi 2021 | 2019–2020 | Oyo | NR | E. coli | DDM | Urine | 49 | [23] |
| Adenipekun 2016 | 2012–2013 | Lagos | NR | E. coli | BMM | Urine, stool | 247 | [24] |
| Adeosun 2019 | 2018–2019 | Osun | NR | K. pnuemoniae | DDM | Urine, wound swab, ear swab, eye swab, other collection sites | 62 | [25] |
| Adesanya 2020 | 2018 | Oyo | Retrospective | K. pneumoniae, E. coli, K. oxytoca, P. mirabilis, E. cloacae, P. vulgaris, Providencia sp. | DDM | Urine, wound swab, tracheal aspirate, sputum, catheter tip swab, pleural fluid, eye swab, pus, ear swab, tissue biopsy, rectal swab, CSF | 114 | [26] |
| Adesina 2019 | NR | Lagos | NR | E. fergusonii | DDM | Wound swab | 3 | [27] |
| Agbo 2015 | 2012–2013 | Enugu | NR | E. coli, Klebsiella sp. | DDM | Urine, stool, sputum wound swab, high vaginal swab | 113 | [28] |
| Akani 2022 | 2022 | Port Harcourt | NR | Salmonella sp., Shigella sp. | DDM | Stool | 10 | [29] |
| Akinlabi 2020 | 2017–2018 | Kano, katsina, Abuja | Cross-sectional | E. coli | DDM | Blood, Ear swab, wound swab, eye swab, urine, stool, high vaginal swab, cerebrospinal fluid | 104 | [30] |
| Akinpelu 2020 | NR | Lagos | NR | Klebsiella sp. | DDM | Urine, blood, sputum | 18 | [31] |
| Akinyemi 2021 | 2015 | Lagos | NR | K. pnuemoniae | DDM | Blood, urine, stool, wound swab, nasal swab | 43 | [32] |
| Akujobi 2013 | NR | Anambra | NR | E. coli | DDM | Urine, semen, wound swab, high vaginal swab, ear swab, sputum | 46 | [33] |
| Aminu 2021 | 2018 | Kano | Cross-sectional | E. coli, K. pneumoniae, E. aerogenes, P. mirabilis, P. vulgaris, E. agglomerans, E. ozaenae, K. oxytoca, E. cloacae, C. freundii, S. odorifera, Salmonella sp. | DDM | Blood, urine, sputum, tracheal aspirate, swabs of other parts | 76 | [34] |
| Anibijuwon 2018 | 2016–2017 | Ekiti, Osun, Oyo | Cross-sectional | E. coli, E. aerogenes, Klebsiella sp., Proteus sp. | DDM | Blood, urine | 59 | [35] |
| Aworh 2019 | 2018–2019 | Abuja | Cross-sectional | E. coli | DDM | Stool | 48 | [36] |
| Bashir 2019 | 2016 | Kano | Cross-sectional | E. coli, Klebsiella sp., Proteus sp. | DDM | Urine, wound/pus, urine catheter, nasal feed tube | 80 | [37] |
| Brinkac 2019 | 2016 | Abuja | NR | Klebsiella sp. | E-test | Blood | 7 | [38] |
| Brown 2017 | 2013–2014 | Ibadan | Cross-sectional | K. pnuemoniae, S. pullorum, Salmonella subsp II | DDM | Blood | 6 | [39] |
| Duru 2020 | 2008–2016 | FCT, Kano | NR | Citrobacter sp., Serratia sp., Pantoea sp., Enterobacter sp., K. pneumoniae, E. coli | DDM | Blood | 160 | [40] |
| Enyinnaya 2022 | NR | Abuja | Cross-sectional | E. coli | DDM, E-test | Blood, urine, wound biopsy/swab, CSF, sputum, aspirates, ear, eye swabs | 400 | [41] |
| Fadeyi 2016 | 2013–2015 | Ilorin | Cross-sectional | K. oxytoca, K. pneumoniae | DDM | Blood, sputum, wound, urine, ear, eye & throat swabs | 50 | [42] |
| Fakorede 2023 | 2021 | Lagos | Cross-sectional | Salmonella sp. | DDM | Blood | 24 | [43] |
| Ibadin 2017 | 2016 | Benin City | NR | Citrobacter sp., E. coli, Klebsiella sp., P. mirabilis, P. vulgaris, Providencia sp., S. marcescens | DDM | Ear swab, eye swab, throat swab, vaginal swabs, wound swab, catheter tip, sputum, stool, urine & aspirates | 258 | [44] |
| Iduh 2020 | NR | Sokoto | NR | E. coli, K. pneumoniae, Salmonella sp. | DDM | Stool | 177 | [45] |
| Iliyasu 2016 | 2011–2014 | Kano | Retrospective | E. coli, K. pneumoniae, P. mirabilis, Enterococcus sp. | – | NR | 17 | [46] |
| Iregbu 2015 | 2010–2012 | Abuja | Retrospective | E. coli, K. pneumoniae | – | CSF | 10 | [47] |
| Isaiah 2011 | NR | Yola | NR | E. coli | DDM | Urine, sputum, blood, stool, high vaginal swabs, wound swabs & Abscess | 350 | [48] |
| Jamal 2022 | 2019 | Abuja | NR | E. coli, K. pneumoniae, S. marcescens, P. mirabilis, M. morganii, C. freundii, E. cloacae | E-test | NR | 200 | [49] |
| John 2023 | NR | Cross River | NR | P. mirabilis, Klebsiella sp., E. coli, Providencia sp., Enterobacter sp., Citrobacter sp. | DDM | Wound swab, throat swab | 37 | [50] |
| Kankara 2022 | 2018 | Katsina | Prospective | E. coli, K. pneumoniae, E. aerogenes, P. mirabilis, | DDM | Urine, wound swabs, hand swabs, sputum, stool, catheter swab | 15 | [51] |
| Kingsley 2013 | 2011–2012 | Uyo | Cross-sectional | E. coli, K. pneumoniae, Enterobacter sp, P. mirabilis, S. typhi | DDM | Blood | 57 | [52] |
| Makanjuola 2018 | 2014 | Ibadan | NR | Klebsiella sp., E. coli, Proteus sp., Enterobacter sp. | DDM | Blood, urine, wound swabs, biopsies, tracheal aspirates | 39 | [53] |
| Medugu 2022 | 2019–2020 | Abuja | Cross-sectional | E. coli | VITEK 2 AST-280/VITEK 2 AST-281 cards | Stool, CSF, aspirate, endocervical swab, wound swab, blood, urine | 107 | [54] |
| Mofolorunsho 2021 | 2018 | Anyibga | Cross-sectional | E. coli, K. pneumoniae | DDM | Urine | 78 | [55] |
| Mohammed 2015 | 2014 | Borno | Cross-sectional | K. pneumoniae, E. coli, P. mirabilis, K. oxytoca, M. morganii, C. freundii, S. marcescens, E. aerogenes, K. ozaenae, H. alvei, C. sedlakii | DDM | Blood, urine, cerebrospinal fluid, stool, and swabs of patients with invasive diseases | 225 | [56] |
| Motayo 2013 | NR | Abeokuta | NR | E. coli, K. pneumoniae | DDM | Blood, urine, CSF, genitals, etc. | 21 | [57] |
| Nkup 2022 | 2020 | Jos | Prospective | K. pneumoniae | DDM | Urine, sputum, wound swab | 8 | [58] |
| Nwafia 2019 | 2016–2017 | Enugu | Cross-sectional | E. coli | DDM | Urine, pleural & peritoneal aspirate, blood, wound swabs, CSF | 70 | [59] |
| Oduyebo 2015 | 2013 | Lagos | NR | K. pneumoniae, K. oxytoca, K. ozaenae, E. coli, E. agglomerans, E. aerogenes, E. cloacae, P. mirabilis, P. vulgaris, S. rubidaea, M. morganii, C. freundi, P. rettgeri | DDM | Urine, blood, sputum, wound, pus | 177 | [60] |
| Ojuawo 2020 | 2017 | Ilorin | Prospective | K. oxytoca, K. pneumoniae | DDM | Blood, sputum | 23 | [61] |
| Okpalanwa 2019 | 2018 | Ankpa | Cross-sectional | Salmonella sp. | DDM | Stool | 9 | [62] |
| Oladipo 2021 | NR | Ibadan | NR | K. pneumoniae | DDM | Wound swab | 100 | [63] |
| Oli 2017 | 2016 | Awka | Cross-sectional | E. coli, Salmonella sp. | DDM | Urine | 110 | [64] |
| Oli 2019a | 2017 | Anambra | NR | E. coli, K. pneumoniae, K. oxytoca | DDM | Urine, anal swab, wound swab, sputum | 78 | [65] |
| Oli 2019b | NR | Enugu | NR | E. coli, K. pneumoniae, Enterobacter sp, Citrobacter sp., Proteus sp., Serratia sp. | DDM | Urine, stool | 32 | [66] |
| Omoyibo 2018 | 2014 | South-West | Cross-sectional | E. coli, M. morganii, P. mirabilis, S. odorifera, E. cloacae, E. gergoviae | DDM | Wound swab | 19 | [67] |
| Onanuga 2020 | 2019 | Bayelsa | Cross-sectional | E. coli, K. pneumoniae | DDM | Urine | 42 | [68] |
| Onanuga 2019 | 2015 | Port Harcourt | Cross-sectional | E. coli, K. pneumoniae | DDM | Urine | 81 | [69] |
| Onyedibe 2018 | 2014 | Jos | Cross-sectional | E. coli | DDM | Urine, blood, swabs, aspirates | 220 | [70] |
| Oyekale 2022 | 2020–2021 | Ekiti | Cross-sectional | E.coli, K. aerogenes, P. vulgaris, P. mirabilis | DDM | Blood | 20 | [71] |
| Raji 2013 | 2011 | Lagos | Cross-sectional | E. coli, K. pneumoniae, P. mirabilis, E. cloacae | E-test | Urine, blood, skin swab, soft tissue, surgical site, nasal swab, ear swab, body fluids | 96 | [72] |
| Romanus 2009 | 2009 | Enugu | NR | K. pneumoniae | DDM | Urine, blood, sputum | 186 | [73] |
| Shitta 2021 | 2016–2017 | Osun | Cross-sectional | E. coli, Klebsiella sp., P. vulgaris, S. rubidae, Citrobacter sp., Enterobacter sp., S. typhi | DDM | Urine, wound, high vaginal swab, stool, eye swab, ear swab, endocervical swab | 356 | [74] |
| Ugah 2022 | 2019–2021 | Abia, Enugu, Ebonyi, Imo, Anambra | Cross-sectional | E. coli, K. pneumoniae, K. oxytoca, M. morganii, C. freundii, S. marcescens, P. mirabilis, E. cloacae, S. dysenteriae, S. enterica, P. vulgaris, enterocolytica | DDM | Urine, sputum, CSF, stool, blood, etc. | 400 | [75] |
| Uzoamaka 2017 | 2014–2016 | Enugu | NR | E. coli, K. pneumoniae, Proteus sp. | DDM | Sputum | 259 | [76] |
| Yusuf 2014 | NR | Kano | NR | E. coli, K. pneumoniae, Enterobacter spp, Salmonella sp, P. mirabilis, P. vulgaris | DDM | Urine, catheter tip swab, stool, semen, urogenital swab, abscesses | 547 | [77] |
NR, Not reported; DDM, disk diffusion method; BMD, broth microdilution method; E-test, Epsilometer test.
3.3. Pooled prevalence
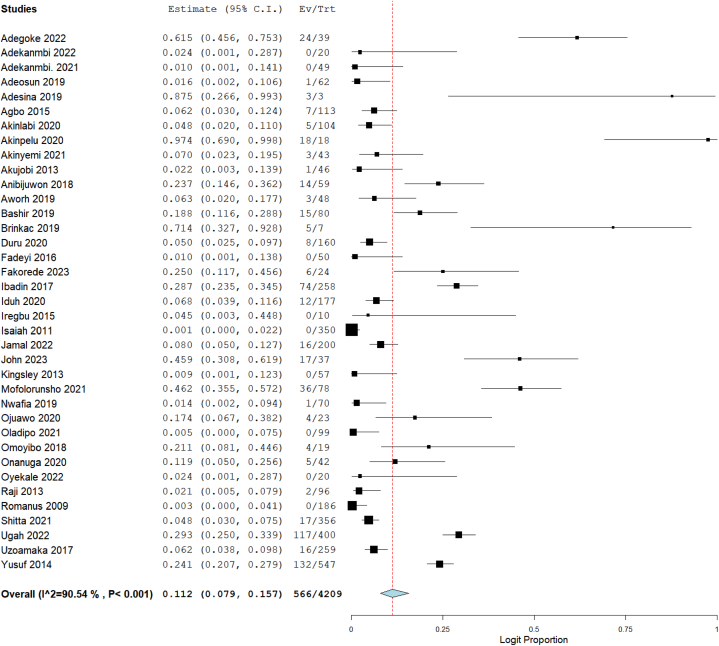
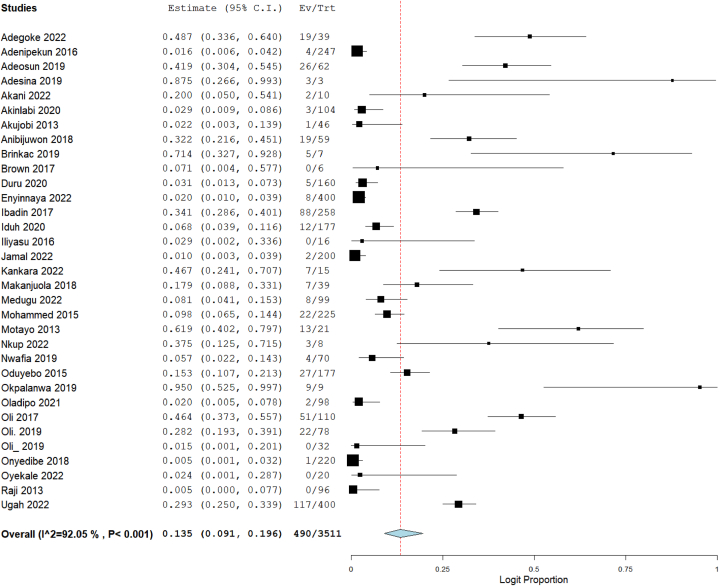
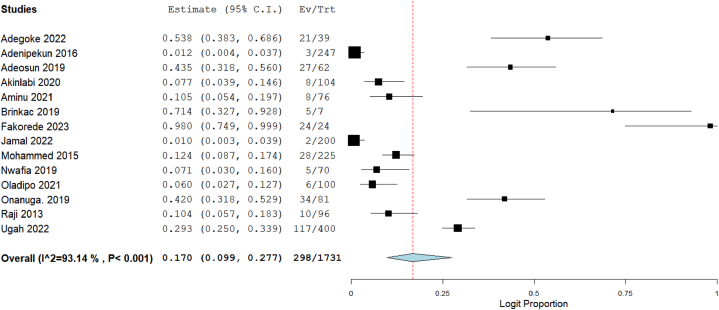
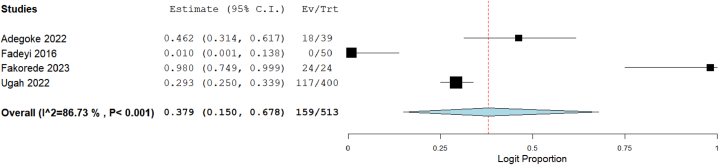
Out of the 57 studies included in this meta-analysis, data on resistance of CRE to imipenem were available in 37 studies, meropenem in 33 studies, ertapenem in 14 studies, and doripenem in 4 studies. The total number of tested Enterobacterales was 4209 for imipenem, 3511 for meropenem, 1731 for ertapenem, and 513 for doripenem. Using the random-effect model to calculate summary estimates, the aggregated prevalence estimate for imipenem resistance among CRE was 11.2 % (95 % CI: 7.9–15.7). Meropenem resistance had an estimate of 13.5 % (95 % CI: 9.1–19.6), while ertapenem and doripenem were estimated at 17.0 % (95 % CI: 9.9–27.7) and 37.9 % (95 % CI: 15.0–67.8), respectively (Fig. 2, Fig. 3, Fig. 4, Fig. 5). Notably, high heterogeneity (I2>85 %, p < 0.001) was observed for all the estimates.
Fig. 2.
Forest plot illustrating the pooled prevalence of imipenem resistance in CRE.
Fig. 3.
Forest plot illustrating the pooled prevalence of meropenem resistance in CRE.
Fig. 4.
Forest plot illustrating the pooled prevalence of ertapenem resistance in CRE.
Fig. 5.
Forest plot illustrating the pooled prevalence of doripenem resistance in CRE.
3.4. Prevalence of CRE in various subgroups
The resistance rates of the examined Enterobacterales against imipenem, meropenem, ertapenem, and doripenem antibiotics underwent additional scrutiny, considering variables such as the study period, geographical region, and study design. Further, analyses based on specific genera of CRE were performed. These comprehensive analyses aimed to offer a thorough perspective on resistance rates and investigate the observed heterogeneity. Forest plots for all subgroup analyses are available in the supplementary document (File S3).
3.4.1. Prevalence of imipenem resistance in CRE in various subgroups
Table 2 provides a summary of the imipenem resistance prevalence in CRE based on various analyzed variables. The subgroup analysis included studies from the six geopolitical zones of Nigeria, with the South-West region being the most represented (n = 13). South-South region exhibited the highest resistance rate at 28.4 % (95 % CI: 14.0–49.2), while the North-East region had the lowest at 0.1 % (95 % CI: 0.0–2.2). The majority of studies (n = 16) were conducted between 2016 and 2020, accounting for a resistance of 13.1 % (95 % CI: 8.2–20.3). Nonetheless, the highest CRE resistance rate (25.0 %, 95 % CI: 11.7–45.6) was observed in the 2021–2023 period. However, the estimate was contributed by only one study. Regarding study design, the analyzed studies were predominantly cross-sectional studies (n = 16). The highest resistance rate was observed in a prospective study (17.4 %, 95 % CI: 6.7–38.2), followed by cross-sectional studies (12.1 %, 95 % CI: 6.9–20.4). While various parameters indicated high heterogeneity, studies conducted in the 2011–2015 period demonstrated moderate heterogeneity (I2 = 52.26 %).
Table 2.
Prevalence of imipenem resistance in CRE in different subgroups.
| Subgroup | Number of studies | Prevalence (%) | 95 % CI | Heterogeneity test |
||
|---|---|---|---|---|---|---|
| Q | I2 (%) | p-value | ||||
| Region | ||||||
| South-South | 5 | 28.4 | 14.0–49.2 | 33.08 | 87.91 | <0.001 |
| South-West | 13 | 9.6 | 4.2–20.6 | 72.60 | 83.47 | <0.001 |
| South-East | 6 | 4.6 | 1.3–14.6 | 83.01 | 93.98 | <0.001 |
| North-Central | 7 | 15.1 | 5.0–37.4 | 64.33 | 90.67 | <0.001 |
| North-West | 3 | 15.3 | 7.2–29.6 | 22.24 | 91.01 | <0.001 |
| North-East | 1 | 0.1 | 0.0–2.2 | NA | NA | NA |
| Overall | 35 | 12.0 | 8.4–16.8 | 347.63 | 90.22 | <0.001 |
| Study period | ||||||
| 2006–2010 | 1 | 0.3 | 0.0–4.1 | NA | NA | NA |
| 2011–2015 | 7 | 5.3 | 2.4–11.2 | 12.57 | 52.26 | 0.05 |
| 2016–2020 | 16 | 13.1 | 8.2–20.3 | 166.37 | 90.98 | <0.001 |
| 2021–2023 | 1 | 25.0 | 11.7–45.6 | NA | NA | NA |
| Overall | 25 | 10.0 | 6.5–15.1 | 220.50 | 89.12 | <0.001 |
| Study design | ||||||
| Cross-sectional | 16 | 12.1 | 6.9–20.4 | 170.56 | 91.21 | <0.001 |
| Retrospective | 1 | 4.5 | 0.3–44.8 | NA | NA | NA |
| Prospective | 1 | 17.4 | 6.7–38.2 | NA | NA | NA |
| Overall | 18 | 12.2 | 7.2–19.8 | 172.61 | 90.15 | <0.001 |
3.4.2. Prevalence of meropenem resistance in CRE in various subgroups
The subgroup analysis on meropenem resistance included studies from all the six geopolitical zones of the country, with the South-West being the most represented (n = 11) (Table 3). The South-South region exhibited the highest pooled resistance rate at 37.2 % (95 % CI: 26.0–50.0), while the lowest was from the North-East (9.8 %, 95 % CI: 6.5–14.4). Studies conducted from 2016 to 2020 were the most abundant (n = 14), constituting the period with the highest pooled resistance rate to meropenem antibiotics at 24.7 % (95 % CI: 17.1–34.1). Cross-sectional studies were the most common in terms of study design (n = 14). A substantial meropenem resistance rate of 43.5 % (95 % CI: 25.2–63.8) was estimated for the prospective study design. A moderate heterogeneity was observed for studies conducted in the South-South region (51.38 %), and no heterogeneity was found for studies in the prospective study design.
Table 3.
Prevalence of meropenem resistance in CRE in different subgroups.
| Subgroup | Number of studies | Prevalence (%) | 95 % CI | Heterogeneity test |
||
|---|---|---|---|---|---|---|
| Q | I2 (%) | p-value | ||||
| Region | ||||||
| South-South | 3 | 37.2 | 26.0–50.0 | 4.11 | 51.38 | 0.128 |
| South-West | 11 | 14.3 | 6.5–28.6 | 98.24 | 89.82 | <0.001 |
| South-East | 6 | 18.9 | 10.3–32.1 | 42.07 | 88.12 | <0.001 |
| North-Central | 7 | 11.5 | 2.6–38.5 | 68.62 | 91.26 | <0.001 |
| North-West | 3 | 13.1 | 2.0–53.1 | 18.38 | 89.12 | <0.001 |
| North-East | 1 | 9.8 | 6.5–14.4 | NA | NA | NA |
| Overall | 31 | 15.0 | 10.2–21.6 | 361.94 | 91.71 | <0.001 |
| Study period | ||||||
| 2011–2015 | 8 | 5.3 | 2.4–11.2 | 37.98 | 81.57 | <0.001 |
| 2016–2020 | 14 | 24.7 | 17.1–34.1 | 114.55 | 88.65 | <0.001 |
| 2021–2023 | 1 | 20.0 | 5.0–54.1 | NA | NA | NA |
| Overall | 23 | 16.0 | 10.9–23.0 | 234.94 | 90.64 | <0.001 |
| Study design | ||||||
| Cross-sectional | 14 | 11.3 | 5.8–21.0 | 206.01 | 93.69 | <0.001 |
| Retrospective | 1 | 2.9 | 0.2–33.6 | NA | NA | NA |
| Prospective | 2 | 43.5 | 25.2–63.8 | 0.18 | 0 | 0.673 |
| Overall | 17 | 13.2 | 7.3–22.8 | 213.49 | 92.51 | <0.001 |
3.4.3. Prevalence of ertapenem resistance in CRE in various subgroups
The South-West region contributed more studies (n = 5) to the ertapenem subgroup analysis, with the highest pooled resistance rate observed for the South-South at 46.5 % (95 % CI: 35.5–57.8) (Table 4). Further, there were more studies conducted in the 2016–2020 study period (n = 7). However, the highest resistance rate of 98 % (95 % CI: 74.9–99.9) was in the 2021–2023 study period. It is worthy of note, however, that this high resistance rate was contributed by a single study. Except for studies conducted in the South-South region, high heterogeneity (>90 %) was observed in the variables assessed for ertapenem resistance.
Table 4.
Prevalence of ertapenem resistance in CRE in different subgroups.
| Subgroup | Number of studies | Prevalence (%) | 95 % CI | Heterogeneity test |
||
|---|---|---|---|---|---|---|
| Q | I2 (%) | p-value | ||||
| Region | ||||||
| South-South | 2 | 46.5 | 35.5–57.8 | 1.49 | 32.64 | 0.223 |
| South-West | 5 | 18.4 | 4.1–54.2 | 75.24 | 94.68 | <0.001 |
| South-East | 2 | 15.9 | 3.5–49.5 | 12.43 | 91.96 | <0.001 |
| North-Central | 2 | 13.5 | 0.1–97.2 | 25.21 | 96.03 | <0.001 |
| North-West | 1 | 10.5 | 5.4–19.7 | NA | NA | NA |
| North-East | 1 | 12.4 | 8.7–17.4 | NA | NA | NA |
| Overall | 13 | 18.1 | 10.4–29.7 | 176.73 | 93.21 | <0.001 |
| Study period | ||||||
| 2011–2015 | 4 | 10.7 | 3.2–30.1 | 60.89 | 95.07 | <0.001 |
| 2016–2020 | 7 | 15.4 | 7.1–30.2 | 76.48 | 92.15 | <0.001 |
| 2021–2023 | 1 | 98.0 | 74.9–99.9 | NA | NA | NA |
| Overall | 12 | 16.2 | 9.0–27.3 | 158.26 | 93.05 | <0.001 |
3.4.4. Prevalence of carbapenem resistant organisms
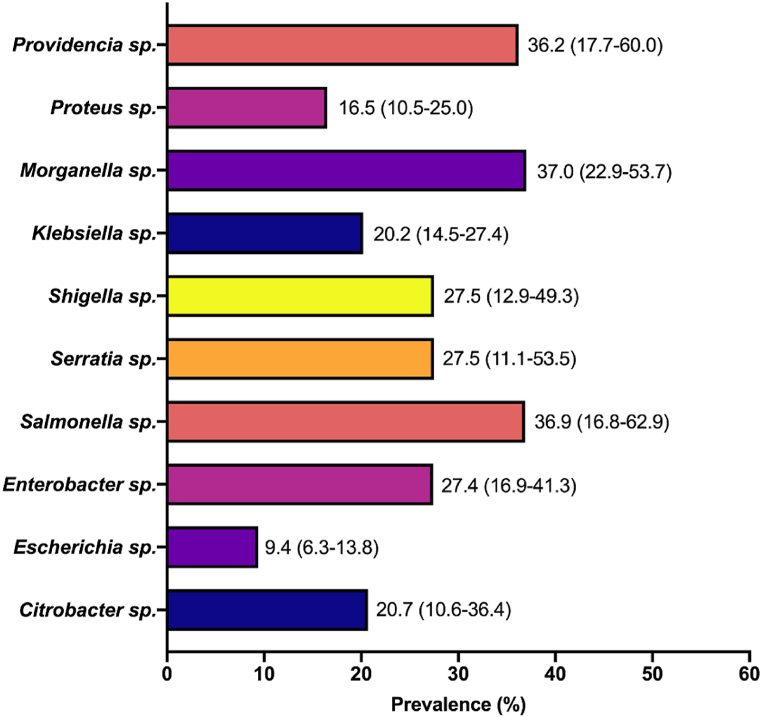
The prevalence of specific genera of CRE to at least one carbapenem antibiotic was also evaluated. The estimates revealed Morganella sp. as the most resistant CRE (37.0 % [95 % CI: 22.9–53.7]) followed by Salmonella sp. (36.9 % [95 % CI: 16.8–62.9]). In contrast, Escherichia sp. was the least resistant (9.4 % [95 % CI: 6.3–13.8]) among the CRE analyzed (Fig. 6).
Fig. 6.
Prevalence of specific CRE genera. Values in parenthesis are 95 % confidence intervals.
3.5. Evaluation of study quality and assessment of publication bias
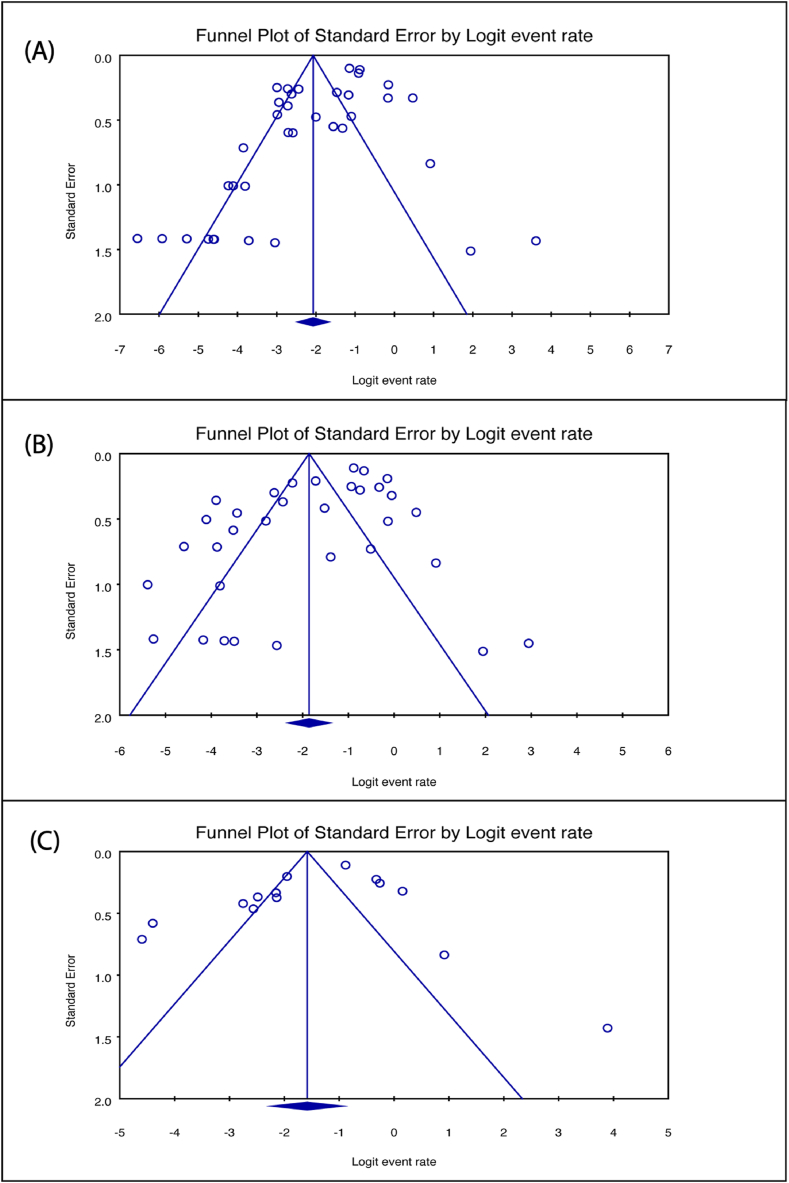
The evaluation of methodological quality using the JBI tool indicated that the included studies demonstrated good quality (File S4). Funnel plots were generated to scrutinize potential publication bias, and their asymmetry, as depicted in Fig. 7, suggested the likelihood of publication bias. Further investigation into funnel plot asymmetry was conducted using Egger's regression test. Some tests yielded significant p-values for studies contributing to the estimates of imipenem (p = 0.0039) (Fig. 7A), meropenem (p = 0.0166) (Fig. 7B), and ertapenem (p = 0.2559) (Fig. 7C) resistance. To address potential biases, a Trim and Fill analysis was performed, with adjusted values revealing changes in resistance estimates: imipenem from 11.2 % to 15.6 % (95 % CI: 11.2–21.3), meropenem from 13.5 % to 17.0 % (95 % CI: 11.7–24.1), and ertapenem from 17.0 % to 14.6 % (95 % CI: 8.3–24.5) (File S5).
Fig. 7.
Funnel plot of publication bias assessment for (A) imipenem [Egger's test, p = 0.0039] (B) meropenem [Egger's test, p = 0.0166] and (C) ertapenem [Egger's test, p = 0.2559] resistance estimates.
4. Discussion
CRE pose a significant and evolving threat to global public health. These multidrug-resistant bacteria have developed resistance to carbapenem antibiotics, which are considered last-line treatments for many severe bacterial infections. The emergence and spread of CRE raise critical concerns due to their potential to cause difficult-to-treat infections, increased morbidity and mortality rates, and the limited availability of effective antibiotics. Surveillance data indicate a rising prevalence of CRE, with reports from healthcare facilities highlighting its emergence in both community and hospital settings. Understanding the geographical distribution and prevalence rates is crucial for implementing targeted interventions. In this study, we provide an overview of CRE in Nigeria, the most populous African nation.
The studies included in this work were conducted in various locations within Nigeria, representing all regions of the country. However, the studies were predominantly from the South-West region (n = 19) followed by the North central. This observed distribution may be attributed to heightened surveillance and reporting activities in these areas. However, the limited representation of CRE data from other regions of the country does not necessarily indicate a lower prevalence of CRE; rather, it could be indicative of a constrained capacity for molecular epidemiology and surveillance studies or a reduced interest in CRE research. Additionally, it is likely that other infectious diseases such as malaria and HIV/AIDS are prioritized over antimicrobial resistance in those regions. Further, the majority of the studies were conducted after 2015, indicating increased interest in CRE research in recent years, which may have been warranted by the global trends in carbapenem resistance [[78], [79], [80]].
Further, E. coli and K. pneumoniae were the most frequently tested Enterobacterales among the included studies. These bacteria are commonly associated with various infections, especially in the urinary tract, respiratory system, and bloodstream [81,82]. The choice to frequently test E. coli and K. pneumoniae for carbapenem resistance may have been driven by their potential to cause serious infections and their propensity to acquire resistance, which can limit treatment options [81,82]. Moreover, prophages, which are significant vectors of horizontal gene transfer, have been detected in these organisms [83,84]. By integrating into bacterial genomes, prophages can spread resistance genes among bacterial populations, enabling the host bacteria to swiftly acquire new resistance traits under antibiotic pressure or other adverse environmental conditions [84]. This mechanism facilitates the rapid adaptation and survival of bacteria in challenging environments [84]. Thus, monitoring the prevalence of carbapenem resistance in these pathogens is crucial for understanding the scope of the issue and implementing effective infection control measures. It is worth noting that the focus on these specific pathogens does not diminish the importance of monitoring other members of the Enterobacterales for carbapenem resistance, as resistance can emerge in various species within this bacterial group. Moreover, other members, including species of Salmonella, Enterobacter, and Citrobacter, among others, were tested, and the pooled prevalence of carbapenem-resistant Morganella sp. was determined to be the highest among the CRE in this study. Comprehensive surveillance and testing strategies are essential to addressing the broader challenge of antimicrobial resistance and ensuring appropriate patient care.
We were interested in elucidating the overall pooled resistance rates of the Enterobacterales to the major carbapenem antibiotics: imipenem, meropenem, ertapenem and doripenem. Our findings revealed doripenem as the antibiotics with the highest resistance rate (37.9 %) among CRE, followed by ertapenem (17.0 %). Although the lowest resistance rate (11.2 %) was against the imipenem antibiotics, the observed rate is also worthy of concern. The high rates of carbapenem resistance may be attributed to frequent, inappropriate, and inaccurate administration of antimicrobial drugs in empirical treatment, coupled with insufficient infection control practices. The carbapenem resistance rates in this study are higher than reports from other countries, including the reported 3 % in Lebanon [85], 3.4 % in Afghanistan [86], 3.5 % in Belgium [87], and 5.74 % in Malaysia [88]. In contrast to our findings, higher carbapenem resistance rates have been reported in Jordan (41.2 %) [89] and Egypt (54.1 %) [90]. The observed variation may be attributed to differences in the methods employed for antibiotic susceptibility testing, the characteristics of the target population, sample types, the variety and quantity of bacterial isolates, disparities in antibiotic use policies, and variations in geographical locations.
In this study, we found the carbapenem resistance rates to vary among the regions of the country. Notably, the highest resistance rate to imipenem (28.4 %), meropenem (37.2 %) and ertapenem (46.5 %) were observed for the South-South region of Nigeria. The reason for the observed high carbapenem resistance in this region is unclear, but it could be linked to the misuse of antibiotics [91]. In contrast, much lower carbapenem resistance rates were observed for the North-East region of the country. This observation may be attributed to the limited number of reports from the region, as only one study contributed to the estimate from the region. Additionally, our analyses indicate a progressive increase in resistance to carbapenem antibiotics over the years in Nigeria. CRE have become a growing concern in Nigeria, mirroring global trends in antimicrobial resistance. The emergence and spread of CRE in the country may have been influenced by various factors, including antibiotic misuse, inadequate infection control practices, and challenges in healthcare infrastructure. The epidemiology of CRE in Nigeria is characterized by an increasing prevalence, affecting both community and hospital settings. Studies suggest that CRE strains have been isolated from clinical specimens, including blood, urine, and wound cultures, indicating the diverse range of infections associated with these resistant bacteria [60,73,74].
Based on the specific members of the Enterobacterales assessed in this study, species of Providencia, Morganella and Salmonella were the most prevalent CRE, with prevalence hovering around 37 %. Previous report in Japan recorded low proportion of Providencia stuartii (1.3 %) and Providencia rettgeri (0.5 %) among the isolated Enterobacterales strains [92]. Moreover, there has been a growing focus on less common carbapenem-resistant CRE species, such as P. rettgeri and K. oxytoca, which are emerging as significant clinical concerns [93,94]. Similarly, in a different report, 9 Morganella morganii were found among 312 carbapenem-resistant Gram-negative bacteria from India [95]. Hence, addressing the notably high prevalence rate observed in this study is essential. In contrast, although E. coli was one of the most frequently tested Enterobacterales in this study, Escherichia sp. was the least prevalent CRE (9.4 %). Nonetheless, the observed rate is lower than the documented rates of carbapenem-resistant E. coli in many other parts of the world, including the 11 % in the United States, 31 % in Singapore, 22 % in Thailand, 18.7 % in Korea and 29 % in Canada [95].
CRE are of significant epidemiological concern due to their association with unfavorable outcomes and their ability to rapidly disseminate within healthcare systems [96]. Previous investigations have consistently demonstrated that CRE infections are linked to a higher mortality rate compared to non-CRE infections. For instance, a study conducted in China revealed a 30-day mortality rate of 65.4 % for CRE bloodstream infections, in contrast to a significantly lower rate of 17.2 % for non-CRE bloodstream infections. They attributed this heightened mortality to host conditions such as endogenous infections and invasive surgical procedures [97]. Furthermore, 30-day mortality rates for CRE infections are also reported to be higher when compared to infections caused by other pathogens exhibiting a broad spectrum of resistance mechanisms [98].
5. Strengths and limitations
This study has its strengths and limitations. To our knowledge, this is the first meta-analysis evaluating the prevalence of CRE from human samples in Nigeria. Because we did not apply any filter for the year in which the studies were performed or published, we believe our report is robust, accommodating all relevant published data. We were, however, unable to perform elaborate analysis on resistance rates to the doripenem antibiotics due to the limited number of available studies.
6. Conclusion
A comprehensive overview of the status of CRE derived from human samples in Nigeria was provided in this study. Resistance to the major carbapenems (imipenem, meropenem, ertapenem, doripenem) were identified, and ranged from 11.2 % for imipenem to 37.9 % for doripenem antibiotics. Our analyses indicated a progressive increase in resistance to carbapenem antibiotics over the years, with the South-South region of the country accounting for higher carbapenem resistance rates. Proactive measures should, therefore, be put in place to check these rising rates of carbapenem resistance in the country to prevent the likely proliferation to other nations. A comprehensive management approach, including antimicrobial stewardship programs, infection prevention and control measures, and surveillance systems should be employed to limit the spread of CRE. Additionally, promoting rational antibiotic use and educating healthcare professionals are essential components of a sustainable strategy.
Future research on CRE should prioritize the development of novel antimicrobial agents and treatment strategies to combat this emerging public health threat. The mechanisms of resistance, transmission dynamics, and genetic evolution of CRE strains should be investigated across different healthcare settings to enhance our understanding of their epidemiology and to ensure the adoption of effective infection control measures. Additionally, exploring the role of environmental reservoirs and potential vectors in CRE spread can provide insights into preventive strategies.
Funding
This research was supported by the Fundamental Research Grant Scheme (FRGS) of the Ministry of Higher Education Malaysia [grant number FRGS/1/2021/SKK06/USM/02/2].
Ethical approval
Not required.
Ethics statement
Not applicable.
Data availability
All data accessed and analyzed in this study are available in the article and its supplementary materials.
CRediT authorship contribution statement
Ahmad Adebayo Irekeola: Writing – review & editing, Writing – original draft, Project administration, Methodology, Investigation, Formal analysis, Conceptualization. Rafidah Hanim Shueb: Writing – review & editing, Validation, Supervision, Project administration, Methodology, Funding acquisition, Conceptualization. Engku Nur Syafirah Engku Abd Rahman: Writing – review & editing, Methodology, Investigation. Hafeez Abiola Afolabi: Writing – original draft, Validation, Methodology, Investigation. Yusuf Wada: Writing – review & editing, Validation, Methodology, Investigation. Abdirahman Hussein Elmi: Writing – review & editing, Methodology, Investigation. Muath Abdu Hakami: Writing – review & editing, Methodology, Investigation. Sfeeah Mofareah Alghzwani: Writing – review & editing, Methodology, Investigation. Osman AE. Elnoubi: Writing – review & editing, Methodology, Investigation. Ahmad A. Alshehri: Writing – review & editing, Supervision, Project administration, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are thankful to the authors who provided additional information when contacted regarding their articles.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e34926.
Contributor Information
Rafidah Hanim Shueb, Email: hanimshueb@gmail.com.
Ahmad A. Alshehri, Email: aaalshehri@nu.edu.sa.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Murray C.J.L., Ikuta K.S., Sharara F., Swetschinski L., Aguilar G.R., Gray A., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet (N. Am. Ed.) 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO . 2021 2021. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report. [Google Scholar]
- 3.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 4.Tompkins K., van Duin D. Treatment for carbapenem-resistant Enterobacterales infections: recent advances and future directions. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:2053–2068. doi: 10.1007/s10096-021-04296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma J., Song X., Li M., Yu Z., Cheng W., Yu Z., et al. Global spread of carbapenem-resistant Enterobacteriaceae: epidemiological features, resistance mechanisms, detection and therapy. Microbiol. Res. 2023;266 doi: 10.1016/j.micres.2022.127249. [DOI] [PubMed] [Google Scholar]
- 6.Aurilio C., Sansone P., Barbarisi M., Pota V., Giaccari L.G., Coppolino F., et al. Mechanisms of action of carbapenem resistance. Antibiotics. 2022;11:421. doi: 10.3390/antibiotics11030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papp-Wallace K.M., Endimiani A., Taracila M.A., Bonomo R.A. Carbapenems: past, present, and future. Antimicrob. Agents Chemother. 2011;55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Duin D., Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 2017;8:460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordmann P., Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 2014;20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 10.Nordmann P., Naas T., Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011;17:1791. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmed N., Tahir K., Aslam S., Cheema S.M., Rabaan A.A., Turkistani S.A., et al. Heavy metal (arsenic) induced antibiotic resistance among extended-spectrum β-lactamase (ESBL) producing bacteria of nosocomial origin. Pharmaceuticals. 2022;15 doi: 10.3390/ph15111426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed N., Khan M., Saleem W., Karobari M.I., Mohamed R.N., Heboyan A., et al. Evaluation of Bi-lateral Co-infections and antibiotic resistance rates among COVID-19 patients. Antibiotics (Basel) 2022;11 doi: 10.3390/antibiotics11020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed N., Habib S., Muzzammil M., Rabaan A.A., Turkistani S.A., Garout M., et al. Prevalence of bacterial pathogens among symptomatic-SARS-CoV-2 PCR-negative patients. Microorganisms. 2022;10 doi: 10.3390/microorganisms10101978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Logan L.K., Weinstein R.A. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irek E.O., Amupitan A.A., Aboderin A.O., Obadare T.O. A systematic review of healthcare-associated infections in Africa: an antimicrobial resistance perspective. Afr J Lab Med. 2018;7:1–9. doi: 10.4102/ajlm.v7i2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munn Z., Moola S., Lisy K., Riitano D., Tufanaru C. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and cumulative incidence data. Int J Evid Based Healthc. 2015;13:147–153. doi: 10.1097/XEB.0000000000000054. [DOI] [PubMed] [Google Scholar]
- 18.Irekeola A.A., Shueb R.H., E A.R.E.N.S., Wada Y., Abdul Rahman Z., Ahmad S., et al. Prevalence of nasopharyngeal carcinoma in patients with dermatomyositis: a systematic review and meta-analysis. Cancers. 2021;13 doi: 10.3390/cancers13081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins J.P.T., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 20.Irekeola A.A., Ear E.N.S., Wada Y., Mohamud R., Lazim N.M., Yean C.Y., et al. Prevalence of EBV infection in 1157 diseased cohorts in Nigeria: a systematic review and meta-analysis. Indian J. Med. Microbiol. 2022;40:420–426. doi: 10.1016/j.ijmmb.2022.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Adegoke A.A., Ikott W.E., Okoh A.I. Carbapenem resistance associated with coliuria among outpatient and hospitalised urology patients. New Microbes New Infect. 2022;48 doi: 10.1016/j.nmni.2022.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adekanmbi A.O., Akinlabi O.C., Usidamen S., Olaposi A.V., Olaniyan A.B. High burden of ESBL- producing Klebsiella spp., Proteus mirabilis, Enterobacter cloacae and Pseudomonas aeruginosa in diagnosed cases of urinary tract infection in a Nigerian Teaching Hospital. Acta Microbiol. Immunol. Hung. 2022;69:127–134. doi: 10.1556/030.2022.01747. [DOI] [PubMed] [Google Scholar]
- 23.Adekanmbi A.O., Usidamen S., Onilude A.A. Molecular characterization of ESBL- producing uropathogenic Escherichia coli recovered from urine samples of patients attending a University Teaching hospital in Nigeria. Acta Microbiol. Immunol. Hung. 2021 doi: 10.1556/030.2021.01380. [DOI] [PubMed] [Google Scholar]
- 24.Adenipekun E.O., Jackson C.R., Ramadan H., Iwalokun B.A., Oyedeji K.S., Frye J.G., et al. Prevalence and multidrug resistance of Escherichia coli from community-acquired infections in Lagos, Nigeria. J Infect Dev Ctries. 2016;10:920–931. doi: 10.3855/jidc.7997. [DOI] [PubMed] [Google Scholar]
- 25.Adeosun I.J., Oladipo E.K., Ajibade O.A., Olotu T.M., Oladipo A.A., Awoyelu E.H., et al. Antibiotic susceptibility of klebsiella pneumoniae isolated from selected tertiary hospitals in Osun state, Nigeria. Iraqi J. Sci. 2019;60:1423–1429. doi: 10.24996/ijs.2019.60.7.2. [DOI] [Google Scholar]
- 26.Adesanya O.A., Igwe H.A. Carbapenem-resistant Enterobacteriaceae (CRE) and gram-negative bacterial infections in south-west Nigeria: a retrospective epidemiological surveillance study. AIMS Public Health. 2020;7:804. doi: 10.3934/publichealth.2020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adesina T., Nwinyi O., De N., Akinnola O., Omonigbehin E. First detection of carbapenem-resistant Escherichia fergusonii strains harbouring beta-lactamase genes from clinical samples. Pathogens. 2019;8 doi: 10.3390/pathogens8040164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agbo E., Eze E. Phenotypic detection of metallo beta lactamases production in imipenem resistant isolates of escherichia coli and klebsiella species isolated from hospital and environmental sources in nsukka, enugu state, Nigeria. J. Biol. Nat. 2015;2:135–141. [Google Scholar]
- 29.Akani N.P., Douglas S.I., Kamani N.C. Characterization of carbapenem-resistant enterobacteriaceae from patients stool in tertiary hospitals, port harcourt, rivers state, Nigeria. J Adv Microbiol. 2022:29–36. [Google Scholar]
- 30.Akinlabi A., Oluwadun A., Alli O.A.T., Oluremi A.S., Webber M.A., Ogbolu D.O. Role of extended spectrum beta lactamases in cephalosporin and carbapenem resistance in Escherichia coli from inpatients and outpatients in Nigeria. J. Clin. Diagn. Res. 2020;14 [Google Scholar]
- 31.Akinpelu S., Ajayi A., Smith S.I., Adeleye A.I. Efflux pump activity, biofilm formation and antibiotic resistance profile of Klebsiella spp. isolated from clinical samples at Lagos University Teaching Hospital. BMC Res. Notes. 2020;13:258. doi: 10.1186/s13104-020-05105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akinyemi K.O., Abegunrin R.O., Iwalokun B.A., Fakorede C.O., Makarewicz O., Neubauer H., et al. The emergence of Klebsiella pneumoniae with reduced susceptibility against third generation cephalosporins and carbapenems in lagos hospitals, Nigeria. Antibiotics (Basel) 2021;10 doi: 10.3390/antibiotics10020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akujobi C.N., Ezeanya C.C. Emergence of carbapenem resistance among extended-spectrum beta-lactamase isolate of Escherichia coli from clinical specimens in a tertiary hospital, Nigeria. Int. J. Microbiol. Res. 2013;5:367–370. [Google Scholar]
- 34.Aminu A., Daneji I.M., Yusuf M.A., Jalo R.I., Tsiga-Ahmed F.I., Yahaya M., et al. Carbapenem-resistant Enterobacteriaceae infections among patients admitted to intensive care units in Kano, Nigeria. Sahel Med. J. 2021;24:1. [Google Scholar]
- 35.Anibijuwon I.I., Gbala I.D., Adebisi O.O. Carbapenem-resistant enterobacteriaceae among in-patients of tertiary hospitals in southwest, Nigeria. Not. Sci. Biol. 2018;10:310–317. [Google Scholar]
- 36.Aworh M.K., Kwaga J., Okolocha E., Mba N., Thakur S. Prevalence and risk factors for multi-drug resistant Escherichia coli among poultry workers in the Federal Capital Territory, Abuja, Nigeria. PLoS One. 2019;14 doi: 10.1371/journal.pone.0225379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bashir A., Garba I., Aliero A.A., Kibiya A., Abubakar M.H., Ntulume I., et al. Superbugs-related prolonged admissions in three tertiary hospitals, Kano State, Nigeria. Pan Afr Med J. 2019;32:166. doi: 10.11604/pamj.2019.32.166.18481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brinkac L.M., White R., D'Souza R., Nguyen K., Obaro S.K., Fouts D.E. Emergence of New Delhi metallo-β-lactamase (NDM-5) in Klebsiella quasipneumoniae from neonates in a Nigerian hospital. mSphere. 2019;4 doi: 10.1128/mSphere.00685-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown B., Dada-Adegbola H., Trippe C., Olopade O. Prevalence and etiology of bacteremia in febrile children with sickle cell disease at a Nigeria tertiary hospital. Mediterr J Hematol Infect Dis. 2017;9 doi: 10.4084/MJHID.2017.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duru C., Olanipekun G., Odili V., Kocmich N., Rezac A., Ajose T.O., et al. Molecular characterization of invasive Enterobacteriaceae from pediatric patients in Central and Northwestern Nigeria. PLoS One. 2020;15 doi: 10.1371/journal.pone.0230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enyinnaya S., Iregbu K., Jamal W., Rotimi V. Antibiotic susceptibility profiles and detection of genes mediating extended-spectrum beta-lactamase (Esbl) production in escherichia coli isolates from National Hospital, Abuja. Niger. J. Clin. Pract. 2022;25:1216–1220. doi: 10.4103/njcp.njcp_1390_21. [DOI] [PubMed] [Google Scholar]
- 42.Fadeyi A., Zumuk C.P., Raheem R.A., Nwabuisi C., Desalu O.O. Prevalence and antibiotic susceptibility pattern of ESBL producing klebsiellae isolated from clinical specimens in a Nigerian tertiary hospital. Afr J Infect Dis. 2016;10:32–37. doi: 10.4314/ajid.v10i1.7. [DOI] [Google Scholar]
- 43.Fakorede C.O., Amisu K.O., Saki M., Akinyemi K.O. Co-existence of extended-spectrum beta-lactamases bla(CTX-M-9) and bla(CTX-M-15) genes in Salmonella species isolated from febrile and diarrhoeagenic patients in Lagos, Nigeria: a cross-sectional study. Eur. J. Med. Res. 2023;28 doi: 10.1186/s40001-022-00960-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ibadin E.E., Omoregie R., Igbarumah I.O., Anogie N.A., Ogefere H.O. Prevalence of extended spectrum β-lactamase, Ampc β-lactamase and metallo-β-lactamase enzymes among clinical isolates recovered from patients with urinary tract infections in Benin city, Nigeria. Int J Enteric Pathog. 2017;5:85–91. [Google Scholar]
- 45.Iduh M.U., Mohammed K., Garba M.K., Nataala S.U., Ashcroft O.F., Abdulkadir A. Prevalence of carbapenem-resistance among enterobacteriaceae isolated from patients attending uduth sokoto, North west Nigeria. Int J Trop Dis Health. 2020;41:55–64. [Google Scholar]
- 46.Iliyasu G., Daiyab F.M., Tiamiyu A.B., Abubakar S., Habib Z.G., Sarki A.M., et al. Nosocomial infections and resistance pattern of common bacterial isolates in an intensive care unit of a tertiary hospital in Nigeria: a 4-year review. J. Crit. Care. 2016;34:116–120. doi: 10.1016/j.jcrc.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 47.Iregbu K.C., Abdullahi N. Profiles of acute bacterial meningitis isolates in children in National Hospital, Abuja. Niger. Med. J. 2015;56:297–300. doi: 10.4103/0300-1652.169749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Isaiah I.N., Nche B.T., Nwagu I.G., Nwagu I.I. Incidence of temonera, sulphuhydryl variables and cefotaximase genes associated with beta-lactamase producing escherichia coli in clinical isolates. N. Am. J. Med. Sci. 2011;3:557–561. doi: 10.4297/najms.2011.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jamal W., Iregbu K., Fadhli A., Khodakhast F., Nwajiobi-Princewill P., Medugu N., et al. A point-prevalence survey of carbapenem-resistant Enterobacteriaceae in two different cities in Kuwait and Nigeria. Afr. J. Clin. Exp. Microbiol. 2022;23:358–368. [Google Scholar]
- 50.John G.E., Okpo E.A., Edeghor U.O., Omang P.A., Odok E.A., Odok A.O. GC-MS analysis and antibacterial susceptibility pattern of crude extracts of Gongronema latifolium and Allium sativum on selected multidrug-resistant clinical isolates. Malays J Microbiol. 2023;19:182–190. doi: 10.21161/mjm.220078. [DOI] [Google Scholar]
- 51.Kankara A.S., Dutsinma U.A., Yusuf I. Antimicrobial activities of piperacillin-tazobactam plus oxyimino- cephalosporins and gentamicin against extensively drug-resistant non-carbapenemase-producing enterobacteriaceae isolated from immunocompromised patients in a Nigerian hospital. Mediterranean Journal of Infection Microbes and Antimicrobials. 2022;11 doi: 10.4274/mjima.galenos.2022.2021.18. [DOI] [Google Scholar]
- 52.Kingsley O.C., Ifeanyi A.O., Edet A.E., Smart O.C. Bacteriologic profile and antibiotics susceptibility pattern of suspected septicaemic patients in Uyo, Nigeria. Res. J. Med. Sci. 2013;7:35–39. doi: 10.3923/rjmsci.2013.35.39. [DOI] [Google Scholar]
- 53.Makanjuola O.B., Fayemiwo S.A., Okesola A.O., Gbaja A., Ogunleye V.A., Kehinde A.O., et al. Pattern of multidrug resistant bacteria associated with intensive care unit infections in ibadan, Nigeria. Ann. Ib. Postgrad. Med. 2018;16:162–169. [PMC free article] [PubMed] [Google Scholar]
- 54.Medugu N., Aworh M.K., Iregbu K., Nwajiobi-Princewill P., Abdulraheem K., Hull D.M., et al. Molecular characterization of multi drug resistant Escherichia coli isolates at a tertiary hospital in Abuja, Nigeria. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-19289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mofolorunsho K.C., Ocheni H.O., Aminu R.F., Omatola C.A., Olowonibi O.O. Prevalence and antimicrobial susceptibility of extended-spectrum beta lactamases-producing Escherichia coli and Klebsiella pneumoniae isolated in selected hospitals of Anyigba, Nigeria. Afr. Health Sci. 2021;21:505–512. doi: 10.4314/ahs.v21i2.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohammed Y., Zailani S.B., Onipede A.O. Characterization of KPC, NDM and VIM type carbapenem resistance enterobacteriaceae from North eastern, Nigeria. J. Biosci. Med. 2015;3:100. [Google Scholar]
- 57.Motayo B.O., Akinduti P.A., Adeyakinu F.A., Okerentugba P.O., Nwanze J.C., Onoh C.C., et al. Antibiogram and plasmid profiling of carbapenemase and extended spectrum Beta-lactamase (ESBL) producing Escherichia coli and Klebsiella pneumoniae in Abeokuta, South western, Nigeria. Afr. Health Sci. 2013;13:1091–1097. doi: 10.4314/ahs.v13i4.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nkup J., Joseph S., Agabi Y., David V., Hashimu Z., Madubulum C., David A., Otobo U., Goyil G., Cirfat N., Anejo-Okopi J., David S. Molecular detection of carbapenem resistant Klebsiella pneumoniae isolated from clinical specimens in Jos, Nigeria. Microbes and Infectious Diseases. 2023;4(2):555–562. doi: 10.21608/mid.2022.145897.1329. [DOI] [Google Scholar]
- 59.Nwafia I.N., Ohanu M.E., Ebede S.O., Ozumba U.C. Molecular detection and antibiotic resistance pattern of extended-spectrum beta-lactamase producing Escherichia coli in a Tertiary Hospital in Enugu, Nigeria. Ann. Clin. Microbiol. Antimicrob. 2019;18:41. doi: 10.1186/s12941-019-0342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oduyebo O.O., Falayi O.M., Oshun P., Ettu A.O. Phenotypic determination of carbapenemase producing enterobacteriaceae isolates from clinical specimens at a tertiary hospital in Lagos, Nigeria. Niger. Postgrad. Med. J. 2015;22:223–227. doi: 10.4103/1117-1936.173973. [DOI] [PubMed] [Google Scholar]
- 61.Ojuawo O.B., Desalu O.O., Fawibe A.E., Ojuawo A.B., Aladesanmi A.O., Opeyemi C.M., et al. Clinical and microbiological profile of adult inpatients with community acquired pneumonia in Ilorin, North Central, Nigeria. Afr. Health Sci. 2020;20:1655–1668. doi: 10.4314/ahs.v20i4.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okpalanwa C.F., Anyanwu M.U., Momoh M.A., Nnamani P.O., Attama A.A. Generic Salmonella in asymptomatic adult volunteers: occurrence, antibiogram, extended-spectrum β-lactamase production and carbapenem resistance. Not. Sci. Biol. 2019;11:383–390. doi: 10.15835/NSB11310436. [DOI] [Google Scholar]
- 63.Oladipo E.K., Awoyelu E.H., Adeosun I.J., Ayandele A.A. Antibacterial susceptibility of clinical isolates of Klebsiella pneumoniae in Nigeria to carbapenems. Iraqi J. Sci. 2021;62:396–401. doi: 10.24996/ijs.2021.62.2.5. [DOI] [Google Scholar]
- 64.Oli A.N., Akabueze V.B., Ezeudu C.E., Eleje G.U., Ejiofor O.S., Ezebialu I.U., et al. Bacteriology and antibiogram of urinary tract infection among female patients in a tertiary health facility in south eastern Nigeria. Open Microbiol. J. 2017;11:292–300. doi: 10.2174/1874285801711010292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oli A.N., Itumo C.J., Okam P.C., Ezebialu I.U., Okeke K.N., Ifezulike C.C., et al. Carbapenem-resistant enterobacteriaceae posing a dilemma in effective healthcare delivery. Antibiotics-Basel. 2019;8 doi: 10.3390/antibiotics8040156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oli A.N., Ogbuagu V.I., Ejikeugwu C.P., Iroha I.R., Ugwu M.C., Ofomata C.M., et al. Multi-antibiotic resistance and factors affecting carriage of extended spectrum β-lactamase-producing enterobacteriaceae in pediatric population of enugu metropolis, Nigeria. Med. Sci. 2019;7 doi: 10.3390/medsci7110104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Omoyibo E.E., Oladele A.O., Ibrahim M.H., Adekunle O.T. Antibiotic susceptibility of wound swab isolates in a tertiary hospital in Southwest Nigeria. Ann. Afr. Med. 2018;17:110–116. doi: 10.4103/aam.aam_22_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Onanuga A., Eboh D.D., Odetoyin B., Adamu O.J. Detection of ESBLs and NDM-1 genes among urinary Escherichia coli and Klebsiella pneumoniae from healthy students in Niger delta university, amassoma, bayelsa state, Nigeria. Pan African Medical Journal One Health. 2020;2 doi: 10.11604/pamj-oh.2020.2.12.23588. [DOI] [Google Scholar]
- 69.Onanuga A., Vincent C.H., Eboh D.D. Carbapenem resistance among extended spectrum beta-lactamases producing Escherichia coli and Klebsiella pneumoniae isolates from patents with urinary tract infections in port-harcourt, Nigeria. Nigerian Journal of Pharmaceutical and Applied Science Research. 2019;8:16–23. [Google Scholar]
- 70.Onyedibe K.I., Shobowale E.O., Okolo M.O., Iroezindu M.O., Afolaranmi T.O., Nwaokorie F.O., et al. Low prevalence of carbapenem resistance in clinical isolates of extended spectrum beta lactamase (ESBL) producing Escherichia coli in North Central, Nigeria. Adv. Infect. Dis. 2018;8:109–120. [Google Scholar]
- 71.Oyekale O.T., Ojo B.O., Olajide A.T., Oyekale O.I. Bacteriological profile and antibiogram of blood culture isolates from bloodstream infections in a rural tertiary hospital in Nigeria. Afr J Lab Med. 2022;11:1807. doi: 10.4102/ajlm.v11i1.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raji M.A., Jamal W., Ojemhen O., Rotimi V.O. Point-surveillance of antibiotic resistance in enterobacteriaceae isolates from patients in a lagos teaching hospital, Nigeria. J Infect Public Health. 2013;6:431–437. doi: 10.1016/j.jiph.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 73.Romanus I.I., Egwu O.A., Ngozi A.T., Chidiebube N.A., Chika E.P. Extended spectrum beta - lactamase (ESBL) mediated resistance to antibiotics among Klebsiella pneumoniae in Enugu metropolis. Macedonian J. Med. Sci. 2009;2:196–199. doi: 10.3889/MJMS.1857-5773.2009.0052. [DOI] [Google Scholar]
- 74.Shitta G., Makanjuola O., Adefioye O., Olowe O.A. Extended spectrum beta lactamase (Esbl), blatem,blashv and blactx-m, resistance genes in community and healthcare associated gram negative bacteria from osun state, Nigeria. Infect. Disord.: Drug Targets. 2021;21:595–602. doi: 10.2174/1871526520999200729181559. [DOI] [PubMed] [Google Scholar]
- 75.Ugah U.I., Udeani T.K. Prevalence of phenotypic carbapenem-resistant Enterobacterales isolates and their distribution by sex, age groups, state and species in south-east Nigeria. Gomal J. Med. Sci. 2022;20:89–96. doi: 10.46903/gjms/20.02.1127. [DOI] [Google Scholar]
- 76.Uzoamaka M., Ngozi O., Johnbull O.S., Martin O. Bacterial etiology of lower respiratory tract infections and their antimicrobial susceptibility. Am. J. Med. Sci. 2017;354:471–475. doi: 10.1016/j.amjms.2017.06.025. [DOI] [PubMed] [Google Scholar]
- 77.Yusuf I., Arzai A.H., Haruna M., Sharif A.A., Getso M.I. Detection of multi drug resistant bacteria in major hospitals in Kano, North-West, Nigeria. Braz. J. Microbiol. 2014;45:791–798. doi: 10.1590/s1517-83822014000300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chatterjee N., Nirwan P.K., Srivastava S., Rati R., Sharma L., Sharma P., et al. Trends in carbapenem resistance in Pre-COVID and COVID times in a tertiary care hospital in North India. Ann. Clin. Microbiol. Antimicrob. 2023;22:1. doi: 10.1186/s12941-022-00549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jean S.-S., Harnod D., Hsueh P.-R. Global threat of carbapenem-resistant gram-negative bacteria. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.823684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhong H., Chen F., Li Y.-J., Zhao X.-Y., Zhang Z.-L., Gu Z.-C., et al. Global trends and hotspots in research of carbapenem-resistant Enterobacteriaceae (CRE): a bibliometric analysis from 2010 to 2020. Ann. Palliat. Med. June 28, 2021;10(6) doi: 10.21037/apm-21-87. Annals of Palliative Medicine 2021. [DOI] [PubMed] [Google Scholar]
- 81.Chang D., Sharma L., Dela Cruz C.S., Zhang D. Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.750662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jalil M.B., Al Atbee M.Y.N. The prevalence of multiple drug resistance Escherichia coli and Klebsiella pneumoniae isolated from patients with urinary tract infections. J. Clin. Lab. Anal. 2022;36 doi: 10.1002/jcla.24619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bleriot I., Trastoy R., Blasco L., Fernández-Cuenca F., Ambroa A., Fernández-García L., et al. Genomic analysis of 40 prophages located in the genomes of 16 carbapenemase-producing clinical strains of Klebsiella pneumoniae. Microb. Genom. 2020;6 doi: 10.1099/mgen.0.000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pei Z., Liu Y., Chen Y., Pan T., Sun X., Wang H., et al. A universe of human gut-derived bacterial prophages: unveiling the hidden viral players in intestinal microecology. Gut Microb. 2024;16 [Google Scholar]
- 85.Moghnieh R., Araj G.F., Awad L., Daoud Z., Mokhbat J.E., Jisr T., et al. A compilation of antimicrobial susceptibility data from a network of 13 Lebanese hospitals reflecting the national situation during 2015–2016. Antimicrob. Resist. Infect. Control. 2019;8:1–17. doi: 10.1186/s13756-019-0487-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mende K., Beckius M.L., Zera W.C., Onmus-Leone F., Murray C.K., Tribble D.R. US Army Med Dep J; 2017. Low Prevalence of Carbapenem-Resistant Enterobacteriaceae Among Wounded Military Personnel; pp. 12–17. [PMC free article] [PubMed] [Google Scholar]
- 87.Huang T.-D., Berhin C., Bogaerts P., Glupczynski Y., group a multicentre study, Caddrobi J, et al. Prevalence and mechanisms of resistance to carbapenems in Enterobacteriaceae isolates from 24 hospitals in Belgium. J. Antimicrob. Chemother. 2013;68:1832–1837. doi: 10.1093/jac/dkt096. [DOI] [PubMed] [Google Scholar]
- 88.Zaidah A.R., Mohammad N.I., Suraiya S., Harun A. High burden of Carbapenem-resistant Enterobacteriaceae (CRE) fecal carriage at a teaching hospital: cost-effectiveness of screening in low-resource setting. Antimicrob. Resist. Infect. Control. 2017;6:1–6. doi: 10.1186/s13756-017-0200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hammour K.A., Abu-Farha R., Itani R., Karout S., Allan A., Manaseer Q., et al. The prevalence of carbapenem resistance gram negative pathogens in a tertiary teaching hospital in Jordan. BMC Infect. Dis. 2023;23:634. doi: 10.1186/s12879-023-08610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kotb S., Lyman M., Ismail G., Abd El Fattah M., Girgis S.A., Etman A., et al. Epidemiology of carbapenem-resistant enterobacteriaceae in Egyptian intensive care units using national healthcare–associated infections surveillance data, 2011–2017. Antimicrob. Resist. Infect. Control. 2020;9:1–9. doi: 10.1186/s13756-019-0639-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Akande-Sholabi W., Oyesiji E. Antimicrobial stewardship: knowledge, perceptions, and factors associated with antibiotics misuse among consumer's visiting the community pharmacies in a Nigeria Southwestern State. J Pharm Policy Pract. 2023;16:120. doi: 10.1186/s40545-023-00629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takei K., Ogawa M., Sakata R., Kanamori H. Epidemiological characteristics of carbapenem-resistant Enterobacterales in Japan: a nationwide analysis of data from a clinical laboratory center (2016–2022) Pathogens. 2023;12:1246. doi: 10.3390/pathogens12101246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wie S.-H. Clinical significance of Providencia bacteremia or bacteriuria. Korean J Intern Med. 2015;30:167. doi: 10.3904/kjim.2015.30.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rajni E., Jain A., Garg V.K., Sharma R., Vohra R., Jain S.S. Providencia causing urinary tract infections: are we reaching a dead end? Indian J. Crit. Care Med. 2022;26:446. doi: 10.5005/jp-journals-10071-24163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma J., Song X., Li M., Yu Z., Cheng W., Yu Z., et al. Global spread of carbapenem-resistant Enterobacteriaceae: epidemiological features, resistance mechanisms, detection and therapy. Microbiol. Res. 2023;266 doi: 10.1016/j.micres.2022.127249. [DOI] [PubMed] [Google Scholar]
- 96.Van Duin D., Arias C.A., Komarow L., Chen L., Hanson B.M., Weston G., et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect. Dis. 2020;20:731–741. doi: 10.1016/S1473-3099(19)30755-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li X., Ye H. Clinical and mortality risk factors in bloodstream infections with carbapenem-resistant Enterobacteriaceae. Can. J. Infect Dis. Med. Microbiol. 2017;2017 doi: 10.1155/2017/6212910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sabino S., Soares S., Ramos F., Moretti M., Zavascki A.P., Rigatto M.H. A cohort study of the impact of carbapenem-resistant Enterobacteriaceae infections on mortality of patients presenting with sepsis. mSphere. 2019;4:10–1128. doi: 10.1128/mSphere.00052-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data accessed and analyzed in this study are available in the article and its supplementary materials.