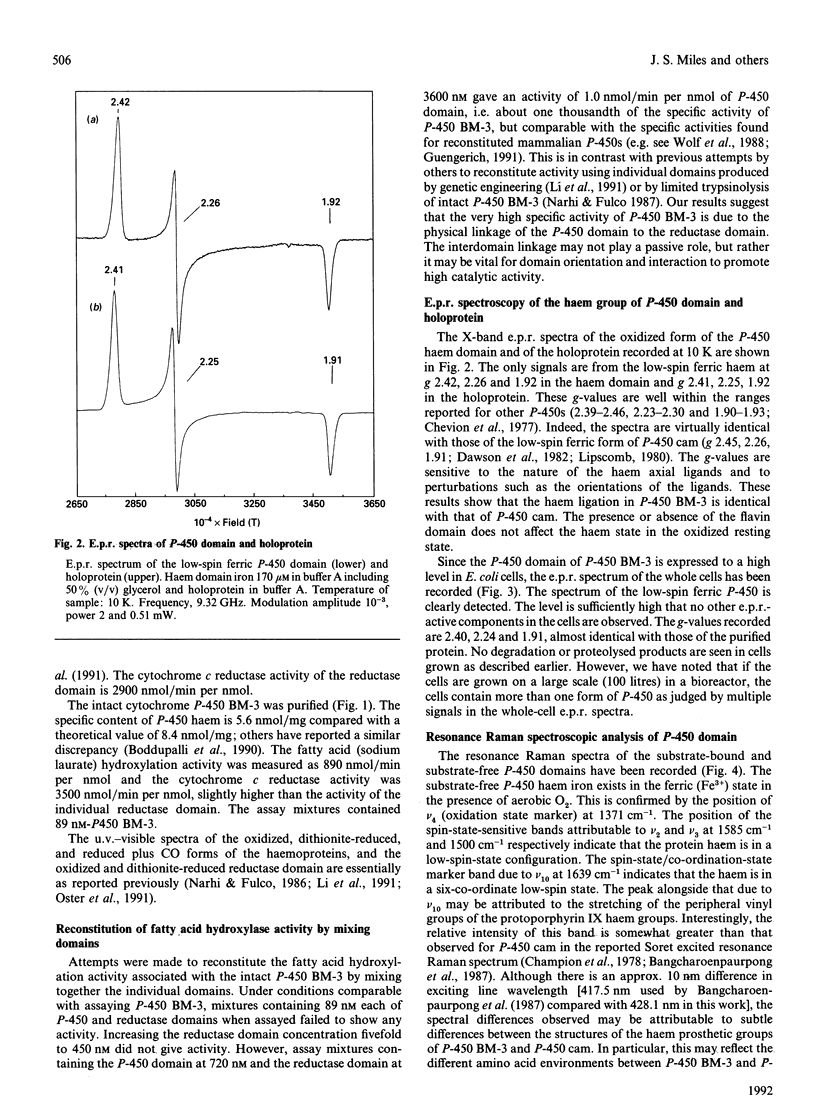
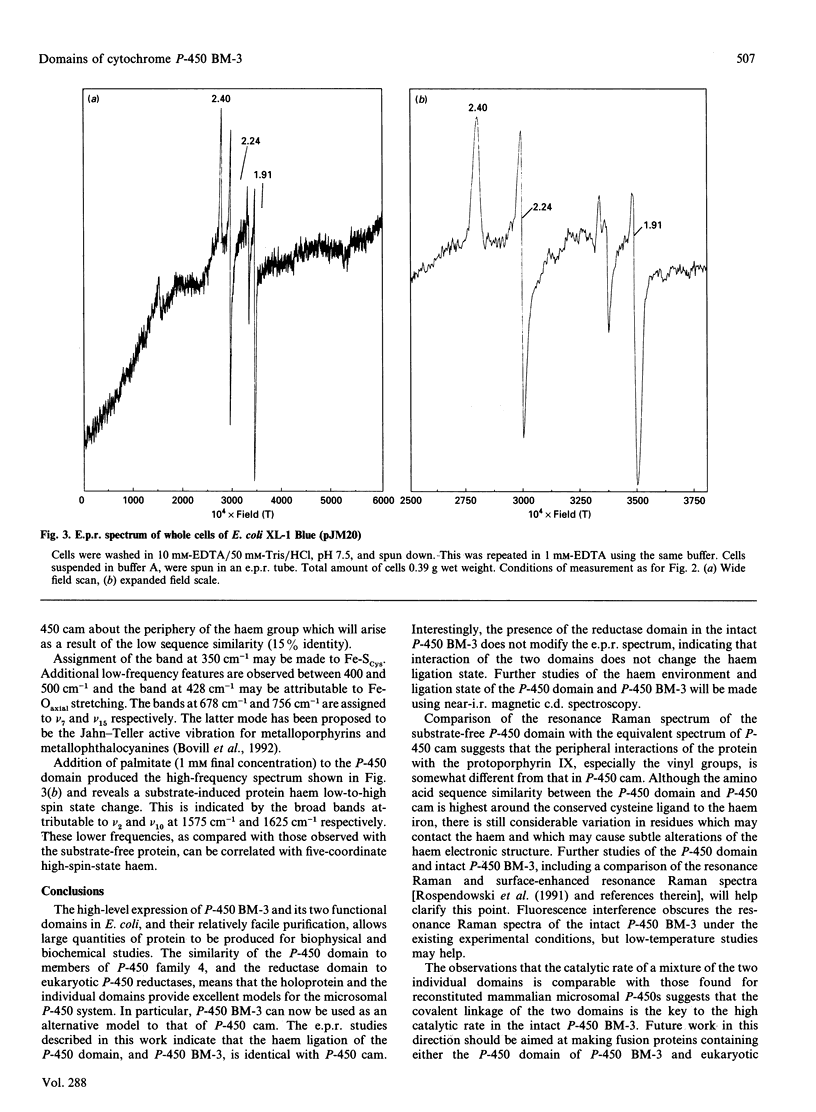
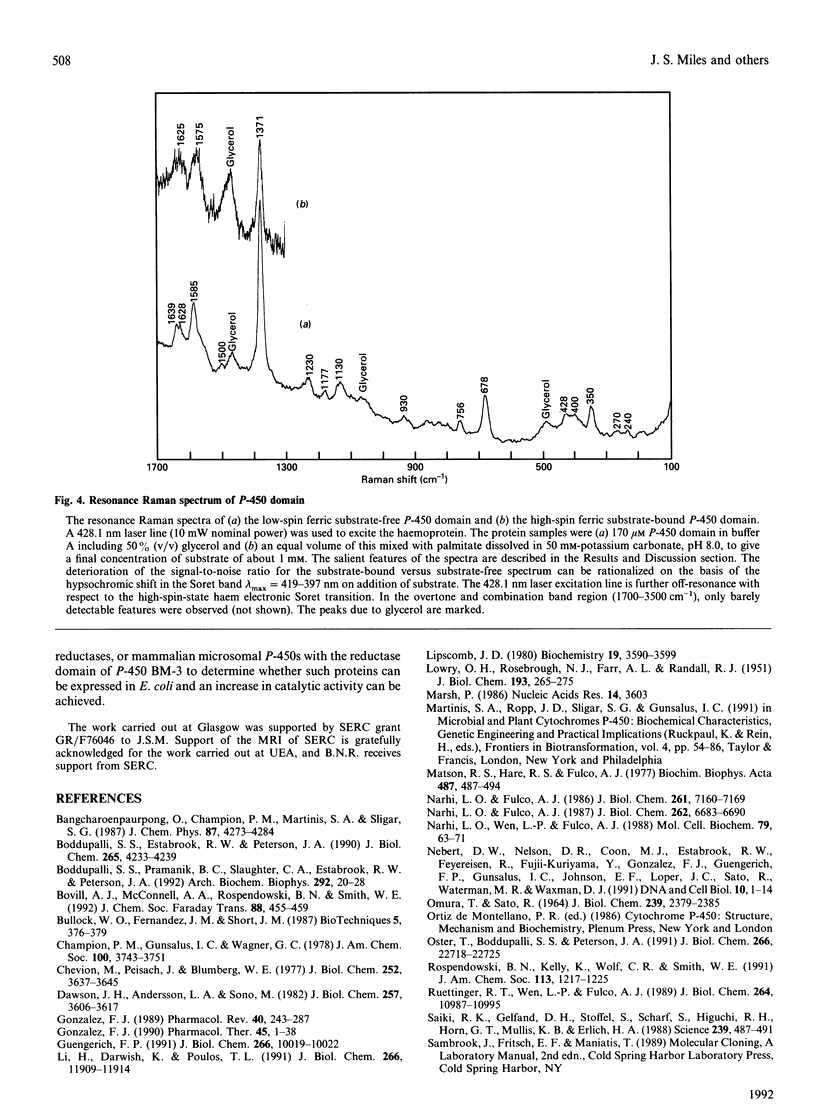
Abstract
1. The gene CYP102 encoding cytochrome P-450 BM-3 and subgenes encoding the cytochrome P-450 and cytochrome P-450 reductase domains have been cloned in Escherichia coli. 2. The protein products of these genes have been overexpressed and purified to homogeneity. 3. The cytochrome P-450 domain is purified in the ferric low-spin state, but is readily converted into the high-spin state by addition of the substrate palmitate (Ks = 1 microM). The cytochrome P-450 reductase domain readily reduces cytochrome c. Mixing the two domains reconstitutes only about one-thousandth of the fatty acid hydroxylase activity associated with the intact cytochrome P-450 BM-3. 4. The X-band e.p.r. spectra of both the cytochrome P-450 domain and intact cytochrome P-450 BM-3 give g-values indicating low-spin ferric haem. The spectra are virtually identical with those of the equivalent form of cytochrome P-450 cam indicating that the haem ligation in cytochrome P-450 BM-3 is identical with that of cytochrome P-450 cam. 5. Resonance Raman spectra of the substrate-free and substrate-bound forms of the cytochrome P-450 domain are given. Spectral differences in comparison with cytochrome P-450 cam may reflect subtle electronic differences between the respective haem environments.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boddupalli S. S., Estabrook R. W., Peterson J. A. Fatty acid monooxygenation by cytochrome P-450BM-3. J Biol Chem. 1990 Mar 15;265(8):4233–4239. [PubMed] [Google Scholar]
- Boddupalli S. S., Pramanik B. C., Slaughter C. A., Estabrook R. W., Peterson J. A. Fatty acid monooxygenation by P450BM-3: product identification and proposed mechanisms for the sequential hydroxylation reactions. Arch Biochem Biophys. 1992 Jan;292(1):20–28. doi: 10.1016/0003-9861(92)90045-x. [DOI] [PubMed] [Google Scholar]
- Chevion M., Peisach J., Blumberg W. E. Imidazole, the ligand trans to mercaptide in ferric cytochrome P-450. An EPR study of proteins and model compounds. J Biol Chem. 1977 Jun 10;252(11):3637–3645. [PubMed] [Google Scholar]
- Dawson J. H., Andersson L. A., Sono M. Spectroscopic investigations of ferric cytochrome P-450-CAM ligand complexes. Identification of the ligand trans to cysteinate in the native enzyme. J Biol Chem. 1982 Apr 10;257(7):3606–3617. [PubMed] [Google Scholar]
- Gonzalez F. J. Molecular genetics of the P-450 superfamily. Pharmacol Ther. 1990;45(1):1–38. doi: 10.1016/0163-7258(90)90006-n. [DOI] [PubMed] [Google Scholar]
- Gonzalez F. J. The molecular biology of cytochrome P450s. Pharmacol Rev. 1988 Dec;40(4):243–288. [PubMed] [Google Scholar]
- Guengerich F. P. Reactions and significance of cytochrome P-450 enzymes. J Biol Chem. 1991 Jun 5;266(16):10019–10022. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Li H. Y., Darwish K., Poulos T. L. Characterization of recombinant Bacillus megaterium cytochrome P-450 BM-3 and its two functional domains. J Biol Chem. 1991 Jun 25;266(18):11909–11914. [PubMed] [Google Scholar]
- Lipscomb J. D. Electron paramagnetic resonance detectable states of cytochrome P-450cam. Biochemistry. 1980 Jul 22;19(15):3590–3599. doi: 10.1021/bi00556a027. [DOI] [PubMed] [Google Scholar]
- Marsh P. Ptac-85, an E. coli vector for expression of non-fusion proteins. Nucleic Acids Res. 1986 Apr 25;14(8):3603–3603. doi: 10.1093/nar/14.8.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson R. S., Hare R. S., Fulco A. J. Characteristics of a cytochrome P-450-dependent fatty acid omega-2 hydroxylase from bacillus megaterium. Biochim Biophys Acta. 1977 Jun 22;487(3):487–494. doi: 10.1016/0005-2760(77)90218-1. [DOI] [PubMed] [Google Scholar]
- Narhi L. O., Fulco A. J. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J Biol Chem. 1986 Jun 5;261(16):7160–7169. [PubMed] [Google Scholar]
- Narhi L. O., Fulco A. J. Identification and characterization of two functional domains in cytochrome P-450BM-3, a catalytically self-sufficient monooxygenase induced by barbiturates in Bacillus megaterium. J Biol Chem. 1987 May 15;262(14):6683–6690. [PubMed] [Google Scholar]
- Narhi L. O., Wen L. P., Fulco A. J. Characterization of the protein expressed in Escherichia coli by a recombinant plasmid containing the Bacillus megaterium cytochrome P-450BM-3 gene. Mol Cell Biochem. 1988 Jan;79(1):63–71. doi: 10.1007/BF00229399. [DOI] [PubMed] [Google Scholar]
- Nebert D. W., Nelson D. R., Coon M. J., Estabrook R. W., Feyereisen R., Fujii-Kuriyama Y., Gonzalez F. J., Guengerich F. P., Gunsalus I. C., Johnson E. F. The P450 superfamily: update on new sequences, gene mapping, and recommended nomenclature. DNA Cell Biol. 1991 Jan-Feb;10(1):1–14. doi: 10.1089/dna.1991.10.1. [DOI] [PubMed] [Google Scholar]
- OMURA T., SATO R. THE CARBON MONOXIDE-BINDING PIGMENT OF LIVER MICROSOMES. II. SOLUBILIZATION, PURIFICATION, AND PROPERTIES. J Biol Chem. 1964 Jul;239:2379–2385. [PubMed] [Google Scholar]
- Oster T., Boddupalli S. S., Peterson J. A. Expression, purification, and properties of the flavoprotein domain of cytochrome P-450BM-3. Evidence for the importance of the amino-terminal region for FMN binding. J Biol Chem. 1991 Nov 25;266(33):22718–22725. [PubMed] [Google Scholar]
- Ruettinger R. T., Wen L. P., Fulco A. J. Coding nucleotide, 5' regulatory, and deduced amino acid sequences of P-450BM-3, a single peptide cytochrome P-450:NADPH-P-450 reductase from Bacillus megaterium. J Biol Chem. 1989 Jul 5;264(19):10987–10995. [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Vermilion J. L., Coon M. J. Identification of the high and low potential flavins of liver microsomal NADPH-cytochrome P-450 reductase. J Biol Chem. 1978 Dec 25;253(24):8812–8819. [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wen L. P., Fulco A. J. Cloning of the gene encoding a catalytically self-sufficient cytochrome P-450 fatty acid monooxygenase induced by barbiturates in Bacillus megaterium and its functional expression and regulation in heterologous (Escherichia coli) and homologous (Bacillus megaterium) hosts. J Biol Chem. 1987 May 15;262(14):6676–6682. [PubMed] [Google Scholar]
- Wolf C. R., Miles J. S., Seilman S., Burke M. D., Rospendowski B. N., Kelly K., Smith W. E. Evidence that the catalytic differences of two structurally homologous forms of cytochrome P-450 relate to their heme environment. Biochemistry. 1988 Mar 8;27(5):1597–1603. doi: 10.1021/bi00405a031. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]