Highlights
-
•
Dimensionality reduction is mandatory for radiomics analyses.
-
•
Test-retest studies allow for a robust dimensionality reduction.
-
•
MR-linacs provide a large amount of data that can be used for radiomic analysis.
-
•
A significant proportion (54 %) of radiomics features extracted from phantom and patient data from a 0.35 T MR-linac are non-reproducible.
Keywords: Radiomics, Robustness, MR-guided radiotherapy
Abstract
Background and purpose
MR-guided radiotherapy adds the precision of magnetic resonance imaging (MRI) to the therapeutic benefits of a linear accelerator. Prior to each therapeutic session, an MRI generates a significant volume of imaging data ripe for analysis. Radiomics stands at the forefront of medical imaging and oncology research, dedicated to mining quantitative imaging attributes to forge predictive models. However, the robustness of these models is often challenged.
Materials and methods
To assess the robustness of feature extraction, we conducted reproducibility studies using a 0.35 T MR-linac system, employing both a specialized phantom and patient-derived images, focusing on cases of pancreatic cancer. We extracted shape-based, first-order and textural features from patient-derived images and only first-order and textural features from phantom-derived images. The impact of the delay between simulation and first fraction images was also assessed with an equivalence test.
Results
From 107 features evaluated, 58 (54 %) were considered as non-reproducible: 18 were uniformly inconsistent across both phantom and patient images, 9 were specific to phantom-based analysis, and 31 to patient-derived data.
Conclusion
Our findings show that a significant proportion of radiomic features extracted from this dual dataset were unreliable. It is essential to discard these non-reproducible elements to refine and enhance radiomic model development, particularly for MR-guided radiotherapy in pancreatic cancer.
1. Introduction
Magnetic Resonance-guided radiotherapy (MRgRT) represents a groundbreaking advancement in the management of cancer, integrating the diagnostic benefits of magnetic resonance imaging (MRI) to the therapeutic power of a linear accelerator. This innovative merger is embodied in the MR-linacs, which utilizes a 0.35 T or 1.5 T MRI scanner to offer unprecedented precision in cancer treatment [1]. The system’s design facilitates meticulous patient positioning through daily image acquisition and enables real-time tracking of the target with continuous cine-MRI during radiation delivery. This dynamic imaging allows for on-the-fly adjustments, pausing the radiation if the target drifts beyond the established safety margins, thereby optimizing treatment accuracy and protecting surrounding healthy tissue.
Radiomics has emerged as a transformative field in medical imaging and oncology, harnessing the power of advanced computing to extract a plethora of quantitative imaging features. These features are the building blocks for sophisticated algorithms that aim to correlate image data with biological characteristics and predict clinical outcome [2], [3]. Radiomics studies should comply with several steps in a typical workflow [4], [5]. One of these steps is to reduce the number of extracted features by removing those that are not robust or those that are redundant for the analysis. This approach, known as dimensionality reduction, reduces the risk of overfitting during model training [6]. This step is crucial for the integrity and applicability of the predictive models generated through radiomic analysis.
In the context of MR-linacs, the large volume of imaging data generated daily offers a fertile ground for radiomic research. The images could also be used to perform delta-radiomics analysis, which represents a comparative longitudinal analysis that evaluates how radiomic features evolve over the course of treatment, providing insights into the biological effects of radiotherapy on tumor and normal tissues.
The focus of this study is to assess the reproducibility of radiomic features derived from two sources: a specialized phantom, and actual patient images from those undergoing treatment for pancreatic cancer using the 0.35 T MRI scanner of an MR-linac system. By examining the consistency of these features, we aim to identify which can be reliably used to enhance the personalization and efficacy of MR-guided radiotherapy. The implications of this research are vast, with the potential to significantly refine radiomic models and ultimately, propel the evolution of patient-specific cancer therapy.
2. Materials and methods
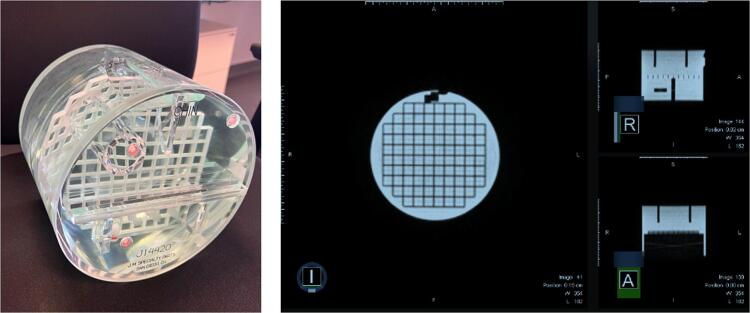
We conducted a detailed investigation into the consistency of radiomic features derived from MR images using a standardized phantom, specifically the American College of Radiology (ACR) phantom, which includes a diverse array of geometric configurations (as displayed in Fig. 1). This phantom is routinely utilized for the calibration and quality assurance of MR imaging systems. In addition, we extended our analysis to a patient cohort undergoing treatment for pancreatic cancer, comparing imaging data from pre-treatment simulations to those obtained on the first day of therapy.
Fig. 1.
ACR phantom and images of this phantom with a 0.35 T MRI.
2.1. Patient criteria
Our study encompassed seventy-four patients treated either for borderline or locally advanced pancreatic cancer with Stereotactic MR-guided Adaptive Radiotherapy (SMART). Treatment regimens varied, with three patients receiving a total dose of 40 Gy across five sessions, and the remaining seventy-one receiving 50 Gy in the same number of fractions. Prior to the SMART regimen, all patients had been treated with induction chemotherapy. Crucially, no chemotherapy was administered in the interim between the simulation examination and the initial fraction of SMART treatment. Participation in this study was contingent upon informed consent, with the study's protocol being duly recorded in the Health Data Hub (registration number: #1802) and receiving the endorsement of the COMERE local research committee (ICM-ART 2020/01).
2.2. Image acquisition
Imaging for both the phantom and patient studies was conducted using the standard MRIdian® protocol for pancreatic cancer: a balanced steady-state gradient echo sequence (True FISP) with a flexible torso phased array coil and characterized by a resolution of 1.63 × 1.63 × 3 mm3, a 276 × 276 matrix size, a TR/TE of 3.84/1.62 ms, a flip angle of 60° and a rapid 17-second acquisition period. For the phantom, eleven sessions of five consecutive acquisitions were performed without repositioning the phantom over three weeks, culminating in a total of 55 acquisitions. The phantom was repositioned between each session. For patients, six MRI scans were performed: one for each of the five SMART fractions and an additional simulation scan conducted. This simulation scan was classically performed from 5 to 14 days after the last chemotherapy injection. To avoid motion artefacts, a breath-hold technique at physiological end-expiration was employed during the imaging process.
2.3. Image pre-processing
Prior to the feature extraction, images underwent a series of pre-processing steps designed to mitigate artefacts and enhance the reliability of subsequent feature calculations. These steps, executed using an in-house python code and adhering to IBSI guidelines [7], included bias field correction via the N4 algorithm [8], noise reduction through anisotropic diffusion filtering (settings: number of iterations = 5; kappa = 5; gamma = 3) [9], [10], and image B-spline interpolation to achieve isotropic voxel dimensions of mm3.
2.4. Features extraction
The feature extraction was preceded by the application of a fixed bin width of 7 for the gray-level discretisation, performed using the open source software Pyradiomics v3.0.1 [11]. As all acquired images had a minimum and maximum value of [0 455] which can be considered like normalized images, this resulting in 65 bins. In total, 107 features were extracted from each image, spanning shape-based metrics, first-order and second-order statistics, as well as advanced textural features via Gray Level Co-occurrence Matrix (GLCM), Gray Level Size Zone Matrix (GLSZM), Gray Level Run Length Matrix (GLRLM), Neighbouring Gray Tone Difference Matrix (NGTDM), and Gray Level Dependence Matrix (GLDM). It is important to note that no filters were used on the images prior to the extraction of these features. Texture features were aggregated using the 3DAverage method (ITBB). For the phantom study, the region of interest (ROI) was automatically outlined using custom in-house code in Matlab and included the entire phantom. For the cohort of pancreatic cancer patients, feature extraction was performed on the gross tumor volume (GTV), which was identified by the treating radiation oncologist on the simulation MRI and then rigidly aligned with the MRI from the first fraction of treatment. If necessary, the radiation oncologist could adjust the GTV contour on the first fraction MRI.
Feature extraction commenced with the application of a consistent bin width of 7 for gray-level discretization, facilitated by the open-source software Pyradiomics v3.0.1. Given that all images were within a value range of [0, 455]—effectively normalized—this process resulted in the formation of 65 bins. Subsequently, 107 features were extracted from each image, encompassing shape-based metrics, first-order and second-order statistics, and advanced textural features. These textural features included analysis through Gray Level Co-occurrence Matrix (GLCM), Gray Level Size Zone Matrix (GLSZM), Gray Level Run Length Matrix (GLRLM), Neighbouring Gray Tone Difference Matrix (NGTDM), and Gray Level Dependence Matrix (GLDM). It's critical to mention that the images underwent no filtration prior to feature extraction. The texture features were compiled using the 3DAverage method (ITBB). For the phantom study, the region of interest (ROI) was precisely defined using bespoke Matlab code and encompassed the entire phantom. In the case of the pancreatic cancer patient cohort, feature extraction was conducted on the gross tumor volume (GTV), which was delineated by the treating radiation oncologist during the simulation MRI and subsequently aligned with the MRI from the initial treatment fraction.
2.5. Statistical analysis
In the phantom study, for each set of five consecutive acquisitions, we calculated both the mean and the coefficient of variation (CoV) for each feature. The CoV, defined as the standard deviation divided by the mean, served as a gauge for the repeatability of the findings. Since the phantom's ROI was automatically segmented, ensuring an identical ROI across all images, shape-based features were excluded from the repeatability assessment. The CoV for each feature was then calculated across all sessions based on each session's average, effectively measuring the phantom's reproducibility. Features were divided into three groups as excellent repeatability (CoV ≤ 5%), good repeatability (5 % < CoV ≤ 10 %) and poor repeatability (10 % < CoV).
For the patient cohort, reproducibility was evaluated by comparing features extracted from the simulation scan and the first treatment fraction. We calculated the intraclass correlation coefficient (ICC) for each feature using Python (version 3.8.18), incorporating the libraries Pandas (version 2.0.3) and Pingouin (version 0.5.4). The ICC is a statistical measure that determines the consistency of measurements by comparing the variance of the same subject to the total variance across all ratings and subjects. Features were subsequently categorized based on their ICC values: excellent reproducibility (ICC ≥ 0.90), good reproducibility (0.75 ≤ ICC < 0.90), and poor reproducibility (ICC ≤ 0.75). The influence of the delay between the simulation MRI and the first fraction MRI was assessed by comparing the mean variation of each feature between two groups of patients based on the median delay between the two MRI. In addition, equivalence tests were performed to evaluate the similarity of feature values between these groups. This comprehensive analysis is poised to contribute significantly to the precision and effectiveness of radiomic research, particularly in the context of MR-guided radiotherapy for pancreatic cancer.
3. Results
3.1. Analysis of phantom-derived radiomic features
For each imaging session of the ACR phantom, a comprehensive suite of 93 radiomic features was extracted from the acquired data. All these features are listed in supplementary material in Table S1. The Fig. 1 provides a representative image of the phantom used in this study as well as its image using 0.35 T MRI. Shape features were deliberately omitted from this phase of analysis due to the focus on other feature categories.
Upon evaluation, 27 of these features (accounting for 29 % of the total) were identified as having suboptimal reproducibility, evidenced by a coefficient of variation (CoV) exceeding 10 % in at least one session or across all sessions cumulatively. Conversely, a significant proportion of the features, 44 in total (47 %), demonstrated high repeatability with a CoV of less than 5 %. Furthermore, 22 features (24 %) displayed good repeatability, falling into the CoV range of greater than 5 % but less than 10 %. The features with poor repeatability are listed in Table 1. The features falling into other categories of repeatability—good, and high—are systematically catalogued in supplementary material in Tables S2 and S3.
Table 1.
List of poorly-repeatable features on phantom images and their coefficient of variation (CoV > 10 %).
| Feature | Coefficient of variation (CoV) |
|---|---|
| original_firstorder_Minimum | 30.7 % |
| original_firstorder_Variance | 10.1 % |
| original_glcm_Autocorrelation | 10.8 % |
| original_glcm_ClusterProminence | 21.0 % |
| original_glcm_ClusterTendency | 11,4% |
| original_glcm_SumSquares | 10.8 % |
| original_gldm_GrayLevelVariance | 10.2 % |
| original_gldm_HighGrayLevelEmphasis | 10.8 % |
| original_gldm_LargeDependenceLowGrayLevelEmphasis | 15.7 % |
| original_gldm_LowGrayLevelEmphasis | 18.5 % |
| original_gldm_SmallDependenceHighGrayLevelEmphasis | 16.1 % |
| original_gldm_SmallDependenceLowGrayLevelEmphasis | 18.4 % |
| original_glrlm_HighGrayLevelRunEmphasis | 10.9 % |
| original_glrlm_LongRunLowGrayLevelEmphasis | 18.6 % |
| original_glrlm_LowGrayLevelRunEmphasis | 19.1 % |
| original_glrlm_ShortRunHighGrayLevelEmphasis | 11.6 % |
| original_glrlm_ShortRunLowGrayLevelEmphasis | 19.4 % |
| original_glszm_HighGrayLevelZoneEmphasis | 12.1 % |
| original_glszm_LargeAreaEmphasis | 10.8 % |
| original_glszm_LargeAreaLowGrayLevelEmphasis | 20.3 % |
| original_glszm_LowGrayLevelZoneEmphasis | 20.3 % |
| original_glszm_SmallAreaHighGrayLevelEmphasis | 12.4 % |
| original_glszm_SmallAreaLowGrayLevelEmphasis | 20.5 % |
| original_glszm_ZoneVariance | 10.8 % |
| original_ngtdm_Complexity | 15.8 % |
| original_ngtdm_Strength | 11.1 % |
| original_glcm_ClusterShade | 16.0 % |
CoV = Coefficient of variation
To illustrate the concept of CoV and its application within our study, the Fig. S1 in supplementary material provides a visual comparison of this metric across four distinct radiomic features. This comparison underscores the variability and potential reliability of each feature in the context of repeated phantom imaging sessions.
3.2. Analysis of the patient-derived radiomic features
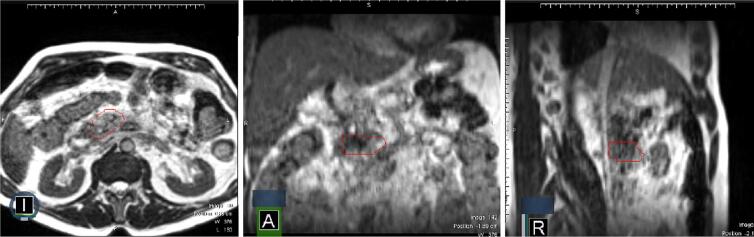
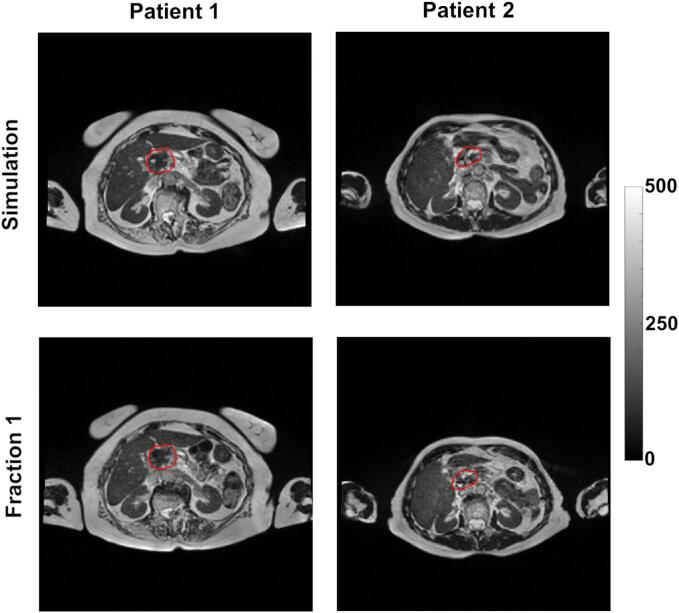
In our analysis of patient-derived data, we meticulously extracted 107 distinct radiomic features from the gross tumor volume (GTV) delineated in each patient's MRI scan, with the full list of these features available in the Table S4 in supplementary material. The Fig. 2 provides a visual representation in 3 plans of a patient's MRI with the GTV clearly marked. The Fig. 3 provides comparison of simulation and first fraction images for two different patients. Out of these features, a significant number—49 features, which equates to 46 %—demonstrated poor reproducibility, as indicated by an intraclass correlation coefficient (ICC) falling below the 75 % threshold. Conversely, we identified 28 features (26 % of the total) that exhibited high reproducibility, with an ICC exceeding 90 %. The remaining 30 features (28 %) showed good reproducibility, with their ICCs ranging between 75 % and 90 %. The features with poor reproducibility are listed in Table 2. The features falling into other categories of reproducibility— good, and high—are systematically catalogued in supplementary material in Tables S5 and S6.
Fig. 2.
Example of patient images with 0.35 T MRI, GTV (ROI) delineated in red.
Fig. 3.
Example of comparison between the simulation and first fraction images for two different patients, after bias field and noise corrections; GTV in red.
Table 2.
List of poorly-reproducible GTV features on simulation vs first fraction images and their interclass correlation (ICC < 75 %).
| Feature | Interclass Correlation (ICC) |
|---|---|
| original_firstorder_RootMeanSquared | 31.6 % |
| original_glcm_JointAverage | 27.9 % |
| original_glcm_Contrast | 11.8 % |
| original_gldm_HighGrayLevelEmphasis | 8.3 % |
| original_firstorder_Range | 26.1 % |
| original_gldm_DependenceNonUniformityNormalized | 60.9 % |
| original_glszm_LargeAreaLowGrayLevelEmphasis | 45.9 % |
| original_glcm_DifferenceEntropy | 66.7 % |
| original_gldm_LargeDependenceHighGrayLevelEmphasis | 29.3 % |
| original_ngtdm_Strength | 28.6 % |
| original_glszm_GrayLevelVariance | 11.0 % |
| original_glcm_SumAverage | 27.9 % |
| original_glszm_ZoneVariance | 74.1 % |
| original_firstorder_InterquartileRange | 51.6 % |
| original_firstorder_Maximum | 23.3 % |
| original_firstorder_90Percentile | 35.0 % |
| original_firstorder_10Percentile | 39.9 % |
| original_firstorder_Variance | 14.5 % |
| original_glcm_ClusterTendency | 16.3 % |
| original_glszm_SmallAreaHighGrayLevelEmphasis | 7.7 % |
| original_glcm_DifferenceVariance | 9.8 % |
| original_glszm_LargeAreaEmphasis | 74.1 % |
| original_glszm_SmallAreaEmphasis | 58.5 % |
| original_gldm_DependenceVariance | 69.3 % |
| original_glcm_SumSquares | 15.2 % |
| original_firstorder_Skewness | 73.3 % |
| original_glszm_ZonePercentage | 73.5 % |
| original_glrlm_GrayLevelVariance | 14.2 % |
| original_glrlm_ShortRunHighGrayLevelEmphasis | 8.3 % |
| original_firstorder_RobustMeanAbsoluteDeviation | 50.0 % |
| original_firstorder_Mean | 31.5 % |
| original_firstorder_Energy | 22.8 % |
| original_firstorder_Median | 31.4 % |
| original_gldm_SmallDependenceHighGrayLevelEmphasis | 5.8 % |
| original_glszm_HighGrayLevelZoneEmphasis | 9.1 % |
| original_glrlm_HighGrayLevelRunEmphasis | 8.4 % |
| original_gldm_GrayLevelVariance | 14.5 % |
| original_glcm_Autocorrelation | 8.0 % |
| original_ngtdm_Contrast | 52.6 % |
| original_gldm_SmallDependenceEmphasis | 69.3 % |
| original_ngtdm_Complexity | 2.9 %% |
| original_glcm_ClusterProminence | 1.3 % |
| original_firstorder_MeanAbsoluteDeviation | 44.3 % |
| original_glcm_ClusterShade | 6.0 % |
| original_glrlm_LongRunHighGrayLevelEmphasis | 8.8 % |
| original_glcm_DifferenceAverage | 37.9 % |
| original_glszm_SizeZoneNonUniformity | 69.0 % |
| original_glszm_SizeZoneNonUniformityNormalized | 54.3 % |
| original_firstorder_TotalEnergy | 22.8 % |
ICC=Interclass correlation
The median delay between the two MRI was 17 days (range 5–33). We did not find any differences of mean feature variation between the 2 groups for the different features, as indicated by the significant results of the equivalence tests conducted, with different ICC results tested. The Table S7 shows 4 examples of mean variation of features exhibiting low reproducibility and 4 examples of features exhibiting high reproducibility between patients having less than 17 days and those having more than 17 days between the two MRI.
3.3. Comparison between phantom and patient features
Fifty-eight features (54 %) were considered as poorly robust on the whole study: 18 in common between phantom and patient data, 9 on the phantom study and 31 on the patient study. These common poorly robust features are outlined in Table 3.
Table 3.
List of common poorly robust features.
| Feature |
|---|
| original_firstorder_Variance |
| original_glcm_Autocorrelation |
| original_glcm_ClusterProminence |
| original_glcm_ClusterTendency |
| original_glcm_SumSquares |
| original_gldm_GrayLevelVariance |
| original_gldm_HighGrayLevelEmphasis |
| original_gldm_SmallDependenceHighGrayLevelEmphasis |
| original_glrlm_HighGrayLevelRunEmphasis |
| original_glrlm_ShortRunHighGrayLevelEmphasis |
| original_glszm_HighGrayLevelZoneEmphasis |
| original_glszm_LargeAreaEmphasis |
| original_glszm_LargeAreaLowGrayLevelEmphasis |
| original_glszm_SmallAreaHighGrayLevelEmphasis |
| original_glszm_ZoneVariance |
| original_ngtdm_Complexity |
| original_ngtdm_Strength |
| original_glcm_ClusterShade |
4. Discussion
In this comprehensive study, we assessed the robustness of radiomic features captured by a 0.35 T MRI scanner integrated within the MRIdian® system, using a standard protocol for pancreatic cancer. The analysis focused on the reproducibility of these features extracted from two distinct sets of images: those obtained from a dedicated phantom and those from patients undergoing treatment. The study objective was to determine the repeatability of various radiomic features, providing a filter to remove those lacking robustness.
Dimensionality reduction is a critical phase in the process of radiomic analysis, aiming toreduce the risk of overfitting in predictive modeling. Overfitting can lead to models that perform exceptionally on training data but fail to generalize to new, unseen data. While machine and deep learning algorithms are commonly employed to perform this reduction, they may not necessarily exclude features that are non-robust under the conditions of acquisition, which is a significant consideration for ensuring the reliability of radiomic studies [12].
Numerous studies have illuminated the profound impact that even minor variations in MRI acquisition parameters can have on the values of radiomic features [13], [14], [15], [16]. Additionally, variations have been noted when identical sequences are deployed on systems from different manufacturers [17], [18], [19]. While post-processing steps have been shown to mitigate some of this variability, they fall short of establishing robustness across all radiomic features [20]. This underscores the necessity of test–retest studies that can identify and exclude features whose variability is attributed not to pathological or therapeutic changes but to inconsistencies in the imaging acquisition parameters themselves [21], [22].
In our analyses, we determined that 54 % of the radiomic features we extracted could be deemed non-reproducible and thus unsuitable for inclusion in subsequent analyses. The approach of dimensionality reduction used in this study is grounded in transparent methodologies, providing an explainable selection of features. This is in stark contrast to the usual opaque nature of artificial intelligence, particularly with deep learning algorithms, where the internal mechanics of feature selection are not always accessible or interpretable.
In our phantom studies, we identified 27 features that were not reproducible, and of these, a significant number were also found to be non-reproducible in the patient image analyses, suggesting a consistency in the feature behavior across different data sets. It was noted that most of the commonly non-reproducible features pertained to texture.. Interestingly, most first-order features that exhibited non-reproducibility in patient data analysis demonstrated good repeatability in phantom data analysis, highlighting a potential discrepancy between phantom and clinical scenarios.
There is a sparse yet growing body of literature exploring the robustness of radiomics features within systems of MR-guided radiotherapy like the MRIdian®.. Our findings align with some of these prior studies, particularly with regard to the robustness of shape-based features and specific textural features [23], [24]. Some of the robust features of the study of Ericsson-Szecsenyi et al. were also robust in our study (for example shape-based features, GLCM sum entropy, GLRLM short-run emphasis, GLRLM long-run emphasis, GLRLM run percentage, GLRLM run length non-uniformity) [23].
The application of Stereotactic Adaptive MR-guided Radiotherapy (SMART) for inoperable pancreatic cancers holds substantial promise, demonstrated by the emerging clinical results [25], [26]. However, with the reality that many patients still face the prospect of recurrence, the development of predictive tools is paramount. Various research groups have ventured to develop and propose radiomic-based models with the intent of predicting patient outcomes after SMART.t. Notably, some of these predictive models have achieved promising levels of accuracy, as evidenced by their reported Area Under the Curve (AUC) statistics [27], [28]. However, a common gap in these studies is the absence of a thorough reproducibility analysis of the radiomic features used within these predictive models.
The method of feature selection delineated in our study aims to ensure that only reproducible features are carried forward into the final radiomic models constructed by various machine learning algorithms.
While our study brings to light several important findings, it is not without its limitations. A notable one is the absence of an inter-observer segmentation analysis, a factor known to introduce variability in the delineation of GTV. By not addressing this variability, we acknowledge a potential source of error that could influence the reproducibility of the radiomic features.
Additionally, our investigation was conducted in a single-center setting, utilizing a singular MRIdian® system and a single imaging protocol with the same coil, acquisition parameters and resolution. The results would be different if any of the imaging parameters were different (i.e. coil, field of view, TE/TR, etc.). This would require a new test–retest study in order to select robust features. The results may also differ when applied to other systems or within different institutional protocols. To enhance the applicability of our findings, a multicentric approach involving various MRIdian® systems, preferably across different geographic locations and patient populations, would be instrumental. Such an approach would allow for the comparison of radiomic feature robustness in a broader context, potentially validating the findings and ensuring that the models developed are more universally applicable. Moreover, the evolving field of radiomics in radiotherapy demands ongoing dialogue between technological advancement and clinical application. As such, the continuous integration of newer imaging technologies and updated radiotherapy techniques will necessitate constant re-evaluation of radiomic feature robustness.
In conclusion, the scope of this study serves not only to refine the process of feature selection in radiomics but also to underscore the importance of rigorous validation in the field of medical imaging and oncology. By advancing methodologies that prioritize the reproducibility and reliability of data, we set a precedent for future research that seeks to harness the power of radiomics in the pursuit of personalized medicine.
CRediT authorship contribution statement
Morgan Michalet: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. Gladis Valenzuela: Investigation, Methodology. Pierre Debuire: Data curation. Olivier Riou: Writing – review & editing. David Azria: Writing – review & editing. Stéphanie Nougaret: Writing – review & editing. Marion Tardieu: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2024.100613.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Mutic S., Dempsey J.F. The ViewRay system: magnetic resonance-guided and controlled radiotherapy. Semin Radiat Oncol. 2014;24:196–199. doi: 10.1016/j.semradonc.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Aerts H.J.W.L., Velazquez E.R., Leijenaar R.T.H., Parmar C., Grossmann P., Cavalho S., et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5 doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibault J.-E., Xing L., Giraud P., El Ayachy R., Giraud N., Decazes P., et al. Radiomics: a primer for the radiation oncologist. Cancer/Radiothérapie. 2020;24:403–410. doi: 10.1016/j.canrad.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Michalet M., Azria D., Tardieu M., Tibermacine H., Nougaret S. Radiomics in radiation oncology for gynecological malignancies: a review of literature. Br J Radiol. 2021 doi: 10.1259/bjr.20210032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Timmeren J.E., Cester D., Tanadini-Lang S., Alkadhi H., Baessler B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging. 2020;11:91. doi: 10.1186/s13244-020-00887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayerhoefer M.E., Materka A., Langs G., Häggström I., Szczypiński P., Gibbs P., et al. Introduction to radiomics. J Nucl Med. 2020;61:488–495. doi: 10.2967/jnumed.118.222893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zwanenburg A., Vallières M., Abdalah M.A., Aerts H.J.W.L., Andrearczyk V., Apte A., et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tustison N.J., Avants B.B., Cook P.A., Zheng Y., Egan A., Yushkevich P.A., et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010 Jun;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perona P., Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell. 1990;12:629–639. doi: 10.1109/34.56205. [DOI] [Google Scholar]
- 10.Weickert J. In: Theor Found Comput Vis. Kropatsch W., Klette R., Solina F., Albrecht R., editors. ViennaSpringer; 1996. Theoretical foundations of anisotropic diffusion in image processing; pp. 221–236. [Google Scholar]
- 11.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parmar C., Grossmann P., Bussink J., Lambin P., Aerts H.J.W.L. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5 doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayerhoefer M., Szomolanyi P., Jirak D., Berg A., Materka A., Dirisamer A., et al. Effects of magnetic resonance image interpolation on the results of texture-based pattern classification. Invest Radiol. 2009;44:405–411. doi: 10.1097/RLI.0b013e3181a50a66. [DOI] [PubMed] [Google Scholar]
- 14.Ford J., Dogan N., Young L., Yang F. Quantitative radiomics: impact of pulse sequence parameter selection on MRI-based textural features of the brain. Contrast Media Mol Imaging. 2018;2018:1–9. doi: 10.1155/2018/1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bologna M., Corino V., Mainardi L. Technical Note: Virtual phantom analyses for preprocessing evaluation and detection of a robust feature set for MRI-radiomics of the brain. Med Phys. 2019;46 doi: 10.1002/mp.13834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prabhu V., Gillingham N., Babb J., Mali R., Rusinek H., Bruno M., et al. Repeatability, robustness, and reproducibility of texture features on 3 Tesla liver MRI. Clin Imaging. 2022;83 doi: 10.1016/j.clinimag.2022.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Lee J., Steinmann A., Ding Y., Lee H., Owens C., Wang J., et al. Radiomics feature robustness as measured using an MRI phantom. Sci Rep. 2021;11 doi: 10.1038/s41598-021-83593-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buch K., Li B., Qureshi M., Kuno H., Anderson S.W., Sakai O. Quantitative assessment of variation in CT parameters on texture features: pilot study using a nonanatomic phantom. Am J Neuroradiol. 2017;38 doi: 10.3174/ajnr.A5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong O., Yuan J., Zhou Y., Yu S., Cheung K. Longitudinal acquisition repeatability of MRI radiomics features: An ACR MRI phantom study on two MRI scanners using a 3D T1W TSE sequence. Med Phys. 2020;48 doi: 10.1002/mp.14686. [DOI] [PubMed] [Google Scholar]
- 20.Eck B., Chirra P., Muchhala A., Hall S., Bera K., Tiwari P., et al. Prospective evaluation of repeatability and robustness of radiomic descriptors in healthy brain tissue regions in vivo across systematic variations in T2-weighted magnetic resonance imaging acquisition parameters. J Magn Reson Imaging. 2021;54 doi: 10.1002/jmri.27635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limkin E.J., Sun R., Dercle L., Zacharaki E.I., Robert C., Reuzé S., et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol Off J Eur Soc Med Oncol. 2017;28:1191–1206. doi: 10.1093/annonc/mdx034. [DOI] [PubMed] [Google Scholar]
- 22.Bologna M., Tenconi C., Corino V., Annunziata G., Orlandi E., Calareso G., et al. Repeatability and reproducibility of MRI-radiomic features: a phantom experiment on a 1.5 T scanner. Med Phys. 2022;50 doi: 10.1002/mp.16054. [DOI] [PubMed] [Google Scholar]
- 23.Ericsson-Szecsenyi R., Zhang G., Redler G., Feygelman V., Rosenberg S., Latifi K., et al. Robustness assessment of images from a 0.35T scanner of an integrated MRI-Linac: characterization of radiomics features in phantom and patient data. Technol Cancer Res Treat. 2022;21 doi: 10.1177/15330338221099113. 15330338221099113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue C., Yuan J., Poon D.M., Zhou Y., Yang B., Yu S.K., et al. Reliability of MRI radiomics features in MR-guided radiotherapy for prostate cancer: Repeatability, reproducibility, and within-subject agreement. Med Phys. 2021;48:6976–6986. doi: 10.1002/mp.15232. [DOI] [PubMed] [Google Scholar]
- 25.Bordeau K., Michalet M., Keskes A., Valdenaire S., Debuire P., Cantaloube M., et al. Stereotactic MR-guided adaptive radiotherapy for pancreatic tumors: updated results of the Montpellier prospective registry study. Cancer (Basel) 2023 doi: 10.3390/cancers15010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuong M.D., Lee P., Low D.A., Kim J., Mittauer K.E., Bassetti M.F., et al. Stereotactic MR-guided on-table adaptive radiation therapy (SMART) for borderline resectable and locally advanced pancreatic cancer: A multi-center, open-label phase 2 study. Radiother Oncol. 2024;191 doi: 10.1016/j.radonc.2023.110064. [DOI] [PubMed] [Google Scholar]
- 27.Simpson G., Spieler B., Dogan N., Portelance L., Mellon E.A., Kwon D., et al. Predictive value of 0.35 T magnetic resonance imaging radiomic features in stereotactic ablative body radiotherapy of pancreatic cancer: a pilot study. Med Phys. 2020;47:3682–3690. doi: 10.1002/mp.14200. [DOI] [PubMed] [Google Scholar]
- 28.Cusumano D., Boldrini L., Yadav P., Casà C., Lee S.L., Romano A., et al. Delta Radiomics analysis for local control prediction in pancreatic cancer patients treated using magnetic resonance guided radiotherapy. Diagnostics. 2021;11:72. doi: 10.3390/diagnostics11010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.