Abstract
The antibiotic AT2433-B1 belongs to a therapeutically important class of antitumor agents. This natural product contains an indolocarbazole aglycone connected to a unique disaccharide consisting of a methoxyglucose and an amino sugar subunit, 2,4-dideoxy-4-methylamino-l-xylose. The configuration of the amino sugar distinguishes AT2433-B1 from its diastereoisomer iso-AT2433-B1. Here we have investigated the interaction of these two disaccharide indolocarbazole derivatives with different DNA sequences by means of DNase I footprinting and surface plasmon resonance (SPR). Accurate binding measurements performed at 4 and 25°C using the BIAcore SPR method revealed that AT2433-B1 binds considerably more tightly to a hairpin oligomer containing a [CG]4 block than to an oligomer with a central [AT]4 tract. The kinetic analysis shows that the antibiotic dissociates much more slowly from the GC sequence compared to the AT one. Preferential binding of AT2433-B1 to GC-rich sequences in DNA was independently confirmed by DNase I footprinting experiments performed with a 117 bp DNA restriction fragment. The specific binding sequence 5′-AACGCCAG identified from the footprints was then converted into a biotin-labeled DNA hairpin duplex and compound interactions with this specific sequence were characterized by high resolution BIAcore SPR experiments. Such a combined approach provided a detailed understanding of the molecular basis of DNA recognition. The discovery that the glycosyl antibiotic AT2433-B1 preferentially recognizes defined sequences offers novel opportunities for the future design of sequence-specific DNA-reading small molecules.
INTRODUCTION
Indolocarbazole antibiotics represent a therapeutically important category of antitumor agents (1). This series includes the antibiotics rebeccamycin and staurosporine, which essentially target DNA topoisomerase I and protein kinase C, respectively (2,3). Derivatives of these natural products, such as the potent topoisomerase I poison NB-506 (4–6) and the promising kinase inhibitor UCN-01 (7,8), are currently undergoing clinical trials as anticancer agents. Several synthetic analogs are being developed world wide, for example, the compounds J-107088 (topoisomerase I inhibitor; Banyu Pharmaceuticals) (9–11) MCR-47 (topoisomerase I inhibitor; MediChem Research) (12), PKC-412 (kinase inhibitor; Novartis) (13) and SB-218078 (kinase inhibitor; SmithKline Beecham) (14).
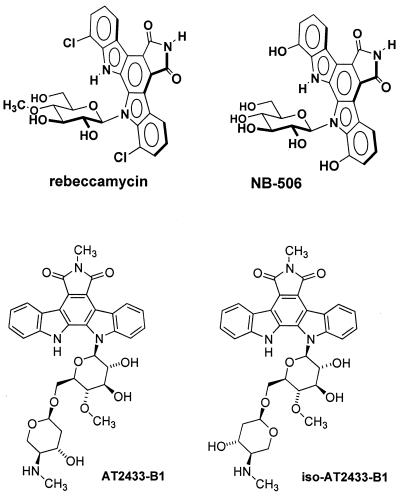
The antitumor antibiotic AT2433-B1 (Fig. 1), originally isolated from culture broth of Actinomadura melliaura, also belongs to the indolocarbazole series (15,16). It contains a unique disaccharide consisting of a methoxyglucose and an amino sugar subunit, 2,4-dideoxy-4-methylamino-l-xylose (17). The mechanism of action of this N-glycosyl indolocarbazole is unknown, but by analogy with rebeccamycin, which contains a methoxyglucose residue (Fig. 1), it was proposed that AT2433-B1 also interacts with DNA as part of its biological action (17). To test this hypothesis, we have developed an approach that uses complementary surface plasmon resonance (SPR) and DNase I footprinting studies to investigate the mechanism of action of AT2433-B1. For comparison, the related indolo[2,3-a]carbazole diglycoside iso-AT2433-B1 (Fig. 1) was also tested. This compound is a diastereoisomer of the natural aminodisaccharide and corresponds to the incorrect structure originally proposed for AT2433-B1 (18). The use of AT2433-B1 and iso-AT2433-B1 provide a unique opportunity to dissect the mechanism of action of these compounds and evaluate the exact influence of the amino deoxysugar residue in the interaction with DNA.
Figure 1.
Structures of the indolocarbazole compounds rebeccamycin, NB-506, AT2433-B1 and iso-AT2433-B1.
MATERIALS AND METHODS
Drugs
Synthesis of the antibiotic AT2433-B1 and its stereoisomer iso-AT2433-B1 have been previously described (17,18). The drugs were dissolved in DMSO at 5 mM and then further diluted with water. The stock solutions of drugs were kept at –20°C and freshly diluted to the desired concentration immediately prior to use.
DNase I footprinting
The 117 bp DNA fragment was prepared by 3′-32P-end-labeling of an EcoRI + PvuII double digest of plasmid pBS (Stratagene) using [α-32P]dATP (3000 Ci mmol–1; Amersham) and AMV reverse transcriptase (Roche). The labeled DNA was purified by gel electrophoresis and resuspended in 10 mM Tris pH 7.0, containing 10 mM NaCl. DNase I footprinting experiments were performed essentially as previously described (19). Reactions were conducted in a total volume of 10 µl. Samples (3 µl) of the labeled DNA fragments were incubated with 5 µl of the buffered solution containing the ligand at appropriate concentration. After 30 min incubation at 37°C to ensure equilibration of the binding reaction, digestion was initiated by addition of 2 µl of a DNase I solution whose concentration was adjusted to yield a final enzyme concentration of ∼0.01 U ml–1 in the reaction mixture. After 3 min the reaction was stopped by freeze drying. Samples were lyophilized and resuspended in 5 µl of an 80% formamide solution containing tracking dyes. The DNA samples were then heated at 90°C for 4 min and chilled in ice for 4 min prior to electrophoresis.
DNA cleavage products were resolved by polyacrylamide gel electrophoresis (PAGE) under denaturating conditions (0.3 mm thick, 8% acrylamide containing 8 M urea). After electrophoresis (∼2.5 h at 60 W, 1600 V in Tris–borate–EDTA buffer solution) gels were soaked in 10% acetic acid for 10 min, transferred to Whatman 3MM paper and dried under vacuum at 80°C. A Molecular Dynamics 425E PhosphorImager and ImageQuant v.3.3 software were used for data analysis. Each resolved band was assigned to a particular bond within the DNA fragments by comparison of its position relative to sequencing standards generated by treatment of the DNA with dimethylsulfate followed by piperidine-induced cleavage at the modified guanine bases in DNA (G-track).
Immobilization of DNA and surface plasmon resonance binding
Three 5′-biotin-labeled DNA hairpins (PAGE purified; Eurogentec) were used in surface plasmon resonance studies (hairpin loop underlined): d(biotin-CAACGCCAGTTTTCTGGCGTTG); d(biotin-CATATATATCCCCATATATATG); and d(biotin-CGCGCGCGTTTTCGCGCGCG).
Samples of hairpin DNA oligomers in HBS-EP buffer at 25 nM concentration were applied to flow cells in streptavidin-derivatized sensor chips (BIAcore SA chips) by direct flow at 2 µl min–1 in a four-channel BIAcore 3000 optical biosensor system. The sensor chips were conditioned with three consecutive 1 min injections of 1 M NaCl in 50 mM NaOH followed by extensive washing with buffer. Nearly the same amounts of all oligomers were immobilized on the surface by non-covalent capture, leaving one of the flow cells blank as a control. Manual injection was used with a flow rate of 2 µl min–1 to achieve long contact times with the surface and to control the amount of DNA bound to the surface. All procedures for binding studies were automated as methods using repetitive cycles of sample injection and regeneration. Steady-state binding analysis was performed with multiple injections of different compound concentrations over the immobilized DNA surfaces for a 10 min period at a flow rate of 20 µl min–1 and 4 or 25°C. Solutions of drug of known concentrations were prepared in filtered and degassed buffer by serial dilutions from stock solution and were injected from 7 mm plastic vials with pierceable plastic crimp caps (BIAcore Inc.).
The instrument response (RU) in the steady-state region is proportional to the amount of bound drug and was determined by linear averaging over a 80 s timespan. The predicted maximum response per bound compound in the steady-state region (RUmax) is determined from the DNA molecular weight, the amount of DNA in the flow cell, the compound molecular weight and the refractive index gradient ratio of the compound and DNA, as previously described (20). In the present case the observed RU values at high concentrations are greater than RUmax, pointing to several binding sites in these DNA sequences. The number of binding sites was determined from Scatchard plots derived from plots of RU/concentration versus RU using linear regression analysis (data not shown). The RUmax value is required to convert the observed response (RU) to the standard binding parameter r (mol drug bound per mol DNA hairpin) using the equation:
r = RU/RUmax
Average fitting of the sensorgrams at the steady-state level was performed with the BIAevaluation 3.0 program. To obtain the affinity constants, the results from the steady-state region were fitted with a multiple equivalent site model using Kaleidagraph for non-linear least squares optimization of the binding parameters with the following equation:
r = n × K × Cfree/(1 + K × Cfree)
where K, the microscopic binding constant, is one variable to fit, r represents mol bound compound per mol DNA hairpin duplex (21), Cfree is the concentration of the compound in equilibrium with the complex and is fixed by the concentration in the flow solution and n is the number of compound binding sites on the DNA duplex and is the second variable to fit. The r values are calculated by the ratio RU/RUmax, where RU is the steady-state response at each concentration and RUmax is the predicted RU for binding of a single compound to the DNA in a flow cell. Global kinetic fits to the sensorgrams to obtain association and dissociation kinetics constants were done using BIAevaluation software and an equivalent site interaction model.
RESULTS
Footprinting studies
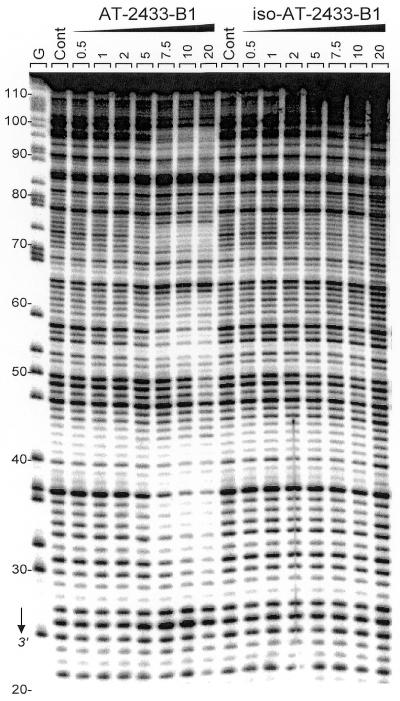
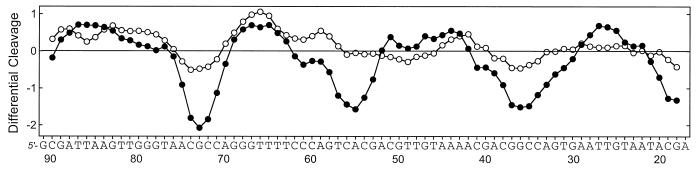
DNase I is a sensitive enzyme for mapping DNA-binding sites of small molecules (22,23). A 32P-labeled DNA restriction fragment of 117 bp was used as substrate and a typical autoradiograph of the sequencing gels used to fractionate the products of partial digestion of the DNA fragment complexed with the two studied compounds is presented in Figure 2. With iso-AT2433-B1 there was very little or no inhibition of DNase I cutting, whereas its regioisomer AT2433-B1 strongly affected cleavage of the DNA substrates by the nuclease. There is no doubt that this antibiotic is a sequence-selective binder. Numerous bands in the drug-containing lanes were weaker than the same bands in the drug-free lane, corresponding to attenuated cleavage, while others displayed relative enhancement of cutting. Band intensities were quantified by densitometry using ImageQuant phosphorimager software. The differential cleavage plots in Figure 3 show that in the presence of 10 µM AT2433-B1 three well-defined regions of attenuated DNA cleavage can be discerned around positions 35, 56 and 73. A fourth binding site can be discerned on the gel in Figure 2 around position 90, but this site, which also corresponds to a GC-rich sequence, lies beyond the area accessible to densitometric analysis. The three footprints coincide with the positions of nucleotide sequences with a high GC content: 5′-CGGCCAG, 5′-GTCACG and 5′-ACGCC.
Figure 2.
Sequence-selective binding. DNase I footprinting gel obtained with the 117 bp PvuII–EcoRI restriction fragment in the presence of graded concentrations of AT2433-B1 and iso-AT2433-B1. The DNA was 3′-end-labeled at the EcoRI site with [α-32P]dATP in the presence of AMV reverse transcriptase. The products of nuclease digestion were resolved on an 8% polyacrylamide gel containing 7 M urea. Control tracks (Cont) contained no drug. Guanine-specific sequence markers obtained by treatment of the DNA with dimethylsulfate followed by piperidine were run in the lane marked G. Numbers on the left side of the gel refer to the standard numbering scheme for the nucleotide sequence of the DNA fragment, as indicated in Figure 3.
Figure 3.
Differential cleavage plots comparing the susceptibility of the 117 bp PvuII–EcoRI restriction fragment to DNase I cutting in the presence of (filled circles) AT2433-B1 and (open circles) iso-AT2433-B1 (10 µM each). Deviation of points towards the lettered sequence (negative values) corresponds to a ligand-protected site and deviation away (positive values) represents enhanced cleavage. Vertical scales are in units of ln(fa) – ln(fc), where fa is fractional cleavage at any bond in the presence of the drug and fc is fractional cleavage of the same bond in the control, given a closely similar extent of digestion in each case. The oligonucleotide sequence selected for the BIAcore experiments is indicated.
The footprints are well resolved with the antibiotic AT2433-B1, whereas the isomeric compound iso-AT2433-B1 has little effect on DNase I cleavage. Only a very weak inhibition of DNase I cleavage can be discerned by densitometry with this compound, again at the GC-rich sequences around positions 35 and 72. Three other DNA restriction fragments were used (data not shown) and in all cases the drugs were found to bind preferentially to GC sites. AT2433-B1 preferentially recognizes sequences such as 5′-CGCC and 5′-GCGCGCG, whereas the sequence selectivity was much reduced with the diastereoisomer, even when using a low temperature (in agreement with the SPR measurements described hereafter).
BIAcore surface plasmon resonance experiments
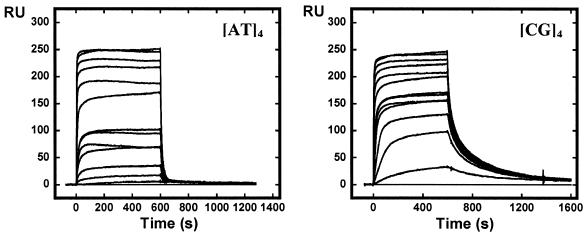
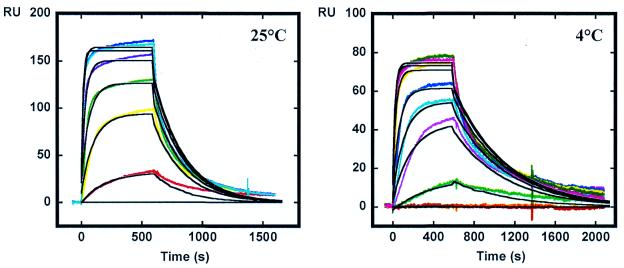
The above DNase I footprinting data indicate that AT2433-B1 interacts preferentially with GC-rich sequences. To further evaluate the drug sequence selectivity, DNA hairpin oligomers with either an alternating [AT]4 or [CG]4 base pair duplex sequence were immobilized on BIAcore SA sensorchips and SPR experiments with AT2433-B1 and its diastereoisomer were conducted at 4 and 25°C. A typical set of sensorgrams (response units versus time) at different drug concentrations for AT2433-B1 binding to the AT and GC oligomers is shown in Figure 4. In all cases the DNA surface was regenerated by buffer flow without additional regeneration agents. As can be seen, AT2433-B1 binds rapidly to both the AT and GC DNA sequences and reaches a stable steady-state plateau, but dissociation rates are quite different depending on the sequence (and temperature).
Figure 4.
SPR sensorgrams for binding of AT2433-B1 to the [AT]4 and [CG]4 DNA hairpin oligomers in HBS-EP buffer at 25°C. The concentration of the unbound ligand in the flow solution varies from 10 nM in the lowest curve to 4 µM in the top curve.
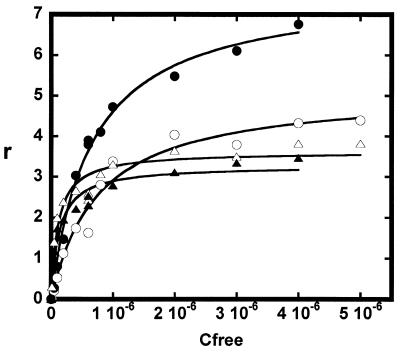
Titrations of the immobilized AT and GC sequence DNA hairpins with AT2433-B1 and iso-AT2433-B1 at 4 and 25°C provided a complete series of sensorgrams, and for each experimental condition the results were converted to plots of r versus Cfree and fitted to determine the binding constants as described in Materials and Methods. The plots obtained with AT2433-B1 are presented in Figure 5. The K values were determined for each set of sensorgrams by non-linear least square fitting of r versus Cfree plots for compound bound to each DNA. Results for AT2433-B1 and iso-AT2433-B1 determined in this manner are collected in Table 1. There is no doubt from these SPR data that the two test compounds bind significantly more tightly to the [CG]4 sequence than to the [AT]4 sequence. For example, at 25°C the ratios KeqGC/KeqAT are 6.8 and 3.6 for AT2433-B1 and iso-AT2433-B1, respectively.
Figure 5.
Binding plots (r versus Cfree) used to determine the affinity constants for AT2433-B1 complexed with the [AT]4 and [CG]4 sequences. To construct these plots, RU values from the steady-state region of the SPR sensorgrams presented in Figure 4 were converted to r (mol drug bound per mol DNA hairpin) and plotted versus the concentration of unbound AT2433-B1 molecules. Circles and triangles refer to the AT and GC sequences and the open and filled symbols correspond to experiments performed at 25 and 4°C, respectively.
Table 1. Binding and dissociation constants for drug binding to the [AT]4 and [CG]4 sequences.
| Sequence | Temperature (°C) | kd (10–3 s–1) | Keq (105 M–1) | n | |
|---|---|---|---|---|---|
| AT2433-B1 | [AT]4 | 25 | a | 13 | 4–5 |
| 4 | 13.7 | 15 | 4–5 | ||
| [CG]4 | 25 | 4.1 | 89 | 3 | |
| 4 | 2.3 | 76 | 3 | ||
| iso-AT2433-B1 | [AT]4 | 25 | a | 1.4 | 6–8 |
| 4 | 9.2 | 2.6 | 6–8 | ||
| [CG]4 | 25 | 56.3 | 5.0 | 4–6 | |
| 4 | 3.2 | 8.5 | 3–4 |
Experiments were performed in HBS-EP buffer.
aDissociation too fast to be measured.
The binding stoichiometry (n values in Table 1) is another important parameter to consider. Considering the length of the hairpin oligonucleotides and the size of the drug molecules, we can estimate that each DNA can offer about three drug intercalation binding sites and this prediction is consistent with the experimental observations for the antibiotic AT2433-B1 interacting with the GC sequence. Higher values (n > 4), as is the case in particular for binding of compound iso-AT2433-B1 to the AT sequence, must reflect the existence of a secondary binding mode, probably an external electrostatic interaction, as we have observed with other cationic DNA ligands (24). Different fitting procedures were used to extract binding parameters with iso-AT2433-B1, but none of them, including a model with two sets of non-equivalent sites, proved better than the multiple equivalent site model (Table 1).
The results clearly indicate that the parent antibiotic AT2433-B1 exhibits a much higher affinity for the GC sequence than its diastereoisomer. At 25°C the GC binding constant (KeqGC) is 18 times higher for AT2433-B1 compared to that measured with iso-AT2433-B1. These observations are in agreement with the footprinting data. Analysis of the kinetic parameters reveals similar differences. Visual inspection of the binding sensorgrams in Figure 4 is sufficient to conclude that AT2433-B1 exhibits slower dissociation rates in the GC interaction compared to the AT sequence. The best fit lines through each experimental plot for the interaction of AT2433-B1 with the alternating GC sequence were obtained by global fitting and are shown in Figure 6. The global equivalent site model provides excellent fits to all of the experimental curves (with very small residuals; data not shown). Similar fits for all plots provided the dissociation kinetics constants listed in Table 1. At 4°C the ratios kdAT/kdGC are 6.2 and 2.9 for AT2433-B1 and iso-AT2433-B1, respectively. In agreement with its larger binding constant, the AT2433-B1 complex with the GC sequence has the lowest kd values at both 4 and 25°C.
Figure 6.
Global fitting of the SPR sensorgrams for the interaction of AT2433-B1 with the [CG]4 sequence at 4 and 25°C. An equivalent site interaction model was used to fit the curves to obtain the association and dissociation kinetic constants.
To deepen the analysis of DNA sequence recognition and further elucidate the mechanism of action of AT2433-B1, additional SPR experiments were performed with the 5′-biotinylated oligomer CAACGCCAGTTTTCTGGCGTTG (hairpin loop underlined) copied directly from the DNase I footprinting experiments (Fig. 2). The differential cleavage plot in Figure 3 clearly shows that the sequence 5′-AACGCCAG (nucleotide positions 69–76) corresponds to a favored binding site for the antibiotic. The SPR experiments were again conducted at 4 and 25°C to compare the kinetic parameters and evaluate the effect of temperature on drug–DNA binding. The equilibrium binding constants collected in Table 2 indicate very clearly that the affinity of AT2433-B1 for the target sequence is considerably higher than that of iso-AT2433-B1. At 25°C, AT2433-B1 binds to AACGCCAG 15 times more tightly than its diastereoisomer. The kinetic parameters also reflect this behavior. As expected, a decrease in temperature from 25 to 4°C affects the kinetics of the drug–DNA interaction, but in both cases the association of AT2433-B1 with the duplex is about five times slower compared to iso-AT2433-B1. At 25°C, the rate of dissociation of AT2433-B1 from the target sequence is about 20 times slower compared to iso-AT2433-B1. It is satisfying to observe that for the two studied compounds the binding constants determined by the ratio ka/kd are very close to the values determined using the steady-state fitting method.
Table 2. Binding and kinetic constants for drug binding to the sequence 5′-AACGCCAG copied from the footprinting experiments.
| AT2433-B1 | iso-AT2433-B1 | |||
|---|---|---|---|---|
| Temperature (°C) | 25 | 4 | 25 | 4 |
| ka (104 M s–1) | 18 | 4.6 | 3.3 | 0.8 |
| kd (10–3 s–1) | 9.6 | 4.9 | 49.5 | 4.8 |
| Keq by kinetics (106 M–1)a | 18.8 | 19.2 | 0.7 | 1.7 |
| Keq by steady-state (106 M–1) | 10.7 | 4.8 | 0.7 | 0.9 |
| n | 3 | 3 | 4–5 | 4–5 |
Experiments were performed in HBS-EP buffer.
aka/kd.
The data in Tables 1 and 2 were obtained under identical experimental conditions and can be mutually compared. It can be noted that the Keq values measured at 25°C for the interaction of AT2433-B1 with the sequence 5′-AACGCCAG derived from the footprinting experiments is slightly higher than that measured with the [CG]4 sequence. Obviously these two sequences provide preferred binding sites for the antibiotic.
DISCUSSION
This is the first time the mechanism of action of the antibiotic AT2433-B1 has been investigated. The results reported here indicate unambiguously that this natural product binds strongly to DNA, which can therefore be considered as a potential target through which the drug exerts its cytotoxic activity. The selective interaction with GC-rich sequences may play a role in the mode of action of this compound. Upon binding to GC sites, the drug is likely to interfere with DNA-reactive enzymes such as polymerases and transcription factors. However, this compound does not stabilize topoisomerase I–DNA covalent complexes (M.Facompré and C.Bailly, unpublished data).
DNase I footprinting experiments indicate that AT2433-B1 preferentially recognizes GC-rich sequences in DNA, such as 5′-CGCC and 5′-GCGCGCG. In contrast, the diastereoisomer iso-AT2433-B1 exhibits a very weak sequence preference, thus showing that the configuration of the xylose subunit of AT2433-B1 is essential for the DNA interaction. As indicated in the Introduction, this synthetic compound corresponds to the incorrect structure originally proposed for AT2433-B1. The absolute stereochemistry of the amino sugar is 3S,4S in AT2433-B1 and 3R,4R in iso-AT2433-B1 (18). Our data illustrate how important it is to correctly assign the stereochemistry of the glycoside residue. A subtle modification of the amino sugar moiety has a profound impact on the drug–DNA recognition process. The amino group must play a privileged role in establishing contact with DNA. In previous studies with rebeccamycin monoglycoside derivatives we have also shown that the presence of an amino group in a suitable position significantly reinforces DNA interaction (25,26).
The SPR technology not only confirms the initial footprinting observations but provides solid quantitative information. The use of two hairpin oligomers with a [CG]4 or [AT]4 block illustrates the GC selectivity of the antibiotic. Equilibrium binding constants of 13 × 105 and 89 × 105 M–1 were measured at 25°C with the AT and GC sequences, respectively. Not only does AT2433-B1 exhibit a significant preference for the GC target but it binds to DNA with high affinity. For comparison, in a recent SPR study performed with the same AT and GC oligomers under identical experimental conditions we showed that the tumor-active analog NB-506, bearing a glucose residue in place of the disaccharide moiety of AT2433-B1, binds equally well to both AT and GC sequences with Keq values of 0.8–0.9 × 105 M–1 (27). The binding affinity of AT2433-B1 for the GC sequence is 100 times higher than that of NB-506. Therefore, we can safely conclude that the amino sugar residue of AT2433-B1 provides a significant driving force for the interaction of the antibiotic with DNA and that this carbohydrate residue may carry the sequence selectivity. At this point it is interesting to refer to the enediyne antibiotics calicheamicin γI1 and esperamicin A1 (28,29) which both bear an oligosaccharide residue with, respectively, an ethylamino or isopropylamino sugar residue similar to the amino sugar subunit (2,4-dideoxy-4-methylamino-l-xylose) of AT2433-B1. Unlike AT2433-B1, these two compounds induce DNA cleavage. NMR studies have revealed that their carbohydrate locates in the minor groove of the DNA double helix to establish hydrogen bonding interaction with the bases (30). The E-ring amino sugar group of both calicheamicin γI1 and esperamicin A1 is protonated to interact with the facing charged phosphate in the DNA complex (31,32) and removal of this charged amino sugar ring (as in esperamicin C) results in a significant decrease in or a loss of DNA cleavage activity (33). By analogy, it is likely that the sugar portion of AT2433-B1 represents the primary structural determinant for binding and the basic element for GC-rich sequence selectivity. If the amino or hydroxy group on the xylose residue establishes a direct contact with a G·C base pair, one can easily understand that the altered configuration of iso-AT2433-B1 can be detrimental to the DNA interaction. The fact that AT2433-B1 does not recognize a precise sequence but binds preferentially to different types of sequences containing at least four consecutive GC suggests that the antibiotic senses DNA conformation rather than DNA sequence per se. GC-rich sequences, which tend to have a wider minor groove (34,35), may accommodate the extended disaccharide more easily than AT tracts.
An additional oligonucleotide issuing directly from the footprinting experiments was used in the SPR experiments to further characterize the DNA interactions of AT2433-B1 and to elucidate its mechanism of action. We have recently used the same combined approach to better understand the molecular basis of DNA recognition by a series of minor groove binders (36). The work reported here shows that the strategy is also valid for intercalators and fully confirms the validity and feasibility of study of small molecule–DNA interaction by means of SPR combined with DNase I footprinting. With this optimized target sequence, 5′-AACGCCAG, the binding affinity for AT2433-B1 reaches 107 M–1, which corresponds to a very tight interaction for a mono-intercalator, comparable or even superior to the binding constants measured with classical intercalating drugs. For example, SPR experiments performed under exactly the same experimental conditions (HBS-EP buffer at 25°C) with the reference intercalator ethidium bromide gave Keq values of 5.6 × 105 and 2.8 × 105 M–1 for the AT and GC oligomers, respectively (C.Carrasco and C.Bailly, unpublished data). The diastereoisomer binds to the 5′-AACGCCAG sequence with a 15-fold lower affinity compared to AT2433-B1, thus reinforcing the conclusion that the configuration of the amino sugar residue is pivotal to the DNA interaction process. With this oligonucleotide it is interesting to observe that the binding constants determined at 4 and 25°C from the kinetic measurements (ka/kd values in Table 2) are almost identical, whereas the association and dissociation constants vary with temperature. With the AT and GC sequences also, the interaction equilibrium constants show a smaller influence of temperature compared to the dissociation kinetics, which are more sensitive to temperature. At 4°C the ratio KeqGC/KeqAT = 5, whereas kdGC/kdAT = 16.
The antibiotic AT2433-B1 forms kinetically stable complexes with a GC oligomer and dissociates 14 times more slowly than its diastereoisomer at 25°C. This shows the importance of the orientations of the OH and NHCH3 substituents on the terminal sugar residue in kinetically stabilizing the drug–DNA complexes. It is reasonable to speculate that compound iso-AT2433-B1 fails to footprint not because of its weaker binding but essentially because of its short residence time on DNA. The equilibrium binding constant for iso-AT2433-B1 bound to the [CG]4 site is six times higher than that of the tumor-active compound NB-506, but only the later compound gives footprints at GC-rich sites (27). Intercalators that have fast binding kinetics, such as proflavine and ethidium, are known to give small footprints (37), whereas those that exhibit slow kinetics, such as DNA-threading intercalators like acridine 4-carboxamides, produce intense footprints, generally at GC-rich sequences (38). At present it is difficult to fathom what structural features can distinguish the complexes of AT2433-B1 and iso-AT2433-B1 since both ligands, in common with other indolocarbazole derivatives (25,26), are believed to bind with the sugar residue positioned in one DNA groove, most likely the minor groove, where it could present a substantial block to the binding of DNase I in the minor groove. According to a preliminary molecular model, the positioning of the aminomethyl group on the xylose residue of AT2433-B1 in the 4S orientation would allow the antibiotic to make an additional hydrogen bonding interaction with a cytosine carbonyl oxygen in the minor groove. Although such a model is not sufficient to delineate in a precise manner which interactions between the drug and DNA are responsible for selective binding to GC sites, it helps to explain why rebeccamycin derivatives containing a monoglycosyl indolocarbazole were found to bind preferentially to nucleotide sequences containing GpT (ApC) and TpG (CpA) steps (39), whereas the antibiotic AT2433-B1 prefers clusters of GC base pairs. This is again an indication that the origin of the selectivity must reside in the interactions between DNA and the glycosyl chain of the drug. It is tempting to conclude that the GC specificity of the antibiotic AT2433-B1 is conferred by possession and specific configuration of the methylaminoxylose residue per se and that it is its interaction with the DNA base pairs which is the predominant feature.
In conclusion, on the basis of the preferred high affinity binding sites, we have obtained quantitative binding and kinetic data that constitute the first investigation of the DNA-binding capacity of the glycosyl antibiotic AT2433-B1. The use of different DNA binding sequences as well as the comparison between the two diastereoisomers leads to the suggestion that the amino sugar residue is an essential element that governs the DNA recognition process. The methodology employed in this study is useful to comprehend how a novel antibiotic discriminates between different sites on the DNA and we expect that it will prove to be of general interest in exploring a variety of reversible interactions between drugs and nucleic acids.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mrs N. Jouy and the IFR22 for access to the BIAcore 3000 instrumentation. This work was supported by grants from the Ligue Nationale Contre le Cancer (Equipe labellisée La Ligue) (to C.B.) and from the National Institutes of Health (to W.D.W.). W.D.W. was the recipient of an INSERM ‘Poste Orange’ fellowship. This research was also supported by a Marie Curie Fellowship of the European Community Program ‘Improving Human Research Potential and the Socio-economic Knowledge Base’ under contract no. HPMFCT-2000-00701 (to C.C.).
REFERENCES
- 1.Long B.H. and Balasubramanian,B.N. (2000) Non-camptothecin topoisomerase I compounds as potential anticancer agents. Exp. Opin. Ther. Patents, 10, 635–666. [Google Scholar]
- 2.Pindur U., Kim,Y.S. and Mehrabani,F. (1999) Advances in indolo [2,3-a]carbazole chemistry: design synthesis of protein kinase C and topoisomerase I inhibitors. Curr. Med. Chem., 6, 29–69. [PubMed] [Google Scholar]
- 3.Bailly C. (2000) Topoisomerase I poisons and suppressors as anticancer drugs. Curr. Med. Chem., 7, 39–58. [DOI] [PubMed] [Google Scholar]
- 4.Yoshinari T., Matsumoto,M., Arakawa,H., Okada,H., Noguchi,K., Suda,H., Okura,A. and Nishimura,S. (1995) Novel antitumor indolocarbazole compound 6-N-formylamino-12,13-dihydro-1,11-dihydroxy-13-(β-D-glucopyranosyl)-5H-indolo[2,3-a]pyrrolo- [3,4-c]carbazole-5,7-(6H)-dione (NB-506): induction of topoisomerase I-mediated DNA cleavage and mechanisms of cell line-selective cytotoxicity. Cancer Res., 55, 1310–1315. [PubMed] [Google Scholar]
- 5.Urasaki Y., Laco,G., Takebayashi,Y., Bailly,C., Kohlhagen,G. and Pommier,Y. (2001) Use of camptothecin-resistant mammalian cell lines to evaluate the role of topoisomerase I in the antiproliferative activity of the indolocarbazole, NB-506 and its topoisomerase I binding site. Cancer Res., 61, 504–508. [PubMed] [Google Scholar]
- 6.Pilch B., Allemand,E., Facompré,M., Bailly,C., Riou,J.F., Soret,J. and Tazi,J. (2001) Specific inhibition of SR splicing factors phosphorylation, spliceosome assembly and splicing by the anti-tumor drug NB506. Cancer Res., 61, 6876–6884. [PubMed] [Google Scholar]
- 7.Akinaga S., Gomi,K., Morimoto,M., Tamaoki,T. and Okabe,M. (1991) Antitumor activity of UCN-01, a selective inhibitor of protein kinase C, in murine and human tumor models. Cancer Res., 51, 4888–4892. [PubMed] [Google Scholar]
- 8.Gescher A. (1998) Analogs of staurosporine: potential anticancer drugs. Biochem. Pharmacol., 31, 721–728. [DOI] [PubMed] [Google Scholar]
- 9.Arakawa H., Morita,M., Kodera,T., Okura,A., Ohkubo,M., Morishima,H. and Nishimura,S. (1999) In vivo antitumor activity of a novel indolocarbazole compound, J-107088, on murine and human tumors transplanted into mice. Jpn J. Cancer Res., 90, 1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshinari T., Ohkubo,M., Fukasawa,K., Egashira,S., Hara,Y., Matsumoto,M., Nakai,K., Arakawa,H., Morishima,H. and Nishimura,S. (1999) Mode of action of a new indolocarbazole anticancer agent, J-107088, targeting topoisomerase I. Cancer Res., 59, 4271–4275. [PubMed] [Google Scholar]
- 11.Komatani H., Kotani,H., Hara,Y., Nakagawa,R., Matsumoto,M., Arakawa,H. and Nishimura,S. (2001) Identification of breast cancer resistant protein/mitoxantrone resistant/placenta-specific, ATP-binding cassette transporter as a transporter of NB-506 and J-107088, topoisomerase I inhibitors with an indolocarbazole structure. Cancer Res., 61, 2827–2832. [PubMed] [Google Scholar]
- 12.Zembower D.E., Zhang,H., Lineswala,J.P., Kuffel,M.J., Aytes,S.A. and Ames,M.M. (1999) Indolocarbazole poisons of human topoisomerase I: regioisomeric analogues of ED-110. Bioorg. Med. Chem., 9, 145–150. [DOI] [PubMed] [Google Scholar]
- 13.Fabbro D., Ruetz,S., Bodis,S., Pruschy,M., Csermak,K., Man,A., Campochiaro,P., Wood,J., O’Reilly,T. and Meyer,T. (2000) PKC412—a protein kinase inhibitor with a broad therapeutic potential. Anticancer Drug Des., 15, 17–28. [PubMed] [Google Scholar]
- 14.Jackson J.R., Gilmartin,A., Imburgia,C., Winkler,J.D., Marshall,L.A. and Roshak,A. (2000) An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res., 60, 566–572. [PubMed] [Google Scholar]
- 15.Golik J., Doyle,T.W., Krishnan,B. and Matson,J.A. (1989) AT2433-A1, AT2433-A2, AT2433-B1 and AT2433-B2 novel antitumor compounds produced by Actinomadura melliaura. II. Structure determination. J. Antibiot., 42, 1784–1789. [DOI] [PubMed] [Google Scholar]
- 16.Matson J.A., Claridge,C., Bush,J.A., Titus,J., Bradner,W.T., Doyle,T.W., Horan,A.C. and Patel,M. (1989) AT2433-A1, AT2433-A2, AT2433-B1 and AT2433-B2, novel antitumor antibiotic compounds produced by Actinomadura malliaura.J. Antibiot., 42, 1547–1555. [DOI] [PubMed] [Google Scholar]
- 17.Chisholm J.D. and Van Vranken,D.L. (2000) Regiocontrolled synthesis of the antitumor antibiotic AT2433-A1. J. Org. Chem., 65, 7541–7553. [DOI] [PubMed] [Google Scholar]
- 18.Chisholm J.D., Golik,J., Krishnan,B., Matson,J.A. and Van Vranken,D.L. (1999) A caveat in the application of the exciton chirality method to N, N-dialkyl amides. Synthesis and structural revision of AT2433-B1. J. Am. Chem. Soc., 121, 3801–3802. [Google Scholar]
- 19.Bailly C. and Waring,M.J. (1995) Comparison of different footprinting methodologies for detecting binding sites for a small ligand on DNA. J. Biomol. Struct. Dyn., 12, 869–898. [DOI] [PubMed] [Google Scholar]
- 20.Davis T.M. and Wilson,W.D. (2000) Determination of the refractive index increments of small molecules for correction of surface plasmon resonance data. Anal. Biochem., 284, 348–353. [DOI] [PubMed] [Google Scholar]
- 21.Connors K.A. (1987) Binding Constants. Wiley, New York, NY.
- 22.Fox K.R. (1997) DNase I footprinting. In Fox,K.R. (ed.), Drug–DNA Interaction Protocols. Humana Press, Totowa, NJ, Vol. 90, pp. 1–22.
- 23.Fox K.R. and Waring,M.J. (2001) High-resolution footprinting studies of drug–DNA complexes using chemical and enzymic probes. Methods Enzymol., 340, 412–430. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen B., Tardy,C., Bailly,C., Colson,P., Houssier,C., Kumar,A., Boykin,D.W. and Wilson,W.D. (2002) Influence of compound structure on affinity, sequence selectivity and mode of binding to DNA for unfused aromatic dications related to furamidine. Biopolymers, 63, 281–297. [DOI] [PubMed] [Google Scholar]
- 25.Bailly C., Qu,X., Anizon,F., Prudhomme,M., Riou,J.F. and Chaires,J.B. (1999) Enhanced binding to DNA and topoisomerase I inhibition by an analog of the antitumor antibiotic rebeccamycin containing an amino-sugar residue. Mol. Pharmacol., 55, 377–385. [DOI] [PubMed] [Google Scholar]
- 26.Bailly C., Goossens,J.F., Laine,W., Anizon,F., Prudhomme,M., Ren,J. and Chaires,J.B. (2000) Formaldehyde cross-linking of a 2′-aminoglucose rebeccamycin derivative to both A·T and G·C base pairs in DNA. J. Med. Chem., 43, 4711–4720. [DOI] [PubMed] [Google Scholar]
- 27.Carrasco C., Vezin,H., Wilson,W.D., Ren,J., Chaires,J.B. and Bailly,C. (2002) DNA binding properties of the indolocarbazole antitumor drug NB-506. Anticancer Drug Des., 16, in press. [PubMed] [Google Scholar]
- 28.Nicolaou K.C., Dai,W.M., Tsay,S.C., Estevez,V.A. and Wrasidlo,W. (1992) Designed enediynes: a new class of DNA-cleaving molecules with potent and selective anticancer activity. Science, 256, 1172–1178. [DOI] [PubMed] [Google Scholar]
- 29.Nicolaou K.C., Smith,A.L. and Yue,E.W. (1993) Chemistry and biology of natural and designed enediynes. Proc. Natl Acad. Sci. USA, 90, 5881–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paloma L.G., Smith,J.A., Chazin,W.J. and Nicolaou,K.C. (1994) Interaction of calicheamicin with duplex DNA: role of the oligosaccharide domain and identification of multiple binding modes. J. Am. Chem. Soc., 116, 3697–3708. [Google Scholar]
- 31.Kumar R.A., Ikemoto,N. and Patel,D.J. (1997) Solution structure of the esperamicin A1–DNA complex. J. Mol. Biol., 265, 173–186. [DOI] [PubMed] [Google Scholar]
- 32.Kumar R.A., Ikemoto,N. and Patel,D.J. (1997) Solution structure of the calicheamicin γ1I–DNA complex. J. Mol. Biol., 265, 187–201. [DOI] [PubMed] [Google Scholar]
- 33.Long B.H., Golik,J., Forenza,S., Ward,B., Rehfuss,R., Dabrowiak,J.C., Catino,J.J., Musial,S.T., Brookshire,K.W. and Doyle,T.W. (1989) Esperamicins, a class of potent antitumor antibiotics: mechanism of action. Proc. Natl Acad. Sci. USA, 86, 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neidle S. (1992) Minor-groove width and accessibility in B-DNA drug and protein complexes. FEBS Lett., 298, 97–99. [DOI] [PubMed] [Google Scholar]
- 35.Wood A.A., Nunn,C.M., Czarny,A., Boykin,D.W. and Neidle,S. (1995) Variability in DNA minor groove width recognised by ligand binding: the crystal structure of a bis-benzimidazole compound bound to the DNA duplex d(CGCGAATTCGCG)2. Nucleic Acids Res., 23, 3678–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson W.D., Wang,L., Tanious,F., Kumar,A., Boykin,D.W., Carrasco,C. and Bailly,C. (2001) BIAcore and DNA footprinting for discovery and development of new DNA targeted therapeutics and reagents. Biacore J., 1, 15–19. [Google Scholar]
- 37.Fox K.R. and Waring,M.J. (1987) Footprinting at low temperature: evidence that ethidium and other simple intercalators can discriminate between different nucleotide sequences. Nucleic Acids Res., 15, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailly C., Denny,W.A., Mellor,L., Wakelin,L.P.G. and Waring,M.J. (1992) Sequence-specificity of the binding of 9-aminoacridine- and amsacrine-4-carboxamides to DNA studied by DNase I footprinting. Biochemistry, 31, 3514–3524. [DOI] [PubMed] [Google Scholar]
- 39.Bailly C., Colson,P., Houssier,C., Rodrigues-Pereira,E., Prudhomme,M. and Waring,M.J. (1998) Recognition of specific sequences in DNA by a topoisomerase I inhibitor derived from the antitumor drug rebeccamycin. Mol. Pharmacol., 53, 77–87. [DOI] [PubMed] [Google Scholar]