Abstract
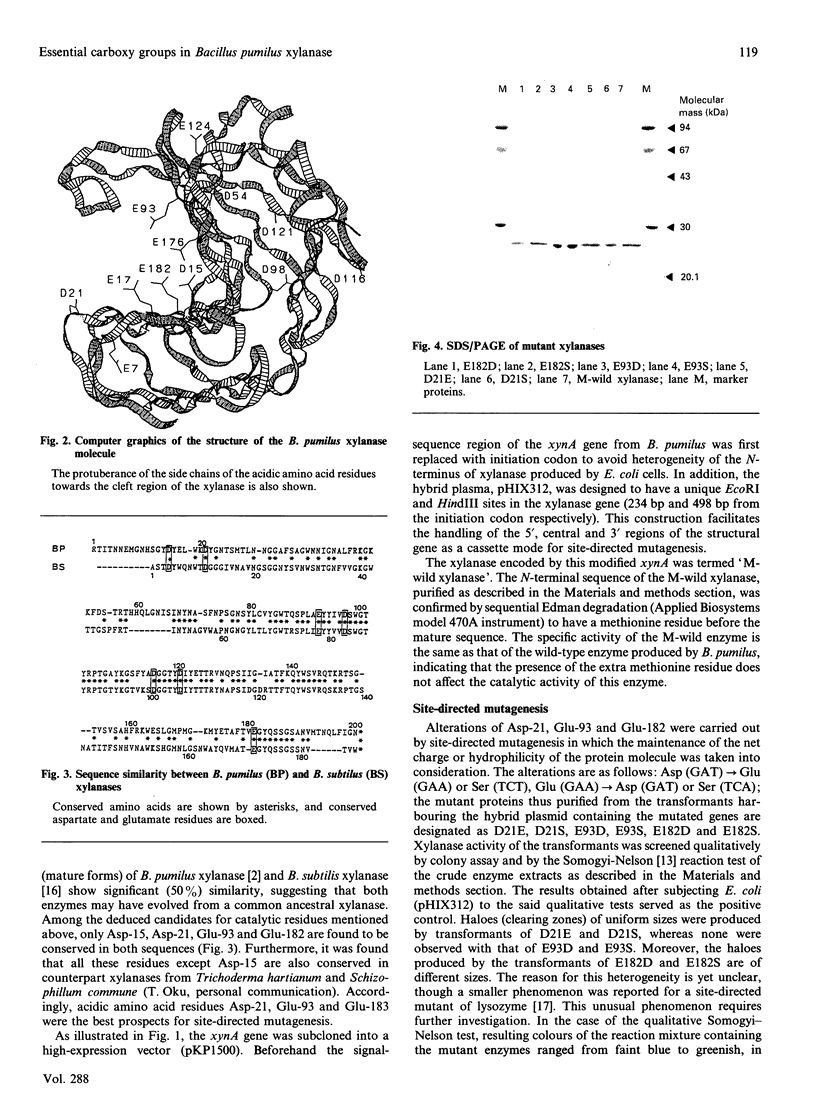
To elucidate the reaction mechanism of xylanase, the identification of amino acids essential for its catalysis is of importance. Studies have indicated the possibility that the reaction mechanism of xylanase is similar to that of hen's egg lysozyme, which involves acidic amino acid residues. On the basis of this assumption, together with the three-dimensional structure of Bacillus pumilus xylanase and its amino acid sequence similarity to other xylanases of different origins, three acidic amino acids, namely Asp-21, Glu-93 and Glu-182, were selected for site-directed mutagenesis. The Asp residue was altered to either Ser or Glu, and the Glu residues to Ser or Asp. The purified mutant xylanases D21E, D21S, E93D, E93S, E182D and E182S showed single protein bands of about 26 kDa on SDS/PAGE. C.d. spectra of these mutant enzymes show no effect on the secondary structure of xylanase, except that of D21E, which shows a little variation. Furthermore, mutations of Glu-93 and Glu-182 resulted in a drastic decrease in the specific activity of xylanase as compared with mutation of Asp-21. On the basis of these results we propose that Glu-93 and Glu-182 are the best candidates for the essential catalytic residues of xylanase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anfinsen C. B., Scheraga H. A. Experimental and theoretical aspects of protein folding. Adv Protein Chem. 1975;29:205–300. doi: 10.1016/s0065-3233(08)60413-1. [DOI] [PubMed] [Google Scholar]
- Bray M. R., Clarke A. J. Essential carboxy groups in xylanase A. Biochem J. 1990 Aug 15;270(1):91–96. doi: 10.1042/bj2700091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. T., Wu C. S., Yang J. T. Circular dichroic analysis of protein conformation: inclusion of the beta-turns. Anal Biochem. 1978 Nov;91(1):13–31. doi: 10.1016/0003-2697(78)90812-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Malcolm B. A., Rosenberg S., Corey M. J., Allen J. S., de Baetselier A., Kirsch J. F. Site-directed mutagenesis of the catalytic residues Asp-52 and Glu-35 of chicken egg white lysozyme. Proc Natl Acad Sci U S A. 1989 Jan;86(1):133–137. doi: 10.1073/pnas.86.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Yasukochi T., Nagatani H., Furuno M., Orita T., Yamada H., Imoto T., Horiuchi T. Construction of a plasmid vector for the regulatable high level expression of eukaryotic genes in Escherichia coli: an application to overproduction of chicken lysozyme. Protein Eng. 1987 Aug-Sep;1(4):327–332. doi: 10.1093/protein/1.4.327. [DOI] [PubMed] [Google Scholar]
- Moriyama H., Hata Y., Yamaguchi H., Sato M., Shinmyo A., Tanaka N., Okada H., Katsube Y. Crystallization and preliminary X-ray studies of Bacillus pumilus IPO xylanase. J Mol Biol. 1987 Jan 5;193(1):237–238. doi: 10.1016/0022-2836(87)90644-9. [DOI] [PubMed] [Google Scholar]
- Panbangred W., Kondo T., Negoro S., Shinmyo A., Okada H. Molecular cloning of the genes for xylan degradation of Bacillus pumilus and their expression in Escherichia coli. Mol Gen Genet. 1983;192(3):335–341. doi: 10.1007/BF00392172. [DOI] [PubMed] [Google Scholar]
- Provencher S. W., Glöckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981 Jan 6;20(1):33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- SMOGYI M. Notes on sugar determination. J Biol Chem. 1952 Mar;195(1):19–23. [PubMed] [Google Scholar]
- Teather R. M., Wood P. J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982 Apr;43(4):777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]