Abstract
Background
Cancer-associated fibroblasts (CAFs) are the prominent cell type in the tumor microenvironment (TME), and CAF subsets have been identified in various tumors. However, how CAFs spatially coordinate other cell populations within the liver TME to promote cancer progression remains unclear.
Methods
We combined multi-region proteomics (6 patients, 24 samples), 10X Genomics Visium spatial transcriptomics (11 patients, 25 samples), and multiplexed imaging (92 patients, 264 samples) technologies to decipher the expression heterogeneity, functional diversity, spatial distribution, colocalization, and interaction of fibroblasts. The newly identified CAF subpopulation was validated by cells isolated from 5 liver cancer patients and in vitro functional assays.
Results
We identified a liver CAF subpopulation, marked by the expression of COL1A2, COL4A1, COL4A2, CTGF, and FSTL1, and named F5-CAF. F5-CAF is preferentially located within and around tumor nests and colocalizes with cancer cells with higher stemness in hepatocellular carcinoma (HCC). Multiplexed staining of 92 patients and the bulk transcriptome of 371 patients demonstrated that the abundance of F5-CAFs in HCC was associated with a worse prognosis. Further in vitro experiments showed that F5-CAFs isolated from liver cancer patients can promote the proliferation and stemness of HCC cells.
Conclusions
We identified a CAF subpopulation F5-CAF in liver cancer, which is associated with cancer stemness and unfavorable prognosis. Our results provide potential mechanisms by which the CAF subset in the TME promotes the development of liver cancer by supporting the survival of cancer stem cells.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13073-024-01367-8.
Keywords: Tumor microenvironment, Cancer-associated fibroblast, Spatial transcriptomics, Cancer stem cell, Liver cancer
Background
Tumor growth is supported by the tumor stroma or the tumor microenvironment (TME), which includes the extracellular matrix (ECM), vasculature, and various non-malignant cells (e.g., myeloid cells, lymphocytes, macrophages, and stromal cells). The TME is the “soil” for tumor growth, providing tumor cells with essential nutrients, growth factors, architectural and mechanical support, and a favorable immune microenvironment [1, 2]. The TME also supports cancer stem cells (CSCs), known as the “seeds” of tumors, whose underlying mechanisms are not fully understood [3]. The distribution of CSCs within tumors is not uniform, suggesting a potential interaction between CSCs and the TME [4]. A better understanding of the TME and its interaction with tumor cells, especially CSCs, is conducive to understanding the mechanism of tumor development and designing corresponding treatment strategies.
Among all types of stromal cells, fibroblasts are the predominant component in the TME [4–7], and cancer-associated fibroblasts (CAFs) play important roles in tumor growth and progression. Multiple CAF subsets, such as ECM-remodeling/myofibroblastic CAFs (myCAF), immunomodulatory CAFs (iCAF), and antigen-presenting CAFs, have been identified in various cancers [8–15]. These CAF subtypes possibly directly communicate with surrounding cells via secreted molecules or cell–cell adhesions or indirectly affect cancer cell behavior by remodeling the ECM [5], thereby exhibiting different functions and clinical significance. Targeting these CAF-driven mechanisms may represent a promising approach for cancer treatment.
Primary liver cancer (PLC) was the sixth most commonly diagnosed cancer and the third leading cause of cancer death worldwide in 2020 [16]. PLC comprises mainly hepatocellular carcinoma (HCC; 75–85% of cases), intrahepatic cholangiocarcinoma (ICC; 10–15%), and combined hepatocellular cholangiocarcinoma (CHC; less than 5%). Despite treatment advances [17], the overall prognosis of PLC remains poor. In liver cancer, through gene tracking in mouse models, it has been confirmed that CAFs in HCC [18] and ICC [19] are mainly derived from hepatic stellate cells. Among them, HAS2 expressed by myCAF and HGF expressed by iCAF can promote ICC growth [19]; and some new CAF subtypes in HCC, such as vascular CAF and lipid processing CAF [20], and more complex and diverse cytokine-mediated cellular crosstalk regulate HCC progression [21].
Although single-cell RNA sequencing (scRNA-seq) has provided crucial initial information on CAF diversity [15, 22–30] the emergence of spatial technologies has revolutionized the TME field and more intuitively revealed the coordinated mechanisms of multiple components. The proximity of CAFs to other cells has been explored in glioblastoma [31], breast cancer [32], rectal cancer [33], head and neck squamous cell carcinoma [34], lung cancer [35], and other cancer types [36]. Recently, we characterized the overall spatial heterogeneity of the PLC TME through spatial transcriptomics [37], and other studies have shown that CAFs in HCC are involved in the spatial organization of SPP1+ macrophages [38]. However, the precise spatial neighborhood of certain CAF phenotypes and what functional changes occur through this spatial relationship to support PLC remains unclear.
In this study, we performed multimodal spatial profiling of human liver cancer samples using proteomics, 10X Genomics Visium spatial transcriptomics (ST), and multiplexed imaging to investigate the role of stromal tissues, especially fibroblasts, in PLC. Through spatial omics analysis, we revealed the spatial heterogeneity and functional diversity of the stroma and fibroblasts in the liver cancer microenvironment. Notably, we identified an F5-CAF subpopulation that preferentially localizes within and around tumor nests. Further multiplexed imaging analysis and characterization of the F5-CAF niche in HCC revealed potential pathways by which CAFs maintain tumor stemness through interactions with CSCs.
Methods
Patients and samples
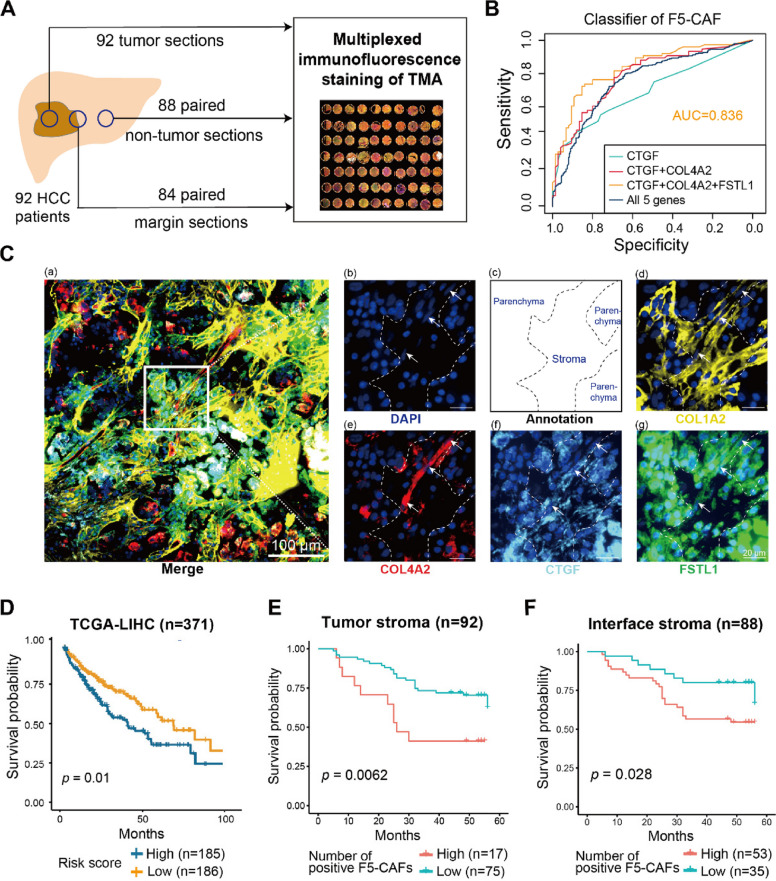
We obtained samples from PLC patients (n = 22) who underwent surgical resection at Eastern Hepatobiliary Surgery Hospital (EHBH). We selected formalin-fixed paraffin-embedded (FFPE) tissue blocks (n = 24) from 6 patients for proteomics analysis, fresh PLC tissues (n = 25) from 11 patients for ST, and fresh tumor tissues from 5 patients for primary cell isolation (Additional file 1: Table S1). All patients were randomly selected and provided informed consent. We provide the clinical information of these patients, where the presence of cirrhosis and the extent of immune cell infiltration in adjacent liver tissues were determined by two experienced pathologists in a single-blind manner based on HE staining.
Two HCC tissue microarrays (TMAs) were purchased from Shanghai Outdo Biotech Company: Cohort 1 (HLivH180Su17), containing cancerous (n = 92) and adjacent normal (n = 88) tissues, and Cohort 2 (HLivH090Su01), containing leading-edge tissues (n = 84). The clinical and pathological data are summarized in Additional file 1: Table S9.
Mass spectrometry (MS)-based proteomics
Matched tumor and adjacent normal tissues (> 5 mm) were cut into 10 µm thick sections and flattened in preprepared sterilized water. Then, the sections were mounted on Leica PEN Membrane Glass Slides (Cat No. 11505189). The deparaffinization of FFPE tissue sections utilized heptan instead of xylene three times for 15 min, followed by 100% ethanol, 95% ethanol, 70% ethanol, and 50% ethanol (3 min each). After only hematoxylin staining, the FFPE tissue sections were sent for laser-capture microdissection (LCM) by a Leica LMD 7000 (Leica Microsystems, Inc., Bannockburn, IL). All stromal tissues from each tissue section were collected by LCM into a 0.2-ml Eppendorf tube and stored at room temperature for further sample preparation, and the remaining parenchymal tissues were collected into 1.5-ml tubes.
The proteins from FFPE tissue tissues used for MS analysis were extracted by a Qproteome FFPE Tissue Kit (Qiagen) according to the manufacturer’s instructions. Note that since tissue sections had already been deparaffinized in the aforementioned H&E staining step, extraction buffer ExB plus supplemented with β-mercaptoethanol could be directly added to the LCM collection tubes, followed by the steps of extraction and cleanup of proteins from FFPE tissues. Finally, the extracted proteins were subjected to MS analysis after tryptic digestion [39]. Protein expression of the stroma regions and remaining parenchyma regions was analyzed using the data-independent acquisition (DIA) mode of a mass spectrometer (Thermo Fisher). Data-dependent acquisition spectrum libraries were constructed before DIA mode to obtain real spectrum libraries.
Quantitative proteomics analysis
The resulting spectra from each fraction were searched separately against the homo_sapiens_uniprot_2021_3_9.fasta (194,557 sequences) database by the search engine Proteome Discoverer 2.2 (PD 2.2, Thermo). The results of the search and identification by PD2.2 software were imported into Spectronaut (version 14.0, Biognosys) software to generate a library. The eligible peptides and product ions were selected from the spectrum by setting peptide and ion pair selection rules to generate a target list [40]. The DIA data were imported, and ion-pair chromatographic peaks were extracted according to the Target List. The ions were matched, and the peak areas were calculated to qualitatively and quantitatively analyze the peptides. iRT was added to the sample to correct the retention time, and the precursor ion Q value cutoff was set to 0.01. The quantitative values were visualized with Bionic Visualizations Proteomaps (https://proteomaps.net/).
Differentially expressed proteins (DEPs; tumor vs. adjacent nontumor, p < 0.05, |logFC|> 1) underlying enrichment analyses. Gene Ontology (GO) and KEGG pathways with adjusted p < 0.05 (Benjamini‒Hochberg method) were considered significantly enriched.
Spatial transcriptomics experiments
Under the guidance of pathologists, we defined the interface as the boundary or transitional region between tumor parenchymal cells and adjacent nontumor parenchymal cells (usually 1–2 mm in distance). The definition of interface areas is distinct from that of the capsule, which refers to the fibrous envelope surrounding the hepatocellular carcinoma tumor itself. Each tissue specimen was embedded in optimal cutting temperature compound medium, immediately frozen in an isopentane slurry made with liquid nitrogen, and finally stored at -80 °C until further processing. Each tissue sample was embedded within 30 min for frozen sectioning after surgical removal. The archives of all patients were collected via the EHBH archive system. Informed consent was obtained from the patients, and all procedures were approved by the ethical committee of EHBH.
Spatial transcriptome experiments were performed according to the user guide of the Visium Spatial Gene Expression Reagent Kit (10X Genomics). Cryosections were mounted onto a spatially barcoded array of 10X Genomics Visium with 10-μm thickness. For processing, the tissue was fixed for 30 min with prechilled methanol at − 20 °C, followed by H&E staining. Slides were finally taken on a Leica SCN400 F whole-slide scanner at 40 × resolution. After capturing ideal tissue morphology information and ensuring that RNA was not degraded (RIN ≥ 7), tissue permeabilization and reverse transcription were immediately conducted by a Visium Spatial Tissue Optimization Kit (10X Genomics). Finally, the library was prepared with second-strand synthesis and denaturation and sequenced by NovaSeq 6000 (Illumina). Each of the spots printed onto the array is 55 μm in diameter and 100 μm from center to center, covering an area of 6.5 × 6.5 mm2.
Basis analysis of the spatial transcriptome
Data processing
The gene-barcode matrices were analyzed with the Seurat pipeline [41] in R. Spots were filtered for a minimum detected gene count of 200 genes. Then, we used the SCTransform function to perform normalization, log-transformation, centering, and scaling across spots, in which percent.mt and nFeature_Spatial were regressed out in a second nonregularized linear regression. Then, the IntegrateData function was used to integrate the expression data from different sections according to 3000 highly variable genes. The integrated data were used for subsequent clustering.
Differential gene expression and correlation with DEPs
Differential expression analysis was performed using the FindAllMarkers function in Seurat (v.4.0) with the following settings: only.pos = TRUE, assay = "SCT", and slot = "data". Genes filtered for an adjusted p value < 0.05 (Wilcoxon rank sum test, Bonferroni method) were regarded as differentially expressed genes (DEGs) and included in the gene set variation analysis (GSVA).
Next, we compared the correlation between protein and mRNA abundance changes. Spots from seven of these 11 patients with similar clinical features and stromal-rich sections were included. Protein‒mRNA pairs were matched with gene IDs. For these protein‒mRNA pairs, we calculated the Spearman correlation of tumor/non-tumor log2(fold change) for DEPs and DEGs.
Expression heterogeneity between ST spots
If ρ is the pairwise Pearson’s correlation coefficient between two expression vectors of highly variable genes, the heterogeneity score can be defined as Pearson’s pairwise distance:
We calculated the heterogeneity score between paired spots from the ST data. Then, hierarchical clustering of Pearson’s pairwise distance showed different functional clusters within tumor stroma (T–S) and non-tumor stroma (N‒S). We also calculated the mean Pearson’s pairwise distances for 100 cases sampled in T–S and N‒S. After 500 bootstrap resampling iterations, we fit the distribution and calculated the significance of the difference between the mean pairwise distance in 2 groups (Wilcoxon rank sum test; ref [42].).
Identification of the major cell types for ST spots
Two methods were used to determine the cell type enriched in each spot: (1) cluster- and marker-based annotation of cell types and (2) estimation of cell purity by deconvoluting cell mixtures from single-cell references by SPOTlight [43].
We combined unsupervised clustering and differential expression to compare the top-ranked DEGs with known cell type-specific expression in the literature. In detail, we input the expression profile of the highly variable genes, principal component analysis (PCA) was performed to project the spots into a low-dimensional space, and uniform manifold approximation and projection (UMAP) was performed for visualization. Clustering was performed using the FindClusters function with a resolution of 0.5 to generate 13 clusters, and DEGs were identified in each cluster. Three of these clusters highly expressed hepatocyte-associated genes, such as ALB, SLC10A1, SLC22A1, CYP2A6, and CYP2C8, and were therefore merged and inferred to be hepatocyte cells. Cancer cells are made up of four clusters and highly expressed GPC3, CDC34, and AFP. Cluster 4 highly expressed MS4A1, CD19, CD3E, CD3G, CD8A, CD8B, and CD28, which means that these spots include T cells and B cells. Other cluster markers highly expressed markers specific for cholangiocytes (KRT19, KRT7), fibroblasts (COL1A2, COL3A1, COL1A1, ACTA2), natural killer cells (NKs; NCAM1) and macrophages (CD68, FCGR2A, CD80, CD86, TGFB1, CD163, MRC1). The remaining three clusters had ambiguous cell types, so they were isolated and reclustered, finally obtaining 4 categories assigned to malignant cholangiocytes, T/B cells, and malignant hepatocytes.
Single-cell references were downloaded from GSE156625 [44], containing broad cell type annotations from 14 liver cancer patients and 1 healthy donor. To detect marker genes, cells were filtered for a minimum detected gene count of 1500 and were randomly sampled from 100 endothelial cells, fibroblasts, lymphocytes, hepatocytes, and myeloid cells. Then, we used the FindAllMarkers function in Seurat (v.4.0) [41] with the following settings: only.pos = TRUE, min.pct = 0.7 and logfc.threshold = 1. Marker genes and single-cell RDS objects were input into the R package SPOTlight [43] to decompose the expression matrix inferring the percentage of each cell type (cell purity) within a spot. When we focused on the specific cell type, we filtered out those spots that did not have the highest percentage of annotated cell type.
Cell type annotation for cohort two: We identified anchors to transfer data from Cohort 1 to Cohort 2 using Seurat’s TransferData function [41]. Each spot obtains a corresponding score for each category, and the category with the highest score is defined as the phenotype of the spot. We examined the cell type annotations in each cohort 2 cluster. We found that the annotation of a small number of cells was inconsistent with that of the majority of cells in the same cluster and corrected their annotation to that of the majority of cells.
Trajectory inference analysis
Using our previously aligned bam files, we first sorted samples by cell barcode. To annotate the spliced and unspliced reads, we used the Velocyto pipeline [45] to generate our loom files. Our steady-state gene-specific velocities were computed and preprocessed following the scVelo python package [46].
Fibroblast subtypes, spatial distribution, and interaction
Identification of fibroblast subtypes
We reclustered fibroblast spots/cells from the ST and scRNA-seq data and then compared their DEGs to match the clusters. For the ST data, 5803 fibroblast-enriched spots were isolated from the broad cell type. Their expression profiles were first combined and the batch effect was corrected using the batch index as a covariate in Seurat [41], then were dimensionally reduced with PCA and clustered at a resolution of 0.5. Similarly, we reclustered 1800 fibroblasts at a resolution of 0.2 from the scRNA-seq data [44] after discarding the cells that expressed fewer than 200 genes or had ambiguous marker genes. We focused on subtypes that were significantly enriched in tumor tissues (Fisher test, p < 0.05).
We mapped clusters in scRNA-seq data to those in ST by scoring single-cell module scores. The list of genes in the module consisted of the top (less than 60) DEGs, and module scores were calculated by three methods (AUCell [47], z score, Garnett [48]). The results of the three methods for assigning single-cell cluster labels to ST clusters were similar, which we showed in Fig. 3D by z scores.
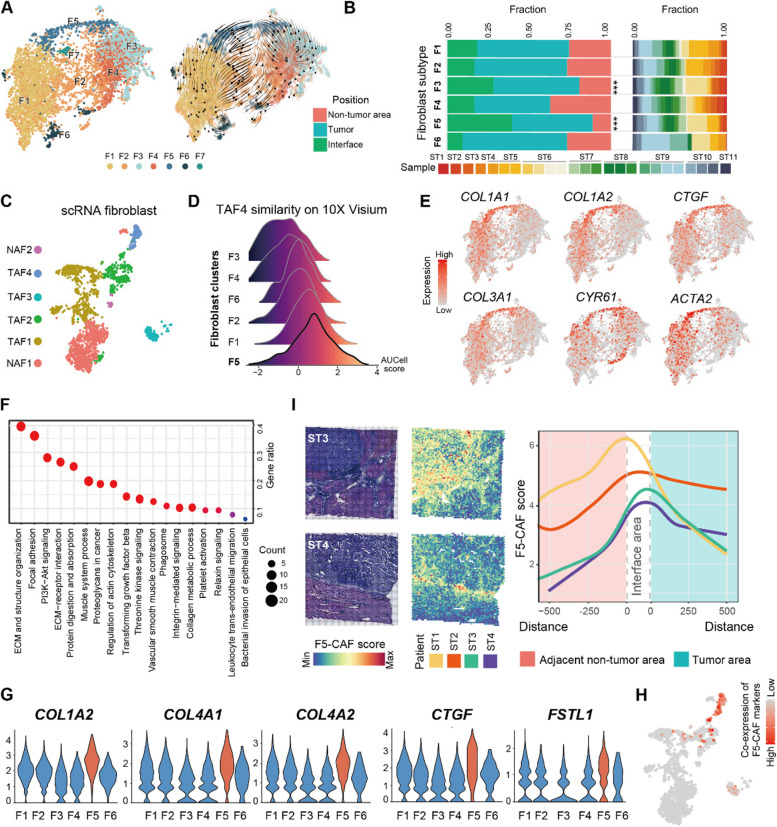
Fig. 3.
A CAF subpopulation in liver cancer and its transcriptional characteristics. A UMAP (left) and trajectory (right) of 5803 fibroblast-enriched spots colored by clusters. B Fibroblast subset composition displayed as percentages at different locations and in individual patients. Stars indicate significant tumor enrichment (Fisher test, p < 0.001). C UMAP of 1836 fibroblasts from the scRNA-seq data colored by clusters. TAF, fibroblasts enriched in the tumor; NAF, other fibroblast subpopulations. D AUCell score matched to TAF4 fibroblasts for each ST cluster. E UMAP feature plots of myofibroblast marker expression in fibroblast-enriched spots. F Gene Ontology (GO) terms for the top 200 upregulated genes in F5 versus other fibroblasts. G The expression of the marker genes of F5. All five genes were significantly highly expressed in F5 (t test, adjusted p < 0.05). H The summed expression of the marker genes in G. Gray indicates that at least one of the 5 marker genes is not expressed, while red reflects a higher combined expression of the marker genes. I The spatial distribution of the F5-CAF score, defined by marker genes in G. Left, representative spatial distribution of the F5-CAF score in two ST tissue sections stained with H&E. Right, line chart of the F5-CAF score by tumor border distance. See also Fig. S3
Marker genes and classification of F5-CAFs
After matching the scRNA-seq and ST data, the common CAF cluster was annotated as F5-CAF because of five marker genes: COL1A2, COL4A1, COL4A2, CTGF, and FSTL1. These five genes were selected from marker genes shared by F5 (from ST data) and TAF4 (from scRNA data). The criteria were q < 0.05 (t test; F5 vs. non-F5 populations), log2(fold change) > 0.5, at least 80% of subtype cells express the gene and other subtypes do not widely express the gene (more than 10% difference).
Based on the expression of the marker genes COL1A2, COL4A1, COL4A2, CTGF, and FSTL1, we employed a random forest model with feature selection to identify the minimal set of genes that could classify F5-CAFs and other fibroblasts. We used this optimized gene set for multiplexed immunofluorescence (mIF) experiments. The ST profile of 80% of the fibroblast-enriched spots was used as the training dataset, and the remaining 20% was used as the test dataset. The classifier was built using a random forest classification method from the randomForest (v.4.7) [49]R package with balanced samples and default parameters. Feature selection was performed by the varImpPlot function to measure variable importance, followed by forward selection to add features. The receiver operation curve (ROC) was plotted by the pROC (v.1.18) [50]R package. Finally, different ROC curves were compared by the roc.test function, and gene combinations with the highest area under the curve (AUC) were selected for F5-CAF classification.
Definition of F5-CAF score and F5-CAF spot
We defined F5-CAF scores as (1) the enrichment score of 5 marker genes of F5-CAFs by ssGSEA [51] and (2) directly summing the normalized expression values of the 5 genes. The results of the two methods are significantly positively correlated. To ensure methodological consistency in module score calculations in subsequent analysis (Fig. 5), we used ssGSEA [51] to calculate the F5-CAF score.
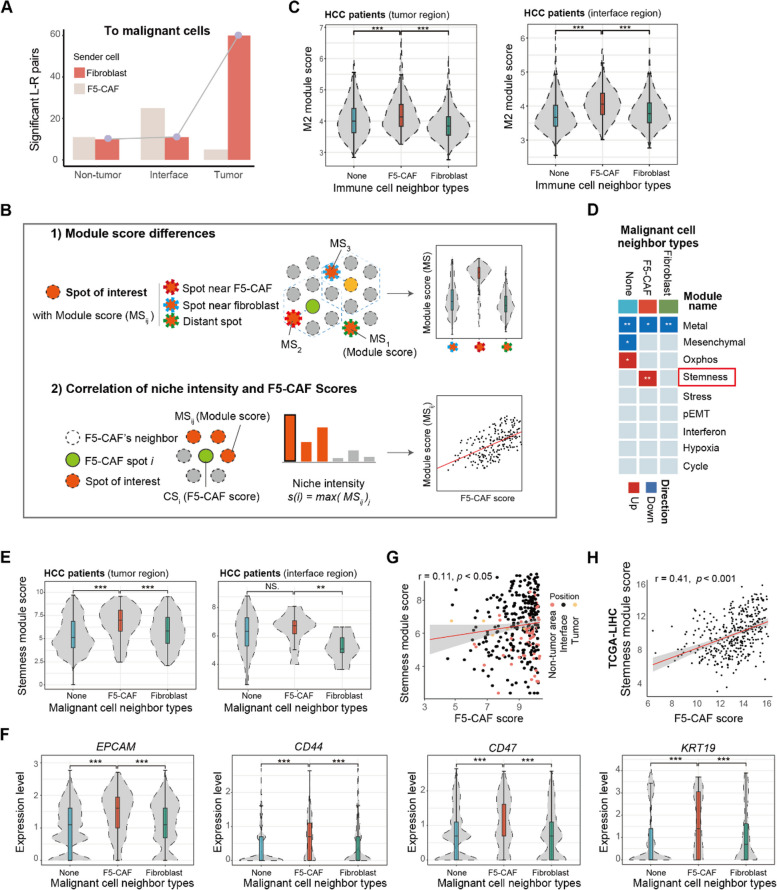
Fig. 5.
Spatial colocalization of F5-CAFs with other cells in the HCC TME. A Significant L–R pairs for F5-CAF/other fibroblast-malignant cells in non-tumor, interface, and tumor areas. Location of malignant cells from malignant spots. B Schematic of the cellular neighborhood. Top, red spots indicate the presence of the cell type of interest and can be divided into three groups (dashed lines in different colors) according to their proximity to fibroblast subpopulations. The dashed hexagon represents a community of fibroblast spots. Bottom, schematic of the calculation of “niche intensity” for each community. C Violin plots of the M2 module score of immune-enriched spots in different groups. ***, p < 0.001 and NS, p > 0.05 by Wilcoxon rank-sum test. D Heatmap of cancer cell states enriched in different groups in the tumor and interface area. The three groups correspond to the definitions in B. The red box marks the significant state in F5-CAF-surrounding malignant spots. Oxphos, oxidative phosphorylation; pEMT, partial epithelial-mesenchymal transition. E Violin plots of the stemness module score of malignant spots in different groups. ***, p < 0.001 and NS, p > 0.05 by Wilcoxon rank-sum test. F Expression of cancer stem cell markers across malignant spots from distinct groups. ***, p < 0.001 and *, p < 0.05 by Wilcoxon rank-sum test. G Pearson correlation between the F5-CAF score and stemness module score in fibroblast-cancer cell communities. The module score was calculated by ssgsea (see the “Methods” section). H Similar to F, for TCGA-LIHC samples. See also Fig. S5 and Fig. S6
We observed some nonfibroblast-enriched spots with high F5-CAF scores, and there were indeed a few fibroblasts in these spots, as confirmed by morphological features in H&E slides. Although these spots had significantly lower numbers of fibroblasts and thus were not previously defined as fibroblast-enriched spots, they were important in spatial analysis because F5-CAF signatures were also present in these spots. Therefore, when we defined F5-CAF spots, we discretized F5-CAF scores using the median F5-CAF score from F5 spots. This approach preserved the spatial localization of F5-CAFs with as few omissions as possible.
Validation of F5-CAFs by mIF staining
The TMA was purchased from Outdo Biotech company (Shanghai, China) with clinical information. MIF was performed using OpalTM chemistry (PerkinElmer, Waltham, USA) with five antibodies against EPCAM (Abcam, ab7504), COL4A2 (Abcam, ab125208), CTGF (Abcam, ab5097), FSTL1 (Abcam, ab71548), and COL1A2 (Abcam, ab96723). Briefly, after deparaffinization, The TMA slides were blocked with antibody diluent for 10 min at room temperature after antigen retrieval buffer. The slides were incubated with the primary antibody for 60 min and subsequently incubated with the HRP-conjugated secondary antibody for 10 min after removing the primary antibody and washing in TBST buffer. Thereafter, the slides were incubated with OpalTM working buffer for 10 min at room temperature and then washed in TBST buffer. 4′,6-diamidino-2-phenylindole (DAPI) was used to stain all nuclei.
Panoramic multispectral scanning of slides was performed by the Tissue-FAXS system (TissueFAXS Spectra, TissueGnostics). Then, we imported the data into Strata-Quest analysis software. We used the spectral library for spectral splitting to obtain a single-channel fluorescence signal, The DAPl channel was used to segmentate and identify the effective nucleus. Each of the individually stained tissue spots in the TMA was utilized to establish the spectral library of the fluorophores to eliminate an interference of cross-fluorescence combined with the spectral unmixing algorithm. For organizational area division, a tissue segmentation algorithm from the software was applied to divide each tissue into parenchymal and stromal areas.
Associations between F5-CAFs and prognosis
The association between F5-CAFs and prognosis was analyzed by The Cancer Genome Atlas (TCGA) gene expression and mIF protein expression data. The expression data and clinical data of 371 liver cancer patients in the TCGA datasets were downloaded from cBioPortal. Considering that the gene combination of COL4A2, CTGF, and FSTL1 best discriminated F5-CAFs, we defined risk scores for these three genes:
where Exp is the expression level of each gene, and β is its regression coefficient obtained from the single-variate Cox model. The TCGA-LIHC patients were divided into high-risk and low-risk groups based on the median risk score.
For the mIF data, we counted stromal cells with positive expression of CTGF, COL4A2, and FSTL1 in tumors as F5-CAFs in HCC. Patients were divided into two groups according to the number of F5-CAFs, and the overall and disease-free survival rates in these two groups were compared using Kaplan‒Meier curves and log-rank tests in the R package survminer and survival (https://github.com/kassambara/survminer).
Spatial distribution of F5-CAFs
To examine how the F5-CAF score was influenced by the cancer cells in the tissue samples, we calculated the correlation of the F5-CAF score expression level and its distance to the tumor border.
Masks of the tumor border annotated by pathologists were created in ImageJ software. The mask covered all pixels considered to belong to this specific area. If p is the pixel, all pixels belonging to the mask will form a set Mt. Then, the coordinates of each spot were extracted from the ST data object using Scanpy (v.1.8.2) [52]. The distance from a spot s to the area t is represented by d(s,t), which is defined as the minimal Euclidean distance from the center of spot s to any pixel p from Mt.
Once distances were determined, for a F5-CAF score of spot s, a tuple (d(s,t), score) was formed. To demonstrate the dependency of the F5-CAF score on the distance to the tumor border, this set of distance-CAF score tuples was then visualized in downstream analysis. To better capture general trends in the data, we used locally weighted scatterplot smoothing (LOWESS) to generate smoothed estimates, which would serve as an approximation of a function f such that score = f(d(s,t)), to be interpreted as if the F5-CAF score value was a function of the distance to the tumor border (statsmodels v.0.31.1 in Python) [53].
Colocalization between F5-CAFs and other cell types
Spatial colocalization analysis was performed in the following two ways:
The NeighborRhood R package [54] was used to check whether the coordinates of the spots were significantly enriched in the same area. We input a neighborhood graph containing spot index pairs for adjacent spots and a data frame with spot index and spot phenotype as defined above. Colocalization scores between and within cell phenotypes were calculated for each spot with its neighbors. Then, colocalization scores were compared to a random distribution with shuffling labels using individual one-tailed permutation tests.
A new strategy for cell state colocalization:
Cell-state enrichment scores for each spot were calculated by UCell [55], singscore [56], and ssGSEA [51]. Module signatures for 9 cancer cell states and 25 immune cell states were obtained from previous studies [57, 58], representing the typical characteristics of different functional cell subtypes.
First, we compared cell-state scores among three locations: spots near CAFs, spots near fibroblasts, and distant spots (Wilcoxon rank-sum test). The test results were integrated by robust rank aggregation (RRA; https://github.com/chuiqin/irGSEA/), and the significantly different cell states are marked with asterisks in the heatmap.
Second, we defined “Niche intensity” s(i) for each CAF spot i to represent the intensity of a certain state that occurs around the CAF. The niche intensity s(i) for each CAF spot i was defined as the maximum cell-state score of phenotypes of interest (e.g., cancer, immune cells) observed among adjacent spots j ∈ Mi, where |Mi|⩽6.
The correlation of niche intensity with the F5-CAF score represents the relationship of a specific cell state to the characteristics of F5-CAFs, thereby reflecting the colocalization of F5-CAFs with specific functional cells.
Ligand‒receptor-mediated interactions between F5-CAFs and other cell types
CellphoneDB Statistical Analysis v.2.0 [59] was used to conduct receptor‒ligand analysis between different spot phenotypes as previously described [60]. We input the count matrix from ST data into CellphoneDB and performed the analysis separately for adjacent, interface, and tumor areas. F5-CAF-specific and top significant interactions compared to other fibroblasts were visualized.
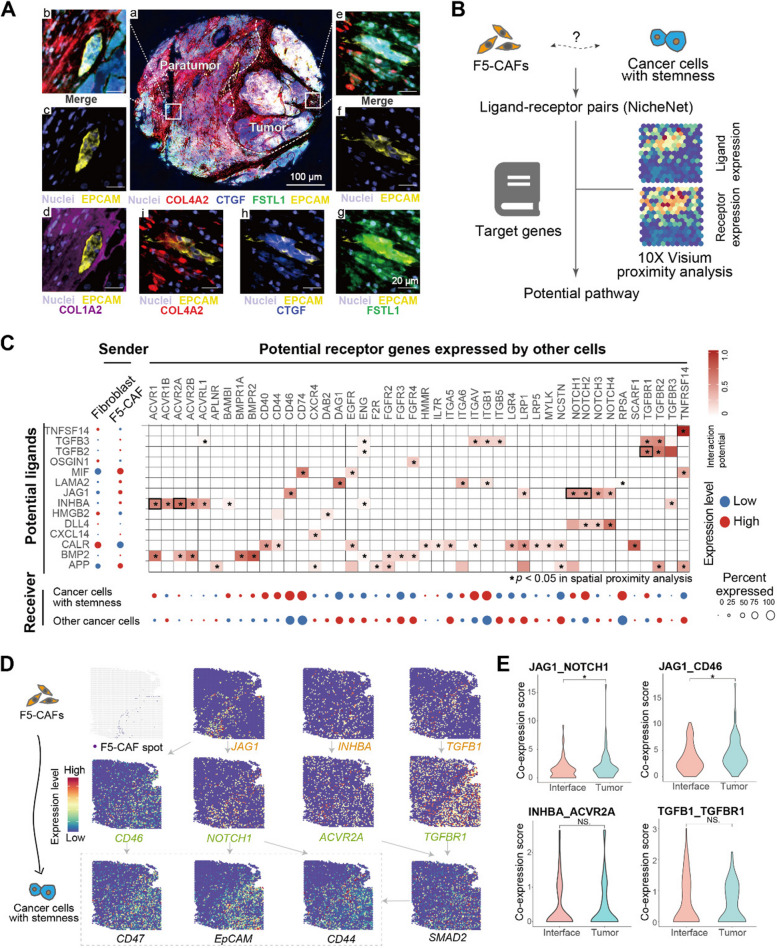
Ligands that regulate CSCs were predicted with NicheNet [61]. DEGs of F5-CAFs and their surrounding CSC spots were input to screen the ligands expressed in CAFs and the corresponding receptors expressed in CSCs as potential interactions. CSC spots were selected based on the median of the stemness scores of cancer cell spots around the F5-CAFs. A total of 65 spots with three scoring algorithms exceeding the threshold were identified as CSC spots (1.48% of all cancer cell spots, consistent with the expected proportion of CSCs). Ligand activity ranking was performed according to the previously obtained differentially expressed gene sets. The top 40 ligands were used to obtain downstream activated target genes.
Next, we analyzed the spatial proximity between ligands and receptors [62]. The average of all ligand‒receptor pairs on each slide was first calculated by averaging the ligand and receptor expression between each F5-CAF spot and its six nearest neighbors and then taking the average of the 6 spots. This calculation for each ligand‒receptor pair was then performed on 1000 randomized permutations of spot identities while preserving the total number of spots per replicate to generate a null distribution for each patient. The p value was calculated by the number of randomized permutations that exceeded the true average.
Isolation of F5-CAFs and related in vitro experiments
Isolation and cell culture of CAFs
CAFs were isolated from patient tumor tissues, which were stored in 5 mL of MACS Tissue Storage Solution (Miltenyi) and transferred to the laboratory at low temperature. Tumor tissues were washed three times in phosphate buffered-saline (PBS) supplemented with 3% penicillin and streptomycin. Next, the tissues were excised into approximately 1 mm3 pieces and then digested with 0.1% collagenase/dispase (Roche), 0.01% hyaluronidase (Yuanye), and 0.002% deoxyribonuclease (DNase) I (Roche) at 37 °C for 1.5 h until the single cells were approximately 80% confluent. After enzyme digestion, the tissue suspension was filtered through a 70 μm cell screen to remove undigested tissues.
The cell filtrate was collected into a new tube and centrifuged at 1000 rpm for 5 min. The supernatant was discarded, and the cell pellets were resuspended in DMEM/F-12 (Corning) supplemented with 20% fetal bovine serum (FBS) and 1% penicillin and streptomycin, plated in 10 cm culture dishes, and maintained at 37 °C and 5% CO2. When cells were attached for 48 h, they were washed with PBS to remove nonadherent cells, and half of the medium was replaced with fresh DMEM/F-12 supplemented with 10% FBS and 1% penicillin and streptomycin. The process was replaced every 3 days until the cells reached 80 ~ 90% confluence and expanded to passage. The first to fifth passages of CAFs were used in these experiments.
Cell culture
The human HCC cell lines Huh7 and Hep3B, the human hepatic stellate cell line LX-2, and the human embryonic kidney cell line HEK293T were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin and maintained at 37 °C and 5% CO2.
Immune cell fluorescence
CAFs, LX2, and HEK293T cells were cultured in 96-well cell culture plates until they were attached and fixed with Immunol Staining Fix Solution (Beyotime) for 10 min, washed three times with PBS, permeabilized with Enhanced Immunostaining Permeabilization Solution (Beyotime) for 10 min, and blocked with QuickBlock Blocking Buffer for Immunol Staining (Beyotime) for 10 min. Then, the cells were incubated with primary antibodies (see below) overnight at 4 °C. Subsequently, the cells were incubated with anti-rabbit IgG (H + L), F(ab')2 Fragment (Alexa Fluor 555 Conjugate, CST) or anti-mouse IgG (H + L) Cross-Adsorbed Secondary Antibody (Alexa Fluor 488, Invitrogen). DAPI was used to stain the nuclei. The F5-CAF ratio in CAFs was determined using a Four Color Multiplex Fluorescent Immunostaining Kit following the manufacturer’s instructions (Absin Bioscience). Images were acquired and analyzed with a LionheartTM FX automated Live Cell Imager (BioTek, USA). The primary antibodies used were alpha-smooth muscle actin (α-SMA) (mouse monoclonal, Boster, BM0002, 1:50), fibronectin (FN) (CST, 26,836, 1:200), COL1A2 (Abcam, ab96723, 1:500), COL4A2 (Abcam, ab125208, 1:1200), CTGF (Abcam, ab5097, 1:700), and FSTL1 (Abcam, ab71548, 1:700).
Supernatant collection of CAFs
CAFs and LX2 cells were seeded in 10 cm dishes in 10 ml of DMEM supplemented with 10% FBS and 1% penicillin and streptomycin until 80 ~ 90% confluence. Thereafter, fresh medium was added, and the conditioned supernatant was collected 48 h later and centrifuged at 2000 rpm for 5 min to remove nonadherent cells and cell debris. All supernatants were frozen at − 80 °C until further use.
Cell proliferation assay
Huh7 and Hep3B cells were seeded at a density of 200 cells/well in 96-well plates. The experimental groups and control group cells were cultured in 200 μL of the aforementioned supernatants and DMEM supplemented with 10% FBS. Then, cell viability was measured by a Cell Counting Kit-8 (CCK-8) (NCM) at 6, 24, 72, and 120 h, and the absorbance of the cells at 450 nm wavelength was detected using the Agilent Synergy H1 Microplate Reader (BioTek, USA).
Coculture experiments and interference assay
Direct and indirect coculture methods were used for the coculture experiments in this study. In brief, CAFs and LX2 cells were mitotically inactivated in DMEM supplemented with 8 μg/mL mitomycin C (Rhawn) at 37 °C with 5% CO2. The cells were incubated for 2 h and washed three times with PBS. Then, they were used as feeder layers for coculture. Subsequently, direct coculture was performed by seeding feeder layer cells (700/well) first and then Huh7 (500/well) or Hep3B (50/well) cells in the upper chamber of 24-well plates. Indirect coculture was performed by seeding Huh7 (500/well) and Hep3B (50/well) cells in the lower chamber and feeder layer cells (700/well) in the upper chamber in a 24-well Transwell apparatus with a 0.4 µm pore size (Corning). Huh7 and Hep3B cells were cultured in a medium supplemented with 10% FBS and 5% calf serum, respectively, for further analysis.
The feeder layers of CAF5 and CAF9 cells were used for the interference assay. Based on the aforementioned coculture experiments, two neutralizing antibodies against discoidin domain receptor 1 (DDR1) (CST, 5583) and COL4A2 (Abcam, ab125208) were used. The antibodies were used at a dilution of 1:2000.
Colony formation assay
Cocultured cells were grown for approximately 10 days, and colonies were fixed with 4% paraformaldehyde and stained with crystal violet for assessment. Colonies containing > 50 cells were counted under a microscope.
Quantitative real-time polymerase chain reaction (RT‒qPCR)
Total RNA was extracted from cultured Huh7 or Hep3B cells using RNAiso Plus (Takara), and the RNA concentration (ng/mL) and purity were measured using a Nanodrop 2000 spectrophotometer (Thermo Scientific, USA). RNAs with OD260/OD280 ratios ranging from 1.8 to 2.1 were used in the following experiments. The FastKing RT Kit (with gDNase) (Tiangen) was used to reverse transcribe the total RNA. RT‒qPCR was performed using FastFire qPCR PreMix (SYBR Green) (Tiangen) according to the manufacturer’s instructions. Glyceraldehyde-3 phosphate dehydrogenase (GAPDH) was used as the reference gene to normalize the mRNA levels. Data were collected with a LightCycler 480 instrument (Roche), and the mRNA levels were analyzed as log2 of the fold difference. The primer sequences are listed in Additional file 1: Table S12.
Statistical analyses were performed using GraphPad Prism 8 software. One-way analysis of variance (ANOVA) and two-way ANOVA were used for multiple comparisons. The error bars in the experiments indicate the standard error of the mean or standard deviation for a minimum of each experiment in triplicate. P < 0.05 was considered statistically significant.
Results
Functional diversity and heterogeneity of the tumor stroma in liver cancer
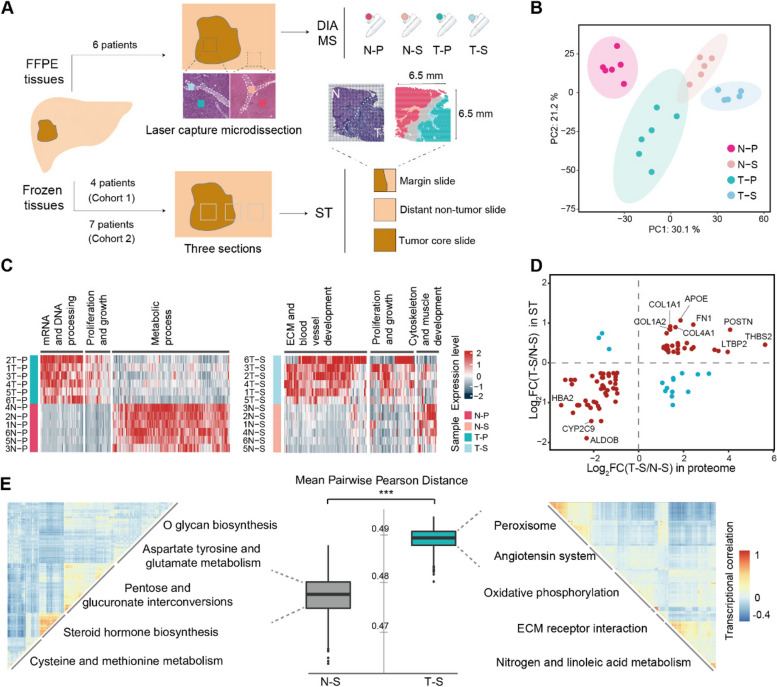
To explore microenvironmental alterations in liver cancer, we first applied MS-based proteomics to investigate global protein changes in the TME. The parenchymal and stromal tissues were extracted via LCM from tumors and corresponding non-tumor areas within rich stromal components which could provide sufficient material for proteomic analysis (Fig. 1A; Additional file 1: Table S1; Additional file 2: Fig. S1A). This process yielded 24 samples, and MS identified 5656 proteins (Additional file 2: Fig. S1B). PCA revealed substantial variation among the four sample groups (Fig. 1B). A total of 997 DEPs were identified between the tumor parenchyma (T-P) and non-tumor parenchyma (N–P) (Fig. 1C, left). The enriched GO terms corresponded to prior studies [63, 64], and were indicative of downregulated metabolism and upregulated proliferation/growth in T–P (Fig. 1C, left; Additional file 1: Table S2). A total of 312 DEPs distinguished the tumor stroma (T-S) from the non-tumor stroma (N‒S) (Fig. 1C, right). The upregulated proteins in T-S (COL11A1, POSTN, EFEMP2, COL4A1, CSPG2, FGL2, SRP14, TGFB1, STAT2, etc.) potentially mediated ECM remodeling, cell adhesion, stromal proliferation, cellular communication and signaling (Additional file 1: Table S3). Pathway enrichment of 312 DEPs revealed increased activity of endocytosis, morphogenesis, wound response, angiogenesis, and integrin pathways in T–S versus N‒S (Additional file 2: Fig. S1C). These results suggest that the stroma also undergoes important functional changes in liver cancer.
Fig. 1.
Functional diversity and heterogeneity of the tumor stroma in human liver cancer revealed by proteomic and ST analysis. A Tissue processing workflow for proteome and spatial transcriptome. ST, 10X Genomics Visium spatial transcriptomics; DIA-MS, data independent collection-mass spectrometry. B Principal component analysis of protein quantification of all samples. Background ellipses indicate 95% confidence intervals. C Differentially expressed proteins in parenchymal cells (left) and stromal cells (right) in different tissues and their corresponding functions. T–P, tumor parenchymal samples; N–P, non-tumor parenchymal samples; T–S, tumor stroma samples; N‒S, non-tumor stroma samples. D Comparison of the degree of differential gene regulation between T–S and N–S in proteomic and transcriptomic data. Red dots are genes with similar changes between the two datasets, and blue dots are genes with opposite changes. E The heterogeneity of the stromal spots in different areas. The boxplot indicates Pearson’s distance, and heatmaps indicate the similarity of the transcriptional profile, clustered by transcriptional correlation. ***, p < 0.001 by Wilcoxon rank sum test. See also Fig. S1
In addition to our previous 21 ST samples from 7 patients (cohort 2), 4 STs from an additional 4 patients were supplemented as cohort 1 (Fig. 1A; Additional file 1: Table S1; ref. [37]). To correspond with the proteome, the T–S and N–S regions for these samples were defined using the same criteria as above. The ST data were converted to pseudo-bulk transcriptomics data to compare the changes in the expression of proteins and mRNAs between T-S and N-S (see Methods). There were 96 genes whose protein and mRNA expression levels were significantly different between T–S and N–S. Most (81 genes) showed consistent up- or down-regulation in the two omics (Fig. 1D) and the fold changes in proteins and mRNAs were positively correlated (Spearman correlation test, r = 0.65, p < 0.0001). Many upregulated genes encode matrix molecules secreted by fibroblasts, which influence cancer in multiple ways (Additional file 1: Table S4; ref. [5]). For example, genes encoding fibril-forming collagens (COL1A1, COL1A2, and COL4A1) ranked among the top 5 upregulated mRNAs and the top 20 upregulated proteins. The fold change for another secreted ECM protein, POSTN, ranked 2nd highest in the proteome and top 5 in the transcriptome. These results suggest that many fibroblast-associated genes upregulated in the tumor stroma remain upregulated after post-transcriptional regulation and may importantly influence hepatocarcinogenesis.
To quantify transcriptomic heterogeneity within the stroma regions, we computed Pearson distances between expression profiles of paired spots (see the “Methods” section). Compared to N‒S, T–S exhibited significantly greater Pearson distances and greater heterogeneity (Wilcoxon rank sum test, p < 0.001; Fig. 1E). Clustering of T‒S spots by Pearson distance yielded more clusters than clustering of N–S spots, indicating more complex and diverse functions of the tumor stroma (Fig. 1E; Additional file 2: Fig. S1D).
Functional diversity of fibroblasts in liver cancer
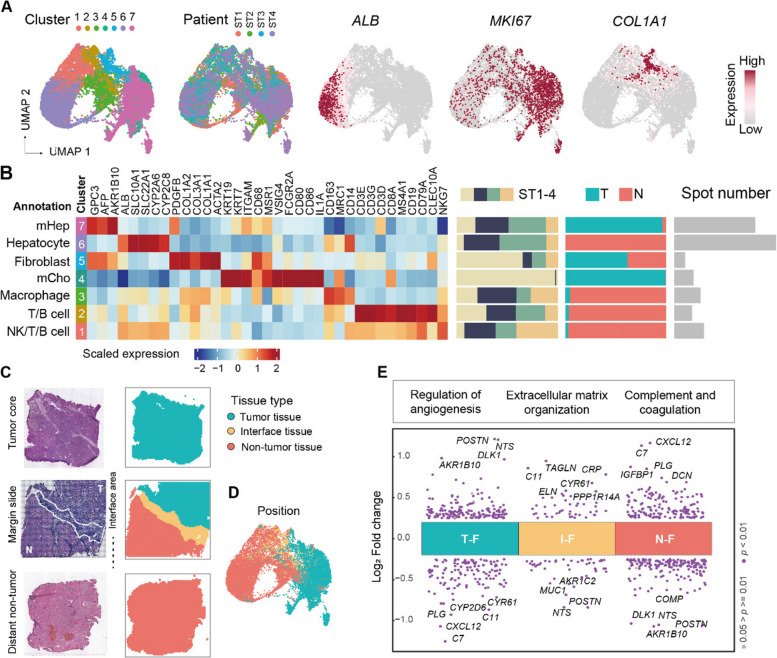
Next, we further investigated the cell composition of PLC using ST data. After integrating four samples from cohort 1 (Additional file 1: Table S1), unsupervised clustering assigned 17,381 spots to clusters, and highly similar clusters were merged, generating 7 robust clusters that were consistently resolved across resolutions (Fig. 2A; see the “Methods” sections). Considering that each spot may contain multiple cells, each cluster's major cell type was determined by (1) known cell type markers and (2) deconvolution estimating cell purity from scRNA-seq data (see the “Methods” section; Additional file 1: Table S5; refs. [44, 65]). The clusters had high expression values of marker genes and a high proportion of major cell types. (Fig. 2B; Additional file 2: Fig. S2A). Normal hepatocytes (ALB+) specifically mapped to non-tumor tissue; malignant cells were preferentially located in tumor areas, including malignant hepatocytes (GPC3+) enriched in HCC patients and malignant cholangiocytes (KRT7+ and KRT19+) enriched in ICC patients; and non-parenchymal cells including fibroblasts (COL1A1+), macrophages (CD68+), and a mixture of NKs, T cells and B cells (Fig. 2B). Having identified the major cell types described above, we next focused our analysis on fibroblasts, which comprise the most prominent stromal component. Cell types from cohort 1 were mapped to 21 ST samples from seven PLC patients in cohort 2 (Additional file 2: Fig. S2B, C; see the “Methods” section; ref. [37]). We totally obtained 6546 fibroblast-enriched spots, and after quality control, the 5803 spots with the highest percentage of fibroblasts were subjected to analysis.
Fig. 2.
Functional diversity and heterogeneity of fibroblasts revealed by ST analysis. A Uniform manifold approximation and projection (UMAP) of ST spots labeled by clusters (left), patients (median), and highlighted by three marker genes (right). B Left, average expression of highly variable genes in each cluster. Right, cluster composition, displayed by individual patient, tumor area, and spot number. mHep, malignant hepatocyte; mCho, malignant cholangiocyte; NK, natural killer; T, tumor; N, non-tumor. C Representative H&E-stained slides (left) and the corresponding spatial location of the spots (right). D UMAP of ST spots labeled by location. E Differential gene expression analysis showing up- and downregulated genes in different locations of fibroblast groups. The functional terms of some upregulated genes in each group are shown in the upper box. T–F, fibroblasts in the tumor area; I–F, fibroblasts in the interface area; N–F, fibroblasts in the non-tumor area. See also Fig. S2
To explore fibroblasts at different spatial locations, all 5803 fibroblast-enriched spots were assigned to three groups by spatial location (Fig. 2C, D): 3160 localized within tumor tissues (T–Fs), 1478 localized in non-tumor tissues (N–Fs) and 1165 localized in interface areas (I–Fs). The expression profiles of all fibroblast-enriched spots were subjected to batch effect correction, and the gene expression in each fibroblast group was compared to that in the other groups. The top differentially expressed genes and pathways are shown in Fig. 2E (Additional file 1: Table S6). T–Fs exhibited elevated expression of POSTN, NTS (participating in metabolism and immune regulation), and DLK1 (a regulator of cell growth) and upregulated angiogenesis- and metabolism-related pathways. N–Fs were enriched for functions such as complement activation and small molecule biosynthetic processes. I–Fs displayed upregulated expression of TAGLN (involved in smooth muscle contraction) and CRP (a biomarker for early inflammation and infection) and were involved in smooth muscle contraction and ECM organization. These findings demonstrate considerable functional divergence among fibroblasts within the liver TME, suggesting significant functional alterations in T–Fs and I–Fs that markedly differ from those in N–Fs.
Identification of a CAF subpopulation with distinct transcriptional profiles
To further investigate the special characteristics of fibroblasts surrounding tumor cells, we explored whether there was a fibroblast subtype specifically enriched in tumor tissues. Re-clustering split 5803 fibroblast-enriched spots into seven subpopulations (F1 ~ F7; Fig. 3A, left; see the “Methods” section). Excluding the minimal cluster F7, the remaining six clusters appeared in most patients. The counted spatial distribution revealed that F1, F2, and F6 were abundant in the tumor/non-tumor areas; F4 was significantly enriched in the non-tumor area; and F3 and F5 were significantly enriched in the tumor/interface area compared to the non-tumor area (Fisher test, p < 0.001; Fig. 3B; Additional file 2: Fig. S3A). The cell trajectory analysis showed that F5 was located terminally with weak differentiation potential (Fig. 3A, right).
Considering that each spot with 55 µm resolution may be a mixture of fibroblasts and a few other types of cells, we then used publicly available scRNA-seq data from another 14 PLC patients to resolve the fibroblast clusters identified by ST (Additional file 2: Fig. S3B; see Methods; ref. [44]). After quality control, 1800 fibroblast cells in the scRNA-seq data were preserved and similarly assigned to six fibroblast subpopulations (Fig. 3C). The numbers of four subpopulations (TAF1–TAF4) were greater in tumor tissues than in non-tumor tissues, while another two subpopulations (NAF1 and NAF2) were more abundant in non-tumor tissues (Additional file 2: Fig. S3C). Fibroblast subpopulations identified by the ST and scRNA-seq datasets were compared using the z score [66] and AUCell score [47]. Analysis initially revealed a similarity between our F5 and TAF4 (Fig. 3D), while F3 was poorly matched with any scRNA-seq cluster (Additional file 2: Fig. S3D, E). Next, a comparison of marker genes and functional characterization suggested closer relatedness between F5 and TAF4. In detail, some collagen genes highly expressed in the tumor stroma proteomic data (see above), namely, COL1A1, COL1A2, COL3A1, and COL4A2, were also highly expressed in TAF4 (Additional file 2: Fig. S3F; Additional file 1: Table S7) and F5 (Fig. 3E; Additional file 1: Table S8). The highly expressed genes of F5 could be enriched for ECM, structural organization, and focal adhesion-related functions (Fig. 3F; refs. [8, 67]), and had a higher myCAF score compared with fibroblast subtypes published in the literature (Additional file 2: Fig. S3G) [12, 19, 68]. However, despite exhibiting elevated CTGF, CYR61, ACTA2, SERPINE1, and COL4A1 expression, F5 also expressed FSTL1 (Additional file 2: Fig. S3H), a marker gene reported as iCAF. Therefore, we believe that it is not completely similar to the recognized myCAF phenotype [4, 68]. We defined the top 5 marker genes for F5 shared with TAF4: COL1A2, COL4A1, COL4A2, CTGF, and FSTL1 (Fig. 3G; Additional file 2: Fig. S3I). When these genes were co-expressed, the unique cells in the scRNA-seq data confirmed that these cells were a single-cell subpopulation (Fig. 3H). Based on these multiple validation methods, we concluded that our F5-CAF corresponded to the single-cell subtype TAF4.
To capture the spatial locations of all F5-CAFs, including those in non-fibroblast-enriched spots, we used a continuous ssGSEA-based F5-CAF score (defined by the expression of COL1A2, COL4A1, COL4A2, CTGF, and FSTL1, see the “Methods” section) instead of discrete cell types. We then performed LOWESS regression to evaluate the association between the F5-CAF score and the distance to the tumor interface. We found that F5-CAF scores were highest in the interface region and decreased further in both tumor regions and adjacent non-tumor regions (Fig. 3I; Additional file 2: Fig. S3J).
F5-CAFs were associated with an unfavorable prognosis in HCC patients
Since HCC is the main type of PLC, we next characterized F5-CAFs mainly in HCC. To validate the existence of the F5-CAF subpopulation in liver cancer at the single-cell level, we performed mIF staining of liver cancer tissues from 92 HCC patients to analyze F5-CAFs (Fig. 4A; Additional file 1: Table S9; see the “Methods” section). The combinations of different marker genes (COL1A2, COL4A1, COL4A2, CTGF, and FSTL1) were used as inputs for random forest models to distinguish F5-CAFs from other fibroblasts (see Methods). The combination of CTGF, COL4A2, and FSTL1 had the lowest number of genes required to accurately identify F5-CAFs (AUC = 0.836; Fig. 4B). Therefore, these three genes and COLIA2, a general marker gene for fibroblasts, were subjected to mIF staining. To avoid interference with gene expression in parenchymal cells, we used CTGF+FSTL1+COL4A2+ cells in the stroma to represent F5-CAFs (Fig. 4C; Additional file 2: Fig. S4A, B; see Methods). To explore the relationship between F5-CAFs and the prognosis of liver cancer patients, we first utilized transcriptional and clinical data from 371 liver patients in TCGA dataset. The risk score was defined for each tumor sample according to the expression levels of F5-CAF markers and was shown to have a significant effect on prognosis (Fig. 4D; see Methods). Then, we counted the number of F5-CAFs in the mIF tissues, with proportions ranging from 0 to 67.6% of all stromal cells in the tumor (Additional file 1: Table S10), and the number of F5-CAFs present in the interface region was proportional to the number in the tumor (Additional file 2: Fig. S4C). Interestingly, the density of F5-CAFs in the interface region was significantly greater than that in the tumor region (Additional file 2: Fig. S4D), consistent with the F5-CAF score results (Fig. 3I) and previous results showing that human HCCs contain fewer fibroblasts than surrounding tissue [68]. We found that abundant F5-CAFs (> 82 cells/mm2) in the tumor stroma were associated with unfavorable survival (Fig. 4E). Similarly, patients with more F5-CAFs (> 22 cells/mm2) in the interface region had shorter survival times (Fig. 4F), while other COL1A2+ fibroblast populations in the stroma region did not reach statistically significant levels in survival analysis (Additional file 2: Fig. S4E, F). Intriguingly, we also observed that the abundance of F5-CAFs in interface and tumor regions was positively correlated with the expression of programmed cell death ligand 1 (PD-L1), which is a ligand of programmed cell death protein 1 (PD-1) and an important immunosuppressive regulator in the TME (Additional file 2: Fig. S4G; Additional file 1: Table S10). Taken together, mIF confirmed the existence of F5-CAFs, and their abundance was associated with a worse prognosis. Given the higher F5-CAF abundance in the interface regions, our results also suggest the potential importance of the interface, which may provide a suitable TME for the growth and invasion of tumor cells.
Fig. 4.
F5-CAFs in HCC patients were associated with an unfavorable prognosis. A The experimental workflow of multiplexed immunofluorescence (mIF) staining of a hepatocellular carcinoma (HCC) tissue microarray (TMA). B Receiver operating characteristic curves of gene combinations predicted by the random forest model. AUC, area under the curve. C Representative composite image of the tumor core tissue by using mIF staining (COL1A2, yellow; COL4A2, red; CTGF, blue; FSTL1, green; and DAPI, dark blue). a, merged image; b, an enlarged subsection of the core highlighted in (a), colored by DAPI nuclear marker with arrows indicating F5-CAF; c, an annotated drawing of the location of parenchymal and stromal tissues; d–g, showing each of the individual markers, together with the DAPI nuclear marker and the autofluorescence signal (pseudocolored black). The spindle cells indicated by the white arrows are fibroblasts that are positive for all five markers. D Overall survival of patients in the TCGA cohort based on the risk score, defined by the expression of F5-CAF markers, stratified by the median value. E Overall survival analysis of HCC patients with a high or low number of F5-CAFs in the tumor stroma, stratified by the best cutoff value. F Overall survival analysis of HCC patients with a high or low number of F5-CAFs in the interface stroma, stratified by the best cutoff value. See also Fig. S4
F5-CAFs are located in niches associated with cancer stemness in HCC
We characterized the cellular neighborhoods surrounding F5-CAFs (termed niches; see Methods) in HCC ST samples. Cell–cell colocalization analysis revealed different fibroblast clusters with different spatial colocalization patterns, with more immune cells, fibroblasts, and cancer cells in the F5-CAF microenvironment than expected by chance (Additional file 2: Fig. S5A, B). In addition, a large number of interactions were also present at the tumor interface, with 3111, 4791, and 3929 significant ligand-receptor pairs (L–R pairs) between cell types in the non-tumor, interface, and tumor areas, respectively (Additional file 2: Fig. S5C). Considering the number of L-R pairs as interaction strength, F5-CAF within the tumor area interacted more strongly with cancer cells than other fibroblasts within the tumor area (Fig. 5A), suggesting greater potential function. They were associated with processes including ECM remodeling (COL1A2: a1b1/a2b1 complex; COL4A2: a1b1/a2b1 complex) and stemness modules (HGF: CD44; FZD7: WNT3), similar phenomena were also seen in scRNA-seq data (Additional file 2: Fig. S5D).
Although cell types surrounding F5-CAF niches can be identified, analyzing fine-grained cell states is challenging due to the limited resolution of 10X Genomics Visium. To address this, we first calculated a continuous module score for each cell state instead of binary labels. We then designed two strategies to analyze colocalization between F5-CAFs and fine-grained cell states (Fig. 5B; see the “Methods” section). First, module scores were compared among spots at different locations: those near F5-CAFs, those near other fibroblasts, and distant spots with no fibroblasts. Second, the maximum module score of spots in the F5-CAF neighborhood was defined as a niche intensity index, and its correlation with the F5-CAF score was subsequently calculated (see the “Methods” section). We applied these strategies to study fine-grained cancer cell states surrounding F5-CAF spots in tumor and interface regions.
Previous studies have reported the immune-excluded TME characterized by macrophages and CAFs [38, 69, 70], and we verified this phenomenon on our data. For 8 HCC patients, F5-CAFs had a higher M2 macrophage score around them both in the tumor and at the interface (Fig. 5C). In scRNA-seq data, we confirmed that more interactions between F5-CAF and M2 macrophages were detected compared with other fibroblasts, included CXCL12: CXCR4; LGALS9: COLEC12/ HAVCR2; ICAM1: aMb2 complex/ aXb2 complex (Additional file 2: Fig. S5E). In the above mIF HCC samples, many F5-CAFs co-localized with CD14+CD206+ M2 macrophages in the interface (Additional file 2: Fig. S5F), which may be related to the expression trend of PD-L1 that we previously observed (Additional file 2: Fig. S4G). For the non-tumor area, we considered the impact of fibrotic background and explored the relationship in fibrotic and non-fibrotic non-tumor areas respectively. The results were not significant (Additional file 2: Fig. S5G). In addition, in our three ICC and CHC patients, we also conducted extended exploration and found obvious co-localization only in the non-tumor areas with fibrosis (Additional file 2: Fig. S5H). Taken together, these results suggest that F5-CAFs may be associated with M2 macrophages and the immunosuppressive TME in HCC patients.
To calculate module scores of 9 published cancer cell states (Additional file 1: Table S11; ref [58].), we defined scores for each malignant spot. Spots around F5-CAFs had stronger stemness module scores than those near other fibroblasts, particularly in tumors (Fig. 5D, 5E; Additional file 2: Fig. S6A). Spots around F5-CAFs expressed higher levels of stemness-related genes in tumors, such as CD44, CD47, KRT19, and EPCAM. (Fig. 5F; Additional file 2: Fig. S6B). We confirmed the correlation between F5-CAF scores and stemness module scores in our cohort (Fig. 5G) and TCGA data (Fig. 5H). Staining for EpCAM, which is generally used to label liver stem cells [71, 72] or liver CSCs [73–75], revealed that EpCAM-positive cells in the tumor stroma were spatially adjacent to F5-CAFs (Fig. 6A), and the number of F5-CAFs was correlated with that of EpCAM-positive cells (Additional file 2: Fig. S6C). In three patients with ICC and CHC, we did not observe differences in the stemness microenvironment of F5-CAF and other fibroblasts, although this requires validation with a larger cohort (Additional file 2: Fig. S6D).
Fig. 6.
Cellular crosstalk between F5-CAFs and HCC CSCs. A Representative composite mIF image of the HCC interface area (EPCAM, yellow; COL4A2, red; CTGF, blue; FSTL1, green; COL1A2, purple and DAPI, weak purple). a, merged image. The dashed curve indicates the boundary of the tumor tissue. b–i, enlarged subsection highlighted in the non-tumor area (b–d) or in the tumor area (e–i) as in (a), showing five merged markers (b, e) or the individual marker(s) in the composite image after spectral unmixing, together with the nuclear marker DAPI (pseudocolored purple) and the autofluorescence signal (pseudocolored black). B Schematic of F5-CAF ligand‒receptor (L–R) analysis in the TME. C Left, average expression of the top ligands predicted by NicheNet across F5-CAFs and other fibroblast subpopulations modulating cancer cells. Middle, heatmap of significant L-R pairs in ST. The black box marks the pathway shown in D. Bottom, average expression of ligand-matched receptors expressed by malignant spots. D Spatial feature plots (ST2 sample) of select ligands expressed by F5-CAFs and cognate receptor expression by cancer cells with stemness. E L–R co-expression scores at different locations. *, p < 0.05 and NS, p > 0.05 by Wilcoxon rank-sum test. See also Fig. S6
To characterize signaling between F5-CAFs and CSCs, we selected malignant spots with higher stemness modules as candidate CSCs and investigated signaling between them and F5-CAFs. We hypothesized that ligands secreted by F5-CAFs interact with CSCs to modulate stemness. Using NicheNet (Fig. 6B), we identified the top predicted L-R pairs with the greatest potential for interaction (heatmap in Fig. 6C). By prioritizing pairs with co-localized expression via spatial proximity analysis (asterisks in Fig. 6C heatmap; see Methods), we identified key stemness pathways, including the NOTCH and TGFβ pathways (black box labeled in Fig. 6C; Additional file 2: Fig. S6E), in which CSC-related genes were expressed (Fig. 6D). Previous studies have reported that Notch ligands expressed by TME cells can promote Notch signaling in tumor cells [76, 77]. Furthermore, we found that the Notch ligand Jag1, which is highly expressed by F5-CAF, binds to Notch receptors on adjacent CSCs, and this co-expression was more pronounced within tumors than at the tumor interface (Fig. 6E; Additional file 2: Fig. S6F). In addition, secretory TGFB and INHBA ligands from F5-CAFs could also interact with TGFBR1 and ACVR2A on CSCs throughout tumors and interface regions (Fig. 6E; Additional file 2: Fig. S6F). The upregulated expression of downstream SMAD2 and SMAD3 can be detected in CSCs near F5-CAF (Additional file 2: Fig. S6G), suggesting that the TGFB1/INHBA-SMAD2/3 signaling axis is involved in the enhancement of stem cell-like properties and the regulation of ECM protein deposition [78]. This phenomenon has been reported in macrophages in pancreatic cancer [79]. Taken together, these results suggest that F5-CAFs maintain liver CSC stemness through multiple pathways.
Patient-derived F5-CAFs promote the in vitro proliferation and stemness of liver cancer cells
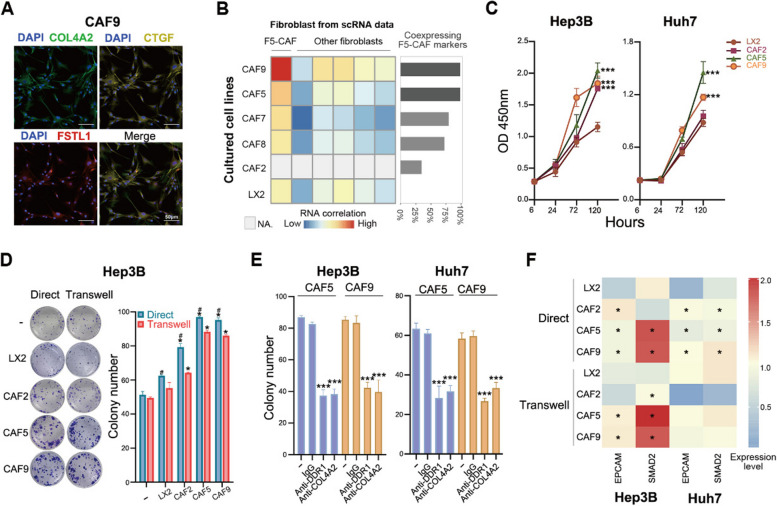
We established five primary CAF lines from resected tumor tissues of five PLC patients (three HCCs and two ICCs; Additional file 1: Table S1). These cells were well attached to the culture plate and exhibited triangular, spindle, star, or spindle shapes with typical fibroblast morphology. All of these cells expressed α smooth muscle actin (α-SMA), fibronectin (FN1), and COL1A2 (Additional file 2: Fig. S7A) and did not express E-cadherin or CD31 (data not shown). Among the five CAF lines, nearly all CAF5 (derived from ICC tissues) and CAF9 (derived from HCC tissues) cells (> 99%) co-expressed CTGF, COL4A2, and FSTL1 (Fig. 7A; Additional file 2: Fig. S7A), the markers of F5-CAFs, consistent with the above results that F5-CAFs exist in both HCC and ICC (Fig. 3B). In contrast, only 35%, 80%, and 73% of cells in CAF2, CAF7 and CAF8, respectively, co-expressed these markers, indicating a mixed cellular composition in these three CAF lines. Concordantly, RNA-seq analysis revealed that the transcriptional profiles of CAF5 and CAF9 more closely resembled that of TAF4 (i.e., F5-CAFs) among the scRNA-seq datasets (Figs. 7B and 3H). Accordingly, these two lines were used as representative F5-CAFs for further experiments.
Fig. 7.
Patient-derived F5-CAFs promote the in vitro proliferation and stemness of liver cancer cells. A Co-expression of the F5-CAF markers (CTGF, COL4A2, and FSTL1) in cultured primary CAF9 cells in mIF staining data. A representative microscopic field of view is shown. B Relationships between cultured primary CAF cells and F5-CAF subtypes in Fig. 3. Correlation of RNA profiles (left) and percentage of cells on mIF data for simultaneous expression of F5-CAF markers as in A (right). Five primary CAF lines were from resected tumor tissues; LX2, human hepatic stellate cell. CAF2 was not included due to insufficient starting cell material for RNA-seq. C CCK-8 experiments showing the effect of the conditioned culture supernatant of CAFs on the growth of liver cancer cells. Note that on day 5, some tumor cells cocultured with CAF9 cells died due to overgrowth. ***, p < 0.001 by ANOVA, compared with LX2. D Colony-formation ability of Hep3B liver cancer cells when cocultured with CAFs directly or in transwells. A representative image of each condition is shown on the left. * and #, p < 0.05, compared with LX2 or Transwell, respectively. E Colony-formation ability of liver cancer cells cocultured directly with CAFs. Culture medium was added without ( −) or with the indicated neutralizing antibody. ***, p < 0.001 by ANOVA, compared with IgG. F Heatmap showing the expression levels of two stemness genes in liver cancer cells after coculture with the indicated CAFs or LX2 cells. *, higher expression level compared with LX2 and p < 0.05. See also Fig. S7
To investigate whether F5-CAFs promote the growth of liver cancer cells, we first cocultured the CAF-conditioned culture supernatant with two types of liver cancer cells in vitro. The conditioned culture supernatant of CAF5 or CAF9 did exhibit a more pronounced effect on stimulating the in vitro growth of liver cancer cells, while the nonspecific CAF cell line CAF2 (exhibiting low CTGF+ and FSTL1+ expression) had a much weaker effect, as compared with the human hepatic stellate cell LX2 (Fig. 7C). Of note, although the CAF5 cells were derived from ICC tissues, they had effects similar to those of CAF9 on HCC cells, consistent with the notion that CAFs in HCC and ICC may have the same origin [20, 68]. Most importantly, compared with LX2 or CAF2 cells, CAF5 and CAF9 cells significantly enhanced the colony formation ability of liver cancer cells (Fig. 7D; Additional file 2: Fig. S7B). When CAF5 or CAF9 cells were directly co-cultured with Hep3B cells, their promoting effect was more pronounced (Fig. 7D). These results indicate that both paracrine and direct contact mechanisms underlie the promoting effect. A recent study showed that activated myofibroblast hepatic stellate cells (myHSCs) promote liver cancer growth by activating discoidin domain receptor 1 (DDR1) [68]. Indeed, when CAF5 or CAF9 cells were co-cultured with liver cancer cells, the addition of a neutralizing antibody against DDR1 or COL4A2, but not a control IgG antibody, inhibited the colony formation of liver cancer cells to some extent (Fig. 7E; Additional file 2: Fig. S7C), indicating that DDR1 may participate in the F5-CAF-mediated promoting effects on liver cancer cells. Finally, we detected the expression of stemness genes in cocultured liver cancer cells. Whether in contact or noncontact culture, CAF5 or CAF9 enhanced the expression of certain stemness genes (Fig. 7F; Additional file 2: Fig. S7D), including EPCAM, CD133, and SMAD2, as mentioned earlier (Fig. 5F; Additional file 2: Fig.S6G). Taken together, these results indicate that F5-CAFs promote the in vitro proliferation and stemness of liver cancer cells.
Discussion
By using spatial multi-omics analysis, we have for the first time comprehensively characterized the tumor stroma and CAFs in liver cancer TME. Our results showed that the tumor stroma of liver cancer was complex and heterogeneous at the proteomic and transcriptional levels, possibly due to the functional diversity and heterogeneity of CAFs in the tumor stroma. More importantly, we identified a CAF subpopulation F5-CAFs in liver cancer that preferentially located within and around tumor nests, marked by the expression of COL1A2, COL4A1, COL4A2, CTGF, and FSTL1, associated with unfavorable prognosis, and colocalization with EpCAM+ CSCs. Thus, our results provide detailed molecular information to explain how CAFs in the TME or the “soil” directly interact with and support the survival of CSCs or the “seeds” of liver cancer.
By comparing F5-CAFs with major CAF subsets reported in previous studies, we found that F5-CAFs express several marker genes identified in myCAFs, including COL1A2 and COL4A1 in human colorectal cancer or lung cancer [4, 80, 81], and CTGF in mouse breast cancer [82]. However, F5-CAFs also highly express FSTL1, which is also expressed in iCAFs in human PDAC [12], and the other marker gene, COL4A2, has not been reported as a marker gene in any CAF subset. Thus, F5-CAFs in liver cancer are similar to but not exactly the same as myCAFs.
The tumor-promoting activity of CAFs or CAF subsets has mainly been investigated in previous studies by single-cell techniques utilizing mouse model systems or in vitro cell culture systems [7]. CAFs are the major producers of ECM components and various other secreted factors, which may induce interactions between CAFs and cancer cells or other cells that promote tumor growth and progression [7]. In liver cancer, CAFs can be isolated from fresh HCC clinical samples and cultured in vitro. Cultured CAFs can secrete HGF [83] or CLCF1 [21], which can regulate liver tumor-initiating cells or enhance the self-renewal ability of HCC cells in cell culture and promote the progression of HCC in mouse models. CAFs could also express FSTL1, which is one of the F5-CAF markers identified in this study, and could promote HCC stemness and metastasis in a preclinical mouse model [84]. In addition, type I collagen, which is enriched in activated myHSCs, can promote proliferation and tumor development via increased stiffness and TAZ activation in peritumoral hepatocytes and via DDR1 activation in established tumors [68]. Taken together, these in vitro culture or animal model studies suggest that CAFs can promote the progression of liver cancer through a variety of mechanisms, consistent with our ST analysis of human liver cancer in the present study. Because ST preserves the native architecture within tissues, our ST and mIF data from clinical samples in this study provide spatial evidence that F5-CAFs interact directly with HCC CSCs to maintain cancer stemness. We also extended the spatial analysis to the ICC or CHC sample, but we acknowledge that the limited ICC/CHC sample size is a limitation of our study. Future studies with larger sample sizes are needed to further elucidate the TME underlying ICC/CHC and validate our findings. In addition, the influences from CAF also have certain heterogeneity in different cancer cell lines, which may be valuable to for future studies. Given the higher F5-CAF abundance in the interface regions, our results also suggest the potential importance of the interface, which may provide a suitable TME for the growth and invasion of tumor cells.
The identification of F5-CAFs in human liver cancer and the discovery of the interactions between F5-CAFs and their neighboring cells can not only help to elucidate the underlying mechanisms of liver cancer development but also make F5-CAFs potential therapeutic targets. Either F5-CAFs or their marker genes can be targets of liver cancer therapy. Indeed, in a preclinical mouse model, blockade of FSTL1 reduced the malignancy and metastasis of HCC, eradicated liver CSCs, and prolonged the survival period [84]. Another gene, CTGF, is a multifunctional signal regulator involved in promoting the occurrence and development of cancer. At present, CTGF has been employed in cancer research with a number of molecules targeting it [85]. As CSCs are endowed with multiple treatment resistance capabilities [86] and F5-CAFs are directly involved in maintaining liver cancer stemness, treatment targeting F5- CAFs may weaken the survival of liver CSCs, as indicated in our in vitro results (Fig. 7E; Additional file 2: Fig. S7C). Furthermore, our results showed that the density of F5-CAFs in the tumor stroma was positively correlated with the expression of PD-L1 in HCC. Therefore, combined treatment targeting both F5-CAFs and PD-L1 may have a synergistic antitumor effect, especially in patients with more F5-CAFs.
Conclusions
In this study, by using spatial multiomics analysis, we have comprehensively characterized the tumor stroma and CAFs in the TME of liver cancer. We identified a CAF subpopulation F5-CAF in HCC, which is marked by the expression of COL1A2, COL4A1, COL4A2, CTGF, and FSTL1, and is associated with cancer stemness and unfavorable prognosis. Our results provide potential mechanisms by which the CAF subset in the TME promotes the development of liver cancer by supporting the survival of CSCs.
Supplementary Information
Additional file 1: Table S1. Clinical and pathological characteristics of each patient for proteomics and 10X Visium analysis and in vitro CAF culture. Table S2. Significantly differentially expressed proteinsbetween tumor and non-tumoral parenchymal cells. Table S3. Significantly differentially expressed proteinsbetween tumor and non-tumor stroma. Table S4. Expressed extracellular matrixproteins detected by proteomics and 10X Visium in tumor and non-tumoral stroma. Table S5. Signature genes of different cell types. Table S6. Differentially expressed genes in fibroblasts of the tumor, non-tumor and interface area. Table S7. Differential expression genes of fibroblast clusters in scRNA-seq data. Table S8. Differential expression genesof fibroblast subtypes in 10X Visium. Table S9. Clinical and pathological characteristics of hepatocellular carcinomapatients for multiplexed immunofluorescenceanalysis. Table S10. The expression level of PD-L1 and CTLA4 and the number of cells with positive expression of the indicated genes in each tissue spot. Table S11. Signature genes of different cancer cell states. Table S12. Primer sequences for RT-qPCR.
Additional file 2: Fig S1. Proteomic landscape and transcriptomic heterogeneity in parenchymal and stromal regions. Fig S2. Cell type annotations in the two cohorts. Fig S3. Identification of fibroblasts enriched in the tumorfrom the scRNA-seq data and the integration of fibroblast features in ST. Fig S4. Overall survival analysis for HCC patients. Fig S5. Spatial distribution and interaction of F5-CAF and M2 macrophage. Fig S6. Interactions of F5-CAF and cancer cells with high stemness. Fig S7. Identification of CAFs and the tumor-promoting effect of F5-CAFs.
Acknowledgements
We thank Zhixuan Tang and Hong Li group for helpful discussions.
Abbreviations
- TME
Tumor microenvironment
- ECM
Extracellular matrix
- CSC
Cancer stem cell
- CAF
Cancer-associated fibroblast
- PLC
Primary liver cancer
- MS
Mass spectrometry
- ST
10X Genomics Visium spatial transcriptomics
Authors’ contributions
Y.H. and H.L. designed the research work. Y.H., H.L., H.W., and L.C. supervised the research work. S.J., Y.H., D.L., N.F., and H.L. wrote the manuscript. S.J., H.W., and D.L. performed the bioinformatics analyses with assistance from H.L., P.L., and H.W. D.L. and N.F. performed experiments with assistance from H.D. and M.Q. H.D. provided FFPE samples and performed pathological analysis. B. Y. and F. F. provided fresh liver cancer samples and clinical data.
Funding
This work was supported by the National Key R&D Program of China (2021YFF1200900 to H.L.), the National Natural Science Foundation of China (82372729 and 81972644 to Y.H., T2122018 and 32170680 to H.L., 91859205, 81830054 and 81988101 to H.W., U21A20376 to L.C.), the National Science and Technology Major Project of China (2018ZX10723204 to Y.H), CAS Youth Innovation Promotion Association (2018307 to H.L.), Shanghai Municipal Science and Technology Major Project (to H.L.), the National Science Foundation of Shanghai (21XD1404600, 21JC1406600, and 22140901000 to L.C.) and Key Project of Jiading District Health Construction Commission of Shanghai (2020-ZD-01 to F. F.), the Basic Medical Research Foundation of the Naval Medical University (2021MS15 and 2022yx005 to F. F.). H.L. was supported by the development fund for Shanghai talents.
the National Key R&D Program of China,2021YFF1200900,Hong Li,the National Natural Science Foundation of China,81972644,Yu-fei He,82372729,Yu-fei He,T2122018,Hong Li,32170680,Hong Li,91859205,Hong-yang Wang,81830054,Hong-yang Wang,81988101,Hong-yang Wang,U21A20376,Lei Chen,the National Science and Technology Major Project of China,2018ZX10723204,Yu-fei He,CAS Youth Innovation Promotion Association,2018307,Hong Li,the National Science Foundation of Shanghai,21XD1404600,Lei Chen,21JC1406600,Lei Chen,22140901000,Lei Chen,Key Project of Jiading District Health Construction Commission of Shanghai,2020-ZD-01,Feiling Feng,the Basic Medical Research Foundation of the Naval Medical University,2021MS15,Feiling Feng,2022yx005,Feiling Feng
Availability of data and materials
Spatial transcriptomics data are available at National Genomics Data Center (raw data: https://ngdc.cncb.ac.cn/gsa-human/browse/HRA000437 [87]; and processed data: https://ngdc.cncb.ac.cn/omix/release/OMIX006870) [88]. RNA-seq data for CAFs are accessible at https://ngdc.cncb.ac.cn/gsa-human/browse/HRA007936 [87]. The scRNA-seq data were obtained from the Gene Expression Omnibus at GSE156625 [44]. The TCGA-LIHC data analyzed in this study can be downloaded from cBioPortal (http://www.cbioportal.org/) [89]. The proteome data have been deposited to the ProteomeXchange Consortium (dataset identifier: PXD054095) via the iProX partner repository (https://www.iprox.cn/) under Project ID: IPX0009295000 [90]. In detail the results data and code can be directly obtained from our Github website: https://github.com/LihongLab/LiverCancer_ST_fibroblast [91]. All the data needed to evaluate the conclusions in the paper are presented in the paper and/or the Supplementary Information. All methods are described in the manuscript.
Declarations
Declarations
Ethics approval and consent to participate.
All patients provided informed consent. The use of clinical samples and all of the procedures were approved by the EHBH's Ethics Committee (No. EHBHKY2018-1–001). The study conformed to the principles of the Helsinki Declaration.
Consent for publication
All authors approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Si-yu Jing, Dan Liu, Na Feng and Hui Dong contributed equally to this work.
Contributor Information
Lei Chen, Email: chenlei@smmu.edu.cn.
Hong-yang Wang, Email: hywangk@vip.sina.com.
Hong Li, Email: lihong01@sinh.ac.cn.
Yu-fei He, Email: yfhe@sibcb.ac.cn.
References
- 1.Brassart-Pasco S, et al. Tumor Microenvironment: Extracellular Matrix Alterations Influence Tumor Progression. Front Oncol. 2020;10:397. 10.3389/fonc.2020.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lv B, et al. Immunotherapy: Reshape the Tumor Immune Microenvironment. Front Immunol. 2022;13: 844142. 10.3389/fimmu.2022.844142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaggianesi M, et al. Messing Up the Cancer Stem Cell Chemoresistance Mechanisms Supported by Tumor Microenvironment. Front Oncol. 2021;11: 702642. 10.3389/fonc.2021.702642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavie D, et al. Cancer-associated fibroblasts in the single-cell era. Nat Cancer. 2022;3(7):793–807. 10.1038/s43018-022-00411-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox TR. The matrix in cancer. Nat Rev Cancer. 2021;21(4):217–38. 10.1038/s41568-020-00329-7 [DOI] [PubMed] [Google Scholar]
- 6.Sahai E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20(3):174–86. 10.1038/s41568-019-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, McAndrews KM, Kalluri R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Rev Clin Oncol. 2021;18(12):792–804. 10.1038/s41571-021-00546-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartoschek M, et al. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat Commun. 2018;9(1):5150. 10.1038/s41467-018-07582-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kieffer Y, et al. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov. 2020;10(9):1330–51. 10.1158/2159-8290.CD-19-1384 [DOI] [PubMed] [Google Scholar]
- 10.Friedman G, et al. Cancer-associated fibroblast compositions change with breast cancer progression linking the ratio of S100A4(+) and PDPN(+) CAFs to clinical outcome. Nat Cancer. 2020;1(7):692–708. 10.1038/s43018-020-0082-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu SZ, et al. Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. Embo j. 2020;39(19): e104063. 10.15252/embj.2019104063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elyada E, et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019;9(8):1102–23. 10.1158/2159-8290.CD-19-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng J, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29(9):725–38. 10.1038/s41422-019-0195-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Single-cell analysis of pancreatic ductal adenocarcinoma identifies a novel fibroblast subtype associated with poor prognosis but better immunotherapy response. Cell Discov. 2021;7(1):36. 10.1038/s41421-021-00271-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, et al. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J Hepatol. 2020;73(5):1118–30. 10.1016/j.jhep.2020.05.039 [DOI] [PubMed] [Google Scholar]
- 16.Sung H, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 17.Llovet JM, et al. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat Cancer. 2022;3(4):386–401. 10.1038/s43018-022-00357-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang SS, et al. Perivenous Stellate Cells Are the Main Source of Myofibroblasts and Cancer-Associated Fibroblasts Formed After Chronic Liver Injuries. Hepatology. 2021;74(3):1578–94. 10.1002/hep.31848 [DOI] [PubMed] [Google Scholar]
- 19.Affo S, et al. Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell. 2021;39(6):866–882.e11. 10.1016/j.ccell.2021.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu GQ, et al. CD36(+) cancer-associated fibroblasts provide immunosuppressive microenvironment for hepatocellular carcinoma via secretion of macrophage migration inhibitory factor. Cell Discov. 2023;9(1):25. 10.1038/s41421-023-00529-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song M, et al. Cancer-Associated Fibroblast-Mediated Cellular Crosstalk Supports Hepatocellular Carcinoma Progression. Hepatology. 2021;73(5):1717–35. 10.1002/hep.31792 [DOI] [PubMed] [Google Scholar]
- 22.Ramachandran P, et al. Single-cell technologies in hepatology: new insights into liver biology and disease pathogenesis. Nat Rev Gastroenterol Hepatol. 2020;17(8):457–72. 10.1038/s41575-020-0304-x [DOI] [PubMed] [Google Scholar]
- 23.Ma L, et al. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell. 2019;36(4):418–430.e6. 10.1016/j.ccell.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim CJ, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut. 2019;68(5):916–27. 10.1136/gutjnl-2018-316510 [DOI] [PubMed] [Google Scholar]
- 25.Ho DW, et al. Single-cell transcriptomics reveals the landscape of intra-tumoral heterogeneity and stemness-related subpopulations in liver cancer. Cancer Lett. 2019;459:176–85. 10.1016/j.canlet.2019.06.002 [DOI] [PubMed] [Google Scholar]
- 26.Losic B, et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat Commun. 2020;11(1):291. 10.1038/s41467-019-14050-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song G, et al. Global immune characterization of HBV/HCV-related hepatocellular carcinoma identifies macrophage and T-cell subsets associated with disease progression. Cell Discov. 2020;6(1):90. 10.1038/s41421-020-00214-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho DW, et al. Single-cell RNA sequencing shows the immunosuppressive landscape and tumor heterogeneity of HBV-associated hepatocellular carcinoma. Nat Commun. 2021;12(1):3684. 10.1038/s41467-021-24010-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184(2):404–421.e16. 10.1016/j.cell.2020.11.041 [DOI] [PubMed] [Google Scholar]
- 30.Wang H, et al. Integrative single-cell transcriptome analysis reveals a subpopulation of fibroblasts associated with favorable prognosis of liver cancer patients. Transl Oncol. 2021;14(1): 100981. 10.1016/j.tranon.2020.100981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain S, et al. Single-cell RNA sequencing and spatial transcriptomics reveal cancer-associated fibroblasts in glioblastoma with protumoral effects. J Clin Invest. 2023;133(5):e147087. [DOI] [PMC free article] [PubMed]
- 32.Cords L, et al. Cancer-associated fibroblast classification in single-cell and spatial proteomics data. Nat Commun. 2023;14(1):4294. 10.1038/s41467-023-39762-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin P, et al. Cancer-associated fibroblasts undergoing neoadjuvant chemotherapy suppress rectal cancer revealed by single-cell and spatial transcriptomics. Cell Rep Med. 2023;4(10): 101231. 10.1016/j.xcrm.2023.101231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, et al. Spatial and Single-Cell Transcriptomics Reveal a Cancer-Associated Fibroblast Subset in HNSCC That Restricts Infiltration and Antitumor Activity of CD8+ T Cells. Cancer Res. 2024;84(2):258–75. 10.1158/0008-5472.CAN-23-1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, et al. Single-cell and spatial transcriptomics reveal POSTN(+) cancer-associated fibroblasts correlated with immune suppression and tumour progression in non-small cell lung cancer. Clin Transl Med. 2023;13(12): e1515. 10.1002/ctm2.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma C, et al. Pan-cancer spatially resolved single-cell analysis reveals the crosstalk between cancer-associated fibroblasts and tumor microenvironment. Mol Cancer. 2023;22(1):170. 10.1186/s12943-023-01876-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu R, et al. Comprehensive analysis of spatial architecture in primary liver cancer. Sci Adv. 2021;7(51):eabg3750. 10.1126/sciadv.abg3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J Hepatol. 2023;78(4):770–82. 10.1016/j.jhep.2023.01.011 [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell. 2016;166(3):755–65. 10.1016/j.cell.2016.05.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–26. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navarro JF, et al. ST Pipeline: an automated pipeline for spatial mapping of unique transcripts. Bioinformatics. 2017;33(16):2591–3. 10.1093/bioinformatics/btx211 [DOI] [PubMed] [Google Scholar]
- 42.Dittrich FL, Wilmot TJ. Threshold testing of vestibular function. J Laryngol Otol. 1965;79(10):888–92. 10.1017/S0022215100064513 [DOI] [PubMed] [Google Scholar]
- 43.Elosua-Bayes M, et al. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 2021;49(9): e50. 10.1093/nar/gkab043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma A, et al. Onco-fetal Reprogramming of Endothelial Cells Drives Immunosuppressive Macrophages in Hepatocellular Carcinoma. Cell. 2020;183(2):377–394.e21. 10.1016/j.cell.2020.08.040 [DOI] [PubMed] [Google Scholar]
- 45.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560(7719):494–8. 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergen V, et al. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38(12):1408–14. 10.1038/s41587-020-0591-3 [DOI] [PubMed] [Google Scholar]
- 47.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–6. 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pliner HA, Shendure J, Trapnell C. Supervised classification enables rapid annotation of cell atlases. Nat Methods. 2019;16(10):983–6. 10.1038/s41592-019-0535-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2001;2:18–22. [Google Scholar]
- 50.Robin X, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hildebrandt F, et al. Spatial Transcriptomics to define transcriptional patterns of zonation and structural components in the mouse liver. Nat Commun. 2021;12(1):7046. 10.1038/s41467-021-27354-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schapiro D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods. 2017;14(9):873–6. 10.1038/nmeth.4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andreatta M, Carmona SJ. UCell: Robust and scalable single-cell gene signature scoring. Comput Struct Biotechnol J. 2021;19:3796–8. 10.1016/j.csbj.2021.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foroutan M, et al. Single sample scoring of molecular phenotypes. BMC Bioinformatics. 2018;19(1):404. 10.1186/s12859-018-2435-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barkley D, et al. Cancer cell states recur across tumor types and form specific interactions with the tumor microenvironment. Nat Genet. 2022;54(8):1192–201. 10.1038/s41588-022-01141-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miao YR, et al. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv Sci (Weinh). 2020;7(7):1902880. 10.1002/advs.201902880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Efremova M, et al. Cell PhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484–506. 10.1038/s41596-020-0292-x [DOI] [PubMed] [Google Scholar]
- 60.Corridoni D, et al. Single-cell atlas of colonic CD8(+) T cells in ulcerative colitis. Nat Med. 2020;26(9):1480–90. 10.1038/s41591-020-1003-4 [DOI] [PubMed] [Google Scholar]
- 61.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17(2):159–62. 10.1038/s41592-019-0667-5 [DOI] [PubMed] [Google Scholar]
- 62.Ji AL, et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell. 2020;182(2):497–514.e22. 10.1016/j.cell.2020.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang Y, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567(7747):257–61. 10.1038/s41586-019-0987-8 [DOI] [PubMed] [Google Scholar]
- 64.Gao Q, et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell. 2019;179(2):561–577.e22. 10.1016/j.cell.2019.08.052 [DOI] [PubMed] [Google Scholar]
- 65.Bian S, et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018;362(6418):1060–3. 10.1126/science.aao3791 [DOI] [PubMed] [Google Scholar]
- 66.Shekhar K, Menon V. Identification of Cell Types from Single-Cell Transcriptomic Data. Methods Mol Biol. 2019;1935:45–77. 10.1007/978-1-4939-9057-3_4 [DOI] [PubMed] [Google Scholar]
- 67.Wu SZ, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53(9):1334–47. 10.1038/s41588-021-00911-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Filliol A, et al. Opposing roles of hepatic stellate cell subpopulations in hepatocarcinogenesis. Nature. 2022;610(7931):356–65. 10.1038/s41586-022-05289-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qi J, et al. Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat Commun. 2022;13(1):1742. 10.1038/s41467-022-29366-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sathe A, et al. Colorectal Cancer Metastases in the Liver Establish Immunosuppressive Spatial Networking between Tumor-Associated SPP1+ Macrophages and Fibroblasts. Clin Cancer Res. 2023;29(1):244–60. 10.1158/1078-0432.CCR-22-2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huch M, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160(1–2):299–312. 10.1016/j.cell.2014.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aizarani N, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572(7768):199–204. 10.1038/s41586-019-1373-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamashita T, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136(3):1012–24. 10.1053/j.gastro.2008.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamashita T, et al. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology. 2013;57(4):1484–97. 10.1002/hep.26168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matsumoto T, et al. Proliferating EpCAM-Positive Ductal Cells in the Inflamed Liver Give Rise to Hepatocellular Carcinoma. Cancer Res. 2017;77(22):6131–43. 10.1158/0008-5472.CAN-17-1800 [DOI] [PubMed] [Google Scholar]
- 76.Sonoshita M, et al. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. 2011;19(1):125–37. 10.1016/j.ccr.2010.11.008 [DOI] [PubMed] [Google Scholar]
- 77.Cao Z, et al. Angiocrine factors deployed by tumor vascular niche induce B cell lymphoma invasiveness and chemoresistance. Cancer Cell. 2014;25(3):350–65. 10.1016/j.ccr.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abdel Mouti M, Pauklin S. TGFB1/INHBA Homodimer/Nodal-SMAD2/3 Signaling Network: A Pivotal Molecular Target in PDAC Treatment. Mol Ther. 2021;29(3):920–36. 10.1016/j.ymthe.2021.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang B, et al. Macrophage-expressed CD51 promotes cancer stem cell properties via the TGF-β1/smad2/3 axis in pancreatic cancer. Cancer Lett. 2019;459:204–15. 10.1016/j.canlet.2019.06.005 [DOI] [PubMed] [Google Scholar]
- 80.Li H, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49(5):708–18. 10.1038/ng.3818 [DOI] [PubMed] [Google Scholar]
- 81.Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24(8):1277–89. 10.1038/s41591-018-0096-5 [DOI] [PubMed] [Google Scholar]
- 82.Sebastian A, et al. Single-Cell Transcriptomic Analysis of Tumor-Derived Fibroblasts and Normal Tissue-Resident Fibroblasts Reveals Fibroblast Heterogeneity in Breast Cancer. Cancers (Basel). 2020;12(5):1307. [DOI] [PMC free article] [PubMed]
- 83.Lau EY, et al. Cancer-Associated Fibroblasts Regulate Tumor-Initiating Cell Plasticity in Hepatocellular Carcinoma through c-Met/FRA1/HEY1 Signaling. Cell Rep. 2016;15(6):1175–89. 10.1016/j.celrep.2016.04.019 [DOI] [PubMed] [Google Scholar]
- 84.Loh JJ, et al. FSTL1 Secreted by Activated Fibroblasts Promotes Hepatocellular Carcinoma Metastasis and Stemness. Cancer Res. 2021;81(22):5692–705. 10.1158/0008-5472.CAN-20-4226 [DOI] [PubMed] [Google Scholar]
- 85.Shen YW, et al. Targeting CTGF in Cancer: An Emerging Therapeutic Opportunity. Trends Cancer. 2021;7(6):511–24. 10.1016/j.trecan.2020.12.001 [DOI] [PubMed] [Google Scholar]
- 86.Turdo A, et al. Meeting the Challenge of Targeting Cancer Stem Cells. Front Cell Dev Biol. 2019;7:16. 10.3389/fcell.2019.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He Y.F., et.al., Spatial multiomics reveals a subpopulation of fibroblasts associated with cancer stemness in human hepatocellular carcinoma, Genome Sequence Archive, https://ngdc.cncb.ac.cn/gsa-human/.
- 88.He Y.F., et.al., Spatial multiomics reveals a subpopulation of fibroblasts associated with cancer stemness in human hepatocellular carcinoma, Open Archive for Miscellaneous Data, https://ngdc.cncb.ac.cn/omix/.
- 89.Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169(7):1327–1341.e23. 10.1016/j.cell.2017.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma J, et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47(D1):D1211–d1217. 10.1093/nar/gky869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jing S.Y., et.al., Source code for ST_liverCancer project. GitHub; 2023. https://github.com/LihongLab/LiverCancer_ST_fibroblast. (2023)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Clinical and pathological characteristics of each patient for proteomics and 10X Visium analysis and in vitro CAF culture. Table S2. Significantly differentially expressed proteinsbetween tumor and non-tumoral parenchymal cells. Table S3. Significantly differentially expressed proteinsbetween tumor and non-tumor stroma. Table S4. Expressed extracellular matrixproteins detected by proteomics and 10X Visium in tumor and non-tumoral stroma. Table S5. Signature genes of different cell types. Table S6. Differentially expressed genes in fibroblasts of the tumor, non-tumor and interface area. Table S7. Differential expression genes of fibroblast clusters in scRNA-seq data. Table S8. Differential expression genesof fibroblast subtypes in 10X Visium. Table S9. Clinical and pathological characteristics of hepatocellular carcinomapatients for multiplexed immunofluorescenceanalysis. Table S10. The expression level of PD-L1 and CTLA4 and the number of cells with positive expression of the indicated genes in each tissue spot. Table S11. Signature genes of different cancer cell states. Table S12. Primer sequences for RT-qPCR.
Additional file 2: Fig S1. Proteomic landscape and transcriptomic heterogeneity in parenchymal and stromal regions. Fig S2. Cell type annotations in the two cohorts. Fig S3. Identification of fibroblasts enriched in the tumorfrom the scRNA-seq data and the integration of fibroblast features in ST. Fig S4. Overall survival analysis for HCC patients. Fig S5. Spatial distribution and interaction of F5-CAF and M2 macrophage. Fig S6. Interactions of F5-CAF and cancer cells with high stemness. Fig S7. Identification of CAFs and the tumor-promoting effect of F5-CAFs.
Data Availability Statement
Spatial transcriptomics data are available at National Genomics Data Center (raw data: https://ngdc.cncb.ac.cn/gsa-human/browse/HRA000437 [87]; and processed data: https://ngdc.cncb.ac.cn/omix/release/OMIX006870) [88]. RNA-seq data for CAFs are accessible at https://ngdc.cncb.ac.cn/gsa-human/browse/HRA007936 [87]. The scRNA-seq data were obtained from the Gene Expression Omnibus at GSE156625 [44]. The TCGA-LIHC data analyzed in this study can be downloaded from cBioPortal (http://www.cbioportal.org/) [89]. The proteome data have been deposited to the ProteomeXchange Consortium (dataset identifier: PXD054095) via the iProX partner repository (https://www.iprox.cn/) under Project ID: IPX0009295000 [90]. In detail the results data and code can be directly obtained from our Github website: https://github.com/LihongLab/LiverCancer_ST_fibroblast [91]. All the data needed to evaluate the conclusions in the paper are presented in the paper and/or the Supplementary Information. All methods are described in the manuscript.