ABSTRACT
We have sequenced the genome of Kurthia gibsonii strain Hakim RU_BHWE, isolated from sewage water. The assembled genome consists of 2.891 Mb with 58.6883× coverage, presenting an average GC content of 36.60%. This genome includes 8 CRISPR arrays, 3 prophages, 3 antibiotic resistance genes, and 12 virulence factor genes.
KEYWORDS: whole genome, sewage water, multidrug-resistant, Kurthia gibsonii, Bangladesh
ANNOUNCEMENT
Since its discovery in 1883 by Hermann Kurth, Kurthia spp. has been known for its wide environmental distribution and its potential to cause opportunistic infections (1 – 5). Genome sequencing is crucial for understanding the survival, adaptation, and role of Kurthia spp. in antimicrobial resistance (6). Reports of multidrug-resistant Kurthia spp. in humans, animals, food, and the environment underscore the importance of ongoing surveillance through One Health approaches to understand its molecular epidemiology and implement effective public health strategies (1 – 3, 5, 7 – 9).
The research techniques and protocols for this study were approved by the Institute of Biological Science (IBSc) at the University of Rajshahi, Bangladesh, under Memo No. 56/321/IAMEBBC/IBSc. In September 2023, we collected samples of sewage water at the University of Rajshahi (24.3733°N, 88.6049°E), following standard procedures. The water samples were mixed thoroughly, transferred to sterile tubes, and transported to the laboratory. We then inoculated these samples on urinary tract infection agar (HiMedia, India) and incubated them aerobically at 37°C for 18–24 hours (10). Kurthia gibsonii was isolated by streaking the cultures on tryptic soy agar (HiMedia), followed by staining and biochemical tests (11). Antibiogram study of the isolates was performed using the disk diffusion method (12), following Clinical and Laboratory Standards Institute guidelines (13). The strain exhibited resistance to penicillin, amoxicillin, tetracycline, and doxycycline. We cultured the isolated strain in nutrient broth (HiMedia) overnight at 37°C and then extracted its genomic DNA using the Qiagen DNA Mini Kit (QIAGEN, Hilden, Germany). The genomic DNA was enzymatically fragmented using the NEBNext dsDNA Fragmentase Kit (NEB, Massachusetts, USA), and size selection was carried out with solid-phase reversible immobilization beads (14). A sequencing library was prepared using the Nextera DNA Flex Library Preparation Kit (Illumina, San Diego, CA, USA), and the library was sequenced with 2 × 150 paired-end reads on the Illumina NextSeq 2000 platform. Quality checks were performed using FastQC v.0.11.7 (15). Raw paired-end reads (n = 2,450,168) were trimmed using Trimmomatic v.0.39 (16), and genome assembly was conducted using Unicycler v.0.4.9 (17). The annotation of the genome was carried out using PGAP v.3.0 (18). The assembled genome was analyzed for antibiotic resistance genes (ARGs) using CARD v.3.2.4 with RGI v.6.0.2 (19) and ResFinder v.4.1 (20), mobile genetic elements (MGEs) using mobileOG-db (21), virulence factor genes using VFDB with VFanalyzer v.4.0 (22), pathogenicity index using PathogenFinder v.1.1 (23), sequence type using MLST v.2.0 (24), CRISPR arrays using CRISPRimmunity (25), prophages using PHASTER (26), and metabolic functional features using RAST v.2.0 (27). We used default parameters for all tools, unless noted otherwise.
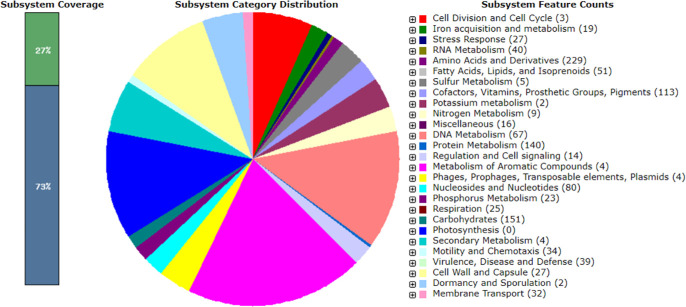
The traits of the draft genomes are documented in Table 1. Notably, 3 ARGs, 12 virulence genes, and 95 MGEs were predicted. MLST classified the genome as sequence type unknown. The genome exhibited eight CRISPR arrays with signature genes (Cas14j, WYL, csa3, cas1, cas2, cas4, cas5, cas7, DEDDh, and cas8c) and three prophages. RAST analysis uncovered 261 subsystems comprising 2,943 genes with 27% coverage (Fig. 1).
TABLE 1.
Genomic traits of the Kurthia strain Hakim RU_BHWE
| Elements | Values |
|---|---|
| Genome size | 2,891,399 bp |
| Genome coverage | 58.6883× |
| G + C content | 36.60% |
| Number of contigs | 104 |
| Contig L50 | 10 |
| Contig N50 | 101,097 bp |
| Total genes | 2,920 |
| Coding sequences | 2,868 |
| Coding genes | 2,836 |
| RNA genes | 52 |
| tRNA genes | 44 |
| rRNAs genes | 3 |
| ncRNAs genes | 5 |
| Pseudo genes | 32 |
| Genes assigned to SEED subsystems | 2,943 |
| Number of subsystems | 261 |
Fig 1.
Metabolic functional features in the assembled genome of the Kurthia gibsonii strain Hakim RU_BHWE in SEED viewer. The 27% coverage indicates the completeness of functional roles within a specific subsystem across different genomes.
ACKNOWLEDGMENTS
Rajshahi University funded this study under Bangladesh’s University Grants Commission (financial year 2023–2024).
M.R.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, and writing (original draft); A.U.-Z.P.: conceptualization, data curation, formal analysis, methodology, software, and writing (original draft); N.Z.: conceptualization, data curation, investigation, and methodology; J.R.: formal analysis, methodology, and software; M.S.A.: formal analysis, methodology, and software; M.U.K.: formal analysis, methodology, and software; S.S.: formal analysis, writing (review and editing), and software; M.H.H.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, resources, supervision, validation, writing (original draft, review, and editing).
Contributor Information
Md. Hakimul Haque, Email: hakim.ahvs@ru.ac.bd.
John J. Dennehy, Department of Biology, Queens College, Queens, New York, USA
DATA AVAILABILITY
The study on Kurthia gibsonii strain Hakim RU_BHWE, conducted using the whole genome sequencing shotgun approach, was submitted to National Center for Biotechnology Information/GenBank, and the assembly was deposited under accession number JBCHWB000000000. The pertinent data, including the original readings, were stored with BioProject accession number PRJNA1102855, BioSample accession number SAMN41030973, and Sequence Read Archive accession number SRR28762083. The specific version mentioned in this document is labeled as JBCHWB000000000.1.
REFERENCES
- 1. Kurth H. 1883. Ueber Bacterium zopfii, eine neue Bakterienart. Berichte der Deutschen Botanischen Gesellschaft 1:97–100. [Google Scholar]
- 2. Ongrádi J, Stercz B, Kövesdi V, Nagy K, Chatlynne L. 2014. Isolation of Kurthia gibsonii from non-gonorrheal urethritis: implications for the pathomechanism upon surveying the literature. Acta Microbiol Immunol Hung 61:79–87. doi: 10.1556/AMicr.61.2014.1.8 [DOI] [PubMed] [Google Scholar]
- 3. Dourou D, Spyrelli ED, Doulgeraki AI, Argyri AA, Grounta A, Nychas G-JE, Chorianopoulos NG, Tassou CC. 2021. Microbiota of chicken breast and thigh fillets stored under different refrigeration temperatures assessed by next-generation sequencing. Foods 10:765. doi: 10.3390/foods10040765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shaw S, Keddie RM. 1983. A numerical taxonomic study of the genus Kurthia with a revised description of Kurthia zopfii and a description of Kurthia gibsonii sp. nov. Syst Appl Microbiol 4:253–276. doi: 10.1016/S0723-2020(83)80054-X [DOI] [PubMed] [Google Scholar]
- 5. Mukhopadhyay BC, Mitra S, Kazi TA, Mandal S, Biswas SR. 2019. Draft genome sequence of cold-tolerant Kurthia gibsonii B83, isolated from spinach leaf. Microbiol Resour Announc 8:e01480-18. doi: 10.1128/MRA.01480-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baker S, Thomson N, Weill FX, Holt KE. 2018. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 360:733–738. doi: 10.1126/science.aar3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gardner GA. 1969. Physiological and morphological characteristics of Kurthia zopfii isolated from meat products. J Appl Bacteriol 32:371–380. doi: 10.1111/j.1365-2672.1969.tb00986.x [DOI] [PubMed] [Google Scholar]
- 8. Sharma BC, Subba R, Saha A. 2012. Kurthia sp, a novel member of phosphate solubilising bacteria from rhizospheric tea soil of Darjeeling hills, India. J of Pharm Bioallied Sci 2:36–39. doi: 10.9790/3008-0233639 [DOI] [Google Scholar]
- 9. Lozica L, Maurić Maljković M, Mazić M, Gottstein Ž. 2022. Kurthia gibsonii, a novel opportunistic pathogen in poultry. Avian Pathol 51:26–33. doi: 10.1080/03079457.2021.1993132 [DOI] [PubMed] [Google Scholar]
- 10. Romance M, Khan MU, Islam MS, Islam MF, Haque MH. 2024. Draft genome sequence of multidrug-resistant Klebsiella pneumoniae Hakim-RU strain isolated from a patient with urinary tract infections in Bangladesh. Microbiol Resour Announc 13:e0008924. doi: 10.1128/mra.00089-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bergey DH, Buchanan RE, Gibbons NE. 1974. Bergey’s Manual of determinative Bacteriology. 8th ed, p 966–1097. American Society for Microbiology, Washington, DC, USA. [Google Scholar]
- 12. Bauer AW, Kirby WMM, Sherris JC, Turck M. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496. doi: 10.1093/ajcp/45.4_ts.493 [DOI] [PubMed] [Google Scholar]
- 13. M100-S32 . 2022. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 14. Liu D, Li Q, Luo J, Huang Q, Zhang Y. 2023. An SPRI beads-based DNA purification strategy for flexibility and cost-effectiveness. BMC Genomics 24:125. doi: 10.1186/s12864-023-09211-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 16. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-LV, Cheng AA, Liu S, et al. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. doi: 10.1093/nar/gkz935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Florensa AF, Kaas RS, Clausen P, Aytan-Aktug D, Aarestrup FM. 2022. ResFinder - an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb Genom 8:000748. doi: 10.1099/mgen.0.000748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown CL, Mullet J, Hindi F, Stoll JE, Gupta S, Choi M, Keenum I, Vikesland P, Pruden A, Zhang L. 2022. mobileOG-dB: a manually curated database of protein families mediating the life cycle of bacterial mobile genetic elements. Appl Environ Microbiol 88:e0099122. doi: 10.1128/aem.00991-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu B, Zheng D, Zhou S, Chen L, Yang J. 2022. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res 50:D912–D917. doi: 10.1093/nar/gkab1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cosentino S, Voldby Larsen M, Møller Aarestrup F, Lund O. 2013. Pathogenfinder - distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 8:e77302. doi: 10.1371/journal.pone.0077302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou F, Yu X, Gan R, Ren K, Chen C, Ren C, Cui M, Liu Y, Gao Y, Wang S, Yin M, Huang T, Huang Z, Zhang F. 2023. CRISPRimmunity: an interactive web server for CRISPR-associated important molecular events and modulators used in geNome edIting tool identifYing. Nucleic Acids Res 51:W93–W107. doi: 10.1093/nar/gkad425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–21. doi: 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The study on Kurthia gibsonii strain Hakim RU_BHWE, conducted using the whole genome sequencing shotgun approach, was submitted to National Center for Biotechnology Information/GenBank, and the assembly was deposited under accession number JBCHWB000000000. The pertinent data, including the original readings, were stored with BioProject accession number PRJNA1102855, BioSample accession number SAMN41030973, and Sequence Read Archive accession number SRR28762083. The specific version mentioned in this document is labeled as JBCHWB000000000.1.