ABSTRACT
House mice, Mus musculus, are highly adapted to anthropogenic spaces. Fecal samples were collected from house mice entering primate enclosure areas at the Duke Lemur Center (Durham, NC, USA). We identified 14 cressdnavirus and 59 microvirus genomes in these mouse feces.
KEYWORDS: microvirus, cressdnavirus, Mus musculus
ANNOUNCEMENT
Invasive rodents alter ecosystem dynamics by outcompeting endemic wildlife for resources and frequently moving between wild and human-inhabited spaces (1 – 5). House mice (Mus musculus), in particular, have become one of the most widespread invasive species, being transported across the planet through human movement and adapting remarkably well to human-altered landscapes (6).
As part of a larger study sampling lemurs, humans, and rodents at the Duke Lemur Center (Durham, NC, USA) under IACUC #A161-21-08, fecal samples were collected in August 2021 from two M. musculus trapped indoors in lemur enclosure areas. Fecal samples were frozen at −80°C until extraction. Once thawed, fecal samples were homogenized with SM buffer [0.1 M NaCl, 50 mM Tris-HCl (pH 7.4)] and centrifuged briefly at 8,000 rpm. Using the Roche High Pure Viral Nucleic Acid Kit (Roche Diagnostics, Germany), DNA was extracted from 200 µL of each supernatant. DNA extract was amplified using the Templiphi kit (GE Healthcare, USA). After library preparation with the Illumina DNA Prep Kit, libraries were sequenced on an Illumina NovaSeq 6000 at the Duke Center for Genomic and Computational Biology yielding 18,080,238 and 23,175,508 paired reads for each of the two samples. Paired-end reads were trimmed using Trimmomatic v0.39 (7) and assembled with MEGAHIT v.1.2.9 (8). Circular contigs were identified by terminal redundancy based on a >10 nt repeat. Diamond v2.1.9 (9) BLASTx was used to identify viral-like sequences against a local NCBI RefSeq viral protein database (release 220). Cenote Taker2 v2.1.5 (10) and VIBRANT v1.2.1 (11) were used to annotate viral genomes. Viral genomes with >98% identity were clustered into virus operational taxonomic units (vOTUs) with SDT v1.2 (12) and used as a reference to map reads with BBMap v38.12 (13). Web-based BLASTn was used to determine the similarity of viruses characterized in this study to known virus genomes. All bioinformatics tools were used with default settings.
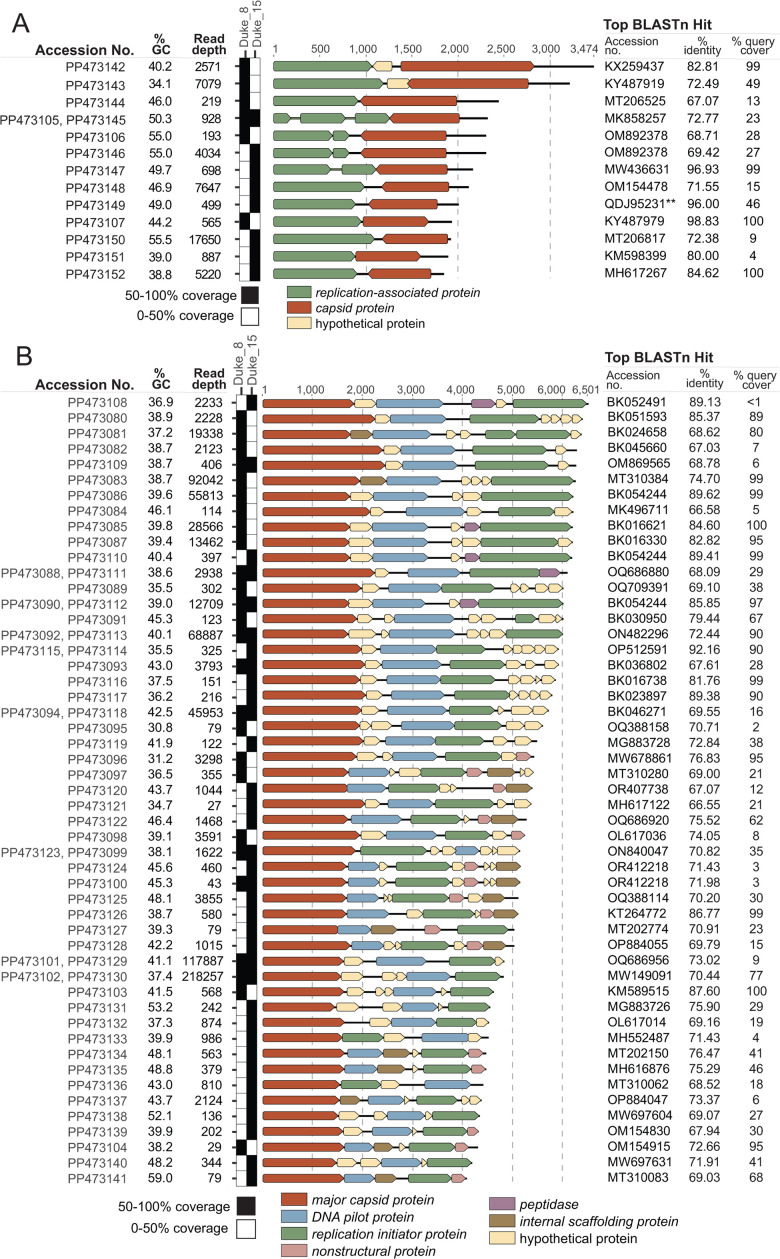
We identified 14 cressdnavirus and 59 microvirus genomes from feces collected from two M. musculus individuals (Duke_8, Duke_15). The Cressdnaviricota phylum is composed of single-stranded DNA viruses infecting an array of eukaryotic hosts including plants, animals, fungi, and protists (14). The 14 cressdnavirus genomes encode a capsid and replication-associated protein, range in length from 1,847 to 3,474 nt, and GC content of 34.1%–55.0% (Fig. 1A). PP473106 and PP473146 share 97.5% similarity and fall within the Smacoviridae family, a group predicted to infect gut archaea (15). PP473147 is a member of the Genomoviridae family, a group of likely fungi-infecting cressdnaviruses (16). The unclassified cressdnavirus genomes (n = 11) identified in this study share similarities with viruses identified from water (n = 5), soil (n = 1), fish tissue (n = 1), damselflies (n = 1), capybara feces (n = 1), and tortoise feces (n = 2).
Fig 1.
(A). Genome organizations of the cressdnaviruses were identified in M. musculus feces in this study. A summary of the accession numbers, GC content, read depth, and top BLASTn hit for each distinct genome is presented. **This BLAST hit represents a Rep protein BLASTp instead of a full genome comparison for PP473149. (B) Genome organizations of the microviruses identified in M. musculus feces in this study. A summary of the accession numbers, GC content, read depth, and top BLASTn hit for each distinct genome is presented. Genomes characterized in this study with >98% nt identity are represented by one row. For (A) and (B), genome coverage plots based on vOTU read mapping depict the presence of all vOTUs across Duke_8 and Duke_15. Black squares represent 50%–100% genome coverage and serve as a high-confidence proxy of vOTU presence.
Microviruses are small, single-stranded DNA bacteriophages (17). The 59 microvirus genomes identified encode at least a major capsid protein, DNA pilot protein, and replication initiator protein, range in length from 4,066 to 6,501 nt, and range in GC content from 30.8%–59.0% (Fig. 1B). The distinct (<98% similarity) microvirus genomes (n = 51) share the highest similarity with microviruses characterized from rodent (M. musculus, Dipodomys merriami, Rattus norvegicus, and Sigmodon arizonae) feces and tissue (n = 8), human samples (n = 17), water (n = 9), ungulate feces (n = 5), soil (n = 3), fish tissue (n = 2), bat feces (n = 2), insects (n = 2), reptile feces (n = 1), avian feces (n = 1), dog feces (n = 1), and airborne particulate (n = 1). The identified microviruses likely infect enterobacteria within M. musculus.
ACKNOWLEDGMENTS
The work described here was supported by TriCEM (Triangle Center for Evolutionary Medicine), Duke Biology, Duke Lemur Center, and Sigma Xi grants awarded to E.N.P. We thank the Duke Lemur Center research team for their help in sample collection.
Contributor Information
Arvind Varsani, Email: arvind.varsani@asu.edu.
Jelle Matthijnssens, Katholieke Universiteit Leuven, Leuven, Belgium .
DATA AVAILABILITY
The sequences of cressdnaviruses and microviruses in this study have been deposited in the NCBI SRA under SRR28214738 and SRR28214739 and GenBank accession numbers PP473080–PP473152.
REFERENCES
- 1. Pittermannová P, Žákovská A, Váňa P, Marková J, Treml F, Černíková L, Budíková M, Bártová E. 2021. Wild small mammals and ticks in Zoos-reservoir of agents with Zoonotic potential? Pathogens 10:777. doi: 10.3390/pathogens10060777doi:34205547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chalkowski K, Lepczyk CA, Zohdy S. 2018. Parasite ecology of invasive species: conceptual framework and new hypotheses. Trends Parasitol 34:655–663. doi: 10.1016/j.pt.2018.05.008 [DOI] [PubMed] [Google Scholar]
- 3. Williams SH, Che X, Garcia JA, Klena JD, Lee B, Muller D, Ulrich W, Corrigan RM, Nichol S, Jain K, Lipkin WI. 2018. Viral diversity of house mice in New York City. mBio 9:e01354-17. doi: 10.1128/mBio.01354-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pankovics P, Boros Á, László Z, Szekeres S, Földvári G, Altan E, Delwart E, Reuter G. 2021. Genome characterization, prevalence and tissue distribution of astrovirus, hepevirus and norovirus among wild and laboratory rats (Rattus norvegicus) and mice (Mus musculus) in Hungary. Infection, Genetics and Evolution 93:104942. doi: 10.1016/j.meegid.2021.104942 [DOI] [PubMed] [Google Scholar]
- 5. Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, Delwart EL. 2011. The fecal viral flora of wild rodents. PLoS Pathog 7:e1002218. doi: 10.1371/journal.ppat.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Witmer GW, Pitt WC. 2012. Invasive rodents in the United States: ecology, impacts, and management
- 7. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033 [DOI] [PubMed] [Google Scholar]
- 9. Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 10. Tisza MJ, Belford AK, Domínguez-Huerta G, Bolduc B, Buck CB. 2021. Cenote-taker 2 democratizes virus discovery and sequence annotation. Virus Evol 7:veaa100. doi: 10.1093/ve/veaa100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kieft K, Zhou Z, Anantharaman K. 2020. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 8:90. doi: 10.1186/s40168-020-00867-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. doi: 10.1371/journal.pone.0108277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bushnell B. 2015. BBMap short-read aligner, and other bioinformatics tools. http://sourceforge net/projects/bbmap/.
- 14. Krupovic M, Varsani A, Kazlauskas D, Breitbart M, Delwart E, Rosario K, Yutin N, Wolf YI, Harrach B, Zerbini FM, Dolja VV, Kuhn JH, Koonin EV. 2020. Cressdnaviricota: a virus phylum unifying seven families of rep-encoding viruses with single-stranded, circular DNA genomes. J Virol 94:e00582-20. doi: 10.1128/JVI.00582-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Díez-Villaseñor C, Rodriguez-Valera F. 2019. CRISPR analysis suggests that small circular single-stranded DNA Smacoviruses infect Archaea instead of humans. Nat Commun 10:294. doi: 10.1038/s41467-018-08167-wdoi:30655519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fiallo-Olivé E, Lett J-M, Martin DP, Roumagnac P, Varsani A, Zerbini FM, Navas-Castillo J. 2021. ICTV virus taxonomy profile: Geminiviridae 2021. J Gen Virol 102:001696. doi: 10.1099/jgv.0.001696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cherwa JE, Fane BA. 2011. Microviridae: microviruses and gokushoviruses. In eLSdoi: 10.1002/047001590X [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences of cressdnaviruses and microviruses in this study have been deposited in the NCBI SRA under SRR28214738 and SRR28214739 and GenBank accession numbers PP473080–PP473152.