ABSTRACT
LBerry, Pembroke, and Zolita are newly isolated bacteriophages that infect Mycobacterium smegmatis mc²155. Based on gene content similarity, LBerry and Pembroke are assigned to cluster A3, and Zolita is assigned to cluster A5. LBerry and Pembroke are 99% identical to Anaysia and Caviar, and Zolita is 99% identical to SydNat.
KEYWORDS: bacteriophage, mycobacteria, genome analysis
ANNOUNCEMENT
The Mycobacterium genus of bacteria includes increasingly antibiotic-resistant human pathogens, such as Mycobacterium tuberculosis and Mycobacterium abscessus (1). With the increasing occurrence of antibiotic-resistant pathogens constituting a global threat to public health, phage therapy has recently been employed as an alternative treatment strategy. Some phages isolated using the nonpathogenic bacterial host M. smegmatis also infect pathogenic Mycobacteria and these phages can potentially be used in phage therapy (2, 3).
LBerry, Pembroke, and Zolita were isolated, purified, and their genomes were annotated through our participation in the SEA PHAGES program (4). Plaque purification, amplification, and production of high-titer lysates were performed as described in the Phage Discovery Guide (5).
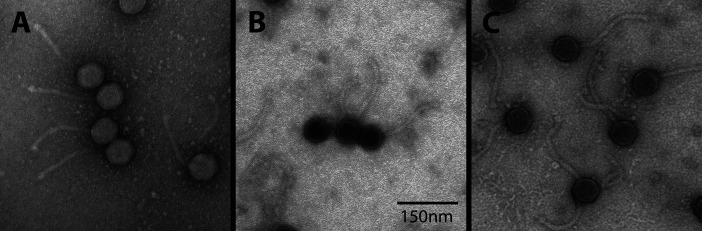
All three phages were isolated from damp grassy soil samples collected in the northeastern US; with LBerry isolated outside of a hotel, Pembroke from a former farm, and Zolita near a flower bed. Each sample was treated with 7H9 liquid medium, filtered (0.2 µm), and inoculated with Mycobacterium smegmatis mc2 155. Samples were incubated at 37°C with shaking for 48 h, then plated on top agar with host bacteria to form plaques. Three rounds of purification were done for LBerry and Pembroke, and four rounds for Zolita. All three phages were determined to have siphovirus morphology via negative-strain transmission electron microscopy (Fig. 1). DNA from each phage was extracted from a high-titer lysate by phenol: chloroform: isoamyl: alcohol extraction (6) and sequenced by the Pittsburgh Bacteriophage Institute (Table 1). Raw reads were verified for accuracy using Consed v29.0 (7) and assembled using Newbler v2.9 (8). All phage genomes have a 3′ single-stranded overhang; the sequences are reported in Table 1 along with genome sizes and GC content for each phage. Based on gene sequence similarities, LBerry and Pembroke were assigned to the A3 cluster while Zolita was assigned to the A5 cluster (9, 10). LBerry and Pembroke are 99% identical to A3 cluster phages Anaysia OP021679 and Caviar ON970623 (11), and Zolita is 99% identical to A5 phage SydNat ON970625.
Fig 1.
Images of (A) LBerry, (B) Pembroke, and (C) Zolita in negative-stained (1% uranyl acetate) taken by a JEOL 200 CX transmission electron microscope. All three phages have siphovirus morphology.
TABLE 1.
Sequencing, genome, and phage characteristics
| Parameter | LBerry | Pembroke | Zolita |
|---|---|---|---|
| Soil sample characteristics | |||
| Collection date | 17 October 2022 | 10 October 2022 | 29 August 2018 |
| Collection location coordinates | 43.058056 N 77.650556 W |
42.076111 N 70.833056 W |
41.843056 N 71.438611 W |
| Phage particle characteristics | |||
| Capsid size (nm) | 68–71 (n = 20) | 59–63 (n = 20) | 67–69 (n = 20) |
| Tail length (nm) | 184–187 (n = 20) | 189–192 (n = 20) | 211–214 (n = 20) |
| Taxonomic identification | |||
| Class | Caudoviricetes | ||
| Genus | Microwolfvirus | Benedictvirus | |
| Species | Unclassified | Benedictvirus Zolita | |
| Sequencing | |||
| Sequencing instrument | Illumina MiSeq v3 reagents | ||
| Library prep kit | TruSeq DNA Nano Prep, S4 Flowcell, v1.5 | NEB Ultra II Library Kit | |
| Number of reads | 100,000 | 100,000 | 552,393 |
| Length of reads (bp) | 150-base single-end reads | ||
| Shotgun coverage (×) | 276 | 280 | 1,523 |
| Phage genome characteristics | |||
| Genome length (bp) | 50,965 | 50,849 | 51,182 |
| 3′ single-stranded overhang sequence | CGGGTGGTAA | CGGGTGGTAA | CGGGAGGTAA |
| GC content (%) | 64.0% | 64.0% | 60.9% |
DNA Master v5.23.6 was used to perform the genome annotations (12). GeneMark v2.5 (13), Glimmer v3.02 (14), and Starterator v.546 (15) were used to determine gene starts. Protein functions were determined using HHpred (PDB, UniProt, Pfam-A v.36, and NCBI v.3.19 databases) (16, 17), BLASTp v.2.14.1 (18), and Phamerator (19). ARAGORN v.1.2.38 (20) and tRNAscan-SE v.2.0 (21) were used to identify tRNAs. Membrane proteins were predicted using TMHMM v.1.0.24 (22) and TOPCONS v.2.0 (23). Unless otherwise stated, default parameters were used for the programs listed.
Cluster A is the largest group of mycobacteriophages, with nearly 800 members. They are genetically diverse (24), and divided into 20 subclusters. LBerry, Pembroke, and Zolita follow the expected synteny of an A cluster phage beginning with a lysis cassette followed by structural proteins, integration proteins, replication/recombination proteins, an immunity repressor, and ending with a series of proteins of unknown function. The presence of immunity repressor and integrase genes in all three phages suggests that these phages could potentially adopt a temperate lifestyle (25). It has been determined that A3 cluster phages are able to infect M. tuberculosis H37Rv (26), indicating that LBerry and Pembroke could be further investigated for application in phage therapy.
ACKNOWLEDGMENTS
This work was completed as part of an undergraduate research project at Providence College. The authors thank the Center for Engaged Learning at Providence College for their support and also thank the North Carolina State University Genomic Sciences Laboratory for sequence assistance, and Graham F. Hatfull and the HHMI SEA-PHAGES program for their support. Electron microscopy work by J.A.D. was conducted at the Marine Biological Laboratory in Woods Hole, MA and is supported by the National Science Foundation EPSCoR Track II Cooperative Agreement (award number 130406). K.C. acknowledges the Walsh fund at Providence College for support.
Contributor Information
Kathleen Cornely, Email: kcornely@providence.edu.
John J. Dennehy, Department of Biology, Queens College, Queens, New York, USA
DATA AVAILABILITY
The genome sequence accession number for LBerry is OR725491 and the SRA accession number is SRX23702564. The genome accession number for Pembroke is OR725495 and the SRA accession number is SRX23702567. The genome accession number for Zolita is MN096372 and the SRA accession number is SRX18224444.
REFERENCES
- 1. Hatfull GF, Hendrix RW. 2011. Bacteriophages and their genomes. Curr Opin Virol 1:298–303. doi: 10.1016/j.coviro.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nobrega FL, Costa AR, Kluskens LD, Azeredo J. 2015. Revisiting phage therapy: new applications for old resources. Trends Microbiol. 23:185–191. doi: 10.1016/j.tim.2015.01.006 [DOI] [PubMed] [Google Scholar]
- 3. Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25:730–733. doi: 10.1038/s41591-019-0437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SCR, et al. 2014. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio 5:e01051-13. doi: 10.1128/mBio.01051-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poxleitner: phage discovery guide. Google scholar. Available from: https://scholar.google.com/scholar_lookup?title=Phage+discovery+guide&publication_year=2018&;. Retrieved 6 Mar 2024.
- 6. Spada S. 2020. Chapter six - methods to purify DNA from extracellular vesicles: focus on exosomes, p 109–118. In Spada S, Galluzzi L (ed), Methods in enzymology. Academic Press. [DOI] [PubMed] [Google Scholar]
- 7. Russell DA. 2018. Sequencing, assembling, and finishing complete bacteriophage genomes, p 109–125. In Clokie MRJ, Kropinski AM, Lavigne R (ed), Bacteriophages: methods and protocols. Vol. 3. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 8. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. doi: 10.1038/nature03959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hatfull GF. 2014. Molecular genetics of mycobacteriophages. Microbiol Spectr 2. doi: 10.1128/microbiolspec.MGM2-0032-2013 [DOI] [PubMed] [Google Scholar]
- 10. Russell DA, Hatfull GF. 2017. PhagesDB: the actinobacteriophage database. Bioinformatics 33:784–786. doi: 10.1093/bioinformatics/btw711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abidin ZU, Aucapina JE, Beauzil S, Berotte CM, Bonsu AO, Burgos GY, Chak STC, Collymore A, Daley ER, Defarias R, et al. 2022. Complete genome sequences of actinobacteriophages Anaysia and Caviar. Microbiol Resour Announc 11:e0094422. doi: 10.1128/mra.00944-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pope WH, Jacobs-Sera D. 2018. Annotation of bacteriophage genome sequences using DNA master: an overview, p 217–229. In Clokie MRJ, Kropinski AM, Lavigne R (ed), Bacteriophages: methods and protocols. Vol. 3. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 13. Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107–1115. doi: 10.1093/nar/26.4.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641. doi: 10.1093/nar/27.23.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pacey M. 2016. Starterator guide. University of Pittsburgh, Pittsburgh, PA. [Google Scholar]
- 16. Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248. doi: 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 19. Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2011. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 12:395. doi: 10.1186/1471-2105-12-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964. doi: 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Möller S, Croning MDR, Apweiler R. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646–653. doi: 10.1093/bioinformatics/17.7.646 [DOI] [PubMed] [Google Scholar]
- 23. Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A. 2015. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43:W401–W407. doi: 10.1093/nar/gkv485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mavrich TN, Hatfull GF. 2019. Evolution of superinfection immunity in cluster A mycobacteriophages. mBio 10:e00971-19. doi: 10.1128/mBio.00971-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mavrich TN, Hatfull GF. 2017. Bacteriophage evolution differs by host, lifestyle and genome. Nat Microbiol 2:17112. doi: 10.1038/nmicrobiol.2017.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guerrero-Bustamante CA, Dedrick RM, Garlena RA, Russell DA, Hatfull GF. 2021. Toward a phage cocktail for tuberculosis: susceptibility and tuberculocidal action of mycobacteriophages against diverse Mycobacterium tuberculosis strains. mBio 12:e00973-21. doi: 10.1128/mBio.00973-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence accession number for LBerry is OR725491 and the SRA accession number is SRX23702564. The genome accession number for Pembroke is OR725495 and the SRA accession number is SRX23702567. The genome accession number for Zolita is MN096372 and the SRA accession number is SRX18224444.