ABSTRACT
The draft genome of a previously documented potential probiotic Weissella cibaria strain GM93m3 from raw goat milk in Nigeria is reported. The total genome size was 2,447,229 with 46 contigs and G+C content of 44.86%.
KEYWORDS: probiotics, Weissella, gut health, food safety
ANNOUNCEMENT
Weissella cibaria, a Gram-positive, rod-shaped, non-motile, lactic acid bacterium, has attracted research interest due to its technological, nutritional, and probiotic potential (1). W. cibaria GM93m3, isolated from raw goat milk collected in June 2019 from dairy farms in Sokoto state, Nigeria (13.0533°N, 5.3223°E), was proposed as a suitable human probiotic candidate in a prior study. The strain showed in vitro the ability to tolerate human gastrointestinal conditions and co-aggregate with pathogens and antioxidant activities (1). Here, we report the draft genome sequence of this W. cibaria GM93m3 strain.
Milk samples were serially diluted in threefolds, inoculated on De Man–Rogosa–Sharpe agar, and incubated anaerobically at 37°C for 24 hours. Distinct colonies were subcultured until pure culture was attained.
High-quality genomic DNA was extracted from pure single colonies of the strain using the Quick-DNA fungal/bacterial miniprep kit (Zymo Research). A genomic library was prepared using the Illumina TruSeq Nano DNA library preparation kit supplied by Illumina Inc. Pair-end sequencing (2 × 150 base pairs) was performed using Illumina NovaSeq 6000 generating 9,224,104 total read counts. The quality of sequence reads was determined using FastQC v0.12.0 (2). Adapter trimming, quality filtering, and per-read quality pruning were done using fastp v0.23.4 (3). Filtered paired-end reads were de novo-assembled using SPAdes v3.15.3 (4). Quality of assemblies was determined using QUAST v4.4 (5), while genome completeness was evaluated using CheckM v1.0.18 (6).
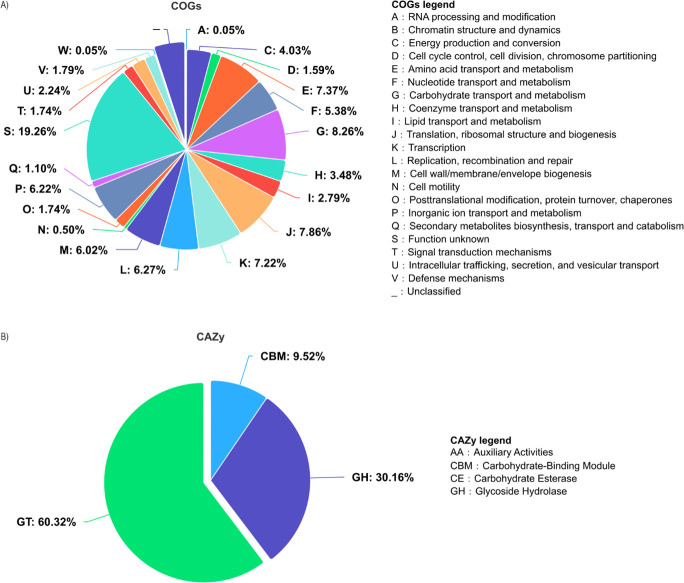
GM93m3 sequence (PRJNA1086807) was assembled into 46 contigs with a genome size of 2,447,229 bp, N50 of 235,178, L50 of 4, G+C content of 44.86%, and coverage of 570.09. CheckM analyses indicated that the genome was 100% complete with no contamination. Species designation of the strain was first determined using 16S rRNA gene extracted from the genome using extractseq version 5.0.0 (7) and blasted against the National Center for Biotechnology Information 16S database (8). The closest hit, which corresponds to W. cibaria strain SRCM103448 (CP035267.1), exhibited an identity similarity of approximately 98%. Further analysis identifying closely similar reference genomes and orthologous average nucleotide identity (ANI) calculated using the Mash/MinHash v2.3 algorithm (9) revealed 98.37% ANI with W. cibaria KACC 11862 (GCA_000193635.2). Functional genome annotation by orthology was performed using eggnog-mapper software v2.1.8 and database v5.0.2 with protein coding sequences (CDSs), translated to proteins before the search identified a total of 2,257 CDSs, 74 transfer RNA genes, 8 ribosomal RNA, and 1 transfer-messenger RNA genes. An overview of clusters of orthologous genes (COGs) functional classification and carbohydrate-active enzymes (CAZy) is presented in Fig. 1 while the genome annotation and 16S rRNA gene are available on Figshare (10). ABRicate v1.0.1 (11) was used to examine plasmids, virulence determinants, and resistance-related genes, with reads aligned to the PlasmidFinder (12), Virulence Factors (13), and Comprehensive Antibiotic Resistance v3.2.9 (14) databases, respectively; however, no hits were detected. The BAGEL v.4.0 webserver (http://bagel4.molgenrug.nl) predicted the absence of genes encoding bacteriocins, which agrees with our previous phenotypic report where the supernatants of strain GM93m3 showed the least antimicrobial activity against selected foodborne pathogens (1). Default parameters were used for all software unless otherwise specified.
Fig 1.
(A) COGs functional classification and (B) CAZy annotated for Weissella cibaria GM93m3 strain.
ACKNOWLEDGMENTS
The authors appreciate the Applied Microbiology International for partly supporting this study through the 2019 Research Support Grant.
M.O.A. performed the conceptualization, data curation, data analysis and visualization, methodology, and writing of the original draft; O.A.O. performed the writing of the original draft; M.S.K. performed the data curation, review, and editing; B.A.O. secured the resources and review and editing; R.A.A. secured the resources and review and editing; C.N.E. performed the conceptualization, resources, methodology, and review and editing.
Contributor Information
Muiz O. Akinyemi, Email: tosin2667@gmail.com.
Rasheed A. Adeleke, Email: rasheed.adeleke@nwu.ac.za.
Chibundu N. Ezekiel, Email: chaugez@gmail.com.
Vanja Klepac-Ceraj, Department of Biological Sciences, Wellesley College, Wellesley, Massachusetts, USA .
DATA AVAILABILITY
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JBBEEN000000000. The version described in this paper is version JBBEEN010000000. The raw data of our study are available in the SRA database of NCBI with accession number SRX23974901.
REFERENCES
- 1. Akinyemi MO, Ogunremi OR, Adeleke RA, Ezekiel CN. 2024. Probiotic potentials of lactic acid bacteria and yeasts from raw goat milk in Nigeria. Probiotics Antimicrob Proteins 16:163–180. doi: 10.1007/s12602-022-10022-w [DOI] [PubMed] [Google Scholar]
- 2. Andrews S. 2010. FastQC: a quality control tool for high throughput sequence. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 3. Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet 16:276–277. doi: 10.1016/s0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- 8. O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–45. doi: 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Akinyemi MO. 2024. Genome annotation of Weissella cibaria GM93m3, a promising probiotic strain from raw goat milk. doi: 10.6084/m9.figshare.25427803.v2. [DOI] [PMC free article] [PubMed]
- 11. Seemann T. Github - Tseemann/Abricate::Mag_Right: mass screening of contigs for antimicrobial and virulence genes. ABRicate. Available from: https://github.com/tseemann/abricate. Retrieved 44 MarMarch 2024. Accessed , 44 MarMarch 2024
- 12. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu B, Zheng D, Zhou S, Chen L, Yang J. 2022. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res 50:D912–D917. doi: 10.1093/nar/gkab1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, Edalatmand A, Petkau A, Syed SA, Tsang KK, et al. 2023. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res 51:D690–D699. doi: 10.1093/nar/gkac920 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Akinyemi MO. 2024. Genome annotation of Weissella cibaria GM93m3, a promising probiotic strain from raw goat milk. doi: 10.6084/m9.figshare.25427803.v2. [DOI] [PMC free article] [PubMed]
Data Availability Statement
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JBBEEN000000000. The version described in this paper is version JBBEEN010000000. The raw data of our study are available in the SRA database of NCBI with accession number SRX23974901.