Abstract
Heme oxygenase-1 (HO-1, HMOX1) degrades heme protecting cells from heme-induced oxidative damage. Beyond its well-established cellular functions, heme has emerged as a stabilizer of G-quadruplexes. These secondary DNA structures interfere with DNA replication. We recently revealed that nuclear HO-1 colocalizes with DNA G-quadruplexes and promotes their removal. Here, we investigate whether HO-1 safeguards cells against replication stress.
Experiments were conducted in control and HMOX1-deficient HEK293T cell lines. Immunostaining unveiled that DNA G-quadruplexes accumulated in the absence of HO-1, the effect that was further enhanced in response to δ-aminolevulinic acid (ALA), a substrate in heme synthesis. This was associated with replication stress, as evidenced by an elevated proportion of stalled forks analyzed by fiber assay. We observed the same effects in hematopoietic stem cells isolated from Hmox1 knockout mice and in a lymphoblastoid cell line from an HMOX1-deficient patient. Interestingly, in the absence of HO-1, the speed of fork progression was higher, and the response to DNA conformational hindrance less stringent, indicating dysfunction of the PARP1-p53-p21 axis. PARP1 activity was not decreased in the absence of HO-1. Instead, we observed that HO-1 deficiency impairs the nuclear import and accumulation of p53, an effect dependent on the removal of excess heme. We also demonstrated that administering ALA is a more specific method for increasing intracellular free heme compared to treatment with hemin, which in turn induces strong lipid peroxidation.
Our results indicate that protection against replication stress is a universal feature of HO-1, presumably contributing to its widely recognized cytoprotective activity.
Keywords: Heme oxygenase-1, Heme, Replication stress, G-quadruplexes, PARP1, Cell cycle
Graphical abstract
Highlights
-
•
Endogenous heme promotes accumulation of G-quadruplexes and induces replication stress.
-
•
By removing excess heme, HO-1 prevents replication stress and improves cell cycle control.
-
•
In the absence of HO-1, replication fork progression can continue despite steric hinderance.
-
•
HO-1 deficiency impairs the PARP1-p53-p21 axis, decreasing p53 nuclear import.
1. Introduction
Heme oxygenase-1 (HO-1, encoded by the HMOX1 gene) is an inducible enzyme that plays a central role in heme catabolism. HO-1 converts heme to biliverdin, carbon monoxide (CO), and iron ion. Due to its ability to remove potentially pro-oxidant heme, HO-1 is widely regarded as an enzyme that protects cells from oxidative damage [1]. In principle, the HO-1 protein localizes in the cytoplasm and anchors to the endoplasmic reticulum, although it was also detected in other cell compartments, including mitochondria [2]. Importantly, HO-1 can translocate to the nucleus, and in some cell types, e.g. hematopoietic stem cells (HSCs), the nuclear form dominates [3].
The role of HO-1 is not limited to its enzymatic activity, and several reports indicate the putative importance of binding to other proteins and nucleic acids [4,5]. HO-1 co-precipitates with proteins involved in replication, DNA damage response (DDR) and DNA repair [5]. One of the possible partners interacting with HO-1 is poly(ADP-ribose) polymerase-1 (PARP1), a highly abundant enzyme that uses NAD+ as a substrate to catalyze poly(ADP-ribosyl)ation (PARylation) [4]. This post-translational modification, involving the covalent binding of highly negatively charged poly(ADP-ribose) (PAR) residues, strongly affects protein functions. Additionally, many proteins (PAR-readers) can bind PAR noncovalently, which may regulate their localization, stability and interactions with other macromolecules [6]. Recognition of DNA damage by PARP1 changes the conformation of its autoinhibitory helical domain (HD), which permits NAD+ binding and converting to PAR [7]. Docking modeling suggests that HO-1 may potentially interact with the HD region to facilitate PARP1 activation [4].
Using a standard chromatin immunoprecipitation assay we were unable to detect binding of HO-1 to DNA [8]. However, the HO-1 protein consists of seven α-helices, which may provide a DNA-binding pocket, and has recently been suggested to regulate gene transcription directly [5]. HO-1 also appears to regulate gene expression through protein-protein interactions with several transcription factors, especially those related to the oxidative stress response. Nuclear HO-1 was shown to bind and stabilize NRF2 and AP-1 transcription factors, thereby increasing the expression of their target genes and improving antioxidant protection [9]. It may also be involved in the type I interferon response or in the regulation of mRNA processing [10]. Thus, direct interaction with other macromolecules is an increasingly appreciated aspect of HO-1 function, but its role is still far from being fully understood.
We have demonstrated that nuclear HO-1 can promote the removal of G-quadruplexes, four-stranded secondary structures composed of guanine-rich nucleic acids. Moreover, using proximity ligation assay (PLA), we proved the colocalization of HO-1 protein with DNA G-quadruplexes in the nucleus of murine HSCs and human HEK293T cells [8].
G-quadruplexes are enriched at telomeres, gene promoters and origins of replication (ORIs), so they can influence both gene transcription and DNA replication [11]. Importantly, G-quadruplexes can form steric hindrances in single-stranded DNA and disrupt the progression of replication forks, which in turn causes replication stress [12]. Indeed, administration of small molecules stabilizing G-quadruplexes slowed down DNA replication and stalled replication forks [13]. An insufficient response to fork stalling can lead to replication fork collapse and the formation of double-strand DNA breaks (DSBs) [14]. As a result, replication stress emerges as an important source of DSBs, significantly impacting genome stability [15]. Similarly, G-quadruplexes have been recognized as significant contributors to DNA damage [16].
The response to replication stress can be regulated by PARP1, which stabilizes stalled forks and is involved in the recruitment of DNA repair proteins [17]. Importantly, PARP1 is also known to bind G-quadruplexes with nanomolar affinities, which in turn enhances its enzymatic activation independently of DNA breaks. PARP1 promotes relaxation of supercoiled DNA and facilitates the unwinding and removal of G-quadruplexes [18].
Importantly, one of the targets of PARP1-mediated PARylation is the tumor suppressor protein p53, which is involved in DNA quality control [19]. In cells where DNA has been damaged, p53 can promote apoptosis, senescence, or transient cell cycle arrest to allow DNA repair [20]. It also associates with active replication forks and plays a central role in the response to replication stress. When fully functional, p53 maintains the appropriate rate of ongoing fork progression and binds to stalled forks to ensure proper restart of replication [19,21]. PARP1 significantly influences p53-DNA binding and regulates p53 trafficking [20]. It is worth noting that p53 can also be directly regulated by heme, the substrate of HO-1, which noncovalently binds to the heme-regulatory motif (HRF) at the molecular ratio 1:1 and accelerates protein degradation [22]. Thus, it can be assumed that the absence of HO-1, which removes excess heme, could reduce the efficiency of p53 pathway.
It has been demonstrated that chemical stabilization of G-quadruplexes may result in replication stress, increased formation of DNA breaks and a high frequency of deletions at the sites of G-quadruplex formation [13,16]. Again, one of the effective stabilizers of G-quadruplexes in cells is heme [8,23,24]. Therefore, it is conceivable that HO-1 may influence key DNA damage response proteins and participates in regulating heme availability for G-quadruplexes, potentially impacting their stability [8]. This implies that HO-1 might provide protection to cells not only against heme-induced oxidative damage but also against replication stress. Our aim was to verify this hypothesis.
2. Materials and methods
HEK293T cells. Cell cultures were performed under standard conditions, at 37 °C in a humidified atmosphere with 5 % CO2. HEK293T cells (kindly obtained from dr. Maciej Wiznerowicz from Wielkopolskie Centrum Onkologii, Poznan, Polska) were cultured in DMEM High-Glucose medium (Biowest) containing 10 % fetal bovine serum (FBS, EurX) and antibiotics (100 IU/mL penicillin and 100 μg/mL streptomycin, Sigma-Aldrich). Cells were routinely cultured in a T-25 flasks (Falcon) and then seeded in 6-well culture plates (Falcon) or 24-well plates (Falcon) onto glass coverslips. In some experiments, cells were incubated with 20 μM hemin (Frontier Scientific), 350 μM δ-aminolevulinic acid hydrochloride (ALA, Sigma-Aldrich), 2.5 mM N-Acetyl-l-cysteine (NAC, Sigma-Aldrich), 500 μM succinylacetone (SA, Sigma-Aldrich); 2 μM pyridostatin hydrochloride (PDS, MedChemTronica), 0.25 μM etoposide (Sigma-Aldrich) for 3 or 24 h. Unless otherwise indicated, experiments were performed in complete medium.
HEK293T cells were modified and characterized as described previously [25]. In short: the HMOX1-deficient HEK293T cell line (KO-HMOX1) was created using CRISPR-Cas9 genome editing with pX330-Pac-Cer plasmid that encodes sgRNA targeting the second exon of human HMOX1 gene. Control cells [WT(mock)] were created using an empty pX330-Pac-Cer vector. CELI mismatch detection assay was performed to evaluate the presence of mutations. HO-1 protein was undetectable in KO-HMOX1 cells both under control conditions and after incubation with hemin [25].
Induced pluripotent stem cells (iPSCs). Murine iPSCs were cultured as described previously [8]. In short: Hmox1-/- iPSCs were reprogrammed from fibroblasts collected from C57BL6 × FVB Hmox1-/- mice and cultured in DMEM High-Glucose medium containing 20 % FBS, 1 % Non-Essential Amino Acids (Life Technologies), 0.1 mM β-mercaptoethanol (Life Technologies), 1000 U/mL leukemia inhibitory factor (LIF, Millipore), and antibiotics (100 IU/mL penicillin and 100 μg/mL streptomycin). Hmox1-/-Hmox2-/- (double knockout, dKO) iPSCs were generated from Hmox1-/- cells using pSpCas9(BB)-2A-Puro plasmid with sgRNA encoding insert targeting Hmox2 (heme oxygenase-2). Then, using lentiviral vectors, constructs providing stable expression of Hmox1 with nuclear localization signal (NLS) or with nuclear export signal (NES) were introduced to Hmox1-/-Hmox2-/- iPSCs [8].
Mouse hematopoietic stem cells (HSCs). Mice were used only for tissue collection, and therefore there was no ethical committee approval required. Mice were housed in individually ventilated cages in specific pathogen-free conditions and had unlimited access to food and water. For the isolation of hematopoietic stem cells, we used C57BL6xFVB Hmox1+/+ or Hmox1-/- mice bred in our facility. Mice were euthanized via CO2 inhalation. Bone marrow was isolated from femurs and tibia, as described earlier [26]. Bones were crushed in a mortar and pestle in 2 % FBS in phosphate buffered saline (PBS, Lonza). Bone marrow isolated from male and female individuals was pooled and collected into tubes, filtered through a cell strainer (100 μm), and centrifuged at 670 g for 10 min at 4oC. Next, the cell pellet was resuspended in an RBC lysis buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) and incubated for 7 min at room temperature. After the dilution of the lysis buffer with 2 % FBS in PBS, the cells were centrifuged and washed with PBS. Finally, the cell pellet was resuspended with 100 μL of 2 % FBS in PBS.
Mouse HSCs (LKS CD150+ CD48−) [26] were sorted using MoFlo XDP FACS sorter (Beckman Coulter) using following clones of antibodies: anti-mouse CD3, clone 17A2; anti-mouse Ly-6G/Ly-6C, clone RB6-8C5; anti-mouse CD11b, clone M1/70; anti-mouse CD45R/B220, clone RA3-6B2; anti-mouse TER-119/erythroid cells, clone Ter-119; anti-mouse-CD150, clone TC15-12F12.2; anti-mouse-CD48, clone HM-48-1 (all Biolegend); anti-mouse-Ly6A/E (Sca1), clone D7; anti-mouse-CD117 (cKit), clone 2B8 (eBioscience). Sorted HSCs were then cultured for 7 days in non-adherent U-shaped 96-well plates (Greiner), ∼2,500 cells/well in standard cell culture conditions. HSC culture medium was StemSpan SFEM (Stem Cell Technologies), supplemented with 20 % of BIT 9500 Serum Substitute (Stem Cell Technologies), murine stem cell factor (mSCF, Peprotech), human thrombopoietin (hTPO, Peprotech), murine interleukin-3 (mIL-3, Peprotech), and human erythropoietin (hEPO, Sigma-Aldrich), all at a concentration of 20 ng/mL.
Patient-derived lymphoblastoid cell line (LCL). Immortalized lymphoblastoid cells were generated as previously described from a patient with genetically confirmed HMOX1 mutation, c.262_268delinsCC (p.Ala88Profs*51) and c.55dupG (p.Glu19Glyfs*14) in trans [27]. LCL cells were characterized by the absence of inducible HO-1 expression. In brief, peripheral blood mononuclear cells (PBMCs) from venous blood samples were incubated with the supernatant of EBV-producing B95-8 marmoset B-lymphoblastoid cells and maintained in RPMI supplemented with l-Glutamine, antibiotics and antimycotics [(100 units/mL of penicillin, 100 μg/mL of streptomycin, and 0.25 μg/mL of amphotericin B (Gibco)] and 20 % FBS at 37 °C and 5 % CO2. The patient's clinical presentation is described in detail elsewhere [27]. He exhibited a severe, multisystem disease with frequent episodes of hyperinflammation irresponsive to immunosuppression, organ failure, and premature death due to pulmonary hemorrhage. Control LCL derived from a healthy donor.
Immunoblotting. Cultured cells were detached with TrypLE in preparation for lysates. Cells were then suspended in cold PBS and centrifuged at 400 g. Pellets were resuspended in Pierce RIPA buffer (Thermo Scientific) with protease inhibitors (complete Protease Inhibitor Cocktail, Merck) and incubated for 5 min at 4 °C with agitation. Lysates were clarified by centrifugation at 8,000 g for 10 min at 4 °C. Protein concentration was determined by a BCA assay kit (Thermofisher) to ensure the equal loading of each sample (10 μg of proteins). Samples were electrophoretically separated in 4–20 % Mini-PROTEAN TGX Precast Protein Gels (BioRad) followed by transfer to a nitrocellulose blotting membrane (BioRad) in transfer buffer (BioRad) with 20 % ethanol. The membranes were blocked with EveryBlot Blocking Buffer (BioRad) for 5 min and incubated with anti-HO-1 (ADI-SPA-896-J, Enzo Life Science), anti-PARP (AF-600-NA, R&D Systems), anti-α-tubulin (T9026, Sigma-Aldrich) primary antibodies overnight at 4 °C. The membranes were then washed three times with Tris-buffered saline with 0.1 % Tween 20 (TBST) and incubated with anti-rabbit-HRP (HO-1), anti-mouse-HRP (α-tubulin), or anti-goat-HRP (PARP) antibodies (Invitrogen) diluted in EveryBlot blocking buffer for 1 h at room temperature. After five washes with TBST, the membrane was incubated with horse-radish peroxidase (HRP) substrate (Bio-Rad) for chemiluminescence detection that was performed on a ChemiDoc MP instrument (Bio-Rad).
MTT reduction assay. Cells (15,000 cells/well) were seeded in a 96-well plate (Falcon) and stimulated with hemin for 24 h at 37°C. After stimulation, cells were cultured with 1 mg/mL MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, Sigma-Aldrich) for ∼1 h at 37°C. Then formazan crystals were dissolved with 50 μL lysis buffer (10 % SDS and 0.6 % acetic acid in DMSO). Absorbance was measured at a wavelength of 570 nm using a TECAN infinite M200 microplate reader (TECAN reader).
Proximity ligation assay (PLA). For in-situ visualization of the colocalization between HO-1 and PARP1 proteins, we followed the Duolink PLA Fluorescence Protocol (Sigma-Aldrich) using the Duolink In Situ Detection Reagents Orange kit (Sigma-Aldrich). Briefly, the cells grown on glass coverslips covered with 1 % Geltrex LDEV-Free Reduced Growth Factor Basement Membrane Matrix (Gibco), were fixed with 4 % electron microscopy-grade, methanol-free paraformaldehyde (Biotum) and permeabilized with 0.2 % PBS-Tx (PBS with 0.2 % Triton X-100) for 10 min at room temperature. Next, cells were blocked in a drop of Blocking Solution (Sigma-Aldrich) at 37°C for 1 h and incubated with primary antibodies [anti-HO-1 (ADI-SPA-896, Enzo), anti-PARP (AF-600-NA, R&D Systems)] diluted 1:100 in Duolink Antibody Diluent (Sigma-Aldrich). After washing in wash buffer A (10 mM Tris, 150 mM NaCl and 0.05 % Tween 20), cells were incubated with secondary antibodies conjugated with PLUS and MINUS probes for 1 h at 37 °C. The following Duolink In Situ PLA secondary antibodies (Sigma-Aldrich) were used: anti-goat PLUS, and anti-rabbit MINUS. Next, the cells were again washed twice in wash buffer A and then incubated with the ligase (diluted 1:40 in ligation buffer) for 30 min at 37 °C. After the next round of washing in wash buffer A, cells were incubated with polymerase (diluted 1:80 in an amplification buffer) for 100 min at 37 °C. Finally, cells were washed in wash buffer B (200 mM Tris and 100 mM NaCl), counterstained with DAPI (0.5 μg/mL, Sigma-Aldrich), mounted in Fluorescence Mounting Medium (Dako), and allowed to dry before imaging. Negative controls were performed using secondary antibodies only. Cells were analyzed with a Zeiss LSM 880 confocal microscope (Zeiss). We imaged single optical sections using the Plan-Neofluar 40 1.30 Oil DIC M27, alpha Plan-Apochromat.
Immunofluorescence staining. Cells grown on glass coverslips covered with 1 % Geltrex LDEV-Free Reduced Growth Factor Basement Membrane Matrix were fixed with ice-cold 80 % methanol (POCh) at −20°C for 10 min or with 4 % Pierce methanol-free formaldehyde (Thermo Scientific) at room temperature for 10 min and followed by two washes with PBS. Cells fixed in formaldehyde were subsequently permeabilized in PBS-Tx at room temperature for 10 min and blocked in 10 % normal donkey serum (Sigma-Aldrich) in 0.1 % PBS-Tx at room temperature for 30 min. Then, the cells were incubated with primary antibodies diluted in 0.1 % PBS-Tx with 1 % donkey serum. Blocking was followed by three PBS washes before incubation with the appropriate secondary antibodies that were diluted 1:400 in 0.1 % PBS-Tx containing 1 % donkey serum at room temperature for one to 3 h. Finally, after washing with PBS and counterstaining the cell nuclei with 0.5 μg/mL DAPI (Sigma-Aldrich) for 10 min at room temperature, the coverslips were mounted in Fluorescence Mounting Medium (Dako) and allowed to dry before imaging. Negative controls with the omission of primary antibodies were performed for each protein. Cells were analyzed using the Axio Observer Z1 microscope (Zeiss) using 63x/1.4 Oil DIC m27 objective or by Leica DMI6000 using 40x/0.75 objective.
For immunocytochemical staining, the following primary antibodies were used: anti-PARP (AF-600-NA, R&D Systems) 1:200; anti-G4 (mouse 1H6 clone, MABE1126, Millipore), anti-HO-1 (ADI-SPA-894, Enzo Life Sciences), anti-phospho-H2AX (Ser139, JBW301 clone, 05–636, EDM Milipore) diluted 1:100, anti-p53 (ab131442, Abcam) diluted 1:200. The following secondary antibodies from Molecular Probes (Invitrogen), diluted 1:400, were used: AlexaFluor 488 anti-rabbit IgG (A21206), AlexaFluor 568 anti-mouse IgG (A10037), AlexaFluor 568 anti-goat IgG (A11057), and AlexaFluor 488 anti-mouse IgG (A21202).
Flow cytometry. Cells for flow cytometry were detached with TrypLE, fixed with 4 % paraformaldehyde in PBS for 10 min, permeabilized with 0.1 % Triton X100 in PBS, and stained with anti-PARP (AF-600-NA, R&D Systems) and then AlexaFluor 568 anti-goat IgG (A11057) antibodies. Fluorescence signal was analyzed using BD LSR Fortessa flow cytometer with excitation at λ = 561 nm and emission at 575/26. Quantitative analysis was performed using the FloJo software.
FRAP analysis. Human PARP1 coding sequence (NCBI Reference Sequence: NM_001618.4) was inserted into a Sleeping Beauty system vector based on the pSBbi-Hyg plasmid (a gift from Eric Kowarz, Addgene plasmid # 60524 [28]) conferring resistance to hygromycin and providing constitutive expression of PARP1 under the control of the UbC promoter, with the Clover green fluorescent protein at the N-terminus. FRAP analysis was performed in living HEK293T cells transfected with PARP1-GFP fusion proteins. A laser-scanning confocal microscope (Zeiss LSM 880) equipped with a 488 nm laser light and a 40 × 1.3 numerical aperture objective was used to perform all photobleaching experiments (bleaching: 488 nm, 150 ms, 5 iterations). Fluorescence recovery was monitored over a 120 s period. The data obtained for recovery were corrected for the background intensity and loss of total fluorescence [29]. The T-half analysis was performed with easyFRAP: https://easyfrap.vmnet.upatras.gr.
ImageStream cytometry. Cells were immunostained using anti-p53 (MA1-7629, Invitrogen) and AlexaFluor 488 anti-rabbit (A21206, Invitrogen) antibodies and co-stained with DAPI (0.5 μg/mL; Sigma-Aldrich) for nuclear imaging. On average, 3,000 events per sample were collected using ImageStream IS100 (Amnis). The spatial relationship between p53 and nuclear images was measured using the ‘Nuclear Translocation’ feature of the IDEAS software package.
Lentivirus production and cell transduction. Lentiviral particles were generated by co-transfection of 3 plasmids into HEK293T cells using Transfection Grade Linear Polyethylenimine Hydrochloride (PEI Max, Polysciences): envelope plasmid (PMD2.G), packaging plasmid (psPAX2) and transfer plasmid (for LivePAR pLV-EF1A-LivePAR-Hygro which was a gift from Robert Sobol (Addgene plasmid #176063) [30], for NAD+ sensors: NS-Grapefruit, NS-Grapefruit-NLS plasmids [31]), which were a gift from Min Hu and Qiuliyang Yu. Forty-eight hours after transfection, lentivirus-containing supernatant was centrifuged at 200 g for 5 min to pellet any cells that were collected during harvesting and the supernatant was filtered through a 0.45 μm PES filter (Braun).
Next, WT(mock), KO-HMOX1 HEK293T cells, or iPSC dKO, NLS and NES were seeded into 6-well plates and after 24 h medium with lentiviral particles (1 mL) was mixed with polybrene (2 mg/mL) and added to the cells. Cells were incubated overnight and then the medium with lentiviral particles was removed and replaced with fresh medium. Next, GFP-positive cells (stable cell lines) were sorted using MoFlo XDP (Beckman Coulter).
Analysis of NAD + levels. The HEK293T cell lines that stably express the FRET sensors (in cytoplasm or nucleus) were seeded at a density of 50,000 cells per well into a 24-well plate. After 48 h of incubation, different compounds were added into the culture medium: 10 mM β-Nicotinamide adenine dinucleotide sodium salt (β-NAD, Sigma-Aldrich), 10 nmol/L FK866 (HY-50876, MedChemExpress), 250 nM etoposide, and 100 μM olaparib. After 24 h incubation cells were washed with DPBS, collected using TrypLE Express (Thermo Scientific) and analyzed using BD LSR Fortessa flow cytometer with excitation at λ = 488 nm and emission at 670/30 nm and 530/30 nm. Quantitative analysis was performed using the FloJo software.
LivePAR Recruitment after laser-induced micro-irradiation. For analysis of PARylation in HEK293T cells, WT(mock) and KO-HMOX1 lines were transfected using lipofectamine 2000 (Invitrogen). On the next day, cells were seeded into μ-Slide 18 Well Glass Bottom (Ibidi) covered with Geltrex.
Stable LivePAR+ iPSC dKO, NLS and NES were seeded into μ-Slide 18 Well Glass Bottom (Ibidi) covered with Geltrex. After the next 24 h, laser micro-irradiation (50 mW laser λ = 405 nm, 95 % of total power, 50 ms) was performed using a Stellaris 5 (Lecia) equipped with HC PL APO CS2 40x/1.30 OIL objective. Time-lapse images of LivePAR (GFP signal) were collected every 1 s during a 1–3 min analysis. Cells stimulated for 1 h before laser irradiation with 100 nM olaparib (inhibitor of PARylation) were used as a control. Quantitative analysis was performed using the Image J software.
PARP1 autoparylation assay. PARP1 autoparylation in the presence of HO-1 was assessed using modified method by Xie and co-workers [32]. Briefly, 10 nM recombinant PARP1 (Active Motif, cat. nr 81037) was incubated with 61.25 nM recombinant HO-1 (R&D Systems, cat. nr 3776-HM-020) in 50 mM Tris-HCL (pH = 8.0), with 2 mM MgCl2, 25 μM biotin-NAD+ (R&D systems, cat. nr 6573/131U), 75 μM NAD+ (Sigma-Aldrich, cat. nr N0632-1G) and 5 μg/mL oligonucleotide which forms (G4, AGGGAGGGCGCTGGGAGGAGGG) or does not form G-quadruplexes (ssDNA, CCAGTTCGTAGTAACCC) in the total volume of 20 μL. The reaction was carried out at room temperature for 1 or 3 min and stopped by adding NuPage LDS sample buffer (Thermo Scientific, cat. nr NP0007). Samples were then separated on 4–15 % MP TGX gel (Bio-Rad, cat. nr 4561083), transferred to nitrocellulose membranes using Trans-Blot Turbo RTA Mini 0.2 μm Nitrocellulose Transfer Kit (Bio-Rad, cat nr. 1704270). Membranes were then blocked with EveryBlot blocking buffer (Bio-Rad, cat. nr 12010020) for 5 min at room temperature and incubated for 1 h with 1:3000 streptavidin-HRP (Thermo Scientific, cat. nr N100). Chemiluminescence was detected with Immobilon Western Chemiluminescent HRP Substrate (Millipore Sigma, cat. nr WBKLS0500) using Chemidoc MP (Bio-Rad). As a negative control we used reaction mix without NAD+. Additionally, specificity of the PARylation detection was confirmed with anti-PAR antibody (R&D Systems, cat. nr 4335-MC-100), (data not shown).
Total heme measurement. Intracellular heme concentration was measured using a fluorescence-based assay [33]. Briefly, cells were detached with TrypLE and suspended in 100 μL 2 M oxalic acid (Sigma-Aldrich) and then split. One sample was heated at 95 °C for 30 min to remove iron from heme. The other solution was treated in the same way but at room temperature throughout to control for the presence of endogenous porphyrins. Fluorescence (wavelengths: excitation 400 nm, emission 662 nm) was measured by Spectra II Microplate Reader (Tecan). Data were normalized to the total protein concentration.
Free heme measurement. For live imaging of cytosolic and nuclear free heme we used HS1 cytosolic and HS1 nuclear plasmids [34] kindly provided by prof. Amit R. Reddit (Georgia Institute of Technology). These constructs consist of a heme-binding domain from cytochrome b562, fused to a pair of fluorescent proteins, EGFP and Katushka 2 (mKATE2). The ratio of EGFP (heme-sensitive) to mKATE2 (heme-insensitive) fluorescence provides a readout of cellular heme independent of sensor concentration [34]. Cells were transfected using polyethyleneimine MAX (PEI MAX, Polysciences) and assay was performed as described previously [8]. Fluorescence was analyzed using LSR Fortessa flow cytometer (Becton Dickinson).
Detection of oxidative stress. We used CellROX Deep Red Reagent for oxidative stress detection (Thermo Fisher Scientific). Cells were cultured in the 12-well plates and stimulated with hemin (20 μM) or ALA (350 μM) for 24 h. Tert-butyl hydroperoxide (TBHP, 200 μM) was added for 30 min as a positive control. To detect reactive oxygen species, CellROX Deep Red reagent (500 nM) was added to each sample and incubated for 30 min at 37 °C. Fluorescence was analyzed using LSR Fortessa flow cytometer (Becton Dickinson).
Lipid Peroxidation. We used the Click-iT Lipid Peroxidation (LAA) Kit for Imaging – Alexa Fluor 488 (C10446, Thermo Fisher Scientific), following the provider's protocol. Cells were cultured in the 24-well plates with round coverslips and stimulated with hemin (20 μM) or ALA (350 μM) for 24 h. Cumene hydroperoxide (CH) served as a positive control. The cells were fixed with 4 % Pierce methanol-free formaldehyde (Thermo Scientific) at room temperature for 10 min and washed twice with PBS. Then, the cells were permeabilized in PBS-Tx at room temperature for 10 min. Lipid peroxidation was detected by incubation of coverslips with 50 μL Click-iT reaction cocktail for 30 min at room temperature. Next, the samples were washed 3 times with PBS and co-stained with DAPI (0.5 μg/mL; Sigma-Aldrich). Fluorescence detection was performed using Leica DM6B fluorescent microscope with PL Fluotar L 20x/0.40 objective.
DNA fiber assay. All procedures were based on the protocol described by Nieminuszczy and colleagues [35]. Cells grown on the 6-well plates were stimulated with nucleotide analogs: 25 mM IdU (5-iodo-2-deoxyuridine, Sigma) and 250 mM CldU (5-chloro-2-deoxyuridine, Sigma), both for 20 min at 37 °C. Next, the cells were washed with PBS, detached with TrypLE, centrifuged, and resuspended in cold Dulbecco's PBS (DPBS) to the concentration of 750,000 cells/mL. Then, the cells were lysed (200 mM Tris-HCl, pH 7.5, 50 mM EDTA, 0.5 % SDS), spread on the glass Superfrost Plus Gold slides (Thermo Scientific) and fixed in acetic acid:methanol (3:1). After incubation in 2.5 N HCl, fixed chromatin slides were incubated in blocking solution (5 % BSA in PBS) and stained with primary antibodies: anti-BrdU rat (BU1/75 (ICR1), ab6326, Abcam) and anti-BrdU mouse (B44, BD, 347580) for 2 h at room temperature; secondary antibodies: Alexa Fluor 488 anti-rat IgG (A21208, Invitrogen) and Alexa Fluor 568 anti-mouse IgG (A10037, Invitrogen) for 1 h at room temperature. Finally, after washing with PBS, the coverslips were mounted in a fluorescence mounting medium (Dako) and allowed to dry before imaging. The LSM 880 AxioObserver (Zeiss) confocal microscope equipped with an EC Plan-Neofluar Plan-Apochromat 63x/1.4 oil DIC M27 objective or Leica DM6B fluorescent microscope with PL Fluotar 63x/1.30 oil objective was used for visualization of DNA fibers. ImageJ software was used to quantify the number and length of DNA fibers from the images.
G4 labeling in DNA fibers. Cells grown on the 12-well plates were stimulated with 10 μM EdU for 30 min. Next, cells were washed with PBS, detached with TrypLE, centrifuged, and resuspended in cold DPBS to the concentration of 650,000 cells/mL. Then, cells were lysed, spread on glass Superfrost Plus Gold (Thermo Scientific) slides and fixed in acetic acid:methanol (3:1) for 10 min. Next, slides were washed in distilled water and twice in 5 % BSA in PBS. EdU staining was performed by incubation for 30 min at room temperature with a Click-iT reaction cocktail (Click-iT EdU Cell Proliferation Kit for Imaging – Alexa Fluor 488, Thermo Fisher Scientific) prepared according to the manufacturer's protocol. After incubation, slides were washed twice in 5 % BSA in PBS and blocked in 10 % donkey serum in PBS-Tx for 30 min at room temperature. The samples were then incubated with anti-G4 primary antibodies (mouse 1H6 clone, Millipore) overnight at 4 °C diluted 1:100 in 1 % donkey serum in PBS-Tx. Finally, after washing with PBS, G-quadruplexes were visualized by secondary antibodies [Alexa Fluor 568 anti-mouse IgG (Thermo Scientific)] diluted 1:400, for 3 h at room temperature. The coverslips were mounted in Fluorescence Mounting Medium (Dako) and fibers were analyzed with a Leica DM6B fluorescent microscope using PL Fluotar 63x/1.30 Oil objective and quantified manually.
Cell cycle and cell culture growth. Cells grown on the 6-well plates were fixed with ice-cold 80 % methanol (POCh) at −20°C for at least 30 min. After washing and centrifuging, cells were stained with FxCyclePI/RNase Staining Solution (Life Technologies). After 15 min of incubation at room temperature samples were analyzed using BD LSR Fortessa (Becton Dickinson) (excitation 488 nm, emission 585 nm). Data were analyzed using FlowJo software (BD Life Sciences) by univariate modeling. To evaluate the cell culture growth rate, cells were seeded in a 12-well plate (150,000 cells/well), collected after 24, 48, and 72 h, and then counted using the BioRad TC20 automated cell counter.
Timelapses – duration of the cell cycle. Cells were seeded (50,000 cells/well) onto a black 24-well plate with a glass bottom (Cellvis). After 24 h, NucSpot Live 650 Nuclear Stain (Biotum) was added to the cells for 30 min at 37°C. Then timelapses were performed using Olympus IX microscope with a Lumencor Spectra X 647 nm fluorescence LED light source for 72 h (image intervals – 15 min). The cell cycle duration (time between metaphase plate of mother and daughter cell) was measured manually using ImageJ software.
Timelapses – p53 nuclear translocation. Cells were seeded (50,000 cells/well) onto a black 24-well plate with a glass bottom (Cellvis). After 24 h, they were transfected with GFP-p53 plasmid, a gift from Tyler Jacks (Addgene plasmid #12091), [36]) by using lipofectamine 2000 and incubated for 24 h. Then 20 μM etoposide was added and timelapses were performed using Olympus IX microscope with a Lumencor Spectra X 647 nm fluorescence LED light source for 3 h (image intervals – 10 min). Nuclear translocation of p53 was calculated by measuring the increase in fluorescence signal in nucleus of single cell (ratio of the last and the first timelapse frame).
Heme depleted serum. Heme-depleted serum was prepared by treating FBS (Biowest) with 1 mM ascorbic acid (Sigma-Aldrich). The mixture was then incubated at 37 °C and mixed at 50 rpm. The depletion of heme was confirmed by measuring the optical absorbance of the serum at 405 nm using a spectrophotometer (Tecan). The optical density is stabilized at around 0.6–0.8 after 7 h. Subsequently, heme depletion was achieved through three rounds of dialysis against cold PBS using 2K MWCO dialysis flasks (Thermo Scientific). The first dialysis was carried out overnight, followed by two more dialysis sessions of 4 h each. Finally, the serum was sterilized by filtering it through 0.2 μm syringe filters (Sarstedt).
Reverse Transcription and Real-Time PCR. Cells were seeded (50,000 cells/well) onto a 12-well plate and treated with 500 μM succinylacetone (SA – Sigma Aldrich) in culture medium with heme depleted serum for 24 h. Additionally, cells were treated with 100 nM olaparib (HY-10162, MedChemExpress) in complete culture medium for 24 h. RNA was isolated using a RNeasy Mini Kit (Qiagen) and reverse-transcribed with a QuantiTect Reverse Transcription Kit (Qiagen) with integrated gDNA removal. The gene expression was assessed on a StepOnePlus thermocycler (Applied Biosystems) with real-time PCR using an SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) and specific primers: EEF2 For – GCG GTC AGC ACA ATG GCA TA, Rev – GAC ATC ACC AAG GGT GTG CAG; CDKN1A (p21) For – AGG ACT CAA CCG TAA TAT CC, Rev – CAG CAG ATC ACC AGA TTA AC.
RNA-seq analysis. We used our previously published data available in the BioProject database, accession no. PRJNA562450 [26].
Statistical Analysis. All experiments were performed in duplicate or in triplicate and were repeated independently at least three times, unless otherwise indicated. Data were analyzed with GraphPad Prism 8.0 software. Tests used in the statistical analysis are listed in the description of the figures. Bar graphs represent mean +SEM; ns – non significant, *-p≤0.05, **-p<0.01, ***- p < 0.001.
Other. The graphical abstract was created with BioRender.com (Agreement number: IV26TQ6OJU).
3. Results
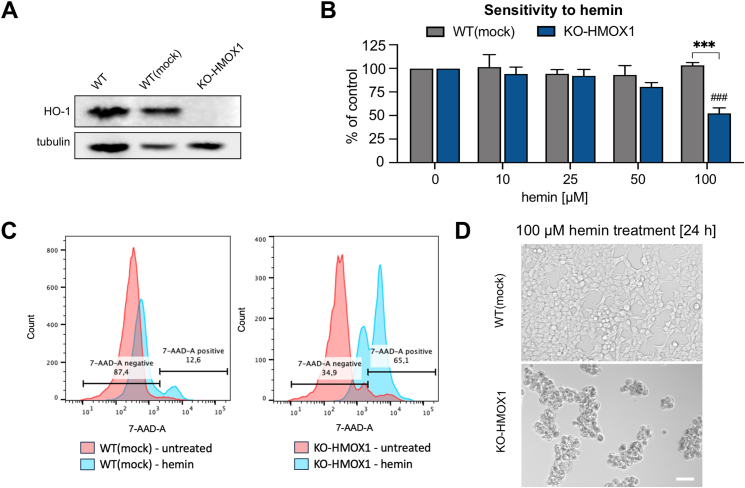
HMOX1-deficient HEK293T cells. In previous study [8], we demonstrated the colocalization of HO-1 protein and DNA G-quadruplexes in the nuclei of immortalized human embryonic kidney HEK293T cells. Here, to investigate the possible effects of G-quadruplex accumulation and the role of HO-1, we employed the HMOX1-deficient HEK293T cell line (KO-HMOX1) created by CRISPR-Cas9 genome editing [25]. Cells treated with empty vector [WT(mock)] were used as a control. Western blot confirmed successful knocking-down of the HMOX1 gene and removal of the HO-1 protein (Fig. 1A). The empty vector did not change HO-1 levels compared to intact cells (WT). As expected, KO-HMOX1 HEK293T cells were more sensitive to hemin treatment than their WT(mock) counterparts, as shown by the results of the MTT reduction assay. Viability of control cells was unaffected even in response to high concentration of hemin (100 μM, 24 h), but was reduced by approximately 50 % in HMOX1-deficient cells (Fig. 1B). Differences in hemin sensitivity were confirmed by cytometric analyzes and cell morphology (Fig. 1C and D).
Fig. 1.
Characterization of HEK293T WT(mock) and KO-HMOX1 cell lines. A) Expression of HO-1 protein in cells cultured under control conditions. Western blot. Tubulin was used as a loading control. B) Effect of hemin (10–100 μM, 24 h) on cell viability. MTT reduction assay. ANOVA. * - KO-HMOX1 vs. WT(mock), # - treated vs. untreated cells. C) Flow cytometer plot for 7-AAD staining after hemin (100 μM, 24 h) treatment and in control (untreated) group. Percentage of 7-AAD negative (range was selected based on comparison to negative control without 7-AAD) and 7-AAD positive cells was indicated for cells after hemin stimulation. D) Effect of hemin (100 μM, 24 h) on cell morphology. Representative images. Bright field, scale bar 200 μm.
HO-1 colocalizes with PARP1 and affects its functions. HO-1 has been reported to co-precipitate with PARP1 [4], a protein involved in both DNA replication and repair, which binds to G-quadruplexes [9,18]. To check whether HO-1 could affect PARP1-dependent pathways, we first compared the expression level of PARP1 protein in WT(mock) and KO-HMOX1 cells. Western blot analysis did not show any differences between cell lines (Fig. 2A). However, a more quantitative assay using flow cytometry-detected immunostaining suggested higher PARP1 protein levels in HMOX1-deficient cells (Fig. 2B). This observation was confirmed by immunofluorescence microscopy, where stronger PARP1 spots were visible in at least some KO-HMOX1 cells (Fig. 2C). Moreover, using PLA we also showed a close colocalization of HO-1 and PARP1 in our HEK293T cells cultured under control conditions (Fig. 2D).
Fig. 2.
The role of HO-1 in the regulation of PARP1 pathway. A) PARP1 protein level. Western blotting (left) and densitometry results (right) of PARP1 in HEK293T cells. Tubulin was used as a loading control. T-test. B) PARP1 protein level. Immunofluorescence staining was analyzed by flow cytometry. T-test. C) PARP1 in HEK293T cells. Representative images of PARP1 intracellular localization (left) and densitometry analysis (right). Mann-Whitney test. Scale bar 20 μm. D) Colocalization (red dots) of HO-1 and PARP1 proteins in WT(mock) cells. Cell nuclei were counterstained with DAPI (blue). Scale bar 10 μm. Proximity ligation assay (PLA). E) Fluorescence recovery after photobleaching (FRAP) analysis of PARP1-GFP using a confocal microscope. Representative images (left) taken at the indicated timepoints (in seconds) and quantitative analysis (right) of T-half of recovery after photobleaching of PARP1-GFP. T-test. Scale bar 20 μm. F) Timelapse analysis of livePAR in WT(mock) and KO-Hmox1 after induction of DNA damage in the cell nucleus. Foci (arrows) in the image demonstrate LivePAR recruitment. G) Recruitment of LivePAR in HEK293T (left) or iPSCs (right) after laser-induced micro-irradiation. H) Effect of β-NAD (10 mM) with and without FK866 (10 nM), etoposide (250 nM), and olaparib (100 nM) on cytoplasmic (left) and nuclear (right) NAD+ levels. The FRET NAD+ sensors were analyzed by flow cytometry. Two-way ANOVA. I) Autoparylation in vitro of recombinant PARP1 in the presence of recombinant HMOX1. The liquid reaction was proceeded with mixture of biotin-NAD+ (25 μM), NAD+ (75 μM) and ssDNA or G4 oligonucleotides (5 μg/mL). Western blotting (right) and densitometry (left) detection with streptavidin-HRP. Reaction mix without NAD+ was used as a negative control. ANOVA. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To test whether HO-1 could have any functional effect on PARP1, we transfected the cells with pEGFP expression plasmid encoding full-length GFP-tagged human PARP1, and then monitored fluorescence recovery after photobleaching (FRAP) [37], induced by pulsing with 488 nm laser light (Fig. 2E). As expected, we observed a rapid increase in the fluorescence intensity of PARP1 at the site of photobleaching, reaching maximum values within 10–20 s. Interestingly, PARP1 accumulated more rapidly in HMOX1-deficient cells (t1/2 = 3.71 s) than in their HO-1 expressing counterparts (t1/2 = 4.78 s, p < 0.05) (Fig. 2E).
In the next step, we used a genetically encoded LivePAR probe for live-cell imaging of PAR accumulation to follow PARP activation in real time in WT(mock) and KO-HMOX1 cells. LivePAR is a PAR-binding fusion protein (PAR binding domain of RNF146) tagged with EGFP [38]. We measured the kinetics of LivePAR recruitment in response to laser micro-irradiation (405 nm, 50 ms) in single cells (Fig. 2F). Probe accumulation reached peak intensity within 1 min. Interestingly, PAR formation at the site of laser micro-irradiation was more intense in HMOX1-deficient HEK293T cells (Fig. 2G). Then, we performed the same measurements using murine induced pluripotent stem cells (iPSCs) lacking Hmox1 and Hmox2 (dKO) or stably expressing either cytoplasmic HO-1 (NES) or nuclear HO-1 (NLS) [8]. The results were similar: PAR formation was more intense in Hmox1-deficient cells, while cytoplasmic or nuclear localization of HO-1 was not significant (Fig. 2G). Hence, it appears that higher PARylation dynamics in the absence of HO-1 is not cell type specific but is a more common feature.
PARP enzymatic activity and PAR accumulation can be modulated by changes in NAD+ levels [38]. Therefore, we analyzed NAD+ availability in both the cytoplasm and nucleus using genetically encoded FRET-based NAD+ sensors targeting appropriate cellular compartments [31]. The sensors report subcellular NAD+ levels as a ratio of the emission intensities at 594 nm and 507 nm (Fig. 2H). Treatment of cells with FK866, an inhibitor of nicotinamide phosphoribosyltransferase-mediated NAD+ synthesis, resulted in a significant reduction in the NAD+ signal, an effect reversed by supplementation of cells with β-NAD, confirming the specificity of NAD+ detection. We observed slightly decreased NAD+ levels in the cytoplasm of KO-HMOX1 HEK293 cells compared to control counterparts. No differences were found in the nucleus (Fig. 2H). Importantly, there were no differences in NAD + levels related to HMOX1 status in cells treated with etoposide (to induce DNA damage response and activate PARP1) or olaparib (PARP1 and PARP2 competitive inhibitor). In both KO-HMOX1 and WT(mock) cells, NAD+ concentrations in the cytoplasm and nucleus after etoposide or olaparib administration were comparable to that in control conditions (Fig. 2H). These results indicate that NAD+ is not a limiting factor influencing PARylation dynamics in HEK293T cells.
Finally, to detect a possible direct effect of HO-1 protein on PARP1 enzymatic activity, we measured autoPARylation of PARP1 protein in vitro in the presence or absence of HO-1 protein (Fig. 2I). We did not observe any effect of HO-1 in such an isolated system with or without dsDNA or G4-forming DNA. Thus, the effect of HO-1 deficiency on PARylation in HEK293T cells appears to be more associated with PARP1 cellular motility, but not with a substrate availability or direct protein-protein interaction modulating PARP1 enzymatic activity.
To sum up, we showed that HO-1 colocalizes with PARP1, and HMOX1 deficiency influences dynamics of laser micro-irradiation-induced PARylation. Next, we checked whether HO-1 affects G-quadruplex accumulation in HEK293T cells and the cellular response to DNA damage.
HO-1 prevents the stabilization of G-quadruplexes. We used immunostaining with specific antibodies to compare the level of G-quadruplexes in the WT(mock) and KO-HMOX1 cell lines. Similarly to murine iPSCs reported in our previous study [8], we detected more G-quadruplexes in HMOX1-deficient HEK293T cells cultured in control conditions (Fig. 3A). Treatment of the cells with pyridostatin (PDS, 2 μM), a well-known stabilizer of G-quadruplexes, significantly elevated the signal, regardless of HMOX1 status. This indirectly indicates that HO-1 is possibly not important in the formation of G-quadruplexes, but rather may reduce their stability by regulating heme levels, the effect not visible in the presence of an exogenous stabilizer.
Fig. 3.
G-quadruplex accumulation and DNA-damage response (DDR) in WT(mock) and KO-HMOX1 cells. A) G-quadruplexes in cells cultured under control conditions or incubated with PDS (2 μM, 24 h). B) DDR measured as γH2AX staining in cells cultured under control conditions or incubated with etoposide (0.25 μM), hemin (20 μM), ALA (350 μM) or PDS (2 μM) for 24 h. Immunocytochemistry and confocal microscopy. Representative images are presented in inserts. Kruskal-Wallis test. * - KO-HMOX1 vs. WT(mock), # - treated vs. untreated cells.
HO-1 protects cells from genotoxic stress. To test whether HO-1 could contribute to the protection of DNA against damage, we used immunocytochemical staining of phosphorylated histone H2AX (γH2AX), as a marker of the DNA damage response (DDR) to DNA double-strand breaks [39]. Under control conditions, KO-HMOX1 cells exhibited more intense γH2AX staining, which may indicate a more pronounced DDR associated with either more severe DNA lesions or less efficient repair of damage (Fig. 3B). Furthermore, exposure of cells to a low concentration of etoposide (0.25 μM), an anticancer drug that generates DSB [40], increased the γH2AX signal, confirming the DDR induction. This effect was significantly stronger in HMOX1-deficient cells (Fig. 3B), indicating that HO-1 may play a role in protecting cells from genotoxic stress.
We supposed that the observed effects of HMOX1 deficiency are related to elevated levels of pro-oxidative heme, which is not sufficiently removed in the absence of a heme-degrading enzyme. Accordingly, we expected that the effect of HMOX1 deficiency would be stronger in cells with elevated levels of intracellular heme. We therefore compared γH2AX staining in WT(mock) and KO-HMOX1 cells exposed to hemin (20 μM) or supplemented with δ-aminolevulinic acid (ALA, 350 μM), a substrate in the heme synthesis pathway (Fig. 3B). Indeed, hemin induced strong DDR, comparable to the response to etoposide. This effect was more pronounced in the absence of HO-1. Importantly, the influence of ALA on DDR was much weaker and observed only in KO-HMOX1 cells. As expected, stabilization of G-quadruplexes by PDS resulted in increased γH2AX staining, regardless of HMOX1 status (Fig. 3B). The obtained results show that HO-1 efficiently protects cells against genotoxic stress induced by increased heme synthesis but is less effective in protecting against hemin.
ALA is a more specific source of intracellular free heme than hemin. To understand the differences between the protective efficacy of HO-1 in response to ALA and hemin, and to test whether the effect on DDR correlates with an increase in intracellular heme levels, we compared the total and free heme pools in WT(mock) and KO-HMOX1 cells treated with hemin or ALA. We chose concentrations of hemin (20 μM) and ALA (350 μM) that did not affect cell viability but increased HO-1 levels in HEK293T cells (Fig. 4A).
Fig. 4.
Effect of hemin and ALA on intracellular heme in HEK293T WT(mock) and KO-HMOX1 cells. A) Expression of HO-1 protein in WT(mock) cells. Immunocytochemistry and fluorescence microscopy. Representative pictures. B) Effect of hemin (20 μM, 24 h) and ALA (350 μM, 24 h) on total heme levels. Oxalic acid assay. ANOVA. C) Free heme in the cytoplasm and nucleus of WT(mock) and KO-HMOX1 cells under control conditions. D) Effect of hemin (20 μM, 3 h) and ALA (350 μM, 24 h) on free heme in the cytoplasm and nucleus of WT(mock) cells. E) Effect of hemin (20 μM, 3 h) and ALA (350 μM, 24 h) on free heme in the cytoplasm and nucleus of KO-HMOX1 cells. Cytoplasmic or nuclear eGFP/mKATE2 fluorescence ratio in cells transfected with reporter plasmids (a lower value indicates a higher concentration of heme). Flow cytometry. ANOVA (B, C), Kruskal-Wallis test (D, E).
We first measured total heme levels using an oxalic acid colorimetric assay. The total heme pool was comparable in WT(mock) and KO-HMOX1 cells cultured under control conditions (Fig. 4B). In response to hemin, total heme levels decreased slightly in control cells, possibly as a result of HMOX1 induction, as such an effect was not visible in the absence of HO-1. In both cell lines, total heme levels did not change significantly in response to ALA (Fig. 4B). However, what may be biologically more important for the expression of heme-regulated genes or the stabilization of G-quadruplexes is the pool of free heme. Therefore, we transfected the cells with eGFP-mKATE2 reporter plasmids and then measured free heme using genetically encoded fluorescent heme sensors localized to the cytoplasm or nucleus. In this system, the green fluorescence of eGFP is quenched by heme, whereas the red fluorescence of mKATE2 is heme-insensitive [34]. A decrease in the green-to-red fluorescence ratio indicates an increase in the free heme level. We compared the levels of heme in cells cultured under control conditions and supplemented with hemin (3 h) as a source of exogenous heme or ALA (24 h) as a substrate for heme synthesis.
Under control conditions, the effect of HMOX1 deficiency on the level of free heme in the cytoplasm was undetectable. However, heme concentration was slightly increased in the nucleus of HMOX1-deficient cells, compared to WT(mock) counterparts (Fig. 4C). Treatment of WT(mock) cells with hemin resulted in a weak, statistically insignificant tendency towards an increase in free heme levels in the cytoplasm and nucleus, as we already reported [8]. The increase in response to ALA was statistically significant (Fig. 4D). In KO-HMOX1 cells, the accumulation of free heme in response to hemin as well as ALA was significant and more pronounced than in WT(mock) cells, in both cytoplasmic and nuclear cell compartments (Fig. 4D). Thus, HO-1 may play a role in regulating the intracellular availability of free heme, although the observed effects are moderate. Worth noting, the influence of hemin and ALA on the DNA damage response does not correlate with the upregulation of intracellular heme.
Hemin, but not ALA, induces strong lipid peroxidation. In the next step, we compared the induction of oxidative stress in WT(mock) and KO-HMOX1 cells after treatment with hemin or ALA. We used a CellROX Deep Red fluorogenic probe designed to measure superoxide and hydroxyl radicals in the cytoplasm (Fig. 5A). The fluorescence was quantified by flow cytometry, 24 h after stimulation. The obtained results were similar in WT(mock) and KO-HMOX1 cells: hemin did not induce the production of radicals in the cytoplasm, whereas ALA caused a relatively weak upregulation, statistically significant in HMOX1-deficient cells. A strong induction in both cell lines was detected in response to tert-butyl hydroperoxide (TBHP), which served as a positive control (Fig. 5A). We also checked the effect of hemin on cytoplasmic ROS production at shorter time points, 3–12 h. Similarly to 24 h, we did not observe an increase in the ROS levels either in WT(mock) or KO-HMOX1 cells (Fig. 5B).
Fig. 5.
Oxidative stress and genotoxic stress in HEK293T cells. A) Effect of hemin (20 μM, 24 h) and ALA (350 μM, 24 h) on ROS generation in WT(mock) and KO-HMOX1 cells. CellROX Deep Red Reagent assay, analyzed by flow cytometry. TBHP (200 μM, 30 min) was used as a positive control. ANOVA. B) Time course of ROS generation after hemin (20 μM) treatment in WT(mock) and KO-HMOX1 cells. C) Representative images of lipid peroxidation in untreated cells and after treatment with hemin, ALA or CH. Scale bar 40 μm. D) Effect of hemin (20 μM, 24 h), ALA (350 μM, 24 h) and hemin (20 μM, 24 h) together with NAC (2.5 mM, 24 h) on lipid peroxidation in WT(mock) and KO-HMOX1 cells. Click-iT Lipid Peroxidation Kit, analyzed using a fluorescence microscope. Cumene hydroperoxide (CH, 100 μM, 24 h) was used as a positive control. ANOVA. E) Time course of lipid peroxidation after hemin (20 μM) treatment in WT(mock) and KO-HMOX1 cells. F) DNA-damage response measured as γH2AX staining in cells exposed to etoposide (0.25 μM) and then cultured without additional stimulation or treated with PDS, hemin alone and hemin together with NAC (2.5 mM) for 24 h. Immunocytochemistry and confocal microscopy. Kruskal-Wallis test. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Additionally, we analyzed the influence of hemin and ALA on lipid peroxidation using the Click-iT kit (Fig. 5C and D). In contrast to cytoplasmic free radical production, lipid peroxidation was strongly enhanced 24 h after stimulation with hemin but not ALA. The effect of hemin was comparable to that of cumene hydroperoxide (CH), a positive control, and completely reversed by antioxidant N-acetylcysteine (NAC, 2.5 mM) supplementation (Fig. 5D). The cell responses to hemin were similar 24 h after stimulation, regardless of HO-1 levels. However, the analysis of earlier time points showed that in WT(mock) cells peroxidation gradually increased reaching a maximum after 24 h, while in cells without HO-1 a very strong signal was visible already after 3 h and persisted throughout the measurement period (Fig. 5E).
Taken together, the effects of ALA and hemin administrations in HEK293T cells were different: ALA slightly upregulated the production of reactive oxygen species in the cytoplasm, while hemin led to strong lipid peroxidation in cell membranes (Fig. 5A–E). These effects were more pronounced in HMOX1-deficient cells.
To understand the potential importance of G-quadruplexes and oxidative stress for hemin-triggered DNA damage response, we induced genotoxic stress by etoposide and at the same time treated the cells with PDS (2 μM), hemin (20 μM) alone or hemin supplemented with NAC (2.5 mM). Analysis of γH2AX staining showed that stabilization of G-quadruplexes by PDS did not increase the etoposide-induced DDR (Fig. 5F). On the other hand, hemin enhanced the response to etoposide. This effect was stronger in the absence of HO-1 and fully reversed in both cell lines by co-incubation with NAC (Fig. 5F). Thus, the increased DDR in hemin-treated cells is oxidative stress-dependent. We expected that the response to hemin-induced lipid peroxidation may strongly interfere with the influence of elevated free heme on G-quadruplex stabilization. Therefore, in further experiments we used ALA supplementation to increase the availability of free heme without strong induction of lipid peroxidation.
Both HMOX1 deficiency and G-quadruplex stabilization lead to replication stress. A direct effect of the stabilization of G-quadruplexes and one of the causes of DNA breaks may be replication stress [12,14]. Therefore, we tested the extent to which administration of PDS or increase in heme levels after ALA treatment affects DNA replication in the presence or absence of HO-1. To this end, we analyzed the progression of replication forks using the fiber assay [35] to directly evaluate the percentage of stalled forks.
We first confirmed that administration of ALA increases free heme levels (Fig. 4D and E) and leads to the accumulation of G-quadruplexes, in both WT(mock) and KO-HMOX1 cells (Fig. 6A and B). Interestingly, under control conditions, the increased levels of G-quadruplexes seen in KO-HMOX1 cells (Fig. 3A) were accompanied by an increase in the fraction of stalled forks in cells undergoing DNA replication (Fig. 6C). A similar increase in stalled forks was found in WT(mock) cells in which G-quadruplexes were stabilized by PDS, whereas in KO-HMOX1 cells replication stress was not further amplified (Fig. 6C). This indicates that both HMOX1 deficiency and G-quadruplex stabilization cause replication stress in HEK293T cells.
Fig. 6.
G-quadruplexes and replication stress in HEK923T cells. A) Effect of treatment with ALA (350 μM, 24 h) on G-quadruplexes in WT(mock) cells. B) Effect of treatment with ALA on G-quadruplexes in KO-HMOX1 cells. Immunocytochemistry and confocal microscopy. Wilcoxon test. C) Effect of PDS (2 μM, 24 h) on DNA replication, assessed as a proportion of fired, ongoing, terminated and stalled replication forks in WT(mock) and KO-HMOX1 cells. Fibers assay. Chi2 test.
Endogenous heme induces replication stress but accelerates fork progression. Induction of heme synthesis by ALA led to a significant increase in the stalled fork proportions in WT(mock) cells (p < 0.01) and tended to further enhance replication stress in KO-HMOX1 cells (p = 0.074). This effect seems to be indeed related to heme synthesis as the opposite trend was observed in cells treated with succinylacetone (SA), a heme synthesis inhibitor (Fig. 7A). Interestingly, despite a higher percentage of stalled forks, the ongoing forks were longer in both WT(mock) and KO-HMOX1 cells after administration of ALA (Fig. 7B). The forks were also longer in untreated HMOX1-deficient cells than in wild-type counterparts. This indicates a higher speed of fork progression in cells with increased synthesis and availability of endogenous heme.
Fig. 7.
Replication stress and G-quadruplex processing in HEK923T cells. A) Effect of ALA (350 μM, 24 h) and succinylacetone (SA, 500 μM, 24 h) on DNA replication, assessed as a proportion of fired, ongoing, terminated and stalled replication forks in WT(mock) and KO-HMOX1 cells. Fibers assay. Chi2 test. B) Length of fibers identified as ongoing forks in untreated cells and after treatment with ALA. Kruskal-Wallis test. C) Representative images of G-quadruplexes (magenta) on DNA fibers (green) of WT(mock) cells. D) Proportion of replication forks with G-quadruplexes detected in the middle or at the end of the fork in untreated cells and after treatment with ALA. Fibers assay. Chi2 test. * - G-quadruplexes in the end of forks, # - G-quadruplexes within the forks. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
HO-1 regulates the response to G-quadruplexes at replication forks. G-quadruplexes can form at replication forks (Fig. 7C). Indeed, we detected them in about 30 % of all forks, with similar frequency in both cell lines cultured in control conditions (28.6 % in WT(mock) cells and 30.3 % in KO-HMOX1 cells, p > 0.4) (Fig. 7D). However, HMOX1 status determined the effects of endogenous heme synthesis on G-quadruplexes within the fork. Namely, administration of ALA did not alter the formation of G-quadruplexes within the forks in WT(mock) cells but increased their frequency to 37.8 % (p < 0.01) in the absence of HO-1 (Fig. 7D).
Moreover, despite the similar frequency of G-quadruplexes at replication forks in cells with and without HO-1, their spatial distribution was different (Fig. 7D). Under control conditions, in WT(mock) cells, 40.4 % of the fork-associated G-quadruplexes were located at the fork ends, indicating fork stalling and replication arrest. After ALA administration and the putative increase in free heme levels, G-quadruplexes did not further accumulate within the forks, but the percentage of forks with G-quadruplexes at their ends increased to 52.4 % (p < 0.01). This suggests that the cells responded to ALA by inhibiting the replication of the DNA strand on which G-quadruplexes had formed. Interestingly, in HMOX1-deficient cells cultured under control conditions, terminal localization of G-quadruplexes was observed in 56.2 % of cases (p < 0.001 in comparison to WT(mock) counterparts). Administration of ALA led to further accumulation of G-quadruplexes within the forks, but here the increase was mainly observed in the middle part (Fig. 7D). This might suggest less restrictive control of the cell cycle in HMOX1-deficient cells after supplementation with ALA, and continuation of DNA replication despite steric hindrance.
HMOX1 deficiency impairs the p53-dependent pathway. A key cell cycle controller is p53, which prevents the occurrence of stalled or collapsed replication forks and blocks the cell cycle in response to DNA damage. Moreover, the balance between p53, its transcriptional target p21 (CDKN1A) and PARP1 allows maintaining a normal speed of replication fork progression [19]. Interestingly, we observed that although p53 protein levels were similar in WT(mock) and KO-HMOX1 cell lines (Fig. 8A), nuclear localization of p53 was reduced in HMOX1-deficient cells (Fig. 8B). To monitor intracellular trafficking of p53, we transfected HEK293T cells with a plasmid encoding a p53-GFP fusion protein to test nuclear translocation of p53 in response to etoposide (20 μM). Time-lapse analysis during the first 3 h after stimulation showed a significant increase in nuclear signal in WT(mock) cells, but not in HMOX1-deficient cells (Fig. 8C).
Fig. 8.
Nuclear translocation of p53. A) Protein level and (B) nuclear translocation of p53 measured by ImageStream. T-test. C) Nuclear translocation of p53-GFP in WT(mock) and KO-HMOX1 HEK293T cells treated with etoposide (20 μM) for 3 h. Timelapses images (right) captured by using fluorescence microscopy. Scale bar 10 μm. D) Nuclear level of p53 in dKO and NLS iPSCs cultured in heme depleted medium with SA (500 μM) for 24 h. ANOVA. E) Expression of p21 (Cdkn1a) in dKO and NLS iPSCs cultured in heme depleted medium with SA (500 μM) or in complete medium with olaparib (100 nM) for 24 h. Kruskal-Wallis test.
The cellular trafficking of p53 may be influenced by PARP1 activity [20] and direct interaction with heme [22]. Our results showed that both PARylation dynamics (Fig. 2G) and free heme availability (Fig. 4C–E) are altered in HMOX1-deficient cells. In the next step, we analyzed the nuclear localization of p53 under heme-deprived conditions, namely in the heme-free medium supplemented with SA to inhibit endogenous heme synthesis. To avoid the effect of HO-2 isoform on heme metabolism, we performed experiments using dKO (without HO-2 and HO-1) and NLS (without HO-2, with nuclear form of HO-1) iPSC. Under control conditions, nuclear localization of p53 was decreased in Hmox1-deficient iPSCs. However, under heme-deprived conditions, the effect of HO-1 disappeared (Fig. 8D). We also measured the expression of Cdkn1a, a direct transcriptional target of p53. Indeed, we found decreased Cdkn1a expression in Hmox1-deficient iPSCs cultured in control medium, but not in heme-deprived conditions (Fig. 8E). In contrast, the effect of HO-1 status was still evident in cells treated with olaparib, inhibitor of PARP1 and PARP2 (Fig. 8E). These results suggest that the effects of HO-1 on p53 pathway are associated mainly with regulation of free heme. If heme levels are low, the presence of HO-1 is irrelevant to p53 transcriptional activity.
HMOX1 deficiency slows down cell culture growth. G-quadruplexes were only present at a minority of replication forks. Therefore, we checked whether the possible influence of ALA on the cell cycle and S-phase arrest is detectable at the cell culture level. A standard cell cycle assay based on the detection of DNA staining with propidium iodide did not show any effect of HMOX1 expression or ALA administration in the highly proliferating HEK293T cell lines (Fig. 9A). Thus, increased formation of G-quadruplexes within replication forks is not sufficient to exert cell cycle arrest in cell cultures. However, monitoring the growth of cell cultures for 48 h and time-lapse image analysis revealed that HMOX1-deficient cells proliferated more slowly (Fig. 9B), confirming our previous observations [25]. Interestingly, administration of ALA did not affect control cells, but additionally attenuated the growth of KO-HMOX1 cell line (Fig. 9C). Furthermore, single-cell division tracking showed that the cell cycle duration is significantly longer in the absence of HO-1 (Fig. 9D). These results indicate that, despite the lack of cell cycle arrest, the proliferation of HMOX1-deficient HEK293T cells is slowed down, independently of the replication fork progression rate.
Fig. 9.
Cell cycle and proliferation of HEK293T cells. A) G1/S/G2 phases of the cell cycle in WT(mock) and KO-HMOX1 cells cultured under control conditions or treated with ALA (250 μM, 24 h). B) Representative images of cells after 12 h, 24 h, 36 h and 48 h. Scale bar 20 μm. C) Number of cells cultured under control conditions or treated with ALA (24 h or 48 h). ANOVA. D) Duration of the cell cycle in WT(mock) and KO-HMOX1 cells cultured under control. Mann-Whitney test.
Protection against replication stress is a universal effect of HO-1. In the last step, we investigated whether the effect of HMOX1 deficiency on the induction of replication stress is specific to immortalized HEK293T cells or is more universal. Previously, we reported [8] that HO-1 protein colocalizes with G-quadruplexes in murine hematopoietic stem cells (HSCs). On the other hand, we have shown that HO-1 deficiency in the bone marrow leads to premature aging of HSCs in Hmox1 knock-out mice [26]. We have also found that this is due to an extrinsic dysfunction of the hematopoietic niche and can be corrected after transplantation to the Hmox1-competent, healthy niche [26]. In contrast, the intrinsic effects of Hmox1 deficiency in HSCs themselves have not been well characterized.
Here, we investigated whether Hmox1 deficiency could intrinsically increase the risk of replication stress in proliferating primary cells. To this end, we isolated hematopoietic stem and progenitor cells (HSPC, Lin−Sca1+Kit+) from the bone marrow of wild-type and Hmox1 knockout mice and cultured them ex-vivo to induce cell cycling. Then, we analyzed the replication forks using the fiber assay. Similarly, to HEK293T cells, Hmox1-deficient HSPCs had a higher proportion of stalled forks (Fig. 10A), indicating their higher susceptibility to replication stress. Also, the mean length of ongoing forks was greater in the absence of HO-1 (Fig. 10B). Interestingly, Hmox1-deficient HSCs isolated from aged animals had increased expression of Parp1 (Fig. 10C), while the expressions of p53-target genes that regulate the cell cycle, such as Cdkn1a (p21) and Plk2, were decreased in comparison to their counterparts in wild-type cells (Fig. 10C). This could suggest some changes in cell cycle regulation or p53 activity, implying a more universal nature of the observed dependencies.
Fig. 10.
Replication stress in primary cells. A) DNA replication, assessed as a proportion of fired, ongoing, terminated and stalled replication forks in HSCs isolated from the bone marrow of WT mice or Hmox1 KO mice and cultured in-vitro for 7 days. Chi2 test. B) Length of fibers identified as ongoing forks in HSCs isolated from the bone marrow of WT mice or KO-Hmox1 mice and cultured in-vitro for 7 days. Fibers assay. T-test, 2 biological repetitions. C) Expression Parp1, Plk2 and Cdkn1a genes in HSCs isolated from the bone marrow of old WT mice or Hmox1 KO mice. RNA-seq, data are presented as FPKM (Fragments Per Kilobase of transcript per Million mapped reads). T-test. D) DNA replication, assessed as a proportion of fired, ongoing, terminated, and stalled replication forks in LCL derived from a healthy donor (WT) and patient carrying HMOX1 mutation (HMOX1-mut). Chi2 test. E) Length of fibers identified as ongoing forks in WT and HMOX1-mut LCL. Fibers assay. T-test.
Finally, we had the opportunity to investigate whether a similar relationship applies to a very rare case of congenital deficiency of HO-1 in humans. We used the lymphoblastoid cell line (LCL) derived from a patient with defective HMOX1 (HMOX1-mut). The previous whole exome-seq analysis [27] identified the mutations (c.55dupG; c.262_268delinsCC in trans), resulting in a frameshift and absence of functional HO-1 protein. We compared these cells with control LCL from a healthy donor. As shown in Fig. 10D, the fraction of stalled forks was increased in HMOX1-mut cells, confirming the replication stress. In accordance with previous models (Fig. 7, Fig. 10B), the mean length of ongoing forks was greater in the absence of correct HO-1 protein (Fig. 10E), which indicates a higher speed of fork progression.
4. Discussion
Replication stress, defined as the slowing or stalling of replication forks, can result from any obstacle that perturbs fork progression, including nucleotide imbalance, DNA breaks, or secondary DNA structures [41]. In this study, we demonstrate that HO-1 deficiency increases the risk of replication stress, manifested by fork stalling, in both immortalized cell lines and primary cells. Experiments conducted in the HEK293T cell line revealed that replication stress is enhanced in response to ALA, a substrate in the heme synthesis pathway. It appears that the direct cause of the increased risk of fork stalling in the absence of HO-1 may be the accumulation of DNA G-quadruplexes stabilized by endogenous heme.
It is known that the accumulation of G-quadruplexes is particularly high in single-stranded DNA during the S phase of the cell cycle [42,43]. When stabilized and left unremoved, G-quadruplexes can stall replication forks, disrupting DNA repair mechanisms, causing DNA damage, and ultimately leading to genomic instability [44]. Additionally, G-quadruplexes are associated with increased susceptibility to oxidative stress because guanine is preferentially oxidized compared to other DNA bases, and guanines within quadruplexes are more sensitive to oxidation than those dispersed [45,46]. It has been shown that a fraction of active replication forks spontaneously form G-quadruplexes at newly unwound DNA [47]. We directly detected the presence of G-quadruplexes at ∼30 % of replication forks in HEK293T cells cultured under control conditions (Fig. 7D). In response to enhanced heme synthesis, the level of fork-associated G-quadruplexes increased, but only in HMOX1-deficient cells. This indicates that HO-1 provides effective protection against replication stress induced by endogenous heme.
The main function of HO-1 is to degrade excess heme [1]. In its FeII and FeIII oxidation states, heme participates in many cellular processes, acting as a ubiquitous cofactor of crucial enzymes (such as cytochromes P450, dioxygenases, or nitric oxide synthases), and as a direct regulator of several transcription factors (such as BACH1, BACH2, NPAS2, REV-ERBα). Heme is known to induce oxidative DNA damage and DNA strand breaks [48]. Importantly, it binds with nanomolar affinity to DNA G-quadruplexes and can stabilize them in living cells [8,49]. At the same time, binding to G-quadruplexes can strongly increase the pro-oxidant properties of heme. Such G-quadruplexes complexed with FeIII-heme, form heme-DNAzymes which utilize hydrogen peroxide for oxidative catalysis [48]. Therefore, the removal of excess heme may not only prevent the stabilization of G-quadruplexes but also the formation of DNAzymes, contributing to the antioxidant effects of HO-1. It is worth noting that PDS, used as a positive control, does not exhibit these properties. While PDS stabilizes G-quadruplexes it does not transform them into DNAzymes.
In our experiments, to increase the availability of cellular free heme, we used hemin (as a source of exogenous heme) and ALA (as a substrate for ALAD in heme synthesis). These are commonly employed approaches [34]. However, our direct side-by-side comparison demonstrates that they have different cellular effects. Administration of ALA increased cytoplasmic and nuclear free heme levels in both control and HMOX1-deficient cell lines. It is worth noting that the increase in free heme level (more effectively induced by ALA) was not reflected in the intensity of the DNA damage response, as measured by γH2AX staining (which was stronger in cells exposed to hemin). This suggests that endogenous free heme itself is not a direct cause of DNA damage.
Hemin increased cellular free heme levels to a statistically significant extent only in HMOX1-deficient cells. Instead, hemin administration resulted in severe lipid peroxidation both in WT(mock) and KO-HMOX1 cells, although this effect appeared faster in the absence of HO-1. This seems not surprising, as hemin can intercalate into the lipid domains, with subsequent release of its free iron, thereby permitting efficient catalysis of lipid oxidation [50]. Simultaneously, hemin caused a strong DNA damage response, comparable to that induced by etoposide and stronger in the absence of HO-1. It is known that products of lipid peroxidation such as reactive aldehydes react directly with DNA bases or generate intermediates which form mutagenic exocyclic adducts [51]. The effect of hemin on DDR was prevented by antioxidant NAC. This indicates that DNA damage caused by hemin is associated with the induction of oxidative stress rather than an increase in free heme levels. Importantly, it also indicates that administration of ALA is a more specific method for increasing the availability of intracellular free heme, while the observed effects of hemin are strongly interfered by the cellular response to oxidative stress.
Similarly, we observed that HMOX1-deficient cells exhibited a stronger response to etoposide-induced genotoxic stress, as evidenced by γH2AX staining. HO-1 has been reported to regulate the DNA damage response by targeting ATM kinase activity [52]. Therefore, our observations may support the assumption that HO-1 influences the activity of DNA repair proteins. However, etoposide also induces pronounced oxidative DNA damage [53,54]. Since the effect of HMOX1 deficiency disappeared in the presence of NAC in our experimental setting, we presume that HO-1 primarily protects HEK293T cells against etoposide-induced oxidative stress.
Endogenous oxidative stress is the main factor causing the accumulation of single-strand DNA breaks, while etoposide, as a topoisomerase inhibitor, also induces double-strand breaks. Both lead to replication stress, depletion of the replication protein-A (RPA) complex, and disruption of cell cycle checkpoints [47,55]. The protein involved in the recognition and repair of both SSBs and DSBs is PARP1 [20].
Binding of PARP-1 to DNA breaks or G-quadruplexes ahead of the replication fork and its subsequent auto-PARylation are key steps in stabilizing stalled forks and restarting replication [18,19]. On the other hand, if de-PARylation is insufficient, PARP1 complexes can themselves form a barrier to replication [56]. Based on docking modeling, it was proposed that HO-1 interacts with the regulatory helical domain (HD) of PARP1 to maintain PARylation. Additionally, HO-1 was suggested to bind to the PARG protein, reducing its dePARylating activity [4]. Therefore, one might expect a decrease in PARylation in the absence of HO-1. However, in our experimental settings, PARP1 activation was not reduced in HO-1-deficient cells; on the contrary, the dynamics of PARylation were even higher (Fig. 2E–G). AutoPARylation analyzed in-vitro using purified proteins did not show any effect of HO-1 on PARP1 activity (Fig. 2I). Therefore, it appears that the increased risk of replication stress that we observed in the absence of HO-1 was not significantly mediated by PARP1 regulation.
In response to DNA damage and replication stress, PARP1 directly interacts with p53 and p21 inhibiting DNA replication [19,20]. Namely, p53 is a substrate for covalent PARylation at the C-terminal domain (CTD) and exhibits a non-covalent, high-affinity interaction with PAR. This non-covalent PARylation decreases sequence-independent DNA binding, resulting in increased sequence-specific transcriptional activity of p53 [20]. Additionally, PARylation impedes the interaction between p53 and the nuclear export receptor CRM1 [19], leading to nuclear accumulation of p53 and upregulation of p21 [19,20].
Interestingly, we observed a reduced accumulation of p53 in the nucleus of KO-HMOX1 and dKO iPS cells (Fig. 8B–D), as well as a decreased expression of its transcriptional targets Plk2 and Cdkn1a (Fig. 10C) in HSCs isolated from aged Hmox1 knockout mice. The downregulation of p53 targets was further confirmed by qRT-PCR (Fig. 8E) and RNA-seq in Hmox1-deficient murine iPSCs, or by qRT-PCR in primary Hmox1-deficient murine fibroblasts (data not shown). Consequently, the PARP1-p53-p21 axis which protects cells against replication stress seems to be dysregulated in the absence of HO-1.
Moreover, the same axis is known to regulate the speed of fork progression [15,16,19,23]. Again, the p53 transcriptional target, p21, controls fork progression, and inhibits DNA synthesis [57]. Experiments in p21-knockdown cells showed fork acceleration, pinpointing p21 as a negative regulator of fork speed [57]. Defects in p53-p21 interplay lead to supra-threshold acceleration of fork elongation [15,16,19,23]. Importantly, accelerated fork progression is a general mechanism triggering replication stress and DNA damage [57,58]. Our experiments indicate that G-quadruplex accumulation is a direct but most likely not the only factor influencing replication in HMOX1-deficient cells. Namely, in the absence of HO-1, we observed a higher progression rate of ongoing forks (Fig. 7, Fig. 10B,E), which can result from impairment of p53-regulated pathway.
Replication arrest, as a reaction to non-canonical DNA structures, is a p53-dependent quality control process regulated mainly by p21 [21]. Another p53 target, PLK2 kinase, controls the G1/S checkpoint [59]. Cells with dysfunctional p53 signaling enter and proceed through S phase regardless of DNA damage [57]. We observed such a progress in replication despite G-quadruplexes localized within the forks in HMOX1-deficient cells (Fig. 7D). This indicates a less efficient replication arrest in response to DNA conformational hindrance in the absence of HO-1.
We did not investigate in detail the molecular mechanism(s) responsible for the observed changes in the p53-dependent pathway. However, our experiments reveal that the main contribution is related to direct regulation of p53 by heme. It has been demonstrated that p53 protein contains a heme-binding CXCP motif and binds heme noncovalently with low micromolar affinity (Kd 1.2 μM) at the molecular ratio 1:1. Heme triggers nuclear export of p53 by unmasking its C-terminal NES sequence, what is followed by accelerated protein degradation through the ubiquitin-proteasome system [22]. It has been reported that inhibition of heme synthesis with succinylacetone led to increased p53 levels. On the contrary, the administration of hemin led to the redistribution of p53 protein in the cell: the level of p53 protein decreased in the nuclear fraction, while increasing in the cytosol. It was accompanied by the decrease in p53-mediated transcription of p21 [22]. We observed a similar relationship when comparing WT(mock) and KO-HMOX1 cells. In the absence of HO-1, the cellular level of p53 protein remained unchanged, but its nuclear localization was reduced (Fig. 8A and B).
Heme-triggered ubiquitylation of p53 [22] affects not only protein degradation but also negatively regulates its nuclear import [60]. In unstressed cells, ubiquitylation of lysines in NLS interferes with binding of p53 to importin-α3. Thus, p53 is trapped in the cytoplasm. In response to stress, the level of p53 ubiquitylation is rapidly reduced and NLS motif becomes competent for efficient recognition of importin-α3, which is critical for nuclear import of p53 [60]. Interestingly, we demonstrated that the nuclear import of p53 induced by the etoposide treatment was strongly diminished in the absence of HO-1 (Fig. 8C).
It should be emphasized that the effect of HO-1 on p53 localization disappeared if we cultured the cells in conditions of low heme availability, namely in a heme-free medium supplemented with a heme synthesis inhibitor (Fig. 8D). This observation was confirmed by the expression of the direct transcriptional target of p53, i.e. p21 (Cdkn1a): expression was reduced in the absence of HO-1 under control conditions, but the effect of HO-1 disappeared in heme-deprived cells (Fig. 8E). In contrast to heme depletion, inhibition of PARylation did not abolish the effects of HO-1. This indicates that the removal of excess heme, which affects the localization of p53, determines the importance of HO-1 for the proper functioning of the p53-p21 pathway.
Generally, if a replication fork encounters a barrier, it stops or disintegrates. Stalled forks are protected from excision by several proteins, including PARP1, and can restart after the hindrance is removed. If the fork is stalled for several hours, it loses the ability to restart, and its disintegration leads to a DNA double-strand break [61]. To restore the correct structure of the replication fork, G-quadruplexes are resolved by helicases, primarily by BRIP1, but also by FANCM, WRN and BLM [62]. It is worth noting that in murine Hmox1-deficient iPSCs and in HSCs isolated from Hmox1 knockout mice, BRIP1 colocalized with G4 and Brip1 expression was strongly upregulated [8].
The cellular response to replication stress involves either activation of replication checkpoints, remodeling of stalled forks and DNA repair to restart replication, or induction of DNA-damage tolerance (DDT) [15]. A form of DDT is translesion DNA synthesis (TLS), which allows to bypass DNA damage [41]. TLS is performed by specialized polymerases that do not have 3'→5′ proofreading properties. This allows for rapid cell proliferation, but at the cost of a greater risk of mutations. However, this prevents the collapse of replication forks and the formation of DNA double-strand breaks [16]. The most important polymerase in TLS is Polθ (encoded by POLQ), suppressed by p53 [21]. Polθ carries out mutagenic replication and often introduces incorrect substitutions. Accordingly, when POLQ is absent, G-quadruplexes cause massive DNA deletions [62]. In the present study, we did not test which of the pathways is active in HEK293T cells, but again in HSCs isolated from Hmox1 knockout mice, Polq expression was strongly upregulated [8], while Western blot showed increased Polθ protein levels in HMOX1-deficient HEK293T cells, especially after treatment with etoposide (data not shown). This may suggest an activation of TLS pathways.
It can be hypothesized that the increased risk of DNA double-strand breaks and TLS activation induced by G-quadruplexes may lead to genome instability, especially in cancer cells, where replication stress is often amplified [63,64]. Indeed, in-silico analysis revealed that 70 % of rearranged genes in leukemia contain a G-quadruplex sequence [65], and rearrangement sites in transcription factor-3 (TCF3), that promote leukemia development, colocalize with regions that can form G-quadruplexes [66]. Another analysis showed that G-quadruplex motifs increase the likelihood of repetitive mutations and may be primary mutagenesis indicators [67].
Interestingly, HO-1 was considered a protein influencing the risk of carcinogenesis and tumor progression, which was mainly attributed to its cytoprotective, pro-angiogenic or immunomodulatory effects [[68], [69], [70]]. Similarly, the genotoxic properties of heme as a pro-oxidant factor have been suggested [71]. In humans, a polymorphism of the HMOX1 promoter has been identified, affecting the level of gene expression [72]. The results of meta-analysis did not show a relationship between these polymorphisms and overall cancer susceptibility [73]. However, less active HMOX1 promoter variants may be associated with a higher incidence of lung adenocarcinoma in male smokers [74] and oral squamous cell carcinoma in betel chewers [75]. Our results suggest that an additional mechanism of HMOX1-dependent protection against tumor induction may involve reducing the risk of replication stress.
Additionally, it is worth noting that replication stress and fork arrest appear to be the main sources of cytosolic DNA. Remnants of DNA generated during DNA repair enter the cytoplasm, and act as a warning signal that regulates the cGAS-STING pathway leading to activation of IRF3 and the induction of type-I interferon response [76]. Interestingly, in the transcriptome of HSCs isolated from the Hmox1 knockout mice, we observed an increase in a panel of genes involved in the interferon response [26]. We suppose that the replication stress in Hmox1-deficient HSCs described in the current study may contribute to such a gene expression profile. We do not expect a similar effect in HEK293T cells, because they lack STING and are unable to induce an interferon reaction [77].
We believe that the protective effect of HO-1 against replication stress is a universal feature because we observed similar results in all models used, including the cells derived from a patient with an HMOX1 frameshift mutation. The cellular effects of HMOX1 mutations in patients are poorly characterized because very few cases have been diagnosed. In our study, the LCL donor patient suffered from hemolytic anemia with elevated inflammation markers throughout childhood, and experienced fatal pulmonary hemorrhage at the age of 5 years. Generally, the lack of HO-1 has more severe consequences in humans than in mice, and usually leads to death in childhood. The lifespan of Hmox1 knockout mice is comparable to that of wild-type mice. However, at the cellular level, replication stress manifested by an increased percentage of stalled forks and increased rate of fork progression, was very similar in cells isolated from Hmox1 knockout mice and HMOX1-mut human patient.
5. Conclusion
We found that HMOX1 deficiency increases the risk not only of oxidative but also of replication stress and, consequently, DNA damage. This is most probably due to the accumulation of DNA G-quadruplexes stabilized by excess heme and an impaired response to replication stress regulated by the PARP1-p53-p21 network. Inefficient removal of heme disrupts the nuclear localization of p53 and the expression of its transcriptional targets, which may explain less stringent replication arrest in response to DNA conformational hindrance. We suppose that protection against replication stress may play a role in the widely recognized cytoprotective activity of HO-1.
Funding
This work was supported by grants from the National Science Centre to A.J. (2015/18/M/NZ3/00387 and 2017/25/B/NZ3/00986) and P.C. (2021/41/N/NZ3/03709). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the paper.
Data availability
Data will be made available on request.
CRediT authorship contribution statement
Patryk Chudy: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Jakub Kochan: Methodology, Conceptualization, Validation. Mateusz Wawro: Methodology, Validation. Phu Nguyen: Investigation. Monika Gorczyca: Investigation. Aliaksandra Varanko: Investigation. Aleksandra Retka: Investigation. Swati Sweta Ghadei: Investigation. Emilija Napieralska: Investigation. Anna Grochot-Przęczek: Conceptualization, Writing – original draft, Writing – review & editing. Krzysztof Szade: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. Lea-Sophie Berendes: Methodology. Julien Park: Methodology. Grzegorz Sokołowski: Investigation. Qiuliyang Yu: Methodology. Alicja Józkowicz: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Witold N. Nowak: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Wojciech Krzeptowski: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing, Visualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Dr. Katarzyna Miękus (Department of General Biochemistry Faculty of Biochemistry, Biophysics and Biotechnology Jagiellonian University) for providing Leica DMI6000 fluorescence microscope. We thank Malopolska Centre of Biotechnology for providing Zeiss LSM 880 confocal microscope and Dr. Paweł Hermanowicz for assist. We thank Ann-Sofie Jemth and Thomas Helleday, Karolinska Institutet, Stockholm, Sweden, for the PARP1 autoparylation protocol. Some of the optical microscopy experiments were performed at the Bioimaging Laboratory, which serves as an imaging core facility at the Faculty of Biochemistry, Biophysics, and Biotechnology, Jagiellonian University.
Contributor Information
Witold N. Nowak, Email: witold.nowak@uj.edu.pl.
Wojciech Krzeptowski, Email: wojciech.krzeptowski@uj.edu.pl.
References
- 1.Dulak J., Deshane J., Jozkowicz A., et al. Heme oxygenase-1 and carbon monoxide in vascular pathobiology: focus on angiogenesis. Circulation. 2008;117(2):231–241. doi: 10.1161/CIRCULATIONAHA.107.698316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn L.L., Midwinter R.G., Ni J., et al. New insights into intracellular locations and functions of heme oxygenase-1. Antioxidants Redox Signal. 2014;20(11):1723–1742. doi: 10.1089/ars.2013.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szade A., Szade K., Mahdi M., et al. The role of heme oxygenase-1 in hematopoietic system and its microenvironment. Cell. Mol. Life Sci. 2021 May;78(10):4639–4651. doi: 10.1007/s00018-021-03803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li C., Wu J., Dong Q., et al. Heme oxygenase-1 impacts oxidative stress-induced neural stem cells senescence by regulating DNA damage repair via PARP-1. SSRN. 2023 Preprint. [Google Scholar]
- 5.Scaffa A., Tollefson G.A., Yao H., et al. Identification of heme oxygenase-1 as a putative DNA-binding protein. Antioxidants. 2022;11(11):2135. doi: 10.3390/antiox11112135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraus W.L. PARPs and ADP-ribosylation: 50 Years … and counting. Mol Cell. 2015;58(6):902–910. doi: 10.1016/j.molcel.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouleau-Turcotte É., Krastev D.B., Pettitt S.J., et al. Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery. Mol Cell. 2022;82(16):2939–2951.e5. doi: 10.1016/j.molcel.2022.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krzeptowski W., Chudy P., Sokołowski G., et al. Proximity ligation assay detection of protein-DNA interactions - is there a link between heme oxygenase-1 and G-quadruplexes? Antioxidants. 2021;10(1):94. doi: 10.3390/antiox10010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang G., Biswasa C., Lin Q.S., et al. Heme oxygenase-1 regulates postnatal lung repair after hyperoxia: role of β-catenin/hnRNPK signaling. Redox Biol. 2013;1(1):234–243. doi: 10.1016/j.redox.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J., Li S., Li C., et al. The non-canonical effects of heme oxygenase-1, a classical fighter against oxidative stress. Redox Biol. 2021;47 doi: 10.1016/j.redox.2021.102170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33(9):2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazouzi A., Velimezi G., Loizou J.I. DNA replication stress: causes, resolution and disease. Exp. Cell Res. 2014;329(1):85–93. doi: 10.1016/j.yexcr.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 13.Zimmer J., Tacconi E.M.C., Folio C., et al. Targeting BRCA1 and BRCA2 deficiencies with G-quadruplex-interacting compounds. Mol Cell. 2016;61(3):449–460. doi: 10.1016/j.molcel.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander J.L., Orr-Weaver T.L. Replication fork instability and the consequences of fork collisions from replication. Genes Dev. 2016;30(20):2241–2252. doi: 10.1101/gad.288142.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saxena S., Zou L. Hallmarks of DNA replication stress. Mol Cell. 2022;82(12):2298–2314. doi: 10.1016/j.molcel.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeman M.K., Cimprich K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16(1):2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraus W.L., Hottiger M.O. PARP-1 and gene regulation: progress and puzzles. Mol. Aspect. Med. 2013;34(6):1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Edwards A.D., Marecki J.C., Byrd A.K., et al. G-Quadruplex loops regulate PARP-1 enzymatic activation. Nucleic Acids Res. 2021;49(1):416–431. doi: 10.1093/nar/gkaa1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindström M.S., Bartek J., Maya-Mendoza A. p53 at the crossroad of DNA replication and ribosome biogenesis stress pathways. Cell Death Differ. 2022;29(5):972–982. doi: 10.1038/s41418-022-00999-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischbach A., Krüger A., Hampp S., et al. The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res. 2018;46(2):804–822. doi: 10.1093/nar/gkx1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy S., Tomaszowski K.H., Luzwick J.W., et al. p53 orchestrates DNA replication restart homeostasis by suppressing mutagenic RAD52 and POLθ pathways. Elife. 2018;7 doi: 10.7554/eLife.31723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen J., Sheng X., Chang Z., et al. Iron metabolism regulates p53 signaling through direct heme-p53 interaction and modulation of p53 localization, stability, and function. Cell Rep. 2014;7(1):180–193. doi: 10.1016/j.celrep.2014.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canesin G., Di Ruscio A., Li M., et al. Scavenging of labile heme by hemopexin is a key checkpoint in cancer growth and metastases. Cell Rep. 2020;32(12) doi: 10.1016/j.celrep.2020.108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canesin G., Muralidharan A.M., Swanson K.D., et al. HO-1 and Heme: G-quadruplex interaction choreograph DNA damage responses and cancer growth. Cells. 2021;10:1801. doi: 10.3390/cells10071801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mucha O., Podkalicka P., Czarnek M., et al. Pharmacological versus genetic inhibition of heme oxygenase-1 - the comparison of metalloporphyrins, shRNA and CRISPR/Cas9 system. Acta Biochim. Pol. 2018;65(2):277–286. doi: 10.18388/abp.2017_2542. [DOI] [PubMed] [Google Scholar]
- 26.Szade K., Zukowska M., Szade A., et al. Heme oxygenase-1 deficiency triggers exhaustion of hematopoietic stem cells. EMBO Rep. 2020;21(2) doi: 10.15252/embr.201947895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berendes L.S., Westhoff P.S., Wittkowski H., et al. Clinical and molecular analysis of a novel variant in heme oxygenase-1 deficiency: unraveling its role in inflammation, heme metabolism, and pulmonary phenotype. Mol Genet Metab Rep. 2023;38 doi: 10.1016/j.ymgmr.2023.101038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowarz E., Löscher D., Marschalek R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015;10(4):647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]
- 29.Rapsomaniki M.A., Kotsantis P., Symeonidou I.E., et al. easyFRAP: an interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics. 2012;28(13):1800–1801. doi: 10.1093/bioinformatics/bts241. [DOI] [PubMed] [Google Scholar]
- 30.Koczor C.A., Haider A.J., Saville K.M., et al. Live cell detection of poly(ADP-ribose) for use in genetic and genotoxic compound screens. Cancers. 2022;14(15):3676. doi: 10.3390/cancers14153676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L., Chen M., Luo M., et al. Ratiometric NAD + sensors reveal subcellular NAD + modulators. ACS Sens. 2023;8(4):1518–1528. doi: 10.1021/acssensors.2c02565. [DOI] [PubMed] [Google Scholar]
- 32.Xie S., Mortusewicz O., Ma H.T., et al. Timeless interacts with PARP-1 to promote homologous recombination repair. Mol. Cell. 2015;60(1):163–176. doi: 10.1016/j.molcel.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Kopacz A., Klóska D., Werner E., et al. A dual role of heme oxygenase-1 in angiotensin II-induced abdominal aortic aneurysm in the normolipidemic mice. Cells. 2021;10(1):163. doi: 10.3390/cells10010163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanna D.A., Harvey R.M., Martinez-Guzman O., et al. Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors. Proc Natl Acad Sci U S A. 2016;113(27):7539–7544. doi: 10.1073/pnas.1523802113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nieminuszczy J., Schwab R.A., Niedzwiedz W. The DNA fibre technique - tracking helicases at work. Methods. 2016;108:92–98. doi: 10.1016/j.ymeth.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 36.Boyd S.D., Tsai K.Y., Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2000;2(9):563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- 37.Mahadevan J., Bowerman S., Luger K. Quantitating repair protein accumulation at DNA lesions: past, present, and future. DNA Repair. 2019;81 doi: 10.1016/j.dnarep.2019.102650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koczor C.A., Saville K.M., Andrews J.F., et al. Temporal dynamics of base excision/single-strand break repair protein complex assembly/disassembly are modulated by the PARP/NAD+/SIRT6 axis. Cell Rep. 2021;37(5) doi: 10.1016/j.celrep.2021.109917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura A., Sedelnikova O.A., Redon C., et al. Techniques for gamma-H2AX detection. Methods Enzymol. 2006;409:236–250. doi: 10.1016/S0076-6879(05)09014-2. [DOI] [PubMed] [Google Scholar]
- 40.Baldwin E.L., Osheroff N. Etoposide, topoisomerase II and cancer. Curr Med Chem Anticancer Agents. 2005;5(4):363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 41.Bai G., Kermi C., Stoy H., et al. HLTF promotes fork reversal, limiting replication stress resistance and preventing multiple mechanisms of unrestrained DNA synthesis. Mol Cell. 2020;78(6):1237–1251.e7. doi: 10.1016/j.molcel.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145(5):678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biffi G., Tannahill D., McCafferty J., et al. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5(3):182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaillard H., García-Muse T., Aguilera A. Replication stress and cancer. Nat. Rev. Cancer. 2015;15(5):276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 45.Kim J.E., Choi S., Yoo J.A., et al. 8-Oxoguanine induces intramolecular DNA damage but free 8-oxoguanine protects intermolecular DNA from oxidative stress. FEBS Lett. 2004;556:104–110. doi: 10.1016/s0014-5793(03)01385-1. [DOI] [PubMed] [Google Scholar]
- 46.Byrd A.K., Zybailov B.L., Maddukuri L., et al. Evidence that G-quadruplex DNA accumulates in the cytoplasm and participates in stress granule assembly in response to oxidative stress. Biol. Chem. 2016;291(34):18041–18057. doi: 10.1074/jbc.M116.718478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee W.T.C., Gupta D., Rothenberg E. Single-molecule imaging of replication fork conflicts at genomic DNA G4 structures in human cells. Methods Enzymol. 2021;661:77–94. doi: 10.1016/bs.mie.2021.08.008. [DOI] [PubMed] [Google Scholar]
- 48.Ibrahim H., Mulyk P., Sen D. DNA G-quadruplexes activate heme for robust catalysis of carbene transfer reactions. ACS Omega. 2019;4(12):15280–15288. doi: 10.1021/acsomega.9b02305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gray L.T., Puig Lombardi E., Verga D., et al. G-Quadruplexes sequester free heme in living cells. Cell Chem. Biol. 2019;26(12):1681–1691.e5. doi: 10.1016/j.chembiol.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Balla G., Jacob H.S., Eaton J.W., et al. Hemin: a possible physiological mediator of low density lipoprotein oxidation and endothelial injury. Arterioscler. Thromb. J. Vasc. Biol. 1991;11(6):1700–1711. doi: 10.1161/01.atv.11.6.1700. [DOI] [PubMed] [Google Scholar]
- 51.Nair U., Bartsch H., Nair J. Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: a review of published adduct types and levels in humans. Free Radic. Biol. Med. 2007;43(8):1109–1120. doi: 10.1016/j.freeradbiomed.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 52.Otterbein L.E., Hedblom A., Harris C., et al. Heme oxygenase-1 and carbon monoxide modulate DNA repair through ataxia-telangiectasia mutated (ATM) protein. Proc Natl Acad Sci U S A. 2011;108(35):14491–14496. doi: 10.1073/pnas.1102295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kagan V.E., Kuzmenko A.I., Tyurina Y.Y., et al. Pro-oxidant and antioxidant mechanisms of etoposide in HL-60 cells: role of myeloperoxidase. Cancer Res. 2001;61(21):7777–7784. [PubMed] [Google Scholar]
- 54.Wang S., Chan S.Y., Deng Y., et al. Oxidative stress induced by Etoposide anti-cancer chemotherapy drives the emergence of tumor-associated bacteria resistance to fluoroquinolones. J. Adv. Res. 2023;S2090–1232(23) doi: 10.1016/j.jare.2023.02.011. 00062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vesela E., Chroma K., Turi Z., et al. Common chemical inductors of replication stress: focus on cell-based studies. Biomolecules. 2017;7(1):19. doi: 10.3390/biom7010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrision D., Gravells P., Thompson R., Bryant H.E., et al. Poly(ADP-Ribose) glycohydrolase (PARG) vs. Poly(ADP-Ribose) polymerase (PARP) – function in genome maintenance and relevance of inhibitors for anti-cancer therapy. Front. Mol. Biosci. 2020;7:191. doi: 10.3389/fmolb.2020.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maya-Mendoza A., Moudry P., Merchut-Maya J.M., et al. High speed of fork progression induces DNA replication stress and genomic instability. Nature. 2018;559(7713):279–284. doi: 10.1038/s41586-018-0261-5. [DOI] [PubMed] [Google Scholar]
- 58.Kanai M., Hanashiro K., Kim S.H., et al. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol. 2007;9(10):1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 59.Matsumoto T., Wang P.Y., Ma W., et al. Polo-like kinases mediate cell survival in mitochondrial dysfunction. Proc Natl Acad Sci. 2009;106(34):14542–14546. doi: 10.1073/pnas.0904229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchenko N.D., Hanel W., Li D., et al. Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha3 binding. Cell Death Differ. 2010;17(2):255–267. doi: 10.1038/cdd.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berti M., Vindigni A. Replication stress: getting back on track. Nat. Struct. Mol. Biol. 2016;23(2):103–109. doi: 10.1038/nsmb.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lerner L.K., Sale J.E. Replication of G Quadruplex DNA. Genes. 2019;10(2):95. doi: 10.3390/genes10020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kosiol N., Juranek S., Brossart P., et al. G-quadruplexes: a promising target for cancer therapy. Mol. Cancer. 2021;20(1):40. doi: 10.1186/s12943-021-01328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Awadasseid A., Ma X., Wu Y., et al. G-quadruplex stabilization via small-molecules as a potential anti-cancer strategy. Biomed. Pharmacother. 2021;139 doi: 10.1016/j.biopha.2021.111550. [DOI] [PubMed] [Google Scholar]
- 65.Katapadi V.K., Nambiar M., Raghavan S.C. Potential G-quadruplex formation at breakpoint regions of chromosomal translocations in cancer may explain their fragility. Genomics. 2012;100(2):72–80. doi: 10.1016/j.ygeno.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 66.Williams J.D., Fleetwood S., Berroyer A., et al. Sites of instability in the human TCF3 (E2A) gene adopt G-quadruplex DNA structures in vitro. Front. Genet. 2015;6:177. doi: 10.3389/fgene.2015.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Georgakopoulos-Soares I., Morganella S., Jain N., et al. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res. 2018;28(9):1264–1271. doi: 10.1101/gr.231688.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Was H., Cichon T., Smolarczyk R., et al. Overexpression of heme oxygenase-1 in murine melanoma: increased proliferation and viability of tumor cells, decreased survival of mice. Am. J. Pathol. 2006;169(6):2181–2198. doi: 10.2353/ajpath.2006.051365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jozkowicz A., Was H., Dulak J. Heme oxygenase-1 in tumors: is it a false friend? Antioxidants Redox Signal. 2007;9(12):2099–2117. doi: 10.1089/ars.2007.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kusienicka A., Bukowska-Strakova K., Cieśla M., et al. Heme oxygenase-1 has a greater effect on melanoma stem cell properties than the expression of melanoma-initiating cell markers. Int. J. Mol. Sci. 2022;23(7):3596. doi: 10.3390/ijms23073596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Glei M., Klenow S., Sauer J., et al. Hemoglobin and hemin induce DNA damage in human colon tumor cells HT29 clone 19A and in primary human colonocytes. Mutat. Res. 2006;594(1–2):162–171. doi: 10.1016/j.mrfmmm.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 72.Taha H., Skrzypek K., Guevara I., et al. Role of heme oxygenase-1 in human endothelial cells: lesson from the promoter allelic variants. Arterioscler. Thromb. Vasc. Biol. 2010;30(8):1634–1641. doi: 10.1161/ATVBAHA.110.207316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang R., Shen J., Yang R., et al. Association between heme oxygenase-1 gene promoter polymorphisms and cancer susceptibility: a meta-analysis. Biomed Rep. 2018 Mar;8(3):241–248. doi: 10.3892/br.2018.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kikuchi A., Yamaya M., Suzuki S., et al. Association of susceptibility to the development of lung adenocarcinoma with the heme oxygenase-1 gene promoter polymorphism. Hum. Genet. 2005;116(5):354–360. doi: 10.1007/s00439-004-1162-2. [DOI] [PubMed] [Google Scholar]
- 75.Chang K.W., Lee T.C., Yeh W.I., et al. Polymorphism in heme oxygenase-1 (HO-1) promoter is related to the risk of oral squamous cell carcinoma occurring on male areca chewers. Br. J. Cancer. 2004:1551–1555. doi: 10.1038/sj.bjc.6602186. 18;91(8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li T., Chen Z.J. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018;215(5):1287–1299. doi: 10.1084/jem.20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reus J.B., Trivino-Soto G.S., Wu L.I., et al. SV40 large T antigen is not responsible for the loss of STING in 293T cells but can inhibit cGAS-STING interferon induction. Viruses. 2020;12(2):137. doi: 10.3390/v12020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.