Abstract
Pre-mRNP complexes were isolated from rat liver nuclei as 40S hnRNP particles, and actin-binding proteins were collected by DNase I affinity chromatography. The bound proteins were analyzed by 2D gel electrophoresis, and the following five hnRNP A/B-type proteins were identified by tandem mass spectrometry: DBP40/CBF-A (CArG binding factor A), a minor hnRNP A2 variant and three minor hnRNP A3 (mBx) variants. DBP40 was chosen for further analysis of the association of actin with the pre-mRNP complex. It was shown in vitro that purified actin binds to recombinant DBP40 suggesting that the interaction between actin and DBP40 is direct in the pre-mRNP particles. The association of actin with DBP40 was further explored in vivo. It was shown in a transfection study that DBP40 appears both in the nucleus and cytoplasm. Microinjection experiments revealed that DBP40 is exported from the nucleus to the cytoplasm. Finally, RNA–protein and protein–protein cross-linking experiments showed that DBP40 interacts with poly(A)+ RNA as well as actin, both in the nucleus and cytoplasm. We propose that actin associated with DBP40, and perhaps with additional hnRNP A/B-type proteins, is transferred from nucleus to cytoplasm bound to mRNA.
INTRODUCTION
Actin is present not only in the cytoplasm but also in the nuclei of somatic cells and oocytes (1). Early studies of actin in amphibian oocytes gave the first clear evidence for actin being a nuclear constituent (2–4). Subsequently, a number of immunocytology and immunoelectron microscopy studies, analyses of permeabilized cells, and in vitro cross-linking experiments provided further strong support for the view that actin is a nuclear component (reviewed in 1). There is likely to be a delicate balance between globular (G) and filamentous (F) actin, the globular form being by far the predominant one in the cell nucleus (4).
Nuclear actin has been tentatively implicated in RNA synthesis. It was shown early on that injection of anti-actin antibodies or actin-binding proteins into amphibian oocyte nuclei blocks transcription of the lampbrush chromosomes but not of ribosomal RNA genes (5). Actin was also reported to be required for efficient transcription in vitro by RNA polymerase II, presumably acting at the pre-initiation level (6). In addition, it has been shown that an anti-actin antibody inhibits in vitro transcription of human respiratory syncytial virus (HRSV) (7). Finally, monomeric actin seems to be required for the chromatin remodeling complex BAF to associate with chromatin during the gene activation process (8). Thus, actin is likely to play a role in the transcription process, but the precise mechanisms are still to be revealed.
Early on, it also seemed attractive to couple nuclear actin to mRNA transport. Actin was recorded in pre-messenger ribonucleoprotein (pre-mRNP, also designated hnRNP) preparations (9), and at the ultrastructural level it was observed in pre-mRNA-containing fibrogranular material in somatic cells (10). Furthermore, direct evidence for a functional role was obtained in a recent microinjection study suggesting that actin is involved in the nucleocytoplasmic transport of retroviral RNAs, and it was also proposed that the transport of cellular mRNAs could be dependent on actin (11). Finally, it has recently been demonstrated in a dipteran insect that actin is bound in a specific manner to at least one of the hnRNP proteins in pre-mRNP particles (12). Evidently, we need more information on the interaction between actin and pre-mRNP particles to also elucidate the specific function(s) of actin in the transport process.
The hnRNP proteins comprise a large number of proteins that are classified into several families and sub-families based on structural/functional motifs (13). In mammals, for example, there are more than 20 major and a large number of minor protein species, designated A1 to U hnRNP proteins (13,14). The most abundant hnRNP proteins belong to the A/B type and exhibit a well defined modular structure. The N-terminal domain is highly conserved and consists of two tandemly repeated, 80–90 amino acid long, RNA-binding domains (RBD). The C-terminal part of the protein is considerably more divergent, is glycine-rich, and is called the auxiliary domain. Therefore, the A/B-type proteins are often referred to as 2xRBD-Gly proteins. These proteins form a steadily expanding family of proteins with a large number of post-transcriptional isoforms as well as extensive post-translational modifications. In addition to a general packing role in RNA processing and transport, a number of specialized functions have been ascribed to the hnRNP A/B proteins. A1 and A2/B1 proteins participate in constitutive (15) as well as in alternative (16) splicing by antagonizing the SR splicing factor ASF/SF2 in the 5′ splice site selection (17,18). A1 has also been implicated in RNA transport as it is shuttling (19) and contains a nuclear export signal (20). A2 is likely to be involved in mRNA transport and mRNA localization in the cytoplasm (21) as well as in translation (22).
In the present study, we have investigated the interaction of actin with pre-mRNP particles in a mammalian system suitable for systematic and large-scale analysis of hnRNP proteins associated with actin. Using 40S hnRNP complexes obtained from fractionated rat liver nuclear extracts, we have isolated actin-binding proteins with DNase I affinity chromatography and further analyzed them by 2D gel electrophoresis combined with mass spectrometry. We have found a specific subset of rat A/B-type hnRNP proteins tightly associated with nuclear actin. These include DBP40/CBF-A (CArG binding factor A), a minor variant of hnRNP A2 and three minor variants of hnRNP A3 (mBx). The interaction of actin with DBP40 and DBP40 with poly(A)+ RNA has been further studied both in vitro and in vivo. It is proposed that the interaction is direct and that the actin–DBP40 complex bound to pre-mRNA in the nucleus is transferred to the cytoplasm bound to mRNA. Finally, as hnRNP A/B proteins are known to enter cytoplasm, it is possible that actin is associated not only with DBP40 but with a specific subset of A/B-type hnRNP proteins in the mRNP particle when delivered to the cytoplasm.
MATERIALS AND METHODS
Antibodies
The monoclonal 3G1 and 1B6 antibodies against Chironomus tentans hnRNP proteins were produced in our laboratory as described (23). The specificity of 3G1 for the C.tentans hrp36 is the same as that of the earlier studied 4F9 antibody (24). The ability of both antibodies to cross-react with a specific set of rat hnRNP proteins is shown in the Results. Monoclonal antibodies 9H10 and 4F4 against hnRNP A1 and hnRNP C1/C2, respectively, were a gift from Dr Gideon Dreyfuss. The monoclonal anti-β actin antibody termed AC-15 was from Sigma. The polyclonal, affinity-purified, anti-actin antibody was produced as described (12).
Plasmids
A cDNA copy of feline DBP40 inserted into pET28d for bacterial expression and into pcDNA3.1 for mammalian expression was kindly donated by Dr Colin Parrish. The bacterial expression vector for human hnRNP A2 (pET9d) was a gift from Dr Adrian Krainer.
Preparation of 40S pre-mRNP fraction
Rat liver 40S hnRNP complexes were obtained after fractionation of nuclear extracts in 15–30% (w/v) sucrose gradients and subsequent pelleting of the material sedimenting in the 40–50S region of the gradient as described (25).
DNase I affinity chromotography
Binding studies with pre-mRNP preparations were carried out using CNBr-activated Sepharose beads to which DNase I was covalently bound as previously described (12). Typically, 30 µl aliquots of the 40S pre-mRNP preparation were incubated with 100 µl of DNase I beads pre-equilibrated in 1× PBS for 30–40 min at 4°C with continuous mixing. Following stringent washes, DNase I beads were resuspended in urea-containing isofocusing sample buffer (ISB) (6 M urea, 0.3% Ampholytes 3/10, 1.25% NP-40) to elute the bound proteins. In parallel, prior to incubation with DNase I beads, 30 µl aliquots of the 40S pre-mRNP fraction were treated with RNaseA at a final concentration of 1 µg/µl for 15 min at room temperature. Following incubation with DNase I beads as described, bound proteins were similarly eluted with ISB buffer after stringent washes. In both cases bound proteins were analyzed by 2D gel electrophoresis.
Binding experiments with purified proteins were performed stepwise. Typically, 100 µl DNase I beads were pre-equilibrated in G-buffer (5 mM Tris pH 7.6, 0.5 mM ATP, 0.1 mM CaCl2 and 0.5 mM DTT) and then saturated with purified G-actin. The excess actin was washed away and the actin/DNase I beads were then subdivided into aliquots and individually incubated with 35S-labeled DBP40 and hnRNP A2 for 30–40 min at 4°C with continuous mixing. The beads were then spun down and washed batchwise six times with 1 ml phosphate buffer saline (PBS) containing 1% NP-40, 0.1% deoxycholate and 1 mM DTT to remove non-specifically bound material. Bound proteins were heat denatured, eluted, resolved by 10% SDS–PAGE and detected by autoradiography.
2D gel electrophoresis
2D gel electrophoresis was performed as described (26). In the first dimension, proteins were resolved on tube gels by non-equilibrium pH-gradient gel electrophoresis (NEPHGE). The tube gels were then carefully extruded from the capillary and equilibrated in DTT-containing Laemmli buffer prior to placing them on the prepared 10% SDS polyacrylamide gel for the second dimension (SDS–PAGE). The separated proteins were analyzed by western blotting or mass spectrometry.
Western blotting
The proteins resolved by 2D gel electrophoresis were transferred onto a PVDF membrane using a semi-dry blotting apparatus as previously described (26). After blocking with 10% (w/v) non-fat dry milk in PBS–Tween 20, each blot was sequentially incubated with monoclonal antibodies 3G1, 1B6 or 9H10, all antibodies diluted 1:500 in 5% (w/v) non-fat dry milk in PBS–Tween 20. Actin was identified with a monoclonal antibody termed AC-15 (Sigma Immunochemicals). The primary antibodies were detected with a goat anti-mouse IgG coupled to horse-radish peroxidase (HRP) (DAKO). In all cases immunodetection was performed using the chemiluminescent method (ECL-Plus, Amersham).
Mass spectrometry
The proteins in the 2D gel were stained with Coomassie blue, and the five major proteins revealed were individually excised after complete destaining of the gel. The proteins were digested within the gel with trypsin, and the generated peptides were collected as described (27). Contaminants and salts were removed from the samples with reverse phase (C18) chromatography desalting columns (ZipTip, Millipore). Normally, 3 µl of sample was applied to the column followed by washing with 20 µl of 0.1% trifluoroacetic acid and elution with 3 µl of 60% acetonitrile containing 1% acetic acid directly into the nano-ES needle.
Mass spectra of the tryptic peptides were recorded on a Voyager DE-Pro matrix-assisted laser desorption/ionization time-of-flight (MALDI–TOF) mass spectrometer (PE Biosystems). Nano-electrospray (nano-ES) collision-induced dissociation (CID) spectra were recorded on a quadrupole TOF tandem mass spectrometer (Micromass). This instrument was equipped with an orthogonal sampling ES-interface (Z-spray, Micromass). Metal-coated nano-ES needles (Protana A/S) were used and opened manually on the stage of a light microscope to give a spraying orifice of ∼5 µm. This resulted in a flow rate of ∼20–50 nl/min when a capillary voltage of 0.8–1.2 kV was applied. Desolvation was facilitated by a counter-current drying gas (nitrogen). The cone voltage was set at 40 V. For the acquisition of CID spectra, the precursor ion isotope cluster was selected with quadrupole 1. The collision voltage was optimized in the range 20–40 V with argon as collision gas. The product ion spectra were recorded on the orthogonal TOF.
Preparation of proteins for protein–protein interaction assays
Purification of native actin. Actin prepared from calf thymus cells as previously described (28), was a gift from Dr Uno Lindberg.
In vitro transcription/translation of 35S-labeled DBP40 and hnRNP A2. The plasmids containing the full-length ORF of DBP40 and hnRNP A2 were used as templates for TNT-coupled in vitro transcription/translation essentially following the manufacturer’s instruction manual (Promega). The proteins were labeled by incorporation of 35S-methionine, electrophoresed in 10% SDS polyacrylamide gels and detected by autoradiography. For purification, the expressed 35S-labeled proteins were affinity-purified by T7-tag antibody agarose (Novagen) as described in the manufacturer’s protocol.
Expression of DBP40 in COS-1 cells
Transfection. Overexpression of DBP40 in COS-1 cells was achieved both by a polyethyleneimine-based protocol as described (29) and with FUGENE following the manufacturer’s instructions (Boehringer).
Preparation of nuclear and cytoplasmic extracts from transfected cells. After transfection, subconfluent COS-1 cells were left in OPTIMEM-1 medium supplemented with PEST and 10% FCS for 24–48 h to allow overexpression of DBP40. Cells were then washed in PBS and resuspended in PBS containing 0.2% NP-40 and 0.1 mM PMSF. The cells were homogenized and spun at 2000 g for 5 min at 4°C. The supernatant constituted the cytosolic extract. The pellet was washed twice in PBS and resuspended in PBS containing 0.1 mg/ml tRNA, sonicated and clarified. The supernatant was retained as the nuclear extract. The extracts were fractionated by SDS–PAGE and analyzed by western blotting using a monoclonal anti-T7 tag antibody.
Microinjection and fluorescence microscopy
Fluorescent labeling of proteins. Histidine-tagged DBP40 was expressed in Escherichia coli and purified by nickel affinity chromatography as already described (30). Aliquots of purified recombinant DBP40 and BSA (Sigma) in PBS were incubated with 2 equivalents of fluorescein-5-maleimide (Pierce) dissolved in dimethylformamide in the dark. The labeling reaction was carried out for 2 h at room temperature with continuous agitation and in the dark. The labeled proteins were subsequently dialyzed overnight against PBS to remove uncoupled fluorescein-5-maleimide and concentrated to ∼2 mg/ml. Protein samples were stored at –20°C in the dark until further use.
Microinjection and fluorescence microscopy. In a first set of experiments, fluorescein-labeled DBP40 (2mg/ml) was co-injected with Texas Red-conjugated dextran (70 kDa; Molecular Probes) into the nucleus of subconfluent COS-1 cells using an Eppendorf microinjector. Immediately, or after a 90 min incubation at 37°C in OPTIMEM-1 medium supplemented with 10% fetal calf serum, the cells were fixed with 4% formaldehyde in PBS for 10 min at room temperature, washed in PBS, and mounted in Mowiol. Images were recorded with a LSM 510 laser scanning microscope (Zeiss LSM 510).
In a second set of microinjection experiments, fluorescein-labeled DBP40 and BSA (2 mg/ml) were injected into separate nuclei of COS-1 cells. After 90 min incubation as above, the cells were fixed and washed (as above) and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. The samples were then washed with PBS and blocked with 5% milk for 20 min at room temperature. Sytox (Molecular Probes) was added to the cells at a final concentration of 0.25 µM to stain the DNA. The samples were washed with PBS, mounted in Mowiol, and examined in the laser scanning microscope as above.
Protein–RNA cross-linking in vivo
COS-1 cells overexpressing DBP40 were irradiated with ultraviolet (UV) light and the protein–RNA cross-links purified by oligo-dT Sepharose as already described (31).
Protein–protein cross-linking in vivo
Proteins in mammalian tissue culture cells overexpressing full-length T7-tagged DBP40 were cross-linked for 20 min at room temperature with the cleavable agent dithiobis-succinimidylpropionate (DSP; Sigma/Aldrich) at a concentration of 0.5 mM in the standard cultivation medium (8). Nuclear and cytosolic extracts were prepared as described above and where appropriate incubated briefly with 8 M urea. The extracts were diluted 10× with PBS, containing 0.2% NP-40 and 1 mM PMSF, and immediately incubated with anti-T7-tag antibody agarose beads for 1 h at 4°C under continuous agitation. Following washes with PBS containing up to 0.1% deoxycholate, the bound proteins were released and the cross-linker cleaved by boiling the sample in 2× Laemmli sample buffer for 10 min. The proteins were resolved by SDS–PAGE and transferred onto a PVDF membrane with a semi-dry blotting apparatus. DBP40 and actin were detected by western blotting using a HRP-conjugated monoclonal antibody against the T7 tag (Novagen) and an affinity-purified polyclonal antibody against actin, respectively.
RESULTS
A specific subset of the hnRNP proteins in pre-mRNP complexes is associated with actin
We chose to study pre-mRNP particles from rat liver nuclei as this source permitted isolation of pre-mRNP in large scale as 40S monoparticles, and the rat major hnRNP proteins have been properly identified (32,33). We isolated 40S monoparticles and fractionated the hnRNP proteins by 2D gel electrophoresis (NEPHGE and SDS–PAGE). A typical pattern of the hnRNP proteins is presented in Figure 1A (top). The major proteins have been indicated in the figure following the nomenclature devised for human proteins (13). The hnRNP proteins A1, A2, B1 are practically identical to the corresponding human proteins (the primary sequence of B2 remains to be determined). The rat hnRNP A3, also known as mBx, is an abundant protein in rat but not in human (32). The hnRNP A3 protein was early recorded in Xenopus laevis (34) and a closely related protein species has been predicted in human (35). Actin is present in the 40S particle preparation as shown by 2D gel electrophoresis and western blot analysis (Fig. 1A, bottom).
Figure 1.
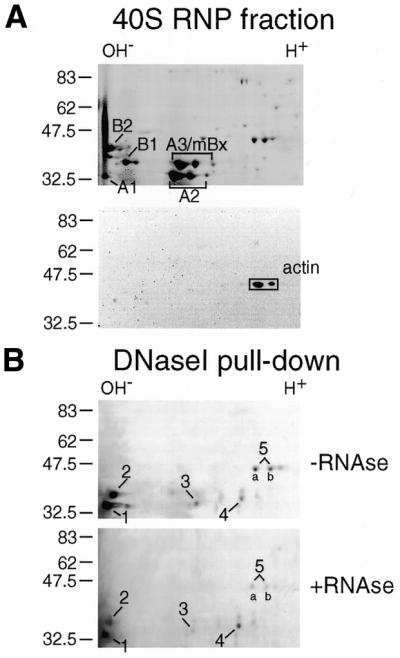
2D electrophoretic analysis of the proteins in 40S pre-mRNP particles and of a subset bound to DNase I–Sepharose. (A) Top, proteins in rat liver 40S hnRNP resolved by 2D gel electrophoresis (NEPHGE/SDS–PAGE). Bottom, western blot analysis of actin in the 40S fraction. (B) 40S pre-mRNP proteins bound to DNase I–Sepharose and subsequently analyzed as in (A). Prior to DNase I–Sepharose chromotography, the sample was split into two portions, one RNase-treated the other non-treated. The gels in (A) and (B) were stained with Coomassie blue. The major spots in (B) were numbered 1–5 and excised for in-gel digestion and subsequent mass spectrometric analysis.
To study whether actin is associated with specific hnRNP proteins in isolated pre-mRNP particles we performed DNase I affinity chromatography, a method based on the high affinity of DNase I for preferentially globular actin (G-actin). DNase I was immobilized on CNBr-activated Sepharose (12), and the DNase I beads were incubated with a 40S hnRNP fraction obtained from rat liver nuclei; beads lacking DNase I were incubated with the RNP fraction in parallel. The experiments were carried out in the presence or absence of exogeneously added RNase A. After incubation, the beads were washed using deoxycholate-containing buffers, and the bound proteins were resolved by 2D gel electrophoresis. Relative to the 40S hnRNP fraction (Fig. 1A), only a small subset of hnRNP proteins remained bound to DNase I–Sepharose (Fig. 1B). Sepharose alone did not bind any proteins (data not shown). The DNase I-bound proteins, designated 1–5 in Figure 1B, display pI values ranging from basic to acidic. The two spots of protein no. 5 (a and b) are assumed to correspond to different variant forms of the protein. Addition of RNase A to the isolated 40S fraction prior to DNase I co-precipitation had the general effect of reducing the amount of proteins resolved by 2D analysis. However the same spots appeared to be present regardless of the RNase A treatment (Fig. 1B, top and bottom). We conclude that actin is associated with a defined subset of hnRNP proteins and that this interaction could be partly dependent on RNA.
Actin was not eluted off the DNase I–Sepharose resin with the urea-containing ISB buffer as shown by 2D gel electrophoresis of the eluted proteins and by western blot analysis using an anti-actin antibody (data not shown). However, when the DNase I–Sepharose beads with adsorbed 40S hnRNP proteins were resuspended in Laemmli SDS-containing loading buffer, actin was detected in the gel (data not shown).
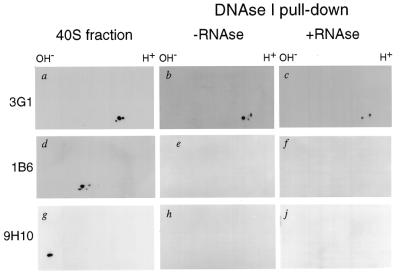
To start characterizing the proteins co-precipitated with DNase I–Sepharose we used three monoclonal antibodies raised against hnRNP proteins: 3G1, 1B6 and 9H10. The 3G1 antibody recognizing the C.tentans hnRNP protein hrp36 (24) detected three spots in the 40S fraction (Fig. 2a), and only two were still present among the proteins pulled-down with DNase I beads (the middle spot is missing) (Fig. 2b and c). The 1B6 antibody made against C.tentans hnRNP proteins (36) cross-reacted with rat major hnRNP A/B proteins in the 40S particles (Fig. 2d), but these proteins were not present in the pull-down fraction (Fig. 2e and f). Finally, the 9H10 antibody, specific for mammalian hnRNP A1 (37), recognized the rat hnRNP A1 in the 40S fraction (Fig. 2g), but this protein did not co-precipitate with actin (Fig. 2h and j). We conclude that only a specific subset of hnRNP proteins can be co-precipitated with actin from a very complex pre-mRNP 40S fraction. Remarkably, the most abundant hnRNP A/B-type proteins do not seem to be among these proteins.
Figure 2.
Western blot analysis of the 40S pre-mRNP proteins and a subset bound to DNase I–Sepharose beads. (a, d and g) Western blot analysis of the rat liver 40S hnRNP proteins using antibodies against C.tentans hrp36 (3G1), C.tentans core hnRNP proteins (1B6) and human hnRNP A1 (9H10), respectively. (b, e and h) Western blot analyses of 40S hnRNP proteins bound to DNase I–Sepharose. The same analysis is reported in (c, f and j), but in this case the 40S fraction was treated with RNase prior to incubation with the DNase I–Sepharose beads.
Identification of the actin-associated hnRNP proteins
Immunodetection using specific monoclonal antibodies did not reveal the nature of the proteins co-precipitated with DNase I. Therefore, the five major protein species (labeled 1–5 in Fig. 1B) were excised from the 2D Coomassie-stained gels. The two tentative variants of protein no. 5 (a and b) were analyzed both individually and together (Fig. 1B). The proteins were in-gel digested with trypsin and subsequently subjected to tandem mass spectrometry. A number of peptides were obtained from each spot, and three from each protein were sequenced (Table 1). Data bank searching allowed complete identification of the following proteins corresponding to the isolated spots: hnRNP A2 (protein no. 1), hnRNP A3 (no. 2), hnRNP A3 (no. 3), hnRNP A3 (no. 4), DBP40/CBF-A (no. 5), all being A/B-type hnRNP proteins. As expected, when the spots 5a and 5b were analyzed individually by tandem mass spectrometry, they turned out to be variants of DBP40/CBF-A. (For further information on the highly homologous proteins DBP40 and CBF-A, see Discussion.)
Table 1. Nano-electrospray tandem mass spectrometry identification of the proteins co-precipitated with DNase I affinity chromatography.
| Spot | Peptide m/z | Charge state | Peptide mass (Da) | Sequence | Identified as |
|---|---|---|---|---|---|
| 1 | 900.01 | 2 | 1798.02 | LFIGGLSFETTEESLR | hnRNP A2 |
| 594.90 | 2 | 1187.80 | IDTIEIITDR | hnRNP A2 | |
| 689.35 | 2 | 1376.70 | GGGGNFGPGPGSNFR | hnRNP A2 | |
| 2 | 955.98 | 2 | 1909.96 | SSGSPYGGGYGSGGGSGGYGSR | hnRNP A3 |
| 584.79 | 2 | 1167.58 | EDTEEYNLR | hnRNP A3 | |
| 625.81 | 2 | 1249.62 | IETIEVM*EDR | hNRNP A3 | |
| 3 | 827.40 | 2 | 1652.80 | GFASVTFDDHDTVDK | hnRNP A3 |
| 625.79 | 2 | 1249.58 | IETIEVM*EDR | hnRNP A3 | |
| 584.72 | 2 | 1167.44 | EDTEEYNLR | hnRNP A3 | |
| 4 | 886.03 | 2 | 1770.06 | LFIGGLSFETTDDSLR | hnRNP A3 |
| 584.73 | 2 | 1167.46 | EDTEEYNLR | hnRNP A3 | |
| 625.79 | 2 | 1249.58 | IETIEVM*EDR | hnRNP A3 | |
| 5 | 752.44 | 2 | 1502.88 | IFVGGLNPEATEEK | DBP40/CBF-A |
| 750.40 | 2 | 1498.80 | EVYQQQQYGSGGR | DBP40/CBF-A | |
| 1098.10 | 2 | 2194.20 | EYFGQFGEIEATELPIDPK | DBP40/CBF-A |
M*, oxidized Met.
DBP40/CBF-A was recorded as two acidic variants of the protein. In the 2D gel, there was also an additional actin-associated protein close to the two DBP40/CBF-A variants, which could represent a third variant.
The actin-associated hnRNP A2 variant (spot 1 in Fig. 1B) behaved differently from the predominant rat hnRNP A2 protein (Fig. 1A). It was highly enriched when DNase I affinity chromatography fractionation was performed, suggesting that we have identified a not yet characterized low abundance variant of hnRNP A2 with a very basic pI.
Three variants of hnRNP A3 with widely different pIs were co-precipitated with DNase I beads from the 40S fraction. The basic variant (spot 2 in Fig. 1B) probably constitutes a novel variant (see major variants in Fig. 1A). The neutral variant (spot 3 in Fig. 1B) has a pI value close to that of the major rat A3 (mBx) variants (Fig. 1A). However, as it is present in such a low amount, it is probably also a minor variant. The acidic variant (spot 4 in Fig. 1B) behaves similarly to the recombinant X.laevis A3 homolog in terms of its acidic pI (data not shown) and is clearly different from the major A3 (mBx) variants (Fig. 1A). Thus, two of the actin-associated A3 variants, and presumably all three, represent minor variants of the hnRNP A3 protein.
In summary, we have identified the major proteins co-precipitated with DNase I–Sepharose beads as being the A/B-type hnRNP protein DBP40/CBF-A, one minor variant of A2 and three minor variants of hnRNP A3.
Actin is directly associated with DBP40
As the DBP40 cDNA sequence has recently been reported by Wang and Parrish (30) and the clone was made available to us, it was feasible to further study the relation between actin and DBP40. Initially, we performed in vitro binding studies with purified actin and DBP40 to establish whether the interaction is direct. T7-tagged DBP40 was translated and 35S-labeled in vitro in a cell-free rabbit reticulocyte lysate and subsequently purified by T7-tag antibody affinity chromatography. In addition, an abundant human hnRNP A2 isoform, used as control in the actin-binding experiments, was expressed and purified in the same way as DBP40. Figure 3A shows an electrophoretic analysis of the 35S-labeled DBP40 and hnRNP A2 proteins used in the binding studies (lanes 1 and 2, respectively).
Figure 3.
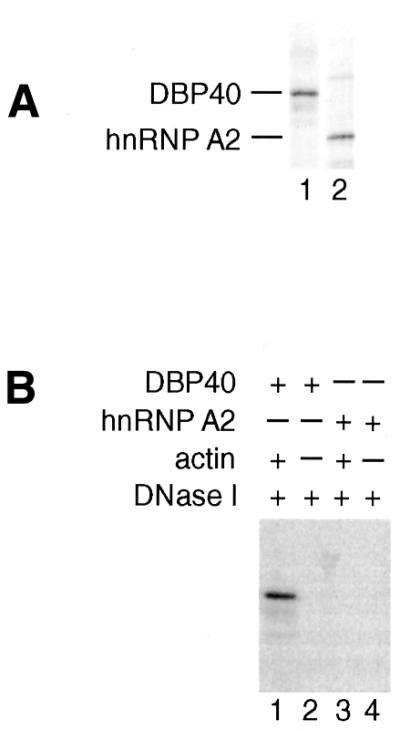
In vitro reconstitution experiments with actin, DBP40 and hnRNP A2. (A) SDS–PAGE of in vitro-translated 35S-labeled DBP40 and hnRNP A2 (lanes 1 and 2, respectively). (B) DNase I affinity chromatography of purified 35S-labeled DBP40 and hnRNP A2 in the presence of unlabeled affinity-purified actin. DBP40 and hnRNP A2 were individually incubated with actin/DNase I beads (lanes 1 and 3) as well as DNase I beads alone as control (lanes 2 and 4). The bound proteins were resolved by SDS–PAGE and visualized by autoradiography. Lane 1, actin plus DBP40; lane 2, no actin plus DBP40; lane 3, actin plus hnRNP A2; lane 4, no actin plus hnRNP A2.
To carry through the binding assay, the purified G-actin was first pre-incubated with DNase I beads pre-equilibrated in low ionic strength G-buffer. Following incubation with purified, 35S-labeled DBP40 or hnRNP A2, the actin/DNase I beads were washed and after heat denaturation the bound proteins were resolved by SDS–PAGE. The gels were fixed and dried, and the proteins detected by autoradiography. Only DBP40 specifically binds actin since DBP40 but not hnRNP A2 was recovered in the bound fraction (Fig. 3B, lanes 1 and 3, respectively). None of the proteins were able to bind to DNase I–Sepharose beads lacking actin (lanes 2 and 4). We conclude that DBP40, but not the abundant hnRNP A2 variant, interacts directly with actin in vitro.
DBP40 is likely to shuttle between nucleus and cytoplasm
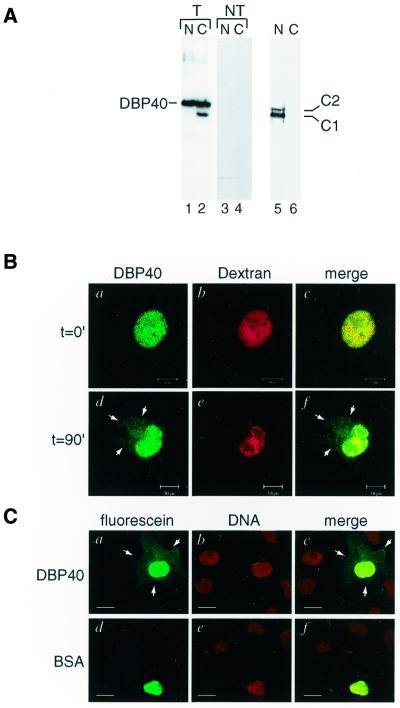
To investigate whether DBP40 is present both in the nucleus and in the cytoplasm, we expressed full-length T7-tagged DBP40 by transfecting sub-confluent COS-1 cells with a DBP40 clone inserted into a plasmid vector for mammalian expression. Cells overexpressing DBP40 were then harvested and extracts prepared from nuclei and cytoplasm. To avoid cytoplasmic contaminations the isolated nuclei were thoroughly washed prior to the preparation of the nuclear extract. The proteins were separated by SDS–PAGE and analyzed by western blotting. Nuclear leakage was excluded by analysis of the distribution of nuclear protein hnRNP C1/C2 (Fig. 4A, lanes 5 and 6). Immunodetection of DBP40 using a monoclonal antibody against the T7 tag indicated that DBP40 is located both in the nucleus and in the cytoplasm (Fig. 4A, lanes 1 and 2, respectively); the second, lower band in the cytoplasmic extract is likely to be a degradation product. As the monoclonal antibody against the T7-tag did not cross-react with any nuclear and cytoplasmic species in the non-transfected cells (Fig. 4A, lanes 3 and 4), it is assumed to be specific for the overexpressed T7-tagged DBP40 protein. Moreover, we have recently raised an antibody against DBP40 and shown that the endogenous protein is distributed both in the nucleus and cytoplasm in non-transfected cells (data not shown). Thus, DBP40 appears both in the nucleus and the cytoplasm.
Figure 4.
Cellular localization and nucleocytoplasmic shuttling of DBP40. (A) Western blot analysis of nuclear (lane 1) and cytoplasmic (lane 2) extracts prepared from transfected (T) COS-1 cells overexpressing DBP40. The DBP40 protein was detected by an anti-T7 tag monoclonal antibody. For comparison, nuclear (lane 3) and cytoplasmic (lane 4) extracts were prepared from non-transfected (NT) COS-1 cells and assayed with the same monoclonal antibody against the T7-tag. As control, the nuclear and cytoplasmic extracts were tested with an antibody to the nuclear protein hnRNP C1/C2 (lanes 5 and 6, respectively). (B) Co-injections of fluorescein-labeled DBP40 and Texas Red-conjugated dextran into nuclei of COS-1 cells. The cells were fixed immediately (t = 0 min) or after incubation for 90 min at 37°C (t = 90 min) and analyzed by confocal microscopy. Bar, 10 µm. (C) Microinjections of fluorescein-labeled DBP40 and BSA into nuclei of COS-1 cells. Following microinjections, the cells were fixed, stained for DNA and examined by confocal microscopy. Bar, 20 µm.
To establish whether DBP40 is exported from the cell nucleus to the cytoplasm, we performed microinjection experiments. Fluorescein-labeled recombinant DBP40 was co-injected with Texas Red-dextran into the nucleus of simian COS-1 cells. The cells were fixed either immediately or after 90 min incubation at 37°C (Fig. 4B). In parallel experiments, DBP40 and albumin, both labeled with fluorescein, were separately injected into simian COS-1 nuclei and incubated for 90 min as above (Fig. 4C). Both experiments showed that DBP40 had been partly transported to the cytoplasm (Fig. 4B and C), whereas both dextran (Fig. 4B) and BSA (Fig. 4C) remained confined to the nucleus. Thus, DBP40 is exported into the cytoplasm. Fluorescein-labeled DBP40 was subsequently injected into the cytoplasm of COS-1 cells. After 15 min incubation at 37°C the protein accumulated in the nucleus, suggesting a fast import kinetics (data not shown). These results, together with the cellular distribution revealed in DBP40-transfected cells, suggest that DBP40 is a shuttling A/B-type hnRNP protein.
DBP40 binds to mRNA in vivo
To determine whether DBP40 also binds to mRNA, cells overexpressing DBP40 were exposed to UV light to promote formation of covalently cross-linked protein–RNA complexes in vivo (19). Nuclear and cytoplasmic extracts were then prepared from UV-irradiated cells, and after SDS treatment the poly(A)+ RNP complexes were bound to oligo-dT Sepharose beads. Following stringent washes, bound RNA–protein complexes were eluted with low ionic strength buffers and then precipitated. The samples were subsequently RNase-treated and resuspended in SDS-loading buffer prior to heat denaturation. The proteins were resolved by SDS–PAGE and analyzed by western blotting. As expected, hnRNP C1/C2, used as control, was cross-linked to nuclear but not cytoplasmic poly(A)+ RNA (Fig. 5, lanes 1 and 2, respectively). DBP40 cross-linked to nuclear poly(A)+ RNA too (Fig. 5, lane 5), but in addition to cytoplasmic poly(A)+ RNA (Fig. 5, lane 6). Neither C1/C2 nor DBP40 proteins co-purified with the RNA if the UV irradiation was omitted (see lanes 3 and 4, and 7 and 8, respectively). We conclude that DBP40 is associated with poly(A)+ RNA both in the nucleus and cytoplasm and appears to accompany the mRNA sequence during the nucleocytoplasmic transport.
Figure 5.
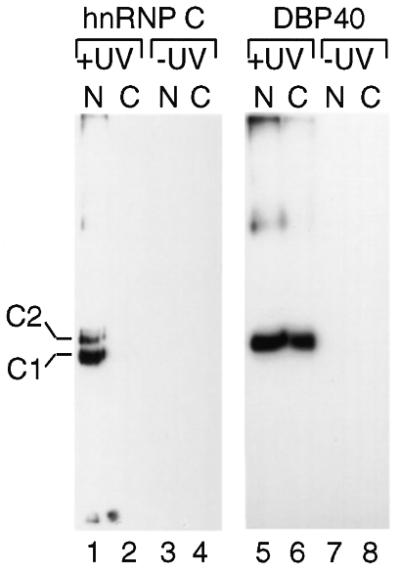
In vivo cross-linking of DBP40 to nuclear and cytoplasmic poly(A)+ RNA. DBP40-transfected cells were UV irradiated, and nuclear (N) and cytoplasmic (C) extracts were prepared, treated with SDS and passed over oligo-dT Sepharose. The bound proteins were analyzed by western blotting using monoclonal antibody 4F4 against hnRNP C1/C2 and T7-tag antibody against DBP40. Non-irradiated cells were studied in parallel.
Actin binds to DBP40 in vivo
To establish whether actin and DBP40 are associated in living cells, transfected COS-1 cells overexpressing T7-tagged DBP40 were treated with the membrane-permeable, cleavable cross-linking reagent DSP (8). Cross-linked nuclear and cytoplasmic extracts were treated with urea to abolish non-cross-linked protein interactions, and DBP40 with associated proteins were immunoprecipitated by anti-T7-tag antibody agarose beads; samples not treated with urea were analyzed in parallel. After cleavage of the cross-linker by boiling, the proteins were resolved by SDS–PAGE, and actin and DBP40 were detected by western blotting using a monoclonal anti-T7-tag antibody and an affinity-purified polyclonal anti-actin antibody, respectively. In the absence of cross-linker, actin was co-precipitated with DBP40 both from a nuclear and from a cytoplasmic extract (Fig. 6A, lanes 1 and 3, respectively), but no actin was recorded after urea treatment (lanes 2 and 4, respectively). The binding of DBP40 to the antibody column was not affected by urea (lanes 2 and 4). In the presence of cross-linker, actin remained partly bound to the DBP40 column even after urea treatment of the nuclear and cytoplasmic extracts (Fig. 6B, lanes 2 and 4, respectively). We conclude that actin and DBP40 are directly bound in vivo and are part of the same complex.
Figure 6.
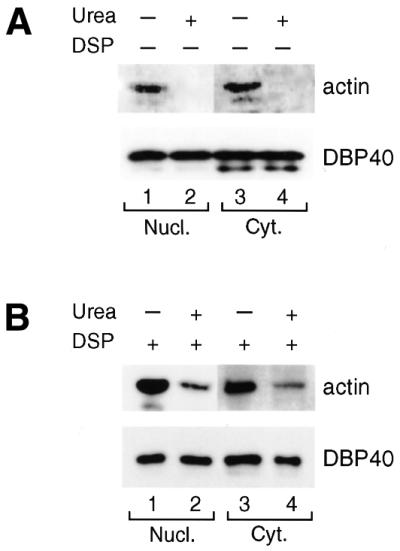
Cross-linking of actin to DBP40 in transfected COS-1 cells. Cultured cells overexpressing DBP40 were treated with DSP and control cells incubated in parallel without DSP. Nuclear and cytoplasmic extracts were prepared from untreated (A) and treated (B) cells. Following urea treatment [lanes 2 and 4 in both (A) and (B)], the samples were subjected to T7-tag antibody agarose immunoprecipitation and bound proteins analyzed by SDS–PAGE and western blotting using an affinity-purified polyclonal antibody against actin and an HRP-conjugated anti-T7-tag monoclonal antibody to detect DBP40.
DISCUSSION
Actin is associated with a subset of hnRNP proteins in pre-mRNP particles
We have now shown that actin is associated with a specific subset of rat liver hnRNP proteins present in a 40S hnRNP particle preparation. The proteins were isolated by DNase I affinity chromatography, fractionated by 2D gel electrophoresis, and identified by tandem mass spectrometry. The following five proteins were recorded: DBP40/CBF-A, a minor hnRNP A2 variant and three minor hnRNP A3 (mBx) variants. These five proteins all belong to the A/B type of hnRNP proteins, which can be resolved by 2D gel electrophoresis into a large number of isoforms (13,14). The diversity among related hnRNP proteins is mainly generated by differential splicing of common RNA precursors but post-translational modifications (phosphorylation, methylation of arginine and glycosylation) contribute as well. Most of the abundant rat hnRNP proteins, including hnRNP A1, B1 and B2 proteins and the major variants of hnRNP A2 and A3, are not part of the DNase I co-precipitated fraction, emphasizing that the actin-associated hnRNP proteins represent a well defined subset of hnRNP proteins.
The A/B-type hnRNP proteins are likely to be bound to pre-mRNA (13), which was directly shown in the present study for DBP40 using UV cross-linking. Such a conclusion is in good agreement with a recent study by Inoue et al. (38), in which it was demonstrated that CBF-A is present in hnRNP particles sedimenting in sucrose gradients up to 200S or more. We conclude that the actin-associated A/B-type hnRNP proteins are likely to be present in pre-mRNP particles and in a specific subset of the 40S hnRNP particles prepared from pre-mRNPs.
In the present study, DBP40/CBF-A appears as two (and possibly three) variants, which are all associated with actin. DBP40 was first described as a feline protein that can bind to parvovirus single-stranded DNA (30), while CBF-A was discovered as a mouse CArG box-binding factor A (39). The feline DBP40 and mouse CBF-A are likely to be homologs with an overall amino acid sequence identity of 92%. DBP40 and CBF-A belongs to a subfamily of highly homologous A/B-type hnRNP proteins, presently comprising 10 members from five species (40). Some of the proteins bind to DNA and are believed to participate in the replication of DNA, e.g. DBP40 (30), or in transcriptional regulation, e.g. CBF-A (39,41,42) and ssDBF (43), while others bind to RNA and are proposed to be involved in post-transcriptional processes, e.g. ABBP1 (44). Some of the proteins might also have dual functions; for example, as mentioned above, the transcriptional regulator CBF-A is associated with RNA in hnRNP particles (38), suggesting that it might also function post-transcriptionally. The fact that the proteins of this subfamily are strikingly similar along their whole lengths and still coupled to widely different cellular processes might suggest that they exert a structural role and perhaps organize the DNA or RNA into well defined higher order structures, a role proposed for A/B-type hnRNP proteins in general (13).
The hnRNP A2 and hnRNP A3 variants associated with actin are low abundance hnRNP proteins and, in particular, the basic variants become highly enriched upon DNase I fractionation. The only functional information available concerns the abundant hnRNP A2 variant. This protein is known to shuttle between the nucleus and the cytoplasm (19) and plays a role in vectorial transport of mRNA in the cytoplasm (21) and enhances cap-dependent translation (22). However, in this study the major A2 variant does not associate with actin, and the minor A2 and A3 variants studied here could exert other functions. In fact, there is good evidence that the variants of a given hnRNP protein play different roles in mRNA metabolism. The polypyrimidine tract binding protein (PTB) and hnRNP D represent two striking examples of this principle. PTB has three different isoforms, which have distinct activities in both alternative splicing and IRES-dependent translation (45). The hnRNP D protein regulates mRNA turnover by acting as an RNA destabilizing protein specifically binding to AU-rich mRNA sequences (46). The four isoforms known show different destabilizing effects on the mRNA (46). We conclude that the minor A2 and A3 variants associated with actin could function in RNA targeting and translation like the abundant hnRNP A2 variant but other roles should also be considered.
The nature of the interaction between actin and the pre-mRNP particle
The DNase I chromatography experiment showed that DBP40 and minor variants of hnRNP A2 and A3 are associated with actin. To start to establish whether the interaction between actin and the hnRNP proteins is direct or indirect, we performed an in vitro reconstitution experiment with purified actin and recombinant DBP40. The result revealed that actin can bind to DBP40, suggesting that actin is interacting directly with DBP40 in the pre-mRNP particle. As DBP40 is also capable of binding RNA, it seems to form a bridge between actin and the pre-mRNA molecule. The corresponding reconstitution experiments could not be carried out with the A2 and A3 variants as cDNA clones for these minor protein variants are not yet available. Therefore, it is still an open question whether actin interacts directly with each one of the five identified, actin-associated hnRNP proteins, or whether it only binds to DBP40, while the other four hnRNP proteins are coupled to DBP40 via protein–protein interactions.
It is also an open question whether actin is in a globular (G) or filamentous (F) form when binding to the pre-mRNP particle. Nuclear actin can appear in individual microfilaments and even as bundles of microfilaments (2), but biochemical studies have shown that by far most nuclear actin appears in a monomeric or short oligomeric state (3,4,47). Furthermore, recent studies with fluorescent phalloidin, a drug specific to filamentous actin, have usually shown that the cytoplasm but not the nucleus is stained in somatic cells (1). Finally, actin-containing Balbiani ring mRNP particles in the nucleus are often not associated with any fibrous structures as demonstrated by immunoelectron microscopy (12). Thus, most of the available evidence suggests that actin associated with pre-mRNP/mRNP particles is in a monomeric or short oligomeric form, a conclusion also drawn by Hofmann et al. (11).
The actin–DBP40 complex presumably enters the cytoplasm bound to mRNA
Similarly to other A/B-type hnRNP proteins, e.g. hnRNP A1 and A2 (19), we show that DBP40 is likely to shuttle between the nucleus and the cytoplasm. The transfection experiment demonstrated that DBP40 appears in both the nucleus and the cytoplasm. Furthermore, the microinjection experiments directly showed that DBP40 is exported from the nucleus to the cytoplasm, i.e. DBP40 is likely to shuttle. The cross-linking experiments revealed that DBP40 is bound to the poly(A)+ RNA as well as to actin both in the nucleus and the cytoplasm. Thus, it seems likely, although it still remains to be proven, that the DBP40–actin complex, along with the associated minor variants of hnRNP A2 and A3, is bound to mRNA during the nucleocytoplasmic transfer.
In the dipteran C.tentans, we have shown that the A/B-type hrp36 protein is bound to actin, and the complex is associated with Balbiani ring pre-mRNA/mRNA from the gene in the cell nucleus to the polysomes in the cytoplasm (12). Thus, the rat DBP40 behaves similarly to the insect hrp36 in the sense that it binds actin in the nucleus and is transported into the cytoplasm coupled to mRNA. However, the mammalian data clearly suggests a more complex interaction between actin and hnRNP proteins in the pre-mRNP/mRNP particles, as actin is associated, directly or indirectly, with at least five different A/B-type proteins. Whether these proteins form a distinct supramolecular entity binding actin or whether they interact with actin as separate molecules remains to be established.
Possible roles of RNP-associated actin
In order to understand the role of actin present in the RNP particles, both nuclear and cytoplasmic functions have to be considered because the actin–DBP40 complex seems to be associated with the RNA throughout the processes of transcription, splicing, transport and translation just like the C.tentans actin–hrp36 complex. Since actin binds to nascent pre-mRNP, it is possible that it mediates packaging of the pre-mRNA into an RNP particle and could thus affect the processing of pre-mRNA. Another possibility would be that actin is involved in intranuclear movement and/or nucleocytoplasmic transport. This latter possibility is supported by a recent study indicating that actin is involved in nuclear export of viral RNAs (11). Furthermore, it has recently been demonstrated that actin contains functional leucine-rich, nuclear export signals, possibly mediating the export of mRNP (48).
In the cytoplasm, the actin–DBP40 complex bound to the mRNA could influence translation, mRNA turnover or mRNA transport, all processes known to be affected by hnRNP proteins (14). A large fraction of mRNA is known to be associated with cytoskeletal structures and in particular with actin microfilaments which may serve a role for RNA transport and/or localization (reviewed in 49). Recently, it has been shown that the cellular microfilament network also plays a direct role in the structure and function of the translation system and perturbations of this network affect protein synthesis (50). Therefore, if the RNA-associated actin proves to be functional in the cytoplasm, either in its G-form or F-form, it could be essential that actin becomes coupled to the mRNA already in the nucleus and is linked to the mRNA entering the cytoplasm. This could demonstrate a requirement for mRNA to undergo a nuclear preparatory event, involving actin and a subset of hnRNP proteins, in order to function properly in the cytoplasm.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Lise-Marie Fjelkestam and Carina Palmberg for technical assistance. The plasmid vectors for bacterial and mammalian expression of DBP40 were kindly donated by Dr Colin R. Parrish and purified actin by Dr Uno Lindberg. We thank Dr Adrian Krainer for supplying the pET9d-hnRNP A2 and Dr Gideon Dreyfuss for providing antibodies against hnRNP A1 and hnRNP C1/C2. The work was supported by the Swedish Research Council (B 5101-879/2001 to B.D.; K 5104-20005891 to T.B.), the Human Frontier Science Program Organization, the Knut and Alice Wallenberg Foundation, and the Ingabritt and Arne Lundberg Foundation. P.P. received Research Fellowships from the European Community and the Blanceflor-Ludovisi Foundation, D.N. a Fellowship from INTAS, and A.G. a Short Term Fellowship from EMBO and a stipend from the Swedish Institute.
REFERENCES
- 1.Rando O.J., Zhao,K. and Crabtree,G.R. (2000) Searching for a function for nuclear actin. Trends Cell Biol., 10, 92–97. [DOI] [PubMed] [Google Scholar]
- 2.Clark T.G. and Rosenbaum,J.L. (1979) An actin filament matrix in hand-isolated nuclei of X. laevis oocytes. Cell, 18, 1101–1108. [DOI] [PubMed] [Google Scholar]
- 3.Clark T.G. and Merriam,R.W. (1977) Diffusible and bound actin in nuclei of Xenopus laevis oocytes. Cell, 12, 883–891. [DOI] [PubMed] [Google Scholar]
- 4.Gounon P. and Karsenti,E. (1981) Involvement of contractile proteins in the changes in consistency of oocyte nucleoplasm of the newt Pleurodeles waltlii. J. Cell Biol., 88, 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheer U., Hinssen,H., Franke,W.W. and Jockusch,B.M. (1984) Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell, 39, 111–122. [DOI] [PubMed] [Google Scholar]
- 6.Egly J.M., Miyamoto,N.G., Moncollin,V. and Chambon,P. (1984) Is actin a transcription initiation factor for RNA polymerase B? EMBO J., 3, 2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke E., Dupuy,L., Wall,C. and Barik,S. (1998) Role of cellular actin in the gene expression and morphogenesis of human respiratory syncytial virus. Virology, 252, 137–148. [DOI] [PubMed] [Google Scholar]
- 8.Zhao K., Wang,W.O., Rando,J., Xue,Y., Swiderek,K., Kuo,A. and Crabtree,G.R. (1998) Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell, 95, 625–636. [DOI] [PubMed] [Google Scholar]
- 9.Brunel C. and Lelay,M.-N. (1979) Two-dimensional analysis of proteins associated with heterogeneous nuclear RNA in various animal cell lines. Eur. J. Biochem., 99, 273–283. [DOI] [PubMed] [Google Scholar]
- 10.Nakayasu H. and Ueda,K. (1985) Association of rapidly-labelled RNAs with actin in nuclear matrix from mouse L5178Y cells. Exp. Cell Res., 160, 319–330. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann W., Reichart,B., Ewald,A., Müller,E., Schmitt,I., Stauber,R.H., Lottspeich,F., Jockusch,B.M., Scheer,U., Hauber,J. and Dabauvalle,M.-C. (2001) Cofactor requirements for nuclear export of Rev Response Element (RRE)- and Constitutive Transport Element (CTE)-containing retroviral RNAs: an unexpected role for actin. J. Cell Biol., 152, 895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Percipalle P., Zhao,J., Pope,B., Weeds,A., Lindberg,U. and Daneholt,B. (2001) Actin bound to the hnRNP protein hrp36 accompanies Balbiani ring mRNA from the gene into polysomes. J. Cell Biol., 153, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Byrd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- 14.Krecic A.M. and Swanson,M.S .(1999) HnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- 15.Sierakowska H., Szer,W., Furdon,P.J. and Kole,R. (1986) Antibodies to hnRNP core proteins inhibit in vitro splicing of human β-globin pre-mRNA. Nucleic Acids Res., 14, 5241–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chabot B., Blanchette,M., Lapierre,I. and La Branche,H. (1997) An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol., 17, 1776–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayeda A. and Krainer,A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- 18.Mayeda A., Munro,S., Caceres,J.F. and Krainer,A.R. (1994) Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J., 13, 5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinol-Roma S. and Dreyfuss,G. (1992) Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature, 355, 730–732. [DOI] [PubMed] [Google Scholar]
- 20.Michael W.M., Choi,M. and Dreyfuss,G. (1995) A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell, 83, 415–422. [DOI] [PubMed] [Google Scholar]
- 21.Hoeck K.S., Kidd,G.J., Carson,J.H. and Smith,R. (1998) hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry, 37, 7021–7029. [DOI] [PubMed] [Google Scholar]
- 22.Kwon S., Barbarese,E. and Carson,J.H. (1999) The cis-acting RNA trafficking signal from myelin basic protein mRNA and its cognate trans-acting ligand hnRNP A2 enhance cap-dependent translation. J. Cell Biol., 147, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun X., Alzhanova-Ericsson,A., Visa,N., Aissouni,Y., Zhao,J. and Daneholt,B. (1998) The hrp23 protein in the Balbiani ring pre-mRNP particles is released just before or at the binding of the particles to the nuclear pore complex. J. Cell Biol., 142, 1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Visa N., Alzhanova-Ericsson,A.T., Sun,X., Kiseleva,E., Björkroth,B., Wurtz,T. and Daneholt,B. (1996) A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell, 84, 253–264. [DOI] [PubMed] [Google Scholar]
- 25.Moraitou M., Patrinou-Georgoula,M. and Guialis,A. (1998) Structural/functional properties of a mammalian multi-component structure containing all major spliceosomal small nuclear ribonucleoprotein particles. Biochem. J., 332, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Farrel P.Z., Goodman,H.M. and O’Farrel,P.H. (1977) High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell, 12, 1133–1142. [DOI] [PubMed] [Google Scholar]
- 27.Oppermann M., Cols,N., Nyman,T., Helin,J., Saarinen,J., Byman,I., Toran,N., Alayia,A.A., Bergman,T., Kalkkinen,N., Gonzalez-Duarte,R. and Jörnvall,H. (2000). Identification of foetal brain proteins by two-dimensional gel electrophoresis and mass spectrometry. Eur. J. Biochem., 267, 4713–4719. [DOI] [PubMed] [Google Scholar]
- 28.Rozycki M., Schutt,C.E. and Lindberg,U. (1991) Affinity chromatography-based purification of profilin:actin. Methods Enzymol., 196, 100–118. [DOI] [PubMed] [Google Scholar]
- 29.Boussif O., Lezoualc’h,F., Zanta,M.A., Mergny,M.D., Scherman,D., Demeneix,B. and Behr,J.-P. (1995). A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA, 92, 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D. and Parrish,C.R. (1999) A heterogeneous nuclear ribonucleoprotein A/B-related protein binds to single-stranded DNA near the 5′ end or within the genome of feline parvovirus and can modify virus replication. J. Virol., 73, 7761–7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinol-Roma S., Adam,S.A., Choi,Y.D. and Dreyfuss,G. (1989) Ultraviolet-induced cross-linking of RNA to proteins in vivo. Methods Enzymol., 180, 410–418. [DOI] [PubMed] [Google Scholar]
- 32.Dangli A., Plomaritoglou,A., Boutou,E., Vassiliadou,N., Moutsopoulos,H.M. and Guialis,A. (1996) Recognition of subsets of the mammalian A/B type core hnRNP polypeptides by novel autoantibodies. Biochem. J., 320, 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plomaritoglou A., Choli-Papadopoulou,T. and Guialis,A. (2000) Molecular characterization of a murine, major A/B-type hnRNP protein: mBx. Biochim. Biophys. Acta, 1490, 54–62. [DOI] [PubMed] [Google Scholar]
- 34.Good P.J., Rebbert,M.L. and Dawid,I.B. (1993) Three new members of the RNP protein family in Xenopus. Nucleic Acids Res., 21, 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takiguchi S., Tokino,T., Imai,T., Tanigami,A., Koyama,K. and Nakamura,Y. (1993) Identification and characterization of a cDNA, which is highly homologous to the ribonucleoprotein gene, from a locus (D10S102) closely linked to MEN2 (multiple endocrine neoplasia type 2). Cytogenet. Cell Genet., 64, 128–130. [DOI] [PubMed] [Google Scholar]
- 36.Wurtz T., Kiseleva,E., Nacheva,G., Alzhanova-Ericsson,A.T., Rosén,A. and Daneholt,B. (1996) Identification of two RNA-binding proteins in Balbiani ring premessenger ribonucleoprotein granules and presence of these proteins in specific subsets of heterogeneous nuclear ribonucleoprotein particles. Mol. Cell. Biol., 16, 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinol-Roma S., Choi,Y.D., Matunis,M.J. and Dreyfuss,G. (1988) Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev., 2, 215–227. [DOI] [PubMed] [Google Scholar]
- 38.Inoue A., Omori,A., Ichinose,S., Takahashi,K.P., Kinoshita,Y. and Mita,S. (2001) S1 proteins C2 and D2 are novel hnRNPs similar to the transcriptional repressor, CArG box motif-binding factor A. Eur. J. Biochem., 268, 3654–3663. [DOI] [PubMed] [Google Scholar]
- 39.Kamada S. and Miwa,T. (1992) A protein binding to CArG box motifs and to single-stranded DNA functions as a transcriptional repressor. Gene, 119, 229–236. [DOI] [PubMed] [Google Scholar]
- 40.Marsich E., Bandiera,A., Tell,G., Scaloni,A. and Manzini,G. (2001) A chicken hnRNP of the A/B family recognizes the single-stranded d(CCCTAA)n telomeric repeated motif. Eur. J. Biochem., 268, 139–148. [DOI] [PubMed] [Google Scholar]
- 41.Bemark M., Olsson,H., Heinegård,D. and Leanderson,T. (1998) Purification and characterization of a protein binding to the SP6 κ promoter. J. Biol. Chem., 273, 18881–18890. [DOI] [PubMed] [Google Scholar]
- 42.Mikheev A.M., Mikheev,A.A., Zhang,Y., Aebersold,R. and Zarbi,H. (2000) CArG binding factor A (CBF-A) is involved in transcriptional regulation of the rat Ha-ras promoter. Nucleic Acids Res., 28, 3762–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smidt M.P., Russchen,B., Snippe,L., Wijnholds,J. and Ab,G. (1995) Cloning and characterization of a nuclear, site specific ssDNA binding protein. Nucleic Acids Res., 23, 2389–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau P.P., Zhu,H.J., Nakamuta,M. and Chan,L. (1997) Cloning of an Apobec-1-binding protein that also interacts with apolipoprotein B mRNA and evidence for its involvement in RNA editing. J. Biol. Chem., 272, 1452–1455. [DOI] [PubMed] [Google Scholar]
- 45.Wollerton M.C., Gooding,C., Robinson,F., Brown,E.C., Jackson,R.J. and Smith,C.W. (2001) Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA, 7, 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loflin P., Chen,C.-Y.A. and Shyu,A.-B. (1999) Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev., 13, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amankwa K.S. and De Boni,U. (1994) Ultrastructural localization of filamentous actin within neuronal interphase nuclei in situ. Exp. Cell Res., 210, 315–325. [DOI] [PubMed] [Google Scholar]
- 48.Wada A., Fukuda,M., Mishima,M. and Nishida,E. (1998) Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J., 17, 1635–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jansen R.P. (1999) RNA–cytoskeletal interactions. FASEB J., 13, 455–466. [PubMed] [Google Scholar]
- 50.Stapulionis R., Kolli,S. and Deutscher,M.P. (1997) Efficient mammalian protein synthesis requires an intact F-actin system. J. Biol. Chem., 272, 24980–24986. [DOI] [PubMed] [Google Scholar]