Abstract
Upon activation by double-stranded RNA in virus-infected cells, the cellular PKR kinase phosphorylates the translation initiation factor eukaryotic initiation factor 2 (eIF2) and thereby inhibits protein synthesis. The γ34.5 and Us11 gene products encoded by herpes simplex virus type 1 (HSV-1) are dedicated to preventing the accumulation of phosphorylated eIF2. While the γ34.5 gene specifies a regulatory subunit for protein phosphatase 1α, the Us11 gene encodes an RNA binding protein that also prevents PKR activation. γ34.5 mutants fail to grow on a variety of human cells as phosphorylated eIF2 accumulates and protein synthesis ceases prior to the completion of the viral life cycle. We demonstrate that expression of a 68-amino-acid fragment of Us11 containing a novel proline-rich basic RNA binding domain allows for sustained protein synthesis and enhanced growth of γ34.5 mutants. Furthermore, this fragment is sufficient to inhibit activation of the cellular PKR kinase in a cell-free system, suggesting that the intrinsic activities of this small fragment, notably RNA binding and ribosome association, may be required to prevent PKR activation.
The innate cellular antiviral response is powerful and multifaceted, involving extensive changes in enzyme activity and cytokine production intended to impede viral replication and spread (22, 38). RNA molecules that are double stranded (dsRNA) or highly structured play a central role in initiating this response in infected cells. These RNA ligands activate numerous latent enzymes which then in turn act on a variety of substrates to effect global changes in cellular physiology. The cellular PKR kinase is a key target that is activated in response to dsRNA. PKR contains two dsRNA binding motifs that in part mediate the dimerization of the enzyme on the dsRNA molecule. Following dimerization, each subunit of the enzyme is thought to phosphorylate the other, thus completing the process of activation and imbuing the enzyme with the ability to phosphorylate other substrates in trans, notably the critical translation initiation factor eukaryotic initiation factor 2 (eIF2) (reviewed in references 6 and 41). Phosphorylation of eIF2 on its α subunit prevents the initiation of protein synthesis and can lead to cell death, thus curtailing viral replication and limiting further propagation of the infection (9, 10, 18, 19). To successfully complete their replicative program and foster efficient dissemination, viruses have evolved a myriad of functions to effectively deal with activated PKR and preclude the cessation of protein synthesis (reviewed in references 17, 18, 22, and 35).
Herpes simplex virus type 1 (HSV-1) is a large DNA virus that latently infects neurons and periodically reinitiates productive growth at epithelial sites, causing blisters, or in the central nervous system, resulting in encephalitis (reviewed in reference 30). HSV-1 contains at least two discrete functions dedicated to preventing the accumulation of phosphorylated eIF2α in infected cells (1, 25). The γ34.5 genes encode a regulatory subunit for the cellular protein phosphatase 1α (13). Thus, the virally modified protein phosphatase 1α holoenzyme is able to reverse the effects of PKR activation by maintaining steady-state levels of active, unphosphorylated eIF2. Deletion of the viral γ34.5 genes creates a host range mutant that is unable to complete its life cycle on a variety of cultured cells as phosphorylated eIF2 accumulates, leading to the premature cessation of protein synthesis at late times postinfection (4, 5). A second viral function that regulates eIF2 phosphorylation was uncovered with the isolation of second-site suppressor mutations that allowed γ34.5 deletion mutants to regain the ability to synthesize proteins and grow on cells that failed to support the replication of the γ34.5 parent mutants (24). These suppressor mutants all expressed the viral Us11 protein at very early times in the infectious cycle (12). Recent studies have demonstrated that Us11 expression prevents the premature cessation of protein synthesis seen for γ34.5 mutants and inhibits PKR activation (2, 25).
Us11 is a basic, 21-kDa RNA binding protein that localizes to the nucleolus, associates with polysomes in infected cells, binds to and regulates the accumulation of at least one viral nonpolyadenylated RNA of unknown function, is incorporated into virions, and reportedly modulates gene expression in a manner akin to that of transactivators encoded by complex retroviruses (8, 31, 32). Structure-function analysis has demonstrated that while both amino- and carboxyl-terminal domains can be packaged into virus particles, the carboxy-terminal 68 amino acids are required to bind RNA, localize to nucleoli, and associate with ribosomes (33). To begin to understand which functions of Us11 may be responsible for regulating protein synthesis and inhibiting PKR activation, we sought to identify the region of Us11 required for these activities. The genetic analysis in this report demonstrates that expression of the Us11 RNA binding domain is necessary and sufficient to overcome the PKR-mediated block to protein synthesis in infected cells. Furthermore, this 68-amino-acid domain effectively prevents PKR activation in vitro, suggesting that the activities ascribed to this domain, notably RNA binding and ribosome association, may be required to prevent PKR activation.
MATERIALS AND METHODS
Plasmids.
All nucleotide numbers are derived from the published sequence of HSV-1 strain 17 (GenBank accession no. X14112). HSV-1 Patton strain DNA was used throughout this study. A universal acceptor vector to express proteins from the full-length α27 promoter and target homologous recombination to the viral thymidine kinase (tk) locus was constructed. This pBluescript II SK(+)-based vector fuses the SalI-EcoRI fragment (nucleotide [nt] 50255 to 47986) from the 5′ region of the tk gene to the full-length α27 promoter (nt 111990 to 113646). Unique HindIII and XbaI sites lie immediately downstream of the α27 promoter. The cloning linker is then followed by the SacI-BamHI fragment (nt 47358 to 45055) from the 3′ region of the tk gene, which also contains a poly(A)+ site. Homologous recombination within the tk locus creates an EcoRI-SacI deletion within the tk (UL23) gene and also affects the synthesis of the UL24 gene product. The full-length Us11 gene was isolated as a HindIII-XbaI fragment following the insertion of a HindIII site immediately after nt 145322 and an XbaI site immediately prior to nt 144714. Us11 gene fragments containing an engineered HindIII site at their 5′ end and an XbaI site at their 3′ terminus were isolated by PCR, and all PCR products were sequenced. The Δ5-87 mutant fuses sequences upstream of the authentic Us11 ATG along with the first four codons (nt 145322 to 145235) to codons 88 to 155 (nt 144985 to 144714). The Δ5-87fs variant, also produced by PCR, is identical to Δ5-87 except for the insertion of an additional cytosine residue between nt 145239 and 145240. This creates a +1 frameshift at the third Us11 codon. Δ88-155 fuses sequences upstream of the authentic Us11 ATG inclusive through codon 87 (nt 145322 to 144986) to a stop codon.
Isolation of recombinant viruses.
Following cotransfection of each individual targeting plasmid with Δ34.5 viral DNA into Vero cells, tk-negative recombinant viruses were isolated by two rounds of plaque purification on 143tk− cells in the presence of bromodeoxyuridine as described in the work of Mulvey et al. (25). Isolation of viral DNA and Southern analysis were performed as described in the work of Mulvey et al. (25).
Cells and viruses. Vero, U373, 143tk−, and 293 cells were from the American Type Culture Collection and were propagated as described previously (25). The HSV-1 Patton strain was used in these studies.
Analysis of total viral protein synthesis.
High-multiplicity-of-infection (MOI) infections and labeling with 35S were performed as described previously (25).
Antibodies.
The Us11 monoclonal antibody was a gift from Richard Roller (31). The polyclonal antibody directed against the Us11 C terminus was a gift from Howard Marsden (16). Following electrophoretic transfer from sodium dodecyl sulfate (SDS)-polyacrylamide gels, Us11 proteins were detected using an ECL kit (Amersham) according to the manufacturer's specifications.
Preparation of S10 extracts and PKR kinase assay.
Twelve confluent 10-cm-diameter dishes of 293 cells were stimulated with 1,000 U of alpha interferon (Hoffmann, La Roche) per ml for 18 h. The cells were washed twice with ice-cold phosphate-buffered saline and once with buffer A (10 mM HEPES-KOH [pH 7.4], 15 mM KCl, 1.5 mM magnesium acetate [Mg(OAc)2], 1 mM dithiothreitol [DTT]) and suspended in an amount of buffer A equivalent to 2.5 packed cell volumes (typically 1.2 ml of packed cells). After swelling for 10 min on ice, the cells were disrupted in a Dounce homogenizer with approximately 30 strokes of a tight-fitting pestle. An amount of buffer B [100 mM HEPES-KOH (pH 7.4), 1.05 M KCl, 35 mM Mg(OAc)2, 10 mM DTT] equivalent to 1/10 of the packed cell volume was added, along with phenylmethylsulfonyl fluoride to a final concentration of 100 μM. The extract was centrifuged at 5,000 × g for 5 min at 4°C, and this supernatant was subsequently spun at 10,000 × g for 10 min at 4°C. The S10 (typically 3 to 5 mg/ml) was quick-frozen in small aliquots and stored at −80°C. Kinase reaction mixtures (25 μl) were assembled on ice and contained 15 μl of S10, 20 μCi of [γ-32P]ATP (6,000 Ci/mmol; NEN), 30 μM ATP, 5 mM Mg(OAc)2, 20 mM HEPES-KOH(pH 7.4), 100 mM KCl, 1.5 mM DTT, and 100 μM phenylmethylsulfonyl fluoride. Purified glutathione S-transferase (GST) fusion proteins were dialyzed into 20 mM HEPES-KOH (pH 7.4)–100 mM KCl–0.5 mM DTT and frozen at −80°C in small aliquots. The reaction mixtures contained reovirus RNA (a gift from A. Shatkin) at 25 ng/ml where indicated. Following incubation at 30°C for 30 min, the reaction mixtures were processed for immunoprecipitation of PKR as described previously (25). PKR phosphorylation was quantified on a PhosphorImager.
RESULTS
Construction of recombinant viruses that express fragments of the Us11 protein.
To determine if a discrete region of Us11 could support enhanced growth of γ34.5 mutants in nonpermissive cultured cells, we engineered recombinant γ34.5 mutant viruses that ectopically express Us11 fragments from a heterologous promoter located within the viral tk locus. This strategy proved useful in our prior studies of the full-length protein (25). Basically, the strong viral promoter from the α27 gene was used to direct synthesis of Us11 fragments at immediate-early times in the viral life cycle (25). This temporal pattern of Us11 expression creates a dominant mutation in the genetic background of a γ34.5-null virus (Δ34.5). The endogenous Us11 allele, located elsewhere in the Us region of the viral genome, is under the control of a stringent late promoter and is not expressed due to the PKR-imposed block of late protein synthesis in Δ34.5-infected cells (4, 5). Sequences flanking this α27-Us11 expression cassette were designed to facilitate homologous recombination within the viral tk genetic locus, resulting in tk-negative viruses.
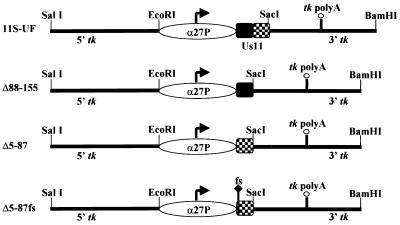
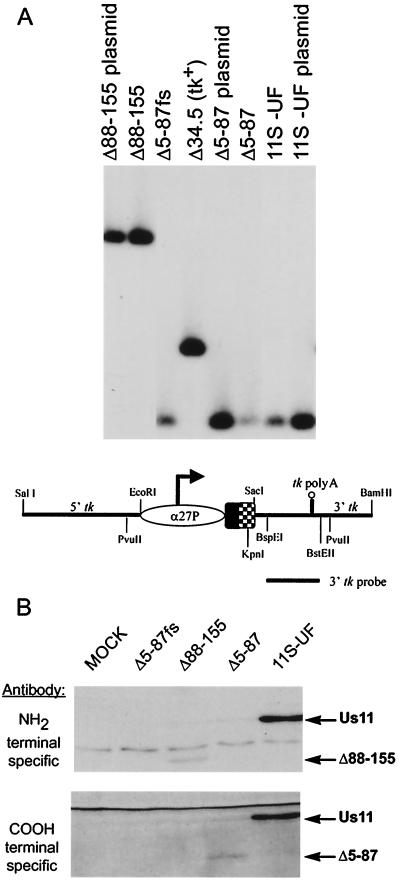
The targeting constructs and the proteins that they are designed to produce are illustrated in Fig. 1. Δ88-155 expresses the amino-terminal 87 amino acids of Us11, while Δ5-87 fuses the amino-terminal 4 amino acids to the carboxyl-terminal amino acids 88 to 155. The fusion of the amino-terminal four amino acids onto the carboxyl-terminal RNA binding domain was necessary to achieve steady-state levels of detectable protein by Western analysis (unpublished data). Δ5-87fs is identical in all respects to Δ5-87 except that it contains a single nucleotide insertion at codon 3 to create a frameshift mutation. 11S-UF produces the wild-type, full-length 155-amino-acid Us11 protein. Analysis of the physical structure of these viruses is presented in Fig. 2A. Southern analysis of the tk locus demonstrates that all of the viruses contain insertions in the tk locus and are homogeneously tk-negative alleles. Moreover, isolates designed to produce Us11 amino-terminal fragments (Δ88-155) can be distinguished from viruses that contain sequences encoding carboxy-terminal sequences (Δ5-87, Δ5-87fs, and 11S-UF) based upon their sensitivity to different restriction endonucleases. The endogenous Us11 locus near the TRs-Us junction was intact and unrearranged (data not shown).
FIG. 1.
Targeting constructs and the Us11-related proteins that they are designed to express. The 5′ and 3′ regions from the HSV-1 tk gene in the illustrated plasmids target homologous recombination to the genomic viral tk locus and create tk-negative recombinant viruses. Transcription from the HSV-1 α27 promoter occurs at immediate-early times postinfection, and the direction of transcription from this promoter is indicated with an arrow. Transcripts are polyadenylated at the poly(A)+ addition site within the 3′ tk region. 11S-UF expresses the full-length, 155-amino-acid Us11 protein from the wild-type Patton strain. The region that encodes the amino-terminal 87 amino acids is shaded, while the region encoding the carboxy-terminal 68 amino acids appears as a checkerboard. Δ88-155 expresses the amino-terminal 87 amino acids, and Δ5-87 fuses the amino-terminal 4 amino acids to the carboxy-terminal 68 residues. Δ5-87fs contains a single nucleotide insertion at codon 3 that results in a +1 frameshift (fs) but is otherwise isogenic to the insert in Δ5-87.
FIG. 2.
(A) Physical analysis of tk-negative recombinant virus genomes. Viral DNA was digested with PvuII and KpnI, fractionated by electrophoresis on 1% agarose gels, transferred to a nylon membrane, and hybridized to a 32P-labeled BspEI-BstEII probe from the 3′ region of the tk gene. The location of the probe and the relevant restriction sites are illustrated for the 11S-UF construct. The Us11 gene segment that encodes the amino-terminal 87 amino acids is shaded, while the segment encoding the carboxyl-terminal 68 amino acids appears as a checkerboard. Recombinant genomes that contain the carboxyl-terminal region of Us11 are sensitive to digestion with KpnI and yield smaller fragments, distinguishing them from recombinant viruses that express only the Us11 amino-terminal 87 amino acids and nonrecombinant viruses that contain an intact tk gene (represented by the Δ34.5 virus). All the plaque-purified, recombinant viruses are homogeneous tk-negative alleles as evidenced by the complete absence of fragments that comigrate with the wild-type tk-positive PvuII fragment contained in the Δ34.5 parent genome. (B) Production of Us11-related proteins by tk-negative recombinant viruses. U373 cells were mock infected or infected at an MOI of 5 with either 11S-UF, Δ5-87, Δ5-87fs, or Δ88-155. At 6 h postinfection, lysates were prepared, and polypeptides were fractionated by SDS-PAGE on 17.5% polyacrylamide gels and electrophoretically transferred to a membrane which was probed with antibodies directed against either the amino or the carboxyl terminus of Us11.
Protein production was documented by analyzing infected cell lysates at immediate-early times postinfection. The 11S-UF virus directed the synthesis of the 21-kDa full-length Us11 protein that cross-reacted with antisera directed against either the amino- or the carboxy-terminal determinants (Fig. 2B). Cells infected with Δ88-155 synthesized a polypeptide consistent with the calculated molecular mass of 9.6 kDa that reacted with a monoclonal antibody directed against the amino terminus of Us11, while the truncated polypeptide encoded by Δ5-87 was visible only with a polyclonal antibody raised against the carboxy terminus of the protein (Fig. 2B). Cells infected with Δ5-87fs did not synthesize detectable amounts of immunoreactive protein. Thus, the recombinant viruses constructed express polypeptides that are of the appropriate size and possess the expected antigenic properties. While the steady-state levels of the isolated amino- and carboxyl-terminal domains are reduced relative to the full-length Us11 protein, the Δ5-87 and Δ88-155 proteins are produced in similar amounts compared to that of full-length Us11.
Expression of the carboxy-terminal 68-amino-acid Us11 RNA binding domain confers a growth advantage upon γ34.5 mutants and enhances protein synthesis.
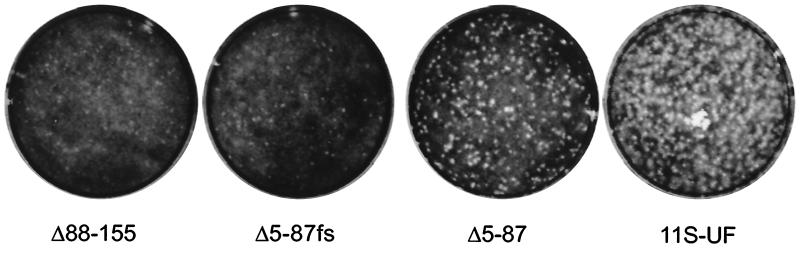
To ascertain the effect of expressing various Us11 protein fragments on the growth of γ34.5 mutants, human U373 glioblastoma cells were infected at a low MOI with 8,000 PFU of each recombinant virus. As γ34.5 mutants cannot sustain late protein synthesis on U373 cells, these cells are nonpermissive for the growth of γ34.5 mutant viruses. At 4.5 days postinfection, the plates were fixed and stained with crystal violet. The 11S-UF virus expresses the full-length Us11 protein and serves as a positive control in this experiment. Figure 3 demonstrates that the 68-amino-acid carboxy-terminal Us11 fragment confers a growth advantage upon γ34.5 mutant viruses as evidenced by the greater numbers of large viral plaques present. The enhanced growth is abrogated by a single nucleotide substitution at codon 3 in the Δ5-87 reading frame. The yield of virus produced on U373 cells infected in parallel at low multiplicity was quantified by determining titers of lysates on permissive Vero cells. Δ5-87 replicates to levels at least 60-fold greater than those of Δ5-87fs. It is important to remember that the steady-state level to which the carboxyl-terminal fragment encoded by Δ5-87 accumulates is markedly reduced relative to that for the full-length Us11 protein (Fig. 2B). 11S-UF, which produces large amounts of full-length Us11 (see Fig. 2B), replicates to levels approximately ninefold greater than those of Δ5-87. This demonstrates that the synthesis of the Δ5-87 protein product is indeed responsible for the observed phenotype. Recombinant viruses that express the amino-terminal 87 amino acids do not display enhanced growth in this assay.
FIG. 3.
A carboxyl-terminal fragment of Us11 that contains the RNA binding domain confers a growth advantage upon a Δ34.5 mutant. Nonpermissive U373 cells (3 × 106 cells) were infected with 8,000 PFU of either 11S-UF, Δ5-87, Δ5-87fs, or Δ88-155. After 4.5 days at 37°C, the cells were fixed with 10% trichloroacetic acid and stained with crystal violet.
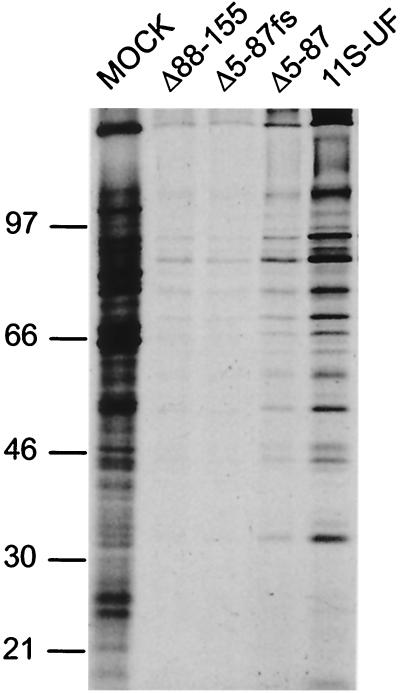
To analyze the regulation of viral protein synthesis, replicate cultures of U373 cells were mock infected or infected with either Δ5-87, Δ5-87fs, Δ88-155, or 11S-UF at a high MOI. At late times postinfection, the cultures were pulse-labeled with 35S-amino acids, and total protein was isolated and fractionated by SDS-polyacrylamide gel electrophoresis (PAGE). Figure 4 demonstrates that cells infected with either 11S-UF or Δ5-87 synthesize substantially more late viral proteins than do cultures infected with the Δ88-155 virus. Although the 11S-UF virus directs even greater levels of late viral protein synthesis, the Δ5-87 gene product is present in markedly smaller quantities relative to the full-length Us11 protein (Fig. 2B, C-terminal sequence-specific antibody), and this will likely influence the efficiency with which protein synthesis is restored. The frameshift mutation introduced into Δ5-87fs abolishes the enhanced levels of protein synthesis, demonstrating that synthesis of the Δ5-87 gene product is required for the increased levels of viral late protein synthesis observed. Cells infected with Δ88-155 are as defective in sustaining late protein synthesis as are those infected with Δ5-87fs. We therefore conclude that expression of the Us11 carboxy-terminal RNA binding domain is necessary and sufficient to overcome the late block to protein synthesis in cells infected with γ34.5 mutant viruses. Finally, to determine if the amino-terminal domain could augment the activity of the carboxyl-terminal RNA binding domain, a mixed infection with recombinant viruses expressing the isolated amino- and carboxyl-terminal domains was performed. The presence of the amino-terminal domain in trans did not enhance the ability of the carboxyl-terminal domain to sustain late viral protein synthesis (data not shown).
FIG. 4.
Expression of the Us11 RNA binding domain is required to overcome the block to protein synthesis in cells infected with Δ34.5 mutants. Nonpermissive U373 cells were mock infected or infected with either Δ88-155, Δ5-87, Δ5-87fs, or 11S-UF at an MOI of 50. Following a 1-h pulse with 35S-amino acids at 10 h postinfection, proteins were solubilized in sample buffer and fractionated by SDS-PAGE on a 12.5% polyacrylamide gel. The fixed, dried gel was subsequently exposed to Kodak XAR film. Numbers at left are molecular masses in kilodaltons.
The 68-amino-acid carboxy-terminal RNA binding domain of Us11 prevents activation of the PKR kinase.
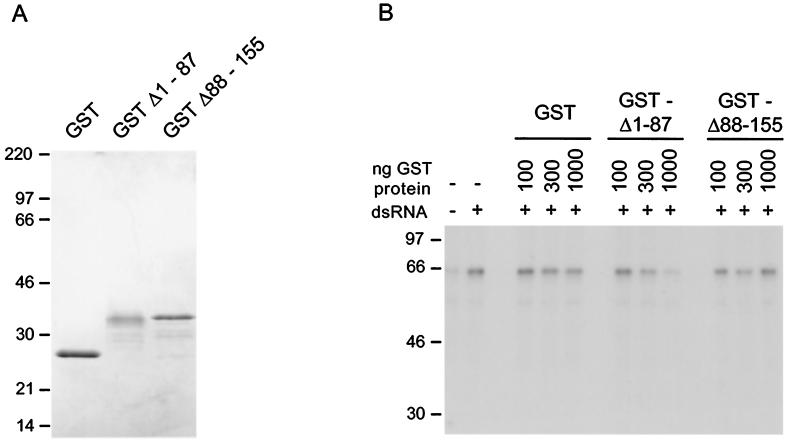
To assess the effect of the 68-amino-acid RNA binding domain on PKR activation, we employed a cell-free system derived from alpha interferon-treated human 293 cells. S10 extracts prepared as described in Materials and Methods served as the source of unactivated PKR. Upon the addition of small amounts of reovirus dsRNA (25 ng/ml), activation of the cellular PKR kinase was evaluated by monitoring its phosphorylation state after immunoprecipitation (Fig. 5B). GST fusion proteins containing either the amino-terminal 87 amino acids or the carboxy-terminal 68 amino acids of Us11 were engineered and purified (Fig. 5A). The Δ1–87 fusion protein contains the RNA binding domain and binds RNA in vitro (33; D. Khoo and I. Mohr, unpublished data). While addition of increasing amounts of purified GST or the GSTΔ88-155 Us11 fusion protein had no measurable effect on PKR activation, addition of increasing amounts of the GSTΔ1-87 Us11 fusion protein resulted in progressive inhibition of PKR activation (Fig. 5B). In multiple experiments, we observed the range of this inhibition to be between three- and sixfold. Thus, a 68-amino-acid Us11 protein fragment that confers a growth advantage upon γ34.5 mutant viruses, precludes the premature cessation of protein synthesis in cells infected with γ34.5 mutants, and binds RNA is necessary and sufficient to prevent activation of the cellular PKR kinase.
FIG. 5.
The 68-amino-acid Us11 RNA binding domain inhibits PKR activation. (A) Two micrograms of purified GST, GSTΔ1-87, or GSTΔ88-155 was subjected to electrophoresis on an SDS-polyacrylamide gel and stained with Coomassie blue. (B) Increasing amounts of either purified GST, GSTΔ1-87, or GSTΔ88-155 were added to 293 S10 extracts. Following the addition of reovirus dsRNA, the reaction mixtures were incubated at 30°C for 30 min. PKR was immunoprecipitated, and the immune complex was fractionated by SDS-PAGE. The gel was fixed and exposed to Kodak XAR film. Numbers at left of each panel indicate molecular masses in kilodaltons.
DISCUSSION
The Us11 gene product specified by HSV is an RNA binding protein that inhibits activation of the cellular PKR kinase (2, 25). Our studies demonstrate that a 68-amino-acid carboxy-terminal protein fragment containing the Us11 RNA binding domain can prevent the premature cessation of protein synthesis that occurs in cells infected with γ34.5 mutants. This same domain can prevent the activation of the cellular PKR kinase in a cell-free system, suggesting that the intrinsic RNA binding activity of this small domain may be intimately involved in precluding PKR activation.
The exact nature of the dsRNA molecule(s) that activates PKR in HSV-infected cells is unknown. It is precisely this ill-defined RNA activator(s) that may constitute the RNA target(s) to which Us11 binds. The Us11 ribonucleoprotein complex could thus sequester these RNAs and prevent PKR activation. The only RNAs reported to interact with the Us11 polypeptide are a nonpolyadenylated viral RNA of unknown function and the 28S and 5.8S rRNAs in the 60S ribosomal subunit of the host (31, 32). While the significance of Us11 recognizing these RNA ligands is not known, it is worth noting that PKR may also associate with ribosomes (42, 43). Us11 could thereby function as a virus-encoded ribosomal protein that prevents PKR activation by structured RNA elements within the ribosomes of HSV-infected cells. Alternatively, tethering PKR to ribosomes via an interaction with Us11 might instead prevent PKR activation by soluble RNA and/or protein activators (20, 28). Such a mechanism has been proposed to explain how ribosomal protein L18 prevents the formation of active PKR dimers (20). Finally, Us11 may have properties of a dsRNA binding protein, as it prevents the activation of PKR mediated by purified reovirus dsRNA.
A physical complex between Us11 and PKR has been observed in infected cells, and this protein-protein interaction may play a role in inhibiting PKR activation (2). Indeed, studies with the vaccinia virus E3L protein have demonstrated a role for both RNA binding and a physical E3L-PKR complex in inhibiting activation of human PKR in Saccharomyces cerevisiae (34). The determinants for a PKR-Us11 interaction could reside either within the RNA binding domain or within the amino-terminal domain. If the amino-terminal domain is involved in contacting PKR, this interaction is not necessary or sufficient to prevent the PKR-mediated inhibition of protein synthesis in infected cells, as Δ88-155 does not support the enhanced growth of Δ34.5 mutants, nor does it inhibit the kinase. A trimeric ribonucleoprotein complex involving a heterodimer between PKR and the Us11 RNA binding domain could also assemble on dsRNA. However, this static complex would be unable to yield an activated kinase molecule. Alternatively, physical association between Us11 and PKR may be required to prevent PKR from interfacing with a variety of protein activators such as PACT or REX (15, 26).
Us11 is the newest member of a class of viral RNA binding proteins, including the influenza virus NS1, vaccinia virus E3L, and reovirus sigma 3 proteins, that can prevent PKR activation. These polypeptides are derived from a diverse group of RNA and DNA viruses, suggesting that their function is evolutionarily quite ancient. However, these proteins share no primary structural homology, raising the possibility that their RNA binding domains are related in three-dimensional space. Structural information exists only for the NS1 protein, which binds multiple RNA ligands, including U6, poly(A), and dsRNA, as a 52-kDa homodimer (3, 21). Three Arg-rich alpha helices within a 73-amino-acid basic domain are thought to dimerize and contact RNA (27, 39). E3L contains a single copy of the consensus dsRNA binding domain (αβββα), as derived from the analysis of the Drosophila Staufen protein, human dsRNA-specific adenine deaminase, PKR, and Escherichia coli RNase III (37). Mutation of conserved residues within this homologous region of E3L significantly reduces dsRNA binding (14). The 41-kDa sigma 3 protein contains a basic 85-amino-acid region at its carboxyl terminus, and several of these residues are absolutely required for binding to dsRNA (7, 23).
The RNA binding domain of Us11 does not contain any sequence elements that are related to characterized RNA recognition motifs from known RNA binding proteins. It is composed of multiple iterations of Arg-X-Pro, where X is often an uncharged polar or acidic amino acid (29, 33, 40). The Us11 protein is conserved between HSV-1 and HSV-2; moreover, different HSV-1 isolates contain variations in the number of repeats, thus accounting for slight differences in the size of the Us11 protein. Roller et al. have proposed that the repetitive Arg-X-Pro domain adopts the conformation of a poly-l-proline II helix, a left-handed single helix characterized by three residues per turn (33). This model aligns all of the basic Arg residues on a single face of the helix. Indeed, modeling such structures predicts that they are capable of recognizing specific sequences in DNA (11). Further investigation of the novel repetitive Arg-X-Pro motif in the 68-amino-acid RNA binding fragment of Us11 will illuminate which residues are required for inhibiting PKR activation and interacting with RNA. This domain may also recognize dsRNA and thus define a new recognition element mediating dsRNA binding. Ultimately, comparing actual structural data for Us11, NS1, E3L, and sigma 3 will yield a more complete understanding of how this seemingly diverse class of proteins recognizes RNA targets and inhibits activation of the cellular PKR kinase.
ACKNOWLEDGMENTS
We thank Rich Roller (University of Iowa), Howard Marsden (MRC Glasgow), and Aaron Shatkin (CABM, Rutgers University) for generously providing reagents for this study (antibodies and reovirus RNA). In addition, we thank David Levy and Bob Schneider for critically reading the manuscript.
J.P. is a predoctoral trainee supported in part by NIH grant 5 T32 AI07180. This work was supported by developmental funds from the Kaplan Cancer Center and a grant from the National Institutes of Health (GM56927) awarded to I.M.
REFERENCES
- 1.Cassady K A, Gross M, Roizman B. The second-site mutation in the herpes simplex virus recombinants lacking the γ134.5 genes precludes shutoff of protein synthesis by blocking the phosphorylation of eIF-2α. J Virol. 1998;72:7005–7011. doi: 10.1128/jvi.72.9.7005-7011.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassady K A, Gross M, Roizman B. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the α subunit of eukaryotic translation initiation factor 2. J Virol. 1998;72:8620–8626. doi: 10.1128/jvi.72.11.8620-8626.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chien C Y, Tejero R, Huang Y, Zimmerman D E, Rios C B, Krug R M, Montelione G T. A novel RNA-binding motif in influenza A virus nonstructural protein 1. Nat Struct Biol. 1997;4:891–895. doi: 10.1038/nsb1197-891. [DOI] [PubMed] [Google Scholar]
- 4.Chou J, Roizman B. The γ34.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc Natl Acad Sci USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou J, Chen J J, Gross M, Roizman B. Association of a M(r) 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with gamma 134.5− mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemens M J. Protein kinases that phosphorylate eIF2 and eIF2B, and their role in eukaryotic cell translational control. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 139–172. [Google Scholar]
- 7.Denzler K L, Jacobs B L. Site-directed mutagenic analysis of reovirus sigma 3 protein binding to dsRNA. Virology. 1994;204:190–199. doi: 10.1006/viro.1994.1523. [DOI] [PubMed] [Google Scholar]
- 8.Diaz J J, Dodon M D, Schaerer-Uthurralt N, Simonin D, Kindbeiter K, Gazzolo L, Madjar J J. Post-transcriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus Us11 protein. Nature (London) 1996;379:273–277. doi: 10.1038/379273a0. [DOI] [PubMed] [Google Scholar]
- 9.Donze O, Dostie J, Sonenberg N. Regulatable expression of the interferon-induced double-stranded RNA dependent protein kinase PKR induces apoptosis and fas receptor expression. Virology. 1999;256:322–329. doi: 10.1006/viro.1999.9618. [DOI] [PubMed] [Google Scholar]
- 10.Gil J, Alcami J, Esteban M. Induction of apoptosis by double-stranded-RNA-dependent protein kinase (PKR) involves the α subunit of eukaryotic translation initiation factor 2 and NF-κB. Mol Cell Biol. 1999;19:4653–4663. doi: 10.1128/mcb.19.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gresh N. Can a polyproline II helical motif be used in the context of sequence selective major groove recognition of B-DNA? A molecular modelling investigation. J Biomol Struct Dyn. 1996;14:255–273. doi: 10.1080/07391102.1996.10508117. [DOI] [PubMed] [Google Scholar]
- 12.He B, Chou J, Brandimarti R, Mohr I, Gluzman Y, Roizman B. Suppression of the phenotype of γ134.5− herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the α47 gene. J Virol. 1997;71:6049–6054. doi: 10.1128/jvi.71.8.6049-6054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He B, Gross M, Roizman B. The gamma (1) 34.5 protein of herpes simplex virus complexes with protein phosphatase 1 alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by the double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho C K, Shuman S. Mutational analysis of the vaccinia virus E3 protein defines amino acid residues involved in E3 binding to double-stranded RNA. J Virol. 1996;70:2611–2614. doi: 10.1128/jvi.70.4.2611-2614.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito T, Yang M, May W S. RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J Biol Chem. 1999;274:15427–15432. doi: 10.1074/jbc.274.22.15427. [DOI] [PubMed] [Google Scholar]
- 16.Johnson P A, Maclean C, Marsden H S, Dalziel R G, Everett R D. The product of gene Us11 of herpes simplex virus type 1 is expressed as a true late gene. J Gen Virol. 1986;67:871–883. doi: 10.1099/0022-1317-67-5-871. [DOI] [PubMed] [Google Scholar]
- 17.Katze M G. Translational control in cells infected with influenza virus and reovirus. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 607–630. [Google Scholar]
- 18.Kaufman R J. Double-stranded RNA-activated protein kinase mediates virus-induced apoptosis: a new role for an old actor. Proc Natl Acad Sci USA. 1999;96:11693–11695. doi: 10.1073/pnas.96.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kibler K V, Shors T, Perkins K B, Zeman C C, Banaszak M P, Biesterfeldt J, Langland J O, Jacobs B L. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. Virology. 1997;71:1992–2003. doi: 10.1128/jvi.71.3.1992-2003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar K U, Srivastava S P, Kaufman R J. Double-stranded RNA-activated protein kinase (PKR) is negatively regulated by 60S ribosomal subunit protein L18. Mol Cell Biol. 1999;19:1116–1125. doi: 10.1128/mcb.19.2.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Lynch P A, Chien C Y, Montelione G T, Krug R M, Berman H M. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol. 1997;4:896–899. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- 22.Mathews M B. Virus cell interactions. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 505–548. [Google Scholar]
- 23.Miller J E, Samuel C E. Proteolytic cleavage of the reovirus sigma 3 protein results in enhanced double-stranded RNA-binding activity: identification of a repeated basic amino acid motif within the C-terminal binding region. J Virol. 1992;66:5347–5356. doi: 10.1128/jvi.66.9.5347-5356.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohr I, Gluzman Y. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 1996;15:4759–4766. [PMC free article] [PubMed] [Google Scholar]
- 25.Mulvey M, Poppers J, Ladd A, Mohr I. A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function. J Virol. 1999;73:3375–3385. doi: 10.1128/jvi.73.4.3375-3385.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel R C, Sen G. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian X Y, Chien C Y, Lu Y, Montelione G T, Krug R M. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA. 1995;1:948–956. [PMC free article] [PubMed] [Google Scholar]
- 28.Raine D A, Jeffrey I W, Clemens M J. Inhibition of the double-stranded RNA-dependent protein kinase by mammalian ribosomes. FEBS Lett. 1998;436:343–348. doi: 10.1016/s0014-5793(98)01163-6. [DOI] [PubMed] [Google Scholar]
- 29.Rixon F J, McGeoch D J. A 3′ coterminal family of mRNA's from the herpes simplex virus type 1 short region: two overlapping reading frames encode unrelated polypeptides one of which has a highly reiterated amino acid sequence. Nucleic Acids Res. 1984;12:2473–2487. doi: 10.1093/nar/12.5.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roizman B R, Sears A E. Herpes simplex viruses and their replication. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 11–68. [Google Scholar]
- 31.Roller R J, Roizman B. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J Virol. 1992;66:3624–3632. doi: 10.1128/jvi.66.6.3624-3632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roller R J, Roizman B. Herpes simplex virus 1 RNA-binding protein US11 negatively regulates the accumulation of a truncated viral mRNA. J Virol. 1991;65:5873–5879. doi: 10.1128/jvi.65.11.5873-5879.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roller R J, Monk L L, Stuart D, Roizman B. Structure and function in the herpes simplex virus 1 RNA-binding protein US11: mapping of the domain required for ribosomal and nucleolar association and RNA binding in vitro. J Virol. 1996;70:2842–2851. doi: 10.1128/jvi.70.5.2842-2851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romano P R, Zhang F, Tan S L, Garcia-Barrio M T, Katze M G, Dever T E, Hinnebusch A C. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol Cell Biol. 1998;18:7304–7316. doi: 10.1128/mcb.18.12.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider R J. Adenovirus and vaccinia virus translational control. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 575–606. [Google Scholar]
- 36.Spector D, Purves F, Roizman B. Role of α-transinducing factor (VP16) in the induction of α genes within the context of viral genomes. J Virol. 1991;65:3504–3513. doi: 10.1128/jvi.65.7.3504-3513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.St. Johnston D, Brown N H, Gall J G, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vilcek J, Sen G. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 39.Wang W, Riedel K, Lynch P, Chien C Y, Montelione G T, Krug R M. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson R J, Vande Woude G F. DNA sequence of an immediate early gene (IE mRNA5) of herpes simplex virus type 1. Nucleic Acids Res. 1982;10:479–491. doi: 10.1093/nar/10.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams B R. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 42.Wu S, Kumar K U, Kaufman R J. Identification and requirement of three ribosome binding domains in dsRNA-dependent protein kinase (PKR) Biochemistry. 1998;37:13816–13826. doi: 10.1021/bi981472h. [DOI] [PubMed] [Google Scholar]
- 43.Zhu S, Romano P R, Wek R C. Ribosome targeting of PKR is mediated by two double-stranded RNA-binding domains and facilitates in vivo phosphorylation of eukaryotic initiation factor-2. J Biol Chem. 1997;272:14434–14441. doi: 10.1074/jbc.272.22.14434. [DOI] [PubMed] [Google Scholar]