Abstract
Actin-related proteins (Arps), which share a basal structure with actin but have distinct functions, have been found in a wide variety of organisms. While their functions are not yet clear, some Arps are localized in the nucleus and are suggested to contribute to the regulation of transcription. An essential gene of Saccharomyces cerevisiae, Act3p/Arp4, encodes the first identified nuclear Arp, which has been shown to bind to core histones in vitro. Here we have analyzed the in vivo function of Act3p/Arp4 on the his4-912δ promoter. Chromatin immunoprecipitation assays show that Act3p/Arp4 is bound to the entire his4-912δ promoter region. Conditional act3/arp4 mutations affect transcription from the his4-912δ promoter, where decreased Act3p/Arp4 binding and a change in nuclease sensitivity of chromatin were observed, showing the involvement of Act3p/Arp4 in the regulation of gene expression through the organization of chromatin structure. Taken together with the presence of Act3p/Arp4 in chromatin remodeling and histone acetyltransferase complexes, it is suggested that Act3p/Arp4 functions in transcriptional regulation to recruit chromatin remodeling and histone acetyltransferase complexes onto chromatin.
INTRODUCTION
Actin-related proteins (Arps), a group of protein families that exhibit moderate sequence similarity among each other and to conventional actin (i.e. muscle actin), have been found in a variety of eukaryotic organisms (1,2). According to the known three-dimensional structure of rabbit muscle actin and sequence comparisons, Arps and conventional actin compose the actin branch within a superfamily of proteins that possess ATPase activity, a superfamily that includes the 70 kDa heat shock cognate protein and hexokinase (3). However, the predicted molecular surfaces of these proteins are quite divergent and thus each protein is thought to be involved in unique functions specific for each family member.
Poch and Winsor (4) proposed a classification of the Arps in Saccharomyces cerevisiae into 10 subfamilies, in which the Arp subfamilies are numbered according to their similarity to conventional actin: Arp1 is most and Arp10 is least similar to actin. Arp subfamilies 1–3 of various organisms have been found and analyzed, showing that these Arps are localized in the cytoplasm and have distinct functions from that of actin isoforms and that each subfamily of Arp has a similar function across eukaryotic phyla (2). On the other hand, in S.cerevisiae the more divergent Arp subfamilies, Arp4–9, are localized in the nucleus (5–8), and also in humans two members of the Arp4 subfamily, hArpNα and hArpNβ/BAF53, are localized in the nucleus (9,10). While functional information on more distantly related Arps than the Arp subfamilies 1–3 is still limited, the information that is available suggests that most of these nuclear Arps function in the organization of chromatin structure.
Saccharomyces cerevisiae Act3p, a member of the Arp4 subfamily, was the first Arp that was demonstrated to be localized in the nucleus (5,6). ACT3/ARP4 is an essential gene coding for a polypeptide of 489 amino acids with a calculated molecular mass of 54.8 kDa (11). Genetic studies suggested that Act3p/Arp4 is involved in transcriptional regulation of, at least, HIS4 and LYS2 and in activity of the his4-912δ promoter (6).
Saccharomyces cerevisiae contains a retrovirus-like transposable element, Ty1, which contains two long terminal repeats, called δ elements, with strong promoter activity. Insertion of Ty1 or a δ element into the 5′-region of genes often causes inactivation of the adjacent gene because of interference or competition between transcriptional signals in the δ element and the native gene promoter (12). Selections for extragenic suppressors of Ty1- or δ element-inactivated genes have identified numerous SPT (Suppressor of Ty) genes and many of them were shown to be involved in transcriptional regulation via effects on chromatin structure (6,12). For example, genes for histones (SPT11 and SPT12) (13), components of the Spt–Ada–Gcn5 acetyltransferase (SAGA) complex (SPT3, SPT7 and SPT20) (14) and TATA-binding factor (SPT15) (15) were identified as SPT genes. Interestingly, act3-3 and act3-4 mutations cause variegated suppression of the δ element-inactivated HIS4 gene (the his4-912δ allele) in cells of an identical genetic background (6), suggesting an epigenetic effect of the mutated Act3p/Arp4 proteins on transcription. However, the molecular mechanisms by which Act3p/Arp4 regulates transcription are not well understood.
Although Act3p/Arp4 shows obvious similarity to conventional actin, it is a unique member of the actin family in terms of the presence of two insertions, I and II (11). Insertion II, consisting of 83 amino acids, is relatively abundant in charged amino acids and is predicted to form a loop-like structure protruding from the surface of the molecule. We previously reported that insertion II interacts with each of the core histones in vitro (16). Additionally, Act3p/Arp4 is present in the NuA4 histone acetyltransferase complex and the Ino80 chromatin remodeling complex purified from cell extracts (17,18).
Here, to investigate the molecular contribution of Act3p/Arp4 to transcriptional regulation in vivo, we have analyzed the association of Act3p/Arp4 with chromatin and the effect of mutations on transcriptional activity of the his4-912δ promoter.
MATERIALS AND METHODS
Yeast strains, media and general methods
Strain MZ3 (MATa pep4-3 trp1 leu2-Δ1 ura3-Δ1) was derived from 20B12 (19). Yeast strains DY2864 (MATa his4-912δ-ADE2 his4-912δ lys2-128δ can1 trp1 ura3 ACT3), DY4285 (MATa his4-912δ -ADE2 lys2-128δ can1 leu2 trp1 ura3 act3-ts26) and DY4519 (MATa his4-912δ -ADE2 lys2-128δ can1 leu2 trp1 ura3 act3-ts12) were used for analyses of chromatin structure. To isolate new temperature-sensitive alleles of ACT3, a plasmid shuffle was performed as described (16,20). DNA sequencing shows that the act3-ts26 and act3-ts12 alleles each have a single amino acid substitution of G187R and G455S, respectively. Protease inhibitors (0.5 µg/ml each antipain, chymostatin, elastatinal, leupeptin and pepstatin A) were added to the buffers described below.
Mobility shift assay of mononucleosomes
The entire coding sequence of Act3p/Arp4 was amplified with a primer set consisting of ACT3-forward and ACT3-reverse (Table 1) by PCR. To construct a plasmid for a glutathione S-transferase (GST)–Act3p/Arp4 fusion protein, by taking advantage of the primer-derived BamHI and EcoRI sites of the amplified PCR products for Act3p/Arp4, the entire coding sequences were inserted into the vector pGEX-2TK (Pharmacia Biotech, Uppsala, Sweden). Escherichia coli (XL-1 blue) was transformed with the constructed plasmid or with a control pGEX-2TK and expression of the fusion protein was induced by addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After further incubation at 30°C for 3 h, cells were harvested by centrifugation and the cell pellet was resuspended in 10 ml of phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3) containing 1 mM DTT and 0.1 mg/ml lysozyme. Cells were lysed and the supernatant was collected. After addition of 500 µl of a slurry of 50% glutathione–Sepharose 4B (Pharmacia Biotech), the fusion protein was bound to the matrix at 4°C for 12 h with shaking. Subsequently the matrix was washed five times with PBS and the fusion protein was recovered in 500 µl of elution buffer (50 mM Tris–HCl, pH 9.5, 20 mM reduced glutathione).
Table 1. Primers used in this work.
Mononucleosomes were reconstructed in vitro with purified core histones of chicken (16) and a 180 bp DNA fragment recovered from BssHII-digested pBluescript SK+ (Stratagene) according to Tatchell and van Holde (21). Mobility shift assays were performed by mixing the reconstituted mononucleosome and the purified GST fusion protein in 10 µl of binding buffer (10 mM HEPES, pH 7.9, 50 mM KCl, 5 mM DTT, 5 mM PMSF, 4 mM MgCl2, 5% glycerol) for 15 min at 37°C and then by electrophoresis on 1.5% agarose gels in 1× TAE (8 mM Tris, 1 mM sodium acetate, 0.4 mM EDTA, pH 8.0). The DNA fragment was detected by Southern blotting probed with the 180 bp fragment labeled with 32P as described below.
Immunoprecipitation of the chromatin fraction containing Act3p/Arp4
Yeast cells were grown in YPD to a concentration of 1.5 × 107 cells/ml. One hundred and fifty milliliters of cells were crosslinked with 1% formaldehyde for 15 min at 26°C. Cells were harvested and suspended in 600 µl of buffer A (0.1 mM Tris–HCl, pH 9.5, 10 mM DTT) and incubated for 15 min at 30°C. Cells were precipitated and washed with buffer B (1.2 M sorbitol, 20 mM HEPES, pH 7.6) and suspended in 1.5 ml of buffer B containing 0.4 mg Zymolyase 100T. Spheroplasts were washed with 20 mM PIPES, pH 6.8, 1 mM MgCl2, 1.2 M sorbitol and then with PBS. Yeast chromatin was prepared from the spheroplasts by incubating in 10 mM HEPES, pH 7.0, 200 mM NaCl, 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton X-100, followed by a wash with 10 mM HEPES, pH 7.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA. The chromatin fraction was isolated following lysis in lysis buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA, 1% SDS and protease inhibitors), sonication of the suspension (10 times for 10 s, resulting fragment size <1 kb) and clarification by centrifugation.
For immunoprecipitation of chromatin containing Act3p/Arp4, 1 ml of clarified chromatin fraction was suspended in 9 ml of Ab-binding buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100) and incubated with 100 ng anti-Act3p/Arp4 antibody (5) for 12 h at 4°C. Twenty microliters bed volume of protein A–Sepharose 4B beads (Amersham Pharmacia, Buckinghamshire, UK) were added and the incubation continued for 3 h. The immunoprecipitates were successively washed for 10 min each with Ab-binding buffer, Ab-binding buffer containing 500 mM NaCl, 10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 1% deoxycholate, 1% NP-40, 0.25 M LiCl and, finally, TE (10 mM Tris–HCl, pH 8.0, 1 mM EDTA). Finally, the precipitates were processed for DNA purification (22) and purified DNAs were dissolved in 40 µl of TE.
PCR analysis of immunoprecipitated DNA
All reactions were carried out in 100 µl volume with final concentrations of 1× PCR buffer (Sigma, St Louis, MO), 0.2 mM dNTPs, 1 µM each primer for fragments I–IV, 10 µl of immunoprecipitated DNA fraction and 0.5 µl of Taq polymerase (Sigma). Identical cycling conditions were used for all primer sets (Table 1). These comprised: an initial denaturation of 96°C for 3 min; 30 cycles of 96°C for 40 s, 54°C for 1 min and 73°C for 1 min. The resulting PCR products were examined by agarose gel electrophoresis.
Indirect end-labeling of micrococcal nuclease-digested chromatin
Yeast nuclei were prepared as described in Weber et al. (5). Nuclei were isolated from 360 ml of a mid-log YPD culture (1.5 × 107 cells/ml) and suspended in ice-cold 20 mM Tris–HCl pH 7.4, 1 M sorbitol, 1 mM CaCl2, 50 mM MgCl2, 5 mM NaCl, 1 mM 2-mercaptoethanol, 0.5 mM spermidine, 30 mM sodium azide and protease inhibitors at a concentration of 100 µg DNA/ml. After preincubation at 37°C for 2 min, micrococcal nuclease (0, 1, 3, 10 and 30 U/ml) was added to 200 µl aliquots of the suspension and chromatin was digested at 37°C for 5 min. The reaction was stopped by adding 20 µl of 250 mM EDTA, 5% SDS, and DNAs were purified from the partially digested chromatin. The purified DNAs were then completely digested with BsaHI. Samples of 1 µg/lane of the processed DNAs were electrophoresed on either 1.25 or 1.75% SeaPlaque GTG agarose gels (FMC BioProducts, Englewood, CO) and transferred to nylon membranes. For indirect end-labeling of the membrane bound DNAs, two DNA fragments consisting of –2116 to –1785 and +41 to +375 (from the start of the ADE2 open reading frame) of the his4-912δ–ADE2 gene were amplified with primer sets for the upstream probe and the downstream probe (Table 1), respectively, and labeled with [α-32P]dCTP (Amersham Pharmacia) by the primer extension method (23). Hybridization and washing were performed according to Church and Gilbert (24) and radioactivity of the labeled DNAs on the membrane was detected.
Restriction endonuclease digestion of chromatin
Nuclei isolated from a mid-log YPD culture (1.5 × 107 cells/ml) were suspended in digestion buffer (20 mM Tris–HCl, pH 7.5, 1 M sorbitol, 5 mM MgCl2, 1 mM CaCl2, 0.5 mM spermidine, 50 mM NaCl, 1 mM 2-mercaptoethanal) or digestion buffer containing 100 mM NaCl at a concentration of 70 ng/µl DNA. Restriction enzymes (10 U) were added to 200 µl aliquots of the suspension and chromatin was digested at 37°C for 1 h. The reaction was stopped by adding 20 µl of 250 mM EDTA, 5% SDS. The detection of a digested fragment containing the his4-912δ region was performed as described above. For the detection of a digested fragment containing the CEN3 region, the 0.7 bp SalI–KpnI fragment from pOS31 that contains the CEN3 DNA sequence (25) was used as probe.
RESULTS
The effect of non-lethal act3/arp4 mutations on transcription
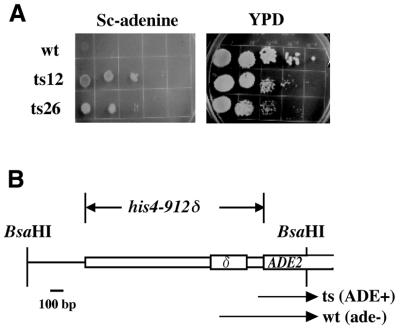
To investigate the involvement of Act3p/Arp4 in transcriptional regulation, the activity of the his4-912δ promoter was tested in two yeast strains possessing conditional act3/arp4 mutations (ts26 and ts12) (16,17). ts26 and ts12 have single amino acid substitutions of G187R and G455S in the genomic ACT3/ARP4 gene, respectively. The his4-912δ–ADE2 allele contains the his4-912δ promoter upstream of the ADE2 gene (Fig. 1B) (6). A wild-type (ACT3/ARP4) strain with the his4-912δ–ADE2 allele does not grow on SC medium lacking adenine (Fig. 1A), due to the δ element inserted in the HIS4 promoter. However, an act3/arp4 mutation (ts26 and ts12) allows a strain with the his4-912δ–ADE2 allele to grow on Sc-Ade medium despite the δ element inserted in the HIS4 promoter (Fig. 1A). These results demonstrate that the act3/arp4 mutations affect the activity of the δ element.
Figure 1.
The effect of non-lethal act3/arp4 mutations on transcriptional activity of the his4-912δ promoter. (A) One hundred microliter samples of YPD medium containing 2.0 × 108 cells/ml ACT3/ARP4 wild-type (wt) or act3/arp4 mutant (ts26 and ts12) strains were washed and suspended in 100 µl of sterile water. Ten-fold dilutions were prepared and 10 µl from each series was spotted onto Sc-Ade or YPD (complete) plates and incubated at 30°C. Incomplete suppression of his4-912δ was observed in ts12 and ts26 strains. (B) Structure of the allele used in this study. The ADE2 promoter is replaced by the his4-912δ promoter where the 334 bp δ element is inserted 162 bp upstream from the original initiating ATG. Arrows show expected transcripts in the strains.
Association of Act3p/Arp4 with the his4-912δ chromatin region in vivo
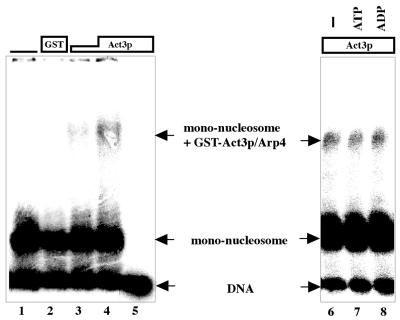
We have previously reported that the insertion II region of Act3p/Arp4 binds to yeast and chicken core histones (16). The binding of recombinant Act3p/Arp4 was checked by mobility shift assay with mononucleosomes reconstituted with chicken core histones and a DNA fragment from a cloning vector, pUC119, according to Tatchell and van Holde (21) (Fig. 2). A complex was detected between recombinant Act3p/Arp4 and mononucleosomes and the amount of complex was dependent on the amount of Act3p/Arp4 added (Fig. 2, lanes 3 and 4). A complex was not detected between mononucleosomes and GST (Fig. 2, lane 2) or between naked DNA and Act3p/Arp4 (Fig. 2, lane 5). These results show that Act3p/Arp4 binds to nucleosomes in vitro. Recently it was shown that recombinant Act3p/Arp4 bound in vitro to nucleosomes reconstructed with core histones from HeLa cells and a 5S ribosomal DNA nucleosome-positioning sequence (17). This shows that binding of Act3p/Arp4 to nucleosomes does not depend on specific DNA sequences.
Figure 2.
In vitro association of Act3p/Arp4 with reconstituted mononucleosomes. Mobility shift assay of GST (lane 2) and recombinant Act3p/Arp4 (GST–Act3p/Arp4) (lane 3–5 and 6–8) was performed with reconstituted mononucleosomes (lanes 1–4 and 6–8) and naked DNA alone (lane 5). Half the amount of GST–Act3p/Arp4 was added in lane 3 as compared with lanes 4 and 5. In lanes 6–8, mobility shift assays with the same amount of GST–Act3p/Arp4 were performed in the absence (lane 6) or presence of 10 mM ATP (lane 7) and 10 mM ADP (lane 8). The positions of the complex consisting of mononucleosome + GST–Act3p/Arp4, mononucleosomes, and naked DNA are shown by arrows.
Actin binds ATP/ADP and nucleotide binding affects intramolecular associations, including actin filament formation. As Act3p/Arp4 can bind ATP (16), we suggested that the binding of ATP analogs could regulate binding of Act3p/Arp4 to the nucleosome. We tested this possibility using a mobility shift assay in the presence of ATP or ADP, which showed that these analogs do not affect binding of Act3p/Arp4 to mononucleosomes (Fig. 2, lanes 6–8). On the other hand, it seems premature to suggest that a modification of Act3p/Arp4 regulates its binding to nucleosomes, because post-translational modification of Arps has not been reported in any organism.
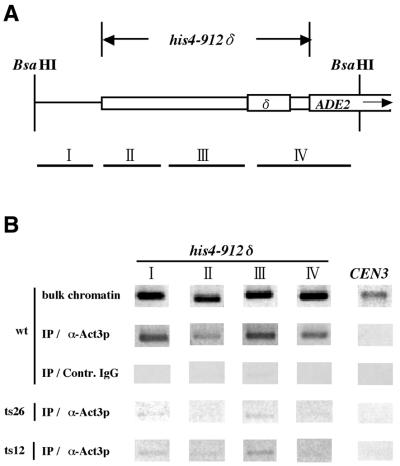
To check the association of Act3p/Arp4 with the his4-912δ promoter region in vivo, we preformed chromatin immunoprecipitation analysis with a crosslinked and shared chromatin fraction. PCR amplification of precipitated DNA with primer sets directed against four parts of the his4-912δ promoter region (Fig. 3A) revealed that an anti-Act3p/Arp4 antibody (5,16), but not mouse immunoglobulin, precipitated chromatin containing each of the four segments of the his4-912δ promoter (I–IV in Fig. 3B) of the strain with wild-type ACT3/ARP4 (wt). This shows that intact Act3p/Arp4 associates with chromatin of the his4-912δ promoter in vivo.
Figure 3.
In vivo association of Act3p/Arp4 with the his4-912δ promoter region of wild-type and act3/arp4 mutants. (A) Act3p/Arp4 was immunoprecipitated (IP/α-Act3p) from crosslinked chromatin from ACT3/ARP4 wild-type (wt) and act3/arp4 mutant (ts26 and ts12) strains. Rabbit immunoglobulin was used as a control (IP/Contr. IgG). DNA purified from the precipitated fractions and input bulk chromatin was analyzed by PCR using primer sets directed against four parts of the his4-912δ promoter region [I–IV in (A) and (B)] and the centromere region of chromosome III [CEN3 in (B)]. (B) The resulting PCR products were resolved on an agarose gel.
Regions I–IV of the his4-912δ promoter region were amplified from the precipitated bulk chromatin with equal efficiency. Taken together with the in vitro experiment with reconstituted nucleosomes containing different DNAs, this suggests that Act3p/Arp4 did not bind to the chromatin in a sequence-specific manner. However, the antibody against Act3p/Arp4 did not precipitate chromatin containing the CEN3 sequence (Fig. 3B). Since the four conserved lysine residues in the H4 tail are required for in vitro binding with Act3p/Arp4 (17), acetylation of histones might contribute to this selectivity.
Decrease in the association of mutated Act3p/Arp4 with chromatin
We next determined the association of Act3p/Arp4 mutants with chromatin of the his4-912δ promoter. These Act3p/Arp4 mutations are outside the region (amino acids 296–399) used to prepare polyclonal antibody and thus the antibody will recognize both wild-type and mutant Act3p/Arp4 proteins with equal affinity. When the chromatin immunoprecipitation assay was performed with strains with a conditional act3/arp4 mutation (ts26 and ts12) grown under permissive conditions, very little chromatin corresponding to regions I–IV was recovered with the anti-Act3p/Arp4 antibody, in contrast to the ACT3/ARP4 strain (Fig. 3B). The chromatin immunoprecipitates from the mutants did not contain the CEN3 region, as was seen for the ACT3/ARP4 strain. This shows that Act3p/Arp4 mutants are defective in associating with the his4-912δ promoter region in vivo.
Micrococcal nuclease sensitivity of the his4-912δ promoter region
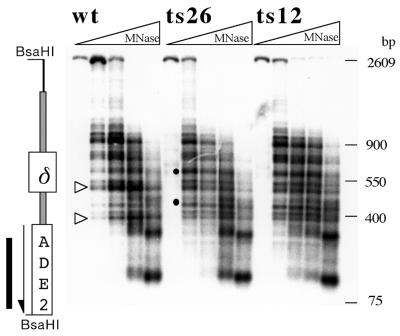
We next examined the effect of Act3p/Arp4 mutations on chromatin structure by analyzing the micrococcal nuclease (MNase) sensitivity of chromatin. Chromatin digested with MNase was analyzed by the indirect end-labeling method and regions that are sensitive to MNase were detected as bands. The experiment shown in Figure 4 examined the chromatin structure of the his4-912δ chromatin region in a strain possessing an intact ACT3/ARP4 gene (wt) and conditional act3/arp4 mutants (ts26 and ts12). Importantly, two of the major MNase cleavage sites observed in the ACT3/ARP4 strain were less abundant in both of the act3/arp4 mutants (Fig. 4, open triangles). Two other cleavage sites (Fig. 4, dots) were more pronounced in both of the act3/arp4 mutants, demonstrating that mutations of Act3p/Arp4 caused a change in chromatin structure in the region examined.
Figure 4.
Chromatin structure around the his4-912δ promoter region of cells possessing an intact ACT3/ARP4 gene and of act3/arp4 mutants. MNase sensitivity of the BsaHI–BsaHI fragment of the his4-912δ promoter region in an ACT3/ARP4 background (wt) and act3/arp4 mutant (ts26 and ts12) strains was analyzed by the indirect end-labeling method using a probe located just downstream of the promoter (solid bar). Chromatin regions that are sensitive to MNase digestion were represented as bands. Positions of the regions correspond to the schematic drawing on the left of the panel. Arrows show the orientation of transcription, open triangles indicate MNase cleavage sites whose intensity was decreased in the mutants and dots show those whose intensity was enhanced in the mutants.
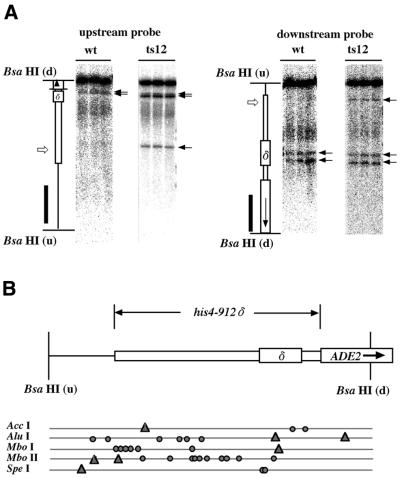
Restriction enzyme sensitivity of the his4-912δ promoter region
To confirm the change in chromatin structure in the mutants, we digested chromatin in isolated nuclei with restriction endonucleases. In Figure 5A chromatin was digested with the restriction enzyme AccI, and AccI sensitivity was detected by the method of indirect end-labeling using a probe located far upstream (Fig. 5A, left) or downstream (Fig. 5A, right) of the promoter (solid bars). There are three AccI sites present in the his4-912δ promoter region, but the most upstream site was not digested in the ACT3/ARP4 strain because of the chromatin structure surrounding the site (Fig. 5A, open arrows on the schematic map). However, the AccI site was digested in the act3/arp4 mutants: ts12 (Fig. 5A) and ts26 (data not shown). This shows that the local chromatin structure surrounding the most upstream AccI site was changed in the act3/arp4 mutants. Reproducible changes in nuclease sensitivity in the his4-912δ promoter region were seen with the five enzymes tested in the act3/arp4 mutants: the results are summarized in Figure 5B. Thus, changes in chromatin structure were detected in various areas in the his4-912δ promoter region, but were not limited to a restricted area.
Figure 5.
Chromatin structure around the his4-912δ promoter region of cells possessing an intact ACT3/ARP4 gene and of act3/arp4 mutants detected by accessibility to a restriction enzyme. (A) AccI sensitivity of the BsaHI–BsaHI fragment surrounding the his4-912δ promoter region in an ACT3/ARP4 background (wt) and an act3/arp4 mutant (ts12) strain were analyzed by the indirect end-labeling method using a probe located far upstream (left) or just downstream (right) of the promoter (solid bars). Positions of the regions are shown on the schematic drawing on the left of each panel. Horizontal arrows indicate observed AccI cleavage sites and open arrows indicate AccI cleavage sites whose intensity was increased in the mutants. Three lanes in each panel correspond to cultures shifted to 26, 30 and 37°C (from left to right) before harvest. (B) The sites where the sensitivity to restriction enzymes was changed in the mutants (triangles) are shown under a map of the his4-912δ promoter region.
Restriction enzyme sensitivity of a centromere region
Centromeric regions in S.cerevisiae have been shown to have distinctive chromatin structures, with positioned nucleosomes emanating from the CDEI, CDEII and CDEIII elements required for centromere function (26). We used the restriction endonuclease MboI to examine nuclease sensitivity of the CEN3 region in isolated nuclei in the ACT3/ARP4 strain and act3/arp4 mutants (ts26 and ts12) (Fig. 6). No reproducible differences were noted, suggesting that act3/arp4 mutations do not affect chromatin structure of centromere regions. This result is consistent with the chromatin immunoprecipitation results (Fig. 3), indicating that Act3p/Arp4 is not associated with the CEN3 centromere region.
Figure 6.
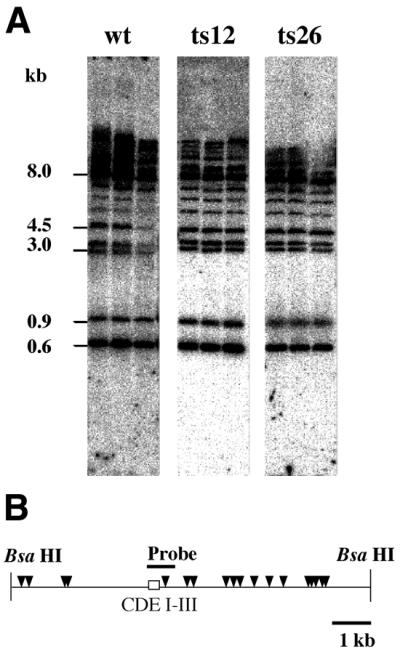
Chromatin structure around the CEN3 centromere region of cells possessing an intact ACT3/ARP4 gene and of act3/arp4 mutants detected by accessibility to a restriction enzyme. (A) MboI sensitivity of the BsaHI–BsaHI fragment surrounding the CEN3 region in an ACT3/ARP4 background (wt) and act3/arp4 mutant (ts26 and ts12) strains were analyzed by Southern blot analysis using a probe located in the CDE I–III region (solid bar). There is no reproducible difference between the wild-type and mutants. (B) A genetic map of the BsaHI–BsaHI fragment surrounding CEN3. The relative positions of the CDE I–III elements are shown by a box and MboI sites are indicated by triangles.
DISCUSSION
We used chromatin immunoprecipitation assays to characterize the association of Act3p/Arp4 with chromatin. We showed that Act3p/Arp4 associates with the his4-912δ promoter region and that act3/arp4 mutations which alter expression from his4-912δ also result in decreased association of Act3p/Arp4 with the promoter. While the insertion II region of Act3p/Arp4 is responsible for binding to core histones in vitro (16), the mutants analyzed here contain amino acid substitutions not within insertion II but in the actin fold. The actin fold is a tertiary structure centered around an ATP/ADP-binding pocket present in actin superfamily members, including Arps (3). In actin, the actin fold undergoes major conformational shifts in response to the 5′ phosphorylation-hydrolysis state of the adenine nucleotide and such shifts are thought to be crucial to the functions of the various actin superfamily members (3,27). Mutations within the actin fold might affect the stability of Act3p/Arp4 and/or cause the formation of irregular Act3p/Arp4 complexes and these changes could affect its association with chromatin.
In mutant cells, in which less of the mutated Act3p/Arp4 is found associated with the his4-912δ chromatin region, changes in chromatin structure of this region are seen, showing the involvement of Act3p/Arp4 in the organization and/or maintenance of chromatin structure. As Act3p/Arp4 interacts with histones through their N-terminal domains, which extend from the nucleosome core (17), it is unlikely that binding of Act3p/Arp4 alters the nucleosome core structure. In addition, while Act3p/Arp4 possesses a putative ATP/ADP-binding motif, this motif is not expected to take part in chromatin remodeling (28), and no sequence motif for any histone modification is found in Act3p/Arp4 (11). Therefore, it is unlikely that Act3p/Arp4 will modulate chromatin structure by itself.
It has been reported that Act3p/Arp4 is a stoichiometric component of the NuA4 histone acetyltransferase complex and Ino80 chromatin remodeling complex (17,18). Taken together with the requirement of Act3p/Arp4 for proper chromatin structure and promoter activity, we suggest that Act3p/Arp4 functions to recruit components of the complexes onto the chromatin for proper chromatin function. On the other hand, while a limited number of complexes involved in chromatin modulation have been characterized so far, actin family proteins, consisting of actin and actin-related proteins, are found in many of them (9,28–31). Thus it is possible that Act3p/Arp4 may recruit other chromatin modifying complexes, in addition to NuA4 and Ino80, to chromatin. Importantly, mutations of Act3p/Arp4 not only affect activity of the his4-912δ promoter, but also activity of some native promoters, including HIS4 and LYS2 (6). Act3p/Arp4 could be involved in the transcriptional regulation of a number of genes by recruiting specific complexes onto chromatin.
While Act3p/Arp4 does not seem to bind nucleosomes in a sequence-specific manner, there is evidence for selective recruitment of complexes involved in the modulation of chromatin structure. Chromatin immunoprecipitation experiments to globally ask what promoters have bound Esa1p, the catalytic subunit of the NuA4 histone acetyltransferase complex that also contains Act3p/Arp4, showed that NuA4 is specifically recruited to ribosomal protein promoters by the DNA-binding factor Rap1 or Abf1 (32). Moreover, depletion of Esa1p leads to a dramatic decrease in acetylation globally (32,33), showing that specific targeting of Esa1p occurs in a background of its global activity on chromatin. Since association of Act3p/Arp4 with chromatin in vitro and in vivo does not depend on specific nucleotide sequences, Act3p/Arp4 could be responsible for such global recruitment of chromatin remodeling and histone acetyltransferase complexes, including the NuA4 complex, rather than for sequence-specific targeting. The global activity of the complexes would serve to maintain a genome-wide balance between active and inactive states of chromatin. At the his4-912δ promoter in the act3/arp4 strains, this balance was altered and activation of the δ element was changed.
On the two promoters, the promoter of the δ element and the native HIS4 promoter responsible for functional Ade2p, the transcriptional effects and changes in chromatin structure caused by act3/arp4 mutations appear to be contradictory. However, recent studies showed that chromatin remodeling is utilized not only for transcriptional activation, but also for transcriptional repression. A chromatin remodeling complex, hSWI/SNF, not only catalyzes the disruption of nucleosome structure, but also the reverse reaction in vitro (34), and a microarray analysis revealed that yeast SWI/SNF acts both as an activator and as a repressor of transcription in vivo (35). It is possible that chromatin remodeling activity is required globally for the maintenance of chromatin fluidity, and its contribution to transcriptional activation or repression depends on the balance between other co-activator or co-repressor activity (36). In the case of the his4-912δ promoter, chromatin remodeling activities would be required for a chromatin structure suitable for both activation of the promoter in the δ element and repression of the HIS4 promoter.
While the association of Act3p/Arp4 with chromatin correlates with correct transcriptional regulation, it is unlikely that the nucleosome-binding property is shared by all nuclear Arps because this insertion II region is specific to Act3p/Arp4 and is not conserved in other Arps. On the other hand, as for Act3p/Arp4, most, if not all, nuclear Arps are expected to be components of chromatin remodeling complexes. Arp7 and Arp9 of S.cerevisiae are components of chromatin remodeling complexes SWI/SNF and RSC (28,29), and a putative human ortholog of Act3p/Arp4, hArpNβ/BAF53 (10), is contained in various protein complexes including the BAF chromatin remodeling and Tip60 histone acetyltransferase complexes (9,30,31). Therefore, the role of nuclear Arps could be more than just recruitment of the complexes and nuclear Arps might also have structural and regulatory functions in these chromatin remodeling complexes. Further analysis of other nuclear Arps with appropriate mutants would be helpful to address their role in transcriptional regulation through changes in chromatin structure.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Eiko Tsuchiya for plasmids. We gratefully acknowledge Dr Ulrike Wintersberger and Irene Görzer for helpful discussions of the results. This work was supported by a Grant-in-Aid for Scientific Research (11760051) from the Ministry of Education, Science, Sport, and Culture, Japan, and by the Kato Memorial Bioscience Foundation.
REFERENCES
- 1.Frankel S. and Mooseker,M.S. (1996) The actin-related proteins. Curr. Opin. Cell Biol., 8, 30–37. [DOI] [PubMed] [Google Scholar]
- 2.Schafer D.A. and Schroer,T.A. (1999) Actin-related proteins. Annu. Rev. Cell. Dev. Biol., 15, 341–363. [DOI] [PubMed] [Google Scholar]
- 3.Holmes K.C., Sander,C. and Valencia,A. (1993) A new ATP-binding fold in actin, hexokinase and Hsc70. Trends Cell Biol., 3, 53–59. [DOI] [PubMed] [Google Scholar]
- 4.Poch O. and Winsor,B. (1997) Who’s who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast, 13, 1053–1058. [DOI] [PubMed] [Google Scholar]
- 5.Weber V., Harata,M., Hauser,H. and Wintersberger,U. (1995) The actin-related protein Act3p of Saccharomyces cerevisiae is located in the nucleus. Mol. Biol. Cell, 6, 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y.W. and Stillman,D.J. (1996) Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev., 10, 604–619. [DOI] [PubMed] [Google Scholar]
- 7.Grava S., Dumoulin,P., Madania,A., Tarassov,I. and Winsor,B. (2000) Functional analysis of six genes from chromosomes XIV and XV of Saccharomyces cerevisiae reveals YOR145c as an essential gene and YNL059c/ARP5 as a strain-dependent essential gene encoding nuclear proteins. Yeast, 11, 1025–1033. [DOI] [PubMed] [Google Scholar]
- 8.Harata M., Oma,Y., Tabuchi,T., Zhang,Y., Stillman,D.J. and Mizuno,S. (2000) Multiple actin-related proteins of Saccharomyces cerevisiae are present in the nucleus. J. Biochem., 128, 665–671. [DOI] [PubMed] [Google Scholar]
- 9.Zhao K., Wang,W., Rando,O.J., Xue,Y., Swiderek,K., Kuo,A. and Crabtree,G.R. (1998) Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell, 95, 625–636. [DOI] [PubMed] [Google Scholar]
- 10.Harata M., Mochizuki,R. and Mizuno S. (1999) Two isoforms of a human actin-related protein show nuclear localization and mutually selective expression between brain and other tissues. Biosci. Biotechnol. Biochem., 63, 917–923. [DOI] [PubMed] [Google Scholar]
- 11.Harata M., Karwan,A. and Wintersberger,U. (1994) An essential gene of Saccharomyces cerevisiae coding for an actin-related protein. Proc. Natl Acad. Sci. USA, 91, 8258–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winston F. and Carlson,M. (1992) Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet., 8, 387–391. [DOI] [PubMed] [Google Scholar]
- 13.Clark-Adams C.D., Norris,D., Osley,M.A., Fassler,J.S. and Winston,F. (1988) Changes in histone gene dosage alter transcription in yeast. Genes Dev., 2, 150–159. [DOI] [PubMed] [Google Scholar]
- 14.Grant P.A., Duggan,L., Côté,J., Roberts,S.M., Brownell,J.E., Candau,R., Ohba,R., Owen-Hughes,T., Allis,C.D., Winston,F., Berger,S.L. and Workman,J.L. (1997) Yeast Gcn5 functions in two multisubunit complexes: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev., 11, 1640–1650. [DOI] [PubMed] [Google Scholar]
- 15.Eisenmann D.M., Dollard,C. and Winston,F. (1989) SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell, 58, 1183–1191. [DOI] [PubMed] [Google Scholar]
- 16.Harata M., Oma,Y., Mizuno,S., Jiang,Y.W., Stillman,D.J. and Wintersberger,U. (1999) The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol. Biol. Cell, 10, 2595–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galarneau L., Nourani,A., Boudreault,A.A., Zhang,Y., Heliot,L., Allard,S., Savard,J., Lane,W.S., Stillman,D.J. and Côté,J. (2000) Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell, 5, 927–937. [DOI] [PubMed] [Google Scholar]
- 18.Shen X., Mizuguchi,G., Hamiche,A. and Wu,C. (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature, 406, 541–544. [DOI] [PubMed] [Google Scholar]
- 19.Wintersberger U., Kuehne,C. and Karwan,A. (1995) Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast, 11, 929–944. [DOI] [PubMed] [Google Scholar]
- 20.Sikorski R.S. and Boeke,J.D. (1991) In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol., 194, 302–318. [DOI] [PubMed] [Google Scholar]
- 21.Tatchell K. and van Holde,K.E. (1977) Reconstitution of chromatin core particles. Biochemistry, 16, 5295–5303. [DOI] [PubMed] [Google Scholar]
- 22.Orlando V. and Paro,R. (1993) Mapping polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell, 75, 1187–1198. [DOI] [PubMed] [Google Scholar]
- 23.Feinberg A.P. and Vogelstein,B. (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem., 132, 6–13. [DOI] [PubMed] [Google Scholar]
- 24.Church G.M. and Gilbert,W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchiya E., Hosotani,T. and Miyakawa T. (1998) A mutation in NPS/STH, an essential gene encoding component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of S.cerevisiae centromeres. Nucleic Acids Res., 26, 3286–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegemann J.H. and Fleig,U.N. (1993) The centromere of budding yeast. Bioessays, 15, 451–460. [DOI] [PubMed] [Google Scholar]
- 27.Boyer L.A. and Peterson,C.L. (2000) Actin-related proteins (Arps): conformational switches for chromatin-remodeling machines? Bioessays, 22, 666–672. [DOI] [PubMed] [Google Scholar]
- 28.Peterson C.L., Zhao,Y. and Chait,B.T. (1998) Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J. Biol. Chem., 273, 23641–23644. [DOI] [PubMed] [Google Scholar]
- 29.Cairns B.R., Erdjument-Bromage,H., Tempst,P., Winston,F. and Kornberg,R.D. (1998) Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell, 2, 639–651. [DOI] [PubMed] [Google Scholar]
- 30.Ikura T., Ogryzko,V.V., Grigoriev,M., Groisman,R., Wang,J., Horikoshi,M., Scully,R., Qin,J. and Nakatani,Y. (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell, 102, 463–473. [DOI] [PubMed] [Google Scholar]
- 31.Fuchs M., Gerber,J., Drapkin,R., Sif,S., Ikura,T., Ogryzko,V., Lane,W.S., Nakatani,Y. and Livingston,D.M. (2001) The p400 complex is an essential E1A transformation target. Cell, 106, 297–307. [DOI] [PubMed] [Google Scholar]
- 32.Reid J.L., Iyer,V.R., Brown,P.O. and Struhl,K. (2000) Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Eas1 histone acetylase. Mol. Cell, 6, 1297–1307. [DOI] [PubMed] [Google Scholar]
- 33.Vogelauer M., Wu,J., Suka,N. and Grunstein,M. (2000) Global acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- 34. Schnitzler,G., Sif,S. and Kingston,R.E. (1998) Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell, 89, 325–328. [DOI] [PubMed] [Google Scholar]
- 35.Sudarsanam P., Iyer,V.R., Brown,P.O. and Winston,F. (2000) Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H.S. and Dean,D.C. (2001) Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene, 20, 3134–3138. [DOI] [PubMed] [Google Scholar]