Abstract
Purpose
Carbapenem-resistant Enterobacterales (CRE) infection is an urgent threat to human health. This study aimed to develop and validate a novel multiplex real-time PCR (multi-qPCR) assay for the detection of the blaKPC, blaNDM, blaIMP, blaOXA-48-like, and blaVIM genes in CRE isolates and clinical samples, as well as to compare it with three phenotypic methods.
Methods
The reliability and limit of detection (LOD) of the multi-qPCR assay were evaluated. PCR and DNA sequencing were used as the reference methods to identify carbapenemase genes in CRE isolates and clinical samples. The accuracy of the multi-qPCR assay, modified carbapenem inactivation and EDTA-modified carbapenem inactivation method (mCIMandeCIM), carbapenemase inhibitor-based combined disk test (CDT), and colloidal gold-based immunochromatographic test was compared with the reference methods with 182 isolates of CRE. Furthermore, 112 clinical samples were collected to validate the efficacy of this multi-qPCR assay.
Results
The standard deviations (CVs) of intra-assay and inter-assay of the multi-qPCR assay were ≤ 0.53% and ≤ 2.04% for detecting the five major carbapenemase genes, respectively; while the LOD ranged from 2×102 copies/mL to 8×102 copies/mL. PCR and DNA sequencing confirmed 168 out of 182 CRE isolates producing carbapenemase(s): KPC (n = 93), NDM (n = 46), IMP (n = 8), OXA-48-like (n = 14), VIM (n = 1), KPC&NDM (n = 5), and KPC&NDM&IMP (n = 1). The accuracy of mCIMandeCIM, CDT, Colloidal Gold, and the multi-qPCR assay was 96.2%, 89.6%, 100%, and 100% respectively for detecting carbapenemase(s) producers. Moreover, the sensitivity and specificity of the multi-qPCR assay were all 100% for the detection of each carbapenemase gene in clinical samples, compared with PCR and sequencing.
Conclusion
For clinical isolate detection, the multi-qPCR assay is comparable to Colloidal Gold, and superior to mCIMandeCIM and CDT; while for clinical samples detection, it also shows excellent performance. Therefore, the multi-qPCR assay has great potential for clinical diagnosis.
Keywords: carbapenem-resistant Enterobacterales, carbapenemases detection, multiplex real-time PCR assay, mCIM/eCIM, combined disk test, immunochromatographic test
Introduction
Antimicrobial resistance is a worldwide public health threat.1 About 700,000 deaths each year are caused by antimicrobial resistance globally, and the number may rise to 10 million by 2050.2 The World Health Organization (WHO) listed carbapenem-resistant Enterobacterales (CRE) as a critical threat in 2017,3 and similarly, the US Centers for Disease Control and Prevention (CDC) also identified CRE as an urgent threat to human health in 2019.4 According to the China Antimicrobial Resistance Surveillance System (CARSS) and China Antimicrobial Surveillance Network (CHINET), the isolation rate of Enterobacterales exceeded 38% among all isolated bacteria.5,6 Klebsiella pneumoniae, which ranks second in isolation rate, has a resistance rate to carbapenems of 10.0% and 28.9%, reported by CARSS and CHINET respectively. Zheng et al reported that the in-hospital mortality rate is up to 33.5%, while the bacteremia-associated mortality rate is even as high as 43.1% among 664 CRE cases.7 Inadequate initial antimicrobial therapy caused by CRE infection often leads to high mortality rates, as identification of CRE using culture methods typically takes 2 to 3 days.8 Rapid detection of CRE directly from patient samples will enable appropriate antibiotic treatment to occur in the early stages of infection.
The most common resistance mechanism of Enterobacterales to carbapenems is the production of carbapenemases, which mainly include KPC, NDM, VIM, IMP, and OXA-48-like worldwide.9 Most of them are plasmid-encoded, as a consequence these carbapenem resistance genes are easily dissemination through horizontal gene transfer. Another less common mechanism of carbapenem resistance is a combination of expression of extended-spectrum beta-lactamases (ESBLs) or AmpCs and porin loss or overexpression of efflux pumps.10 The former is called carbapenemase-producing CRE (CP-CRE), while the latter is called non-carbapenemase-producing CRE (non-CP-CRE). Currently, except for the antimicrobial susceptibility testing method, almost all methods for detecting CRE are targeted at CP-CRE. The culture-based methods,11 carbapenem hydrolysis-based methods,12 as well as immunological methods,12 require at least bacterial isolation. Compared to the above methods, genotypic methods can directly detect carbapenemase genes from biological samples.10 Especially the multiplex real-time PCR (multi-qPCR) assay used for the detection of carbapenemase genes provides a faster result and timely information, which is beneficial for making appropriate treatment decisions in clinical practice.13,14
The current study aimed to establish and validate a multi-qPCR assay for the detection of the five major carbapenemase genes in carbapenem-resistant Enterobacterales isolates and clinical samples, as well as to compare it with three phenotypic methods.
Materials and Methods
Design of Primers and TaqMan-MGB Probes for Multi-qPCR Assay
Multiple sequence alignments were performed using available sequences of the blaKPC, blaNDM, blaIMP, blaOXA-48-like and blaVIM genes of Enterobacterales. The sequences were searched from Primer-BLAST in NCBI by using the primers of blaNDM, blaIMP, and blaVIM genes, blaOXA-48-like gene, and blaKPC gene reported previously.15–17 According to the alignment results, primers and TaqMan-MGB probes were designed and screened referring to our previous work.18 The Primer3 Software version 0.4.0, and the online IDT oligo analyzer 3.1 tool (https://www.idtdna.com/calc/analyzer) were used. The human β-globin gene (GenBank: AH001475.2) was selected as an internal control to monitor the processing of DNA extraction of clinical samples.18 The sequences and characteristics of the primers and probes are shown in Table S1. All primers and TaqMan-MGB probes used in the study were synthesized by Beijing Ruiboxingke Biotechnology Co., Ltd. (Beijing, China).
Evaluation of Reliability and Limit of Detection (LOD) of Multi-qPCR Assay
The intra-assay variation (repeatability) and inter-assay variation (reproducibility) were carried out to evaluate the reliability of this multi-qPCR assay. The Topo Omni DNA Cloning Kit (Clone Smarter, USA) was used for construction of standard plasmids which contain the target amplicon. Then, the standard plasmids were performed serial dilution. The repeatability was evaluated by analyzing the cycle threshold (Ct) average value, standard deviation (SD), and coefficient of variation (CV) of three replicates of each serial dilution in every single multi-qPCR run. The reproducibility was also evaluated by analyzing the Ct average value, SD, and CV, but of three independent multi-qPCR runs of each serial dilution. The independent runs were performed on different dates by two different technicians.
The lowest concentration of the standard plasmids that could be detected reproducibly was regarded as the LOD of this multi-qPCR assay. Based on the results of the Ct value with ten-fold dilution, LOD was obtained by performing two-fold dilution at the lowest ten-fold dilution concentration where the Ct value could be detected.
Bacterial Isolates and Clinical Samples
A total of 182 retrospectively collected and nonduplicate Enterobacterales isolates including Klebsiella pneumoniae (n = 126), Escherichia coli (n = 30), Enterobacter cloacae complex (n = 10), Klebsiella aerogenes (n = 4), Klebsiella oxytoca (n = 3), and others (n = 9) were collected between January 2017 and January 2022 from Beijing Chao-Yang Hospital. All the isolates were identified by Vitek MALDI-TOF mass spectrometry (bioMérieux, France). CRE was defined as isolates with a minimum inhibitory concentration (MIC) of ≥ 4 µg/mL for imipenem or meropenem (resistant to meropenem instead of imipenem for Morganella morganii, Proteus spp., and Providencia spp)., by broth microdilution (BMD) method based on the interpretative criteria from the 2024 Clinical and Laboratory Standards Institute (CLSI).19
A total of 112 retrospectively collected clinical samples, which were mainly culture-positive for gram-negative bacteria such as Enterobacterales, Acinetobacter baumannii, and Pseudomonas aeruginosa from adult patients between January 2021 and December 2023 from Beijing Chao-Yang Hospital were used to evaluate the multi-qPCR assay. These samples were the remaining samples for routine microbiological examination and included sputum (n = 57), bronchoalveolar lavage fluid (n = 28), blood culture (n = 13), pleural fluid (n = 4), abdominal fluid (n = 4), urine (n = 3), and cerebrospinal fluid (n = 3).
Conventional PCR and Sequencing for Detection of Carbapenemase Genes in Clinical Isolates and Samples
For DNA extraction of bacterial isolates, a 1-µL loopful of isolated colonies was well dispersed in 100 μL deionized water (TIANGEN Biotech, Beijing, China), boiled for 5 min at 100 °C, and centrifuged at 10,625×g for 5 min. The supernatant was diluted 10 times with deionized water as a template. The clinical samples were treated with a sample reagent containing NaOH, N-acetyl-L-cysteine, and isopropanol for 15 min. Then, DNA was extracted from 200 μL of the treated sample using a Nextrator NX-48 system (Genolution, Seoul, Korea) according to the manufacturer’s instructions as a template.
Carbapenemase genes including blaKPC, blaNDM, blaIMP, blaOXA-48-like, and blaVIM genes were each detected by conventional PCR and sequencing. The primers of the above genes used in the study were designed by Jiangsu Macro & Micro-Test Med-Tech Co., Ltd., and listed in Table S2. Briefly, 15 µL of 2×GC buffer containing Mg2+ (Takara Bio Inc., Japan) was mixed with 0.55 µL TaKaRa Taq DNA polymerase for amplifying blaOXA-48-like and blaVIM genes (0.35 µL for blaKPC, blaNDM, and blaIMP genes), 3 µL dNTP, 0.6 µL (10 µM) of forward and reverse primers for amplifying blaKPC, blaOXA-48-like, and blaVIM genes (0.5 µL for blaNDM and blaIMP genes), and DNase/RNase-free deionize water to a final volume of 25 µL. Then, 5 µL of the extracted DNA was added to the mixture. The touchdown PCR program was set as follows: an initial denaturation step at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 30s, annealing at 66–54 °C (decreasing 0.3 °C per cycle) for 30s, elongation at 72 °C for 30s, and a final extension at 72 °C for 7 min.
The PCR products were analyzed by electrophoresis and sequenced bidirectionally using an ABI 3730XL DNA sequencer (Applied Biosystems, USA). The gene sequence with a minimum of 99% sequence identity and 99% coverage threshold was deemed for the same genotype, which was compared with those in the database located at the NCBI blast server (http://blast.ncbi.nlm.nih.gov).
Performance of mCIMandeCIM for Detection of CP-CRE Isolates
The mCIM test was performed using a meropenem disk, which was similar to the CLSI report.19 The details were described as follows. A 1-µL loopful of bacteria for Enterobacterales was emulsified from a blood agar plate cultured overnight in a tube with 2 mL trypticase soy broth (TSB; Becton, Dickinson and Company, USA). A 10–μg meropenem disk (Oxoid, UK) was added after vortexing for 10–15 s, and entirely immersed in the suspension, which was incubated for 4 h ± 15 min at 35 ± 2 °C in ambient air. Immediately following completion of the incubation, a mCIM indicator organism (E. coli ATCC 25922) suspension with turbidity equivalent to ~0.5 McF was inoculated on an Müeller-Hinton II agar (Becton, Dickinson and Company, USA) plate. The meropenem disk was removed after expelling excess liquid from the disk using a 10-µL loop. Then the disk was placed on the MH II agar plate after 3–10 min of inoculation with E. coli ATCC 25922, which was incubated for 18–24 h in an inverted position at 35 ± 2 °C in ambient air. The result was interpreted based on the zone diameter of the meropenem disk and considered positive if 6–15 mm or the presence of pinpoint colonies within a 16–18 mm zone, indeterminate if 16–18 mm, and negative if ≥ 19 mm. A second 2 mL TSB tube containing 5 mM EDTA was used for eCIM test. Other operations were the same as mCIM, and process the mCIM and eCIM tubes in parallel for each isolate. The meropenem disks used in mCIM and eCIM for the same isolate were placed on the same MH II agar plate.
Performance of Combined Disk Test (CDT) for Detection of CP-CRE Isolates
KPC inhibitor APB (3-aminophenyl boronic acid hydrochloride, Sigma-Aldrich, USA) solution (50 mg/mL), and metallic β-lactamase (MBL) inhibitor EDTA (ethylenediaminetetraacetic acid disodium salt dihydrate, Sigma-Aldrich, USA) solution (0.5 M) were previously treated using a 0.22 μm filter membrane (Millipore, Germany) and stored at 4 °C.20,21 The CDT was carried out using four 10-μg imipenem disks (Oxoid, UK), including a disk alone, a disk plus 10 μL of APB for KPC inhibition, a disk plus 2 μL of EDTA for MBL inhibition, and a disk plus both 10 μL of APB and 2 μL of EDTA for simultaneously inhibition of KPC and MBL.22 The four disks were placed onto Müeller-Hinton II agar plates inoculated with a ~0.5 McFarland CRE isolate suspension. The inhibition zones were measured after incubation for 16–18 hours at 35 ± 2 °C in ambient air. An increase of ≥ 5 mm in the inhibition zone diameter of the imipenem disk containing inhibitors (APB, EDTA, or both) in comparison to the imipenem disk alone was suggestive of KPC, MBL, or both carbapenemases production, respectively.
Performance of Immunochromatographic Test (Colloidal Gold) for Detection of CP-CRE Isolates
A full 1-μL inoculation loop of bacteria harvested from Columbia blood agar mixed with extraction buffer preloaded in a 1.5 mL centrifuge tube, vortexed, and 80 μL of this suspension was transferred into the Carbapenemase Detection Kit (Colloidal Gold, not launched yet) manufactured by Jiangsu Macro & Micro-Test Med-Tech Co., Ltd. The result was read after 15 min of incubation at room temperature.
Performance of the Multi-qPCR Assay for Detection of Carbapenemase Genes in Clinical Isolates and Samples
Due to the channel limitations of Applied Biosystems 7500 real-time PCR system (Applied Biosystems, Waltham, MA, USA), the multi-qPCR assay was divided into two sets. Set 1 and Set 2 consisted of blaIMP, blaOXA-48-like, and blaKPC genes, and blaVIM, blaNDM, and human β-globin genes, respectively.
A 25-µL reaction mixture consisted of a 2 µL extracted DNA template, 23 µL primers, probes, DNase/RNase-free deionized water, and PCR master mix. The details are shown in Table S3. The multi-qPCR program was as follows: 95 °C for 3 min, followed by 40 cycles of 95 °C for 15s, and 60 °C for 1 min. Fluorescence signals were collected at 60 °C each cycle of the extension step. The standard plasmids containing the target amplicon were used as positive quality control, and sterile water was used as negative quality control. A Ct ≤38 for clinical isolates and Ct ≤36 for clinical samples is considered positive. If the Ct value falls between 36–38 for clinical samples, the test needs to be repeated.
These clinical samples underwent routine laboratory bacterial culture according to medical orders.
Statistical Analysis
Statistical analysis was performed using VassarStats online software (VassarStats.net). The results of all kinds of tests were compared with conventional PCR and sequencing. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and 95% confidence interval (CI) of all kinds of tests were also calculated.
Results
The Repeatability and Reproducibility of the Multi-qPCR Assay
To evaluate the repeatability and reproducibility, different standard plasmids containing the corresponding target amplicon with different concentrations were detected by the multi-qPCR assay. The results are shown in Table S4. The intra-assay CVs of Ct values for detecting the blaIMP, blaOXA-48-like, blaKPC, blaVIM, and blaNDM genes ranged from 0.23% to 0.53%, 0.11% to 0.47%, 0.28% to 0.48%, 0.22% to 0.52%, and 0.27% to 0.47%, respectively; while the inter-assay CVs of Ct values for detecting these genes ranged from 0.84% to 1.19%, 0.81% to 1.54%, 0.94% to 2.04%, 0.86% to 1.46%, and 0.53% to 1.34%, respectively.
Linear Relationship and LOD of the Multi-qPCR Assay
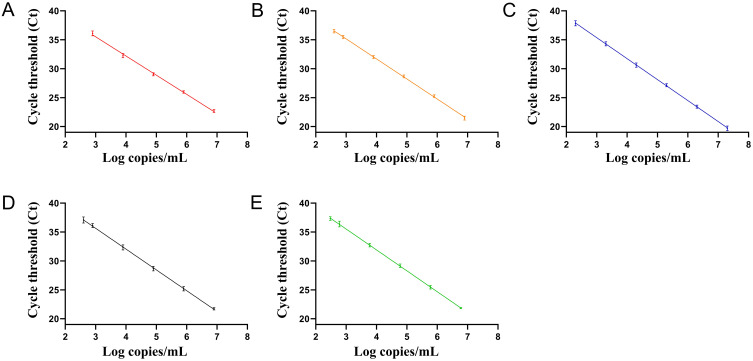
There were good linear relationships between the logarithm concentration of standard plasmids and the Ct value, which are shown in Figure 1. The coefficient of determination R2 for the detection of blaIMP, blaOXA-48-like, blaKPC, blaVIM, and blaNDM genes were 0.9953, 0.9978, 0.9977, 0.9964, and 0.9977, respectively. In addition, the LOD for detecting the blaIMP, blaOXA-48-like, blaKPC, blaVIM, and blaNDM genes were 8×102 copies/mL, 4×102 copies/mL, 2×102 copies/mL, 4×102 copies/mL, and 3×102 copies/mL, respectively.
Figure 1.
Standard curve for the multi-qPCR assay for detection of (A) blaIMP, y = −3.317x+45.48 (R2 = 0.9953), (B) blaOXA-48-like, y = −3.467x+45.56 (R2 = 0.9978), (C) blaKPC, y = −3.625x+46.24 (R2 = 0.9977), (D) blaVIM, y = −3.598x+46.45 (R2 = 0.9964), and (E) blaNDM genes, y = −3.616x+46.38 (R2 = 0.9977).
Conventional PCR and Sequencing for Confirming Carbapenemase Genes in CRE Isolates
The results of conventional PCR and sequencing of 182 CRE isolates are shown in Table 1. Conventional PCR confirmed 168 CRE isolates to be CP-CRE, while 14 isolates were non-CP-CRE due to PCR negative for all detected carbapenemase genes in this study. Among 168 CP-CRE isolates, only one carbapenemase gene was detected in 162 isolates, including 93 blaKPC, 46 blaNDM, 8 blaIMP, 1 blaVIM, and 14 blaOXA-48-like genes; two carbapenemase genes (blaKPC and blaNDM genes) were detected in 5 isolates, and three (blaKPC, blaNDM, and blaIMP genes) were detected in 1 isolate.
Table 1.
PCR and Sequencing, and the Results of mCIMandeCIM, Combined Disk Test, Colloidal Gold, and Multi-qPCR Assay for All Tested CRE Isolates
| Conventional PCR and Sequencing of CRE Isolates (n) | mCIMandeCIM | Combined disk test | Colloidal Gold | Multi-qPCR assay | |||||
|---|---|---|---|---|---|---|---|---|---|
| Serine Carbapenemase | MBL(s) | Not Detected | KPC | MBL | KPC&MBL(s) | Not detected | |||
| KPC (93) | 92 | 1 | 0 | 93 | 0 | 0 | 0 | 93 | 93 |
| NDM (46) | 0 | 46 | 0 | 0 | 45 | 0 | 1 | 46 | 46 |
| IMP (8) | 0 | 8 | 0 | 0 | 8 | 0 | 0 | 8 | 8 |
| VIM (1) | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| OXA-48-like (14) | 14 | 0 | 0 | 0 | 0 | 0 | 14 | 14 | 14 |
| KPC&NDM (5) | 5 | 0 | 0 | 0 | 0 | 5 | 0 | 5 | 5 |
| KPC&NDM&IMP (1) | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| None (14) | 0 | 0 | 14 | 3 | 0 | 1 | 10 | 14 | 14 |
| Total (182) | 112 | 56 | 14 | 96 | 54 | 7 | 25 | 182 | 182 |
| Accuracy | 96.2% | 89.6% | 100% | 100% | |||||
Abbreviation: MBL(s), metallic β-lactamase(s).
Accuracy of mCIMandeCIM, CDT, Colloidal Gold, and Multi-qPCR Assay for Detecting CP-CRE Isolates
mCIMandeCIM can only distinguish between serine carbapenemase and MBL, and cannot further classify them. Compared with conventional PCR and DNA sequencing methods, mCIMandeCIM accurately detected NDM (45/45), IMP (8/8), VIM (1/1) as MBL, and OXA-48-like (14/14) as serine carbapenemase, and 14 non-CP-CRE as none in this study (Table 1). However, one KPC-producing isolate was incorrectly identified as an MBL producer. In addition, all of the six isolates that produced both KPC and MBL(s) were incorrectly identified as serine carbapenemase producers. Therefore, the accuracy of mCIMandeCIM was 96.2%.
CDT is usually able to detect KPC, MBL, and KPC&MBL. It cannot detect OXA-48-like, and further classify MBL. In the study, CDT accurately detected KPC (73/73); IMP (8/8) and VIM (1/1) as MBL; KPC&NDM (5/5) and KPC&NDM&IMP (1/1) as KPC&MBL(s). However, one NDM-producing isolate, and all of 14 OXA-48-like-producing isolates were incorrectly identified as not detected. Moreover, of the 14 non-CP-CRE, 3 isolates were incorrectly identified as KPC producers, and one isolate was incorrectly identified as KPC&MBL(s) producer. Thus, the accuracy of CDT was 89.6%.
Colloidal Gold and multi-qPCR methods can detect all of the five major carbapenemases. In the current study, both Colloidal Gold and multi-qPCR assays accurately detected KPC (73/73), NDM (45/45), IMP (8/8), VIM (1/1), OXA-48-like (14/14), KPC&NDM (5/5), and KPC&NDM&IMP (1/1). Besides, all of the 14 non-CP-CRE isolates were tested negative by both methods. Therefore, the accuracy of the Colloidal Gold and the multi-qPCR assays was all 100%. The sensitivity, specificity, PPV, and NPV were all 100% for both methods for the detection of each carbapenemase gene, which are shown in Table S5.
Carbapenemase Genes Detection Directly from Clinical Samples by Multi-qPCR Assay
To verify the ability of the multi-qPCR assay to detect carbapenemase genes directly from clinical samples, 112 retrospectively collected clinical samples were detected, using conventional PCR and sequencing as reference standards. Among them, 60 samples tested positive, with 2 carbapenemase genes in one sample, and three carbapenemase genes in another sample, while 52 samples tested negative by PCR and sequencing. The results of culture, conventional PCR and sequencing, and the multi-qPCR assay of these clinical samples are shown in Table S6. The culture results of 105 (93.8%) samples were consistent with the conventional PCR and sequencing results. The results of the multi-qPCR assay were completely consistent with the results of PCR and sequencing, with a total detection of 51 blaKPC, 7 blaNDM, 1 blaIMP, 1 blaVIM, and 3 blaOXA-48-like genes. Therefore, the sensitivity, specificity, PPV, and NPV of the multi-qPCR assay were all 100% for the detection of the five major carbapenemase genes in clinical samples (Table 2).
Table 2.
Accuracy of the Multi-qPCR Assay for Detection of Carbapenemase Genes Directly from Clinical Samples
| Conventional PCR and sequencing (n) | Accuracy of multi-qPCR assay (%) | |||
|---|---|---|---|---|
| Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | |
|
blaKPC (51) |
100 (91.3–100) |
100 (92.6–100) |
100 (91.3–100) |
100 (92.6–100) |
|
blaNDM (7) |
100 (56.1–100) |
100 (95.6–100) |
100 (56.1–100) |
100 (95.6–100) |
|
blaIMP (1) |
100 (5.5–100) |
100 (95.8–100) |
100 (5.5–100) |
100 (95.8–100) |
|
blaVIM (1) |
100 (5.5–100) |
100 (95.8–100) |
100 (5.5–100) |
100 (95.8–100) |
|
blaOXA-48-like (3) |
100 (31.0–100) |
100 (95.8–100) |
100 (31.0–100) |
100 (95.8–100) |
Abbreviations: CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value.
We conducted further analysis on the 7 samples with mismatched culture and PCR and sequencing results (Table 3). The inconsistent results were all respiratory samples. Among them, Klebsiella pneumoniae isolate of one sample (S7) was confirmed as a carbapenemase-producing carbapenem-susceptible Enterobacterales by PCR and sequencing. For the other 6 samples, we traced the routine laboratory bacterial culture results of other samples from each patient. Within five days of the date of the sample detected by the multi-qPCR assay, we found that all of the patients of three samples (S11, S28, and S45) were isolated with CRE isolates at least twice with their other samples (same sample type with the sample detected by the multi-qPCR assay). It indicates that CRE isolates are also present in these three samples. However, the patients of the other three samples (S24, S58, and S59) had not been isolated with CRE isolates during hospitalization. It is well known that if the Ct value is too large (>36) then contamination may have occurred during the experiment, but the Ct values of S24, S58, and S59 were 25.0, 32.2, and 27.1, respectively, which were not significantly larger than the Ct values of other samples. It might be because the CRE was a non-dominant isolate.
Table 3.
Analysis on the 7 Samples with Mismatched Culture and PCR and Sequencing Results
| Sample number | Sample type | Culture | Conventional PCR and sequencing of the sample | Multi-qPCR assay of the sample | Conventional PCR and sequencing of the isolates of the sample | Comments |
|---|---|---|---|---|---|---|
| S7 | sp | aba-R; kpn-S | KPC | KPC | KPC (kpn-S) | carbapenemase-producing carbapenem-susceptible Enterobacterales |
| S11 | sp | aba-R; ani | NDM | NDM | None | The patient isolated CRE isolates two times within five days of the date of the sample detected by the multi-qPCR assay |
| S24 | sp | kpn-S; aba-R | OXA-48-like | OXA-48-like | None | CRE may be a non-dominant isolate |
| S28 | sp | eco-S | KPC | KPC | None | The patient isolated CRE isolates two times within five days of the date of the sample detected by the multi-qPCR assay |
| S45 | balf | eco-S | OXA-48-like | OXA-48-like | None | The patient isolated CRE isolates three times within five days of the date of the sample detected by the multi-qPCR assay |
| S58 | balf | aba-R | KPC | KPC | None | CRE may be a non-dominant isolate |
| S59 | sp | pmi-S | NDM | NDM | None | CRE may be a non-dominant isolate |
Abbreviations: sp, sputum; balf, bronchoalveolar lavage fluid; aba, Acinetobacter baumannii; kpn, Klebsiella pneumoniae; ani, Aspergillus niger; eco, Escherichia coli; pmi, Proteus mirabilis; S, susceptible to imipenem and meropenem; R, resistant to imipenem and meropenem (resistant to meropenem instead of imipenem for Morganella morganii, Proteus spp., and Providencia spp.).
Discussion
In this study, a multi-qPCR assay was designed, constructed, and validated for the detection of five major carbapenemase genes. In addition, three phenotypic methods were also assessed and compared with the multi-qPCR assay for carbapenemase detection using a large collection of clinical CRE isolates and evaluated their applicability in clinical settings.
The TaqMan-MGB probes were designed and used in the multi-qPCR assay. Since the existence of MGB (minor groove binder) can remarkably increase the Tm value of the probe,23 the probe can be designed to be shorter, with 13–17 bp in the current study. Compared with the methods reported in the past, the base number of the probes is significantly reduced.24–27 This will reduce the missed detection caused by base mutations in the target gene sequence to a certain extent. Moreover, the TaqMan-MGB probe has a lower background signal. Therefore, the designed probes and primers enable high specificity and sensitivity.
The low CVs of intra-assay (≤ 0.53%) and inter-assay (≤ 2.04%), as well as good linear relationship (R2 ≥ 0.9953) of the multi-qPCR assay indicate that it can provide a repeatable and reproducible quantification of target template DNA. Furthermore, the low LOD range (2–8×102 copies/mL) suggests it has a high sensitivity. These advantages make the multi-qPCR an excellent assay for clinical application.
Correctly detecting the type of carbapenemase produced by Enterobacterales isolates is crucial for clinical treatment, as limited new antibiotics target specific types of carbapenemase.28,29 mCIM/eCIM and inhibitor-based CDT for the detection of carbapenemase in CP-CRE isolates were recommended by CLSI19 and EUCAST,30 respectively. mCIMandeCIM can distinguish between serine carbapenemase (KPC and OXA-48-like) and MBL (NDM, IMP, and VIM), but it cannot further classify them. mCIMandeCIM had very high accuracy in detecting isolates that only produced one type of carbapenemase in the study, which is similar to previously reported results.31,32 However, once the isolate produces both serine carbapenemase and MBL(s), it will make an erroneous judgment that the isolate only produces serine carbapenemase (Table 1). CDT can detect KPC, MBL, and KPC&MBL, but it cannot detect OXA-48-like and further classify MBL. CDT had a high accuracy for differentiating KPC, MBL, or KPC&MBL in this study, which is similar to the report by Li et al.33 However, non-CP-CRE isolates and the isolates producing OXA-48-like had similar results, so CDT could not distinguish them (Table 1).
Although the above two phenotypic methods have high accuracy, both require overnight incubation. The immunochromatographic test, also called Colloidal Gold in the study, only takes 15 minutes to confirm the five major carbapenemases of the isolates. The commercial immunochromatographic tests including RESIST-5 O.O.K.N.V. and CARBA-5 have demonstrated excellent accuracy and clinical value in China.34,35 The Colloidal Gold (not launched yet) manufactured by Jiangsu Macro & Micro-Test Med-Tech Co., Ltd. had excellent results in a multi-center clinical trial. It had a total coincidence rate of 100% (95% CI, 99.18%-100%) with the commercial CARBA-5. Therefore, the current study selected locally produced Colloidal Gold for the detection of carbapenemase in clinical isolates. In this study, it had also shown excellent performance, superior to mCIM&eCIM and CDT (Table 1), with 100% sensitivity and specificity for detection of each of the five major carbapenemases (Table S5).
Although Colloidal Gold can be performed in a short period of time, obtaining highly accurate results still requires bacterial isolation, which may delay appropriate antibiotic treatment to some extent. The multi-qPCR assay can not only detect isolates, but also directly detect clinical samples. In isolate validation, the multi-qPCR assay correctly detected the carbapenemase genes of all isolates, even if there were two or three carbapenemase genes in one isolate (Table 1). Therefore, the sensitivity and specificity were all 100% (Table S5), which indicates that the designed probes and primers, as well as the constructed assay, are suitable and exceedingly good. In clinical sample validation, the results of the multi-qPCR assay were also completely consistent with those of conventional PCR and sequencing (Tables S6 and 2). Even in some respiratory samples, the multi-qPCR assay could provide more resistance information than the culture results (Table 3). It has great potential for application in clinical diagnosis.
In this study, a carbapenemase-producing carbapenem-susceptible Klebsiella pneumoniae was found when clinical samples were detected by the multi-qPCR assay. It suggests that the presence of a very small number of isolates with low or no carbapenemase production in clinical samples. We reviewed the patient’s case and found that carbapenemase-resistant Klebsiella pneumoniae was cultured in the sputum and blood samples after 26 and 28 days, respectively. It indicates that carrying the carbapenemase gene is a potential risk for treatment failure, even if carbapenem antibiotics are initially susceptible. A similar study also supports this viewpoint.36 Early recognition of producers of carbapenemases is very important not only in clinical treatment, but also in controlling the spread of carbapenemase-producing bacteria.37
A detailed comparison describing the main features of each test is shown in Table 4. Compared to the other three phenotypic methods, the biggest advantage of the multi-qPCR assay is that it can detect clinical samples, greatly reducing detection time and providing a basis for the application of appropriate antibiotics. Even so, it cannot be ignored that the three phenotype methods also have their own advantages and application scenarios.
Table 4.
The Main Characteristics of Four Methods for Detecting Carbapenemase
| Parameter | mCIMandeCIM | CDT | Gold Colloidal | Multi-qPCR assay |
|---|---|---|---|---|
| Time (h) | 22–28 | 16–18 | 0.5 | 2 |
| Test principle | Carbapenem hydrolysis and inhibitor-based test | Inhibitor-based test | immunochromatographic test | Multiplex real-time PCR |
| Equipment | No | No | No | Yes |
| Cost | Low | Low | Moderate | High |
| Sample type | Bacterial colonies | Bacterial colonies | Bacterial colonies | Bacterial colonies, clinical samples |
| Result | Serine carbapenemase, MBL(s) | KPC, MBL(s), KPC&MBL(s) | KPC, NDM, IMP, VIM, OXA-48-like | KPC, NDM, IMP, VIM, OXA-48-like |
| Result interpretation | Simple | Simple | Simple | Simple |
| Strengths | simple, low costs | simple, low costs | Fast, simple, moderate costs, differentiation of the big 5 carbapenemases | Relatively fast, simple, differentiation of the big 5 carbapenemases |
| Limitations | Long time-to-result, false identification for producing serine carbapenemase and MBL(s) simultaneously, unable to detect clinical samples | Long time-to-result, unable to detect OXA-48-like, unable to detect clinical samples | unable to detect clinical samples | Expensive, additional equipment required |
The current study has some limitations. First, the number of blaVIM genes was small (n = 1), which resulted in a widened 95% CI for the sensitivity of detecting the blaVIM gene. Second, very rare types of carbapenemase genes (eg blaNMC, blaIMI, and blaSME) were not included, which may provide inaccurate information to clinical practice in the future.
Conclusion
In summary, our new multi-qPCR assay has high accuracy for the detection of carbapenemases in CRE isolates in the study. The multi-qPCR assay is comparable to Colloidal Gold, and superior to mCIMandeCIM and CDT. mCIMandeCIM and CDT offer the chance for easy implementation of carbapenemases detection in routine laboratories, but both require bacterial isolation and overnight incubation, which increases turnaround time. Colloidal Gold offers rapid and cost-effective detection but still requires bacterial isolation. Although there are shortcomings, these three phenotype methods also have their own advantages and application scenarios, which still have great value in clinical application. The designed and constructed multi-qPCR assay, with high reliability and low LOD, can not only detect the five major carbapenemase genes in isolates but also detect them in clinical samples, greatly reducing turnaround time.
Acknowledgments
The authors would like to thank all members of the Department of Infectious Diseases and Clinical Microbiology staff at Beijing Chao-Yang Hospital (Beijing, China) for contributing to this work.
Funding Statement
This research was funded by the National Natural Science Foundation of China (Grant No. 82302551); Beijing Natural Science Foundation (Grant No. 7234369); Beijing Chao-Yang Hospital Science and Technology Innovation Fund (Grant No. 22kcjjyb-19); and the Reform and Development Program of Beijing Institute of Respiratory Medicine (Grant No. Ggyfz202425 and Ggyfz202419).
Data Sharing Statement
All data sets generated or analyzed during this study are included in the article. Further inquiries can be directed to the corresponding author.
Ethical Approval
The study was carried out according to the Declaration of Helsinki and was approved by the Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University (2022-8-24-1). Patient consent was waived due to the retrospective nature of the study. The study maintains patient records and information in a confidential manner. Information in patient records or information collected from the patient is kept in strict confidence following the Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.World Health Organization (WHO). Global antimicrobial resistance surveillance system (GLASS) report: early implementation 2020; 2020. Available from: https://iris.who.int/bitstream/handle/10665/332081/9789240005587-eng.pdf?sequence=1. Accessed 27, July 2024.
- 2.O’Neill J Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Review on Antimicrobial Resistance. 2016. Available from: https://iiif.wellcomecollection.org/file/b28552179_AMR%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations.pdf. Accessed 27, July 2024.
- 3.World Health Organization (WHO). WHO bacterial priority pathogens list, 2024: bacterial pathogens of public health importance, to guide research, development and strategies to prevent and control antimicrobial resistance. 2024. Available from: https://iris.who.int/bitstream/handle/10665/376776/9789240093461-eng.pdf?sequence=1&isAllowed=y. Accessed 27, July 2024.
- 4.US Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States 2019. 2019. Available from: http://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. Accessed 27 July 2024.
- 5.China Antimicrobial Resistance Surveillance System. (CARSS). 2023. Available from: https://www.carss.cn/Report/Details?aId=917. Accessed 27, July 2024.
- 6.China Antimicrobial Surveillance Network (CHINET). 2023; Avialable from: http://www.chinets.com/Data/AntibioticDrugFast. Accessed 27, July 2024.
- 7.Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE network. Antimicrob Agents Chemother. 2018;62(2):e01882–17. doi: 10.1128/AAC.01882-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satlin MJ, Chen L, Patel G, et al. Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant Enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob Agents Chemother. 2017;61(4):e02349–16. doi: 10.1128/AAC.02349-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammoudi Halat D, Ayoub Moubareck C. The current burden of carbapenemases: review of significant properties and dissemination among gram-negative bacteria. Antibiotics (Basel). 2020;9(4):186. doi: 10.3390/antibiotics9040186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caliskan-Aydogan O, Alocilja EC. A review of carbapenem resistance in Enterobacterales and its detection techniques. Microorganisms. 2023;11(6):1491. doi: 10.3390/microorganisms11061491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei M, Wang P, Wang S, Yang C, Gu L. HB&L system for rapid phenotypic detection of clinical carbapenem-resistant Enterobacterales isolates. J Glob Antimicrob Resist. 2021;26:272–278. doi: 10.1016/j.jgar.2021.02.036 [DOI] [PubMed] [Google Scholar]
- 12.Baeza LL, Pfennigwerth N, Greissl C, et al. Comparison of five methods for detection of carbapenemases in Enterobacterales with proposal of a new algorithm. Clin Microbiol Infect. 2019;25(10):1286.e1289–1286.e1215. doi: 10.1016/j.cmi.2019.03.003 [DOI] [PubMed] [Google Scholar]
- 13.Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S. Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother. 2012;67(4):906–909. doi: 10.1093/jac/dkr563 [DOI] [PubMed] [Google Scholar]
- 14.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–495. doi: 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 15.Jing X, Zhou H, Min X, et al. The simplified carbapenem inactivation method (sCIM) for simple and accurate detection of carbapenemase-producing gram-negative bacilli. Front Microbiol. 2018;9:2391. doi: 10.3389/fmicb.2018.02391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi: 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Wang P, Liu J, et al. Molecular detection of Nocardia: development and application of a real-time PCR assay in sputum and bronchoalveolar lavage fluid samples. Eur J Clin Microbiol Infect Dis. 2023;42(7):865–872. doi: 10.1007/s10096-023-04619-4 [DOI] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 34th ed. ed. CLSI supplement M100.. Wayne, PA: CLSI; 2024. [Google Scholar]
- 20.Petropoulou D, Tzanetou K, Syriopoulou VP, Daikos GL, Ganteris G, Malamou-Lada E. Evaluation of imipenem/imipenem+EDTA disk method for detection of metallo-beta-lactamase-producing Klebsiella pneumoniae isolated from blood cultures. Microb Drug Resist. 2006;12(1):39–43. doi: 10.1089/mdr.2006.12.39 [DOI] [PubMed] [Google Scholar]
- 21.Doi Y, Potoski BA, Adams-Haduch JM, Sidjabat HE, Pasculle AW, Paterson DL. Simple disk-based method for detection of Klebsiella pneumoniae carbapenemase-type beta-lactamase by use of a boronic acid compound. J Clin Microbiol. 2008;46(12):4083–4086. doi: 10.1128/JCM.01408-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pournaras S, Zarkotou O, Poulou A, et al. A combined disk test for direct differentiation of carbapenemase-producing Enterobacteriaceae in surveillance rectal swabs. J Clin Microbiol. 2013;51(9):2986–2990. doi: 10.1128/JCM.00901-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutyavin IV, Afonina IA, Mills A, et al. 3’-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28(2):655–661. doi: 10.1093/nar/28.2.655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smiljanic M, Kaase M, Ahmad-Nejad P, Ghebremedhin B. Comparison of in-house and commercial real time-PCR based carbapenemase gene detection methods in Enterobacteriaceae and non-fermenting gram-negative bacterial isolates. Ann Clin Microbiol Antimicrob. 2017;16(1):48. doi: 10.1186/s12941-017-0223-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellington MJ, Findlay J, Hopkins KL, et al. Multicentre evaluation of a real-time PCR assay to detect genes encoding clinically relevant carbapenemases in cultured bacteria. Int J Antimicrob Agents. 2016;47(2):151–154. doi: 10.1016/j.ijantimicag.2015.11.013 [DOI] [PubMed] [Google Scholar]
- 26.Lund M, Petersen MB, Jørgensen AL, Paulmann D, Wang M. Rapid real-time PCR for the detection of IMP, NDM, VIM, KPC and OXA-48 carbapenemase genes in isolates and spiked stool samples. Diagn Microbiol Infect Dis. 2018;92(1):8–12. doi: 10.1016/j.diagmicrobio.2018.04.002 [DOI] [PubMed] [Google Scholar]
- 27.van der Zee A, Roorda L, Bosman G, et al. Multi-centre evaluation of real-time multiplex PCR for detection of carbapenemase genes OXA-48, VIM, IMP, NDM and KPC. BMC Infect Dis. 2014;14(1):27. doi: 10.1186/1471-2334-14-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terreni M, Taccani M, Pregnolato M. New antibiotics for multidrug-resistant bacterial strains: latest research developments and future perspectives. Molecules. 2021;26(9):2671. doi: 10.3390/molecules26092671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. Infectious diseases society of America 2023 guidance on the treatment of antimicrobial resistant gram-negative infections. Clin Infect Dis. 2023;ciad428. doi: 10.1093/cid/ciad428 [DOI] [PubMed] [Google Scholar]
- 30.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. Version 2.01. EUCAST. 2017. Available at: http://www.eucast.org/. [accessed 27, July 2024.].
- 31.Zhu Y, Jia P, Li X, et al. Carbapenemase detection by NG-Test CARBA 5-A rapid immunochromatographic assay in carbapenem-resistant Enterobacterales diagnosis. Ann Transl Med. 2021;9(9):769. doi: 10.21037/atm-20-8216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai YM, Wang S, Chiu HC, Kao CY, Wen LL. Combination of modified carbapenem inactivation method (mCIM) and EDTA-CIM (eCIM) for phenotypic detection of carbapenemase-producing Enterobacteriaceae. BMC Microbiol. 2020;20(1):315. doi: 10.1186/s12866-020-02010-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Li C, Cai X, et al. Performance of modified carbapenem inactivation method and inhibitor-based combined disk test in the detection and distinguishing of carbapenemase producing Enterobacteriaceae. Ann Transl Med. 2019;7(20):566. doi: 10.21037/atm.2019.09.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han R, Guo Y, Peng M, et al. Evaluation of the immunochromatographic NG-Test Carba 5, RESIST-5 O.O.K.N.V. and IMP K-seT for rapid detection of KPC-, NDM-, IMP-, VIM-type, and OXA-48-like carbapenemase among Enterobacterales. Front Microbiol. 2020;11:609856. doi: 10.3389/fmicb.2020.609856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Bai L, Liu J, et al. Parallel validation of the NG-Test Carba 5 and the Xpert Carba-R for detection and characterization of carbapenem-resistant Enterobacterales causing bloodstream infections. J Mol Diagn. 2021;23(8):1007–1014. doi: 10.1016/j.jmoldx.2021.05.009 [DOI] [PubMed] [Google Scholar]
- 36.Hong T, Moland ES, Abdalhamid B, et al. Escherichia coli: development of carbapenem resistance during therapy. Clin Infect Dis. 2005;40(10):e84–86. doi: 10.1086/429822 [DOI] [PubMed] [Google Scholar]
- 37.Pasteran F, Mendez T, Guerriero L, Rapoport M, Corso A. Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J Clin Microbiol. 2009;47(6):1631–1639. doi: 10.1128/JCM.00130-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data sets generated or analyzed during this study are included in the article. Further inquiries can be directed to the corresponding author.