Abstract
The chemical cleavage of linear DNA by Cu(II):thiol combinations was investigated using end-labelled double-stranded restriction fragments. Single-strand cleavage of the target DNA occurred with apparent sequence preference, but there appeared to be no apparent consensus sequence for such preferentially cleaved sites based on studies of three different restriction fragments. For any given restriction fragment, the observed Cu(II):thiol DNA-cleavage sequence preference was found to be independent of the nature of the thiol used. Cleavage of 3'-end-labelled DNA generated fragments bearing 5'-phosphoryl termini, and cleavage of 5'-end-labelled DNA gave rise to fragments bearing 3'-phosphoryl and 3'-'phosphatase-inert' termini in equal proportions. No appreciable amounts of fragments bearing 3'- or 5'-hydroxy termini were detected. This pattern of cleavage products is similar to that observed using the hydroxyl-radical-generating systems, 60Co gamma-irradiation or methidium propyl EDTA:Fe(II), but is dissimilar to that found with the Cu(II):phenanthroline or bleomycin systems. Cu(II):thiol cleavage of DNA fragments containing inserts of repeated dinucleotide sequence showed alternating patterns of relative cleavage intensities within the insert-sequence regions which were not seen in the sequences flanking the insert sequence. The assembled experimental data indicate that a reaction product of the Cu(II):thiol interaction, probably the hydroxyl radical, causes DNA cleavage. The Cu(II):thiol interaction may occur free in solution or via a copper species which is bound non-specifically to DNA. The observed low sequence-dependence of the cleavage reaction probably reflects the nature of the DNA structure itself, the Cu(II):thiol system serving as a structural probe of sequence-dependent structural variations in the DNA structure.
Full text
PDF

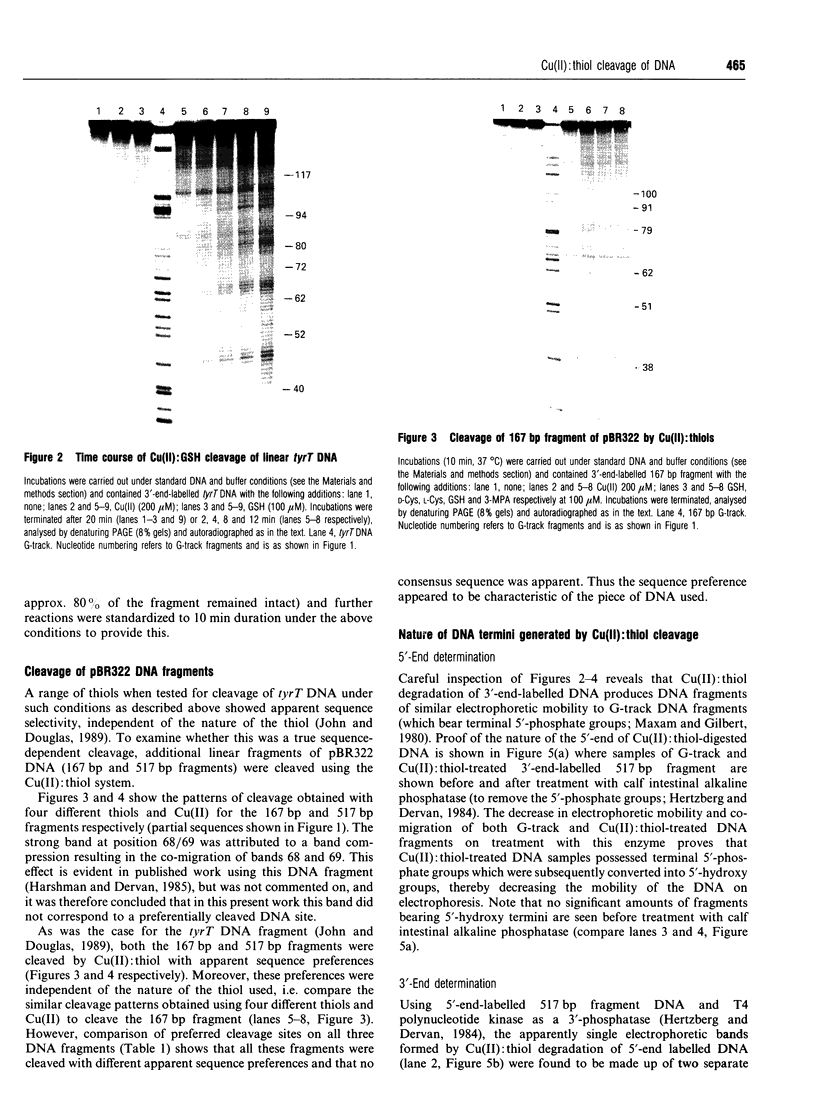
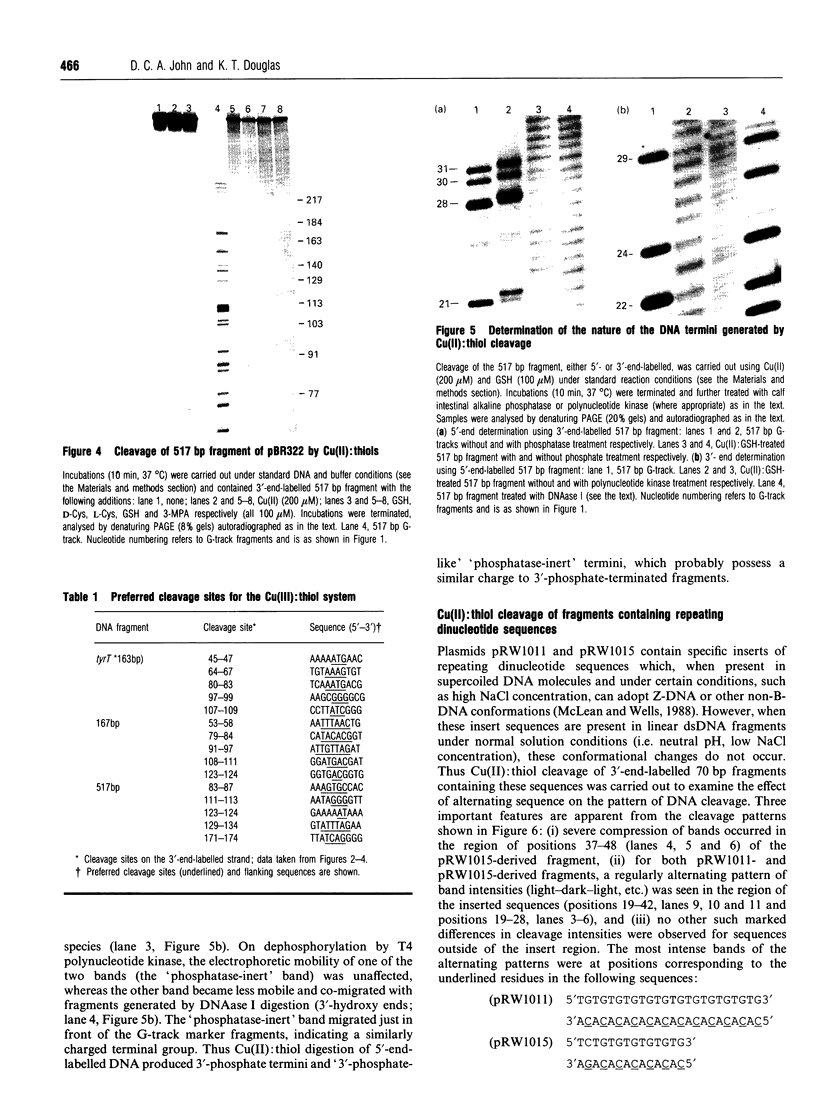


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen C. H., Sigman D. S. Nuclease activity of 1,10-phenanthroline-copper: sequence-specific targeting. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7147–7151. doi: 10.1073/pnas.83.19.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou S. H. DNA- and protein-scission activities of ascorbate in the presence of copper ion and a copper-peptide complex. J Biochem. 1983 Oct;94(4):1259–1267. doi: 10.1093/oxfordjournals.jbchem.a134471. [DOI] [PubMed] [Google Scholar]
- Downey K. M., Que B. G., So A. G. Degradation of DNA by 1,-10-phenanthroline. Biochem Biophys Res Commun. 1980 Mar 13;93(1):264–270. doi: 10.1016/s0006-291x(80)80275-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Dreyer G. B., Dervan P. B. Sequence-specific cleavage of single-stranded DNA: oligodeoxynucleotide-EDTA X Fe(II). Proc Natl Acad Sci U S A. 1985 Feb;82(4):968–972. doi: 10.1073/pnas.82.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Ebright Y. W., Pendergrast P. S., Gunasekera A. Conversion of a helix-turn-helix motif sequence-specific DNA binding protein into a site-specific DNA cleavage agent. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2882–2886. doi: 10.1073/pnas.87.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiel R. J., Beerman T. A., Mark E. H., Datta-Gupta N. DNA strand scission activity of metalloporphyrins. Biochem Biophys Res Commun. 1982 Aug;107(3):1067–1074. doi: 10.1016/0006-291x(82)90630-1. [DOI] [PubMed] [Google Scholar]
- Ford K., Fox K. R., Neidle S., Waring M. J. DNA sequence preferences for an intercalating porphyrin compound revealed by footprinting. Nucleic Acids Res. 1987 Mar 11;15(5):2221–2234. doi: 10.1093/nar/15.5.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman K. D., Dervan P. B. Molecular recognition of B-DNA by Hoechst 33258. Nucleic Acids Res. 1985 Jul 11;13(13):4825–4835. doi: 10.1093/nar/13.13.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- John D. C., Douglas K. T. Apparent sequence preference in cleavage of linear B-DNA by the Cu(II):thiol system. Biochem Biophys Res Commun. 1989 Dec 29;165(3):1235–1242. doi: 10.1016/0006-291x(89)92734-4. [DOI] [PubMed] [Google Scholar]
- Kirshenbaum M. R., Tribolet R., Barton J. K. Rh(DIP)3(3+): a shape-selective metal complex which targets cruciforms. Nucleic Acids Res. 1988 Aug 25;16(16):7943–7960. doi: 10.1093/nar/16.16.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M., Yoon C., Goyne T., Thederahn T., Sigman D. S. Nuclease activity of 1,10-phenanthroline-copper ion: reaction with CGCGAATTCGCG and its complexes with netropsin and EcoRI. Biochemistry. 1986 Nov 18;25(23):7401–7408. doi: 10.1021/bi00371a023. [DOI] [PubMed] [Google Scholar]
- Kłysik J., Stirdivant S. M., Larson J. E., Hart P. A., Wells R. D. Left-handed DNA in restriction fragments and a recombinant plasmid. Nature. 1981 Apr 23;290(5808):672–677. doi: 10.1038/290672a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLean M. J., Wells R. D. The role of DNA sequence in the formation of Z-DNA versus cruciforms in plasmids. J Biol Chem. 1988 May 25;263(15):7370–7377. [PubMed] [Google Scholar]
- Montalvo-Alvarez A. M., Landau I., Baccam D., Chabaud A. G., Ginsburg H. Experimental modifications of the circadian rythm of Plasmodium vinckei petteri following cryopreservation; probable resistance of the merozoïte to thawing. C R Acad Sci III. 1988;307(1):5–10. [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E., Jeppesen C., Buchardt O. Uranyl salts as photochemical agents for cleavage of DNA and probing of protein-DNA contacts. FEBS Lett. 1988 Aug 1;235(1-2):122–124. doi: 10.1016/0014-5793(88)81245-6. [DOI] [PubMed] [Google Scholar]
- Reed C. J., Douglas K. T. Chemical cleavage of plasmid DNA by glutathione in the presence of Cu(II) ions. The Cu(II)-thiol system for DNA strand scission. Biochem J. 1991 May 1;275(Pt 3):601–608. doi: 10.1042/bj2750601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed C. J., Douglas K. T. Single-strand cleavage of DNA by Cu(II) and thiols: a powerful chemical DNA-cleaving system. Biochem Biophys Res Commun. 1989 Aug 15;162(3):1111–1117. doi: 10.1016/0006-291x(89)90788-2. [DOI] [PubMed] [Google Scholar]
- Riddles P. W., Blakeley R. L., Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- Sagripanti J. L., Kraemer K. H. Site-specific oxidative DNA damage at polyguanosines produced by copper plus hydrogen peroxide. J Biol Chem. 1989 Jan 25;264(3):1729–1734. [PubMed] [Google Scholar]
- Shafer G. E., Price M. A., Tullius T. D. Use of the hydroxyl radical and gel electrophoresis to study DNA structure. Electrophoresis. 1989 May-Jun;10(5-6):397–404. doi: 10.1002/elps.1150100518. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Graham D. R., D'Aurora V., Stern A. M. Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline . cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J Biol Chem. 1979 Dec 25;254(24):12269–12272. [PubMed] [Google Scholar]
- Sluka J. P., Horvath S. J., Bruist M. F., Simon M. I., Dervan P. B. Synthesis of a sequence-specific DNA-cleaving peptide. Science. 1987 Nov 20;238(4830):1129–1132. doi: 10.1126/science.3120311. [DOI] [PubMed] [Google Scholar]
- Timsit Y., Vilbois E., Moras D. Base-pairing shift in the major groove of (CA)n tracts by B-DNA crystal structures. Nature. 1991 Nov 14;354(6349):167–170. doi: 10.1038/354167a0. [DOI] [PubMed] [Google Scholar]
- UDENFRIEND S., CLARK C. T., AXELROD J., BRODIE B. B. Ascorbic acid in aromatic hydroxylation. I. A model system for aromatic hydroxylation. J Biol Chem. 1954 Jun;208(2):731–739. [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Kashige N., Kojima M., Nakashima Y. Specificity of nucleotide sequence in DNA cleavage induced by D-glucosamine and D-glucosamine-6-phosphate in the presence of Cu2+. Agric Biol Chem. 1990 Feb;54(2):519–525. [PubMed] [Google Scholar]
- Wilson J. M., Danos O., Grossman M., Raulet D. H., Mulligan R. C. Expression of human adenosine deaminase in mice reconstituted with retrovirus-transduced hematopoietic stem cells. Proc Natl Acad Sci U S A. 1990 Jan;87(1):439–443. doi: 10.1073/pnas.87.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]