Abstract
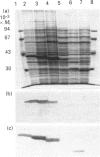
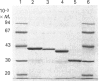
The structural organization of penicillin-binding protein (PBP) 5 was investigated by C-terminal truncation. Compared with other low-M(r) penicillin-interacting proteins, PBP5 carries a C-terminal extension of about 100 amino acids. The sites for introduction of stop codons were chosen on the basis of the established three-dimensional structure of the Streptomyces albus G beta-lactamase [Dideberg, Charlier, Wéry, Dehottay, Dusart, Erpicum, Frère and Ghuysen (1987) Biochem. J. 245, 911-913] and comparative hydrophobic cluster analysis [Gaboriaud, Bissery, Bencheritt and Mornon (1987) FEBS Lett. 224, 149-155]. Two stop codons were introduced at positions Ile-354 or Val-348 to construct an optimized soluble form of PBP5 for crystallization purposes. The newly constructed soluble and enzymically active form (PBP5s353) was isolated by dye-affinity chromatography and gave rise to small crystals. Another two stop codons were introduced at positions Arg-261 or Ala-276 to determine the minimal enzymically active 'core protein'. The truncated form (PBP5s275), missing the entire C-terminal extension, showed unaltered penicillin-binding characteristics and a catalytic-centre activity 40% that of PBP5s353 + 9 using bisacetyl-L-Lys-D-Ala-D-Ala as substrate. This protein, however was more susceptible to proteolytic degradation, which might indicate a role of the C-terminal portion in stabilizing the protein.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bittner M., Kupferer P., Morris C. F. Electrophoretic transfer of proteins and nucleic acids from slab gels to diazobenzyloxymethyl cellulose or nitrocellulose sheets. Anal Biochem. 1980 Mar 1;102(2):459–471. doi: 10.1016/0003-2697(80)90182-7. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Dideberg O., Charlier P., Wéry J. P., Dehottay P., Dusart J., Erpicum T., Frère J. M., Ghuysen J. M. The crystal structure of the beta-lactamase of Streptomyces albus G at 0.3 nm resolution. Biochem J. 1987 Aug 1;245(3):911–913. doi: 10.1042/bj2450911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L. C., Schwarz U., Keck W., Charlier P., Dideberg O., Ghuysen J. M. Properties and crystallization of a genetically engineered, water-soluble derivative of penicillin-binding protein 5 of Escherichia coli K12. Eur J Biochem. 1988 Jan 15;171(1-2):11–16. doi: 10.1111/j.1432-1033.1988.tb13751.x. [DOI] [PubMed] [Google Scholar]
- Frére J. M., Leyh-Bouille M., Ghuysen J. M., Nieto M., Perkins H. R. Exocellular DD-carboxypeptidases-transpeptidases from Streptomyces. Methods Enzymol. 1976;45:610–636. doi: 10.1016/s0076-6879(76)45054-1. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C., Bissery V., Benchetrit T., Mornon J. P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987 Nov 16;224(1):149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Jackson M. E., Pratt J. M. An 18 amino acid amphiphilic helix forms the membrane-anchoring domain of the Escherichia coli penicillin-binding protein 5. Mol Microbiol. 1987 Jul;1(1):23–28. doi: 10.1111/j.1365-2958.1987.tb00522.x. [DOI] [PubMed] [Google Scholar]
- Kelly J. A., Dideberg O., Charlier P., Wery J. P., Libert M., Moews P. C., Knox J. R., Duez C., Fraipont C., Joris B. On the origin of bacterial resistance to penicillin: comparison of a beta-lactamase and a penicillin target. Science. 1986 Mar 21;231(4744):1429–1431. doi: 10.1126/science.3082007. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Meijers J., Peters R., van der Hoek P., van Alphen L. Electrophoretic resolution of the "major outer membrane protein" of Escherichia coli K12 into four bands. FEBS Lett. 1975 Oct 15;58(1):254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Meissner P. S., Sisk W. P., Berman M. L. Bacteriophage lambda cloning system for the construction of directional cDNA libraries. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4171–4175. doi: 10.1073/pnas.84.12.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Palomeque-Messia P., Englebert S., Leyh-Bouille M., Nguyen-Distèche M., Duez C., Houba S., Dideberg O., Van Beeumen J., Ghuysen J. M. Amino acid sequence of the penicillin-binding protein/DD-peptidase of Streptomyces K15. Predicted secondary structures of the low Mr penicillin-binding proteins of class A. Biochem J. 1991 Oct 1;279(Pt 1):223–230. doi: 10.1042/bj2790223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot P. J., Knobler R. L., Buchmeier M. J. Western and dot immunoblotting analysis of viral antigens and antibodies: application to murine hepatitis virus. J Immunol Methods. 1984 Oct 12;73(1):177–188. doi: 10.1016/0022-1759(84)90043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D. J., Strominger J. L. Cleavage of a COOH-terminal hydrophobic region from D-alanine carboxypeptidase, a penicillin-sensitive bacterial membrane enzyme. Characterization of active, water-soluble fragments. J Biol Chem. 1979 Jun 10;254(11):4863–4875. [PubMed] [Google Scholar]
- Waxman D. J., Strominger J. L. Limited proteolysis of the penicillin-sensitive D-alanine carboxypeptidase purified from Bacillus subtilis membranes. Active water-soluble fragments generated by cleavage of a COOH-terminal membrane anchor. J Biol Chem. 1981 Feb 25;256(4):2059–2066. [PubMed] [Google Scholar]
- Waxman D. J., Strominger J. L. Primary structure of the COOH-terminal membranous segment of a penicillin-sensitive enzyme purified from two Bacilli. J Biol Chem. 1981 Feb 25;256(4):2067–2077. [PubMed] [Google Scholar]
- van der Linden M. P., Mottl H., Keck W. Cytoplasmic high-level expression of a soluble, enzymatically active form of the Escherichia coli penicillin-binding protein 5 and purification by dye chromatography. Eur J Biochem. 1992 Feb 15;204(1):197–202. doi: 10.1111/j.1432-1033.1992.tb16624.x. [DOI] [PubMed] [Google Scholar]