Abstract
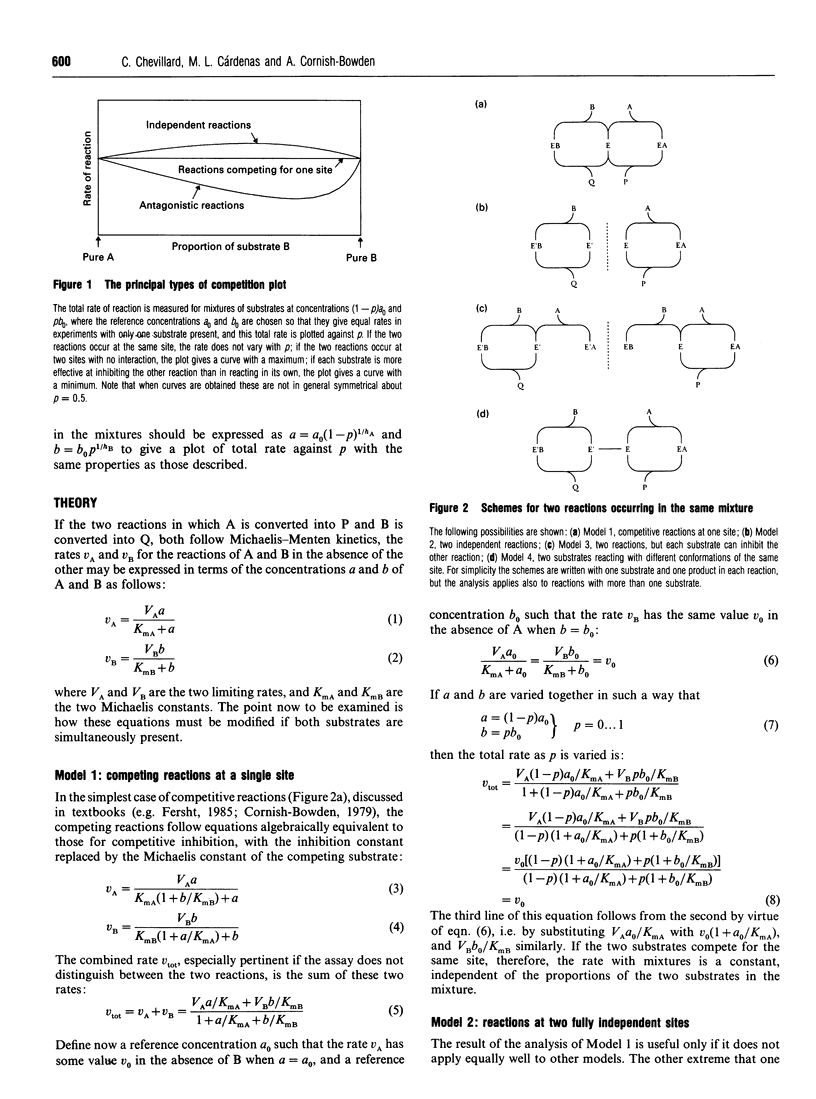
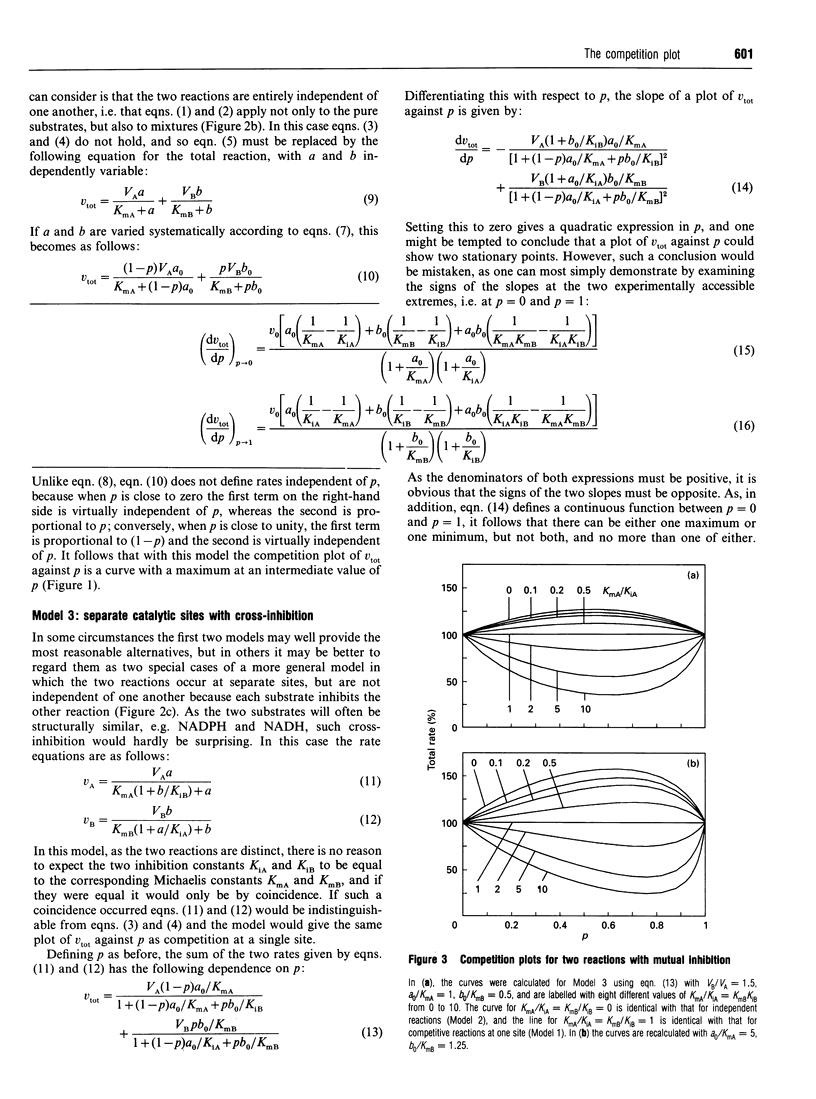
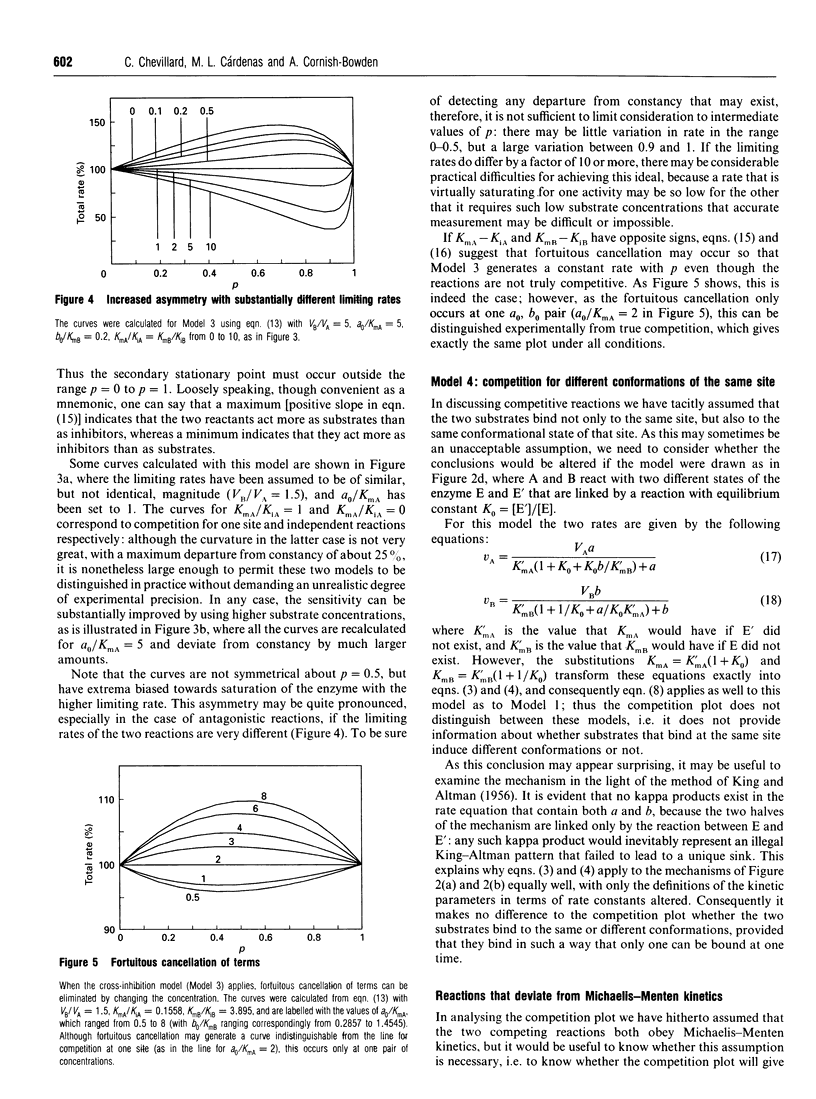
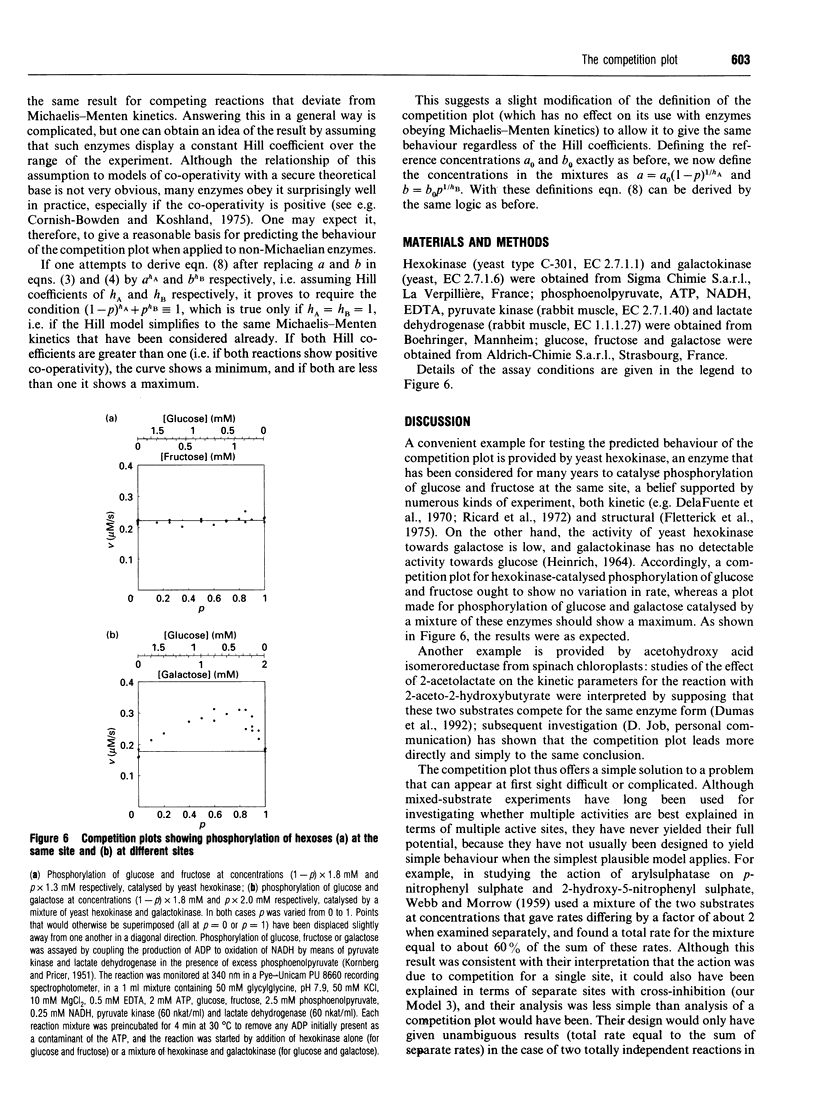
The competition plot is a method for determining whether or not two enzyme-catalysed reactions occur at the same active site. It is a plot of total rate against p, where p varies from 0 to 1 and specifies the concentrations (1-p)a0 and pb0 of two substrates in terms of reference concentrations a0 and b0 chosen so as to give the same rates at p = 0 and p = 1. If the two substrates react at the same site, the competition plot gives a horizontal straight line, i.e. the total rate is independent of p. Independent reactions at two separate sites give a curve with a maximum; separate reactions with cross-inhibition generate curves with either maxima or minima according to whether the Michaelis constants of the two substrates are smaller or larger than their inhibition constants in the other reactions. Although ambiguous results can sometimes arise, experimental strategies exist for avoiding them, for example working as close as possible to the lower of the two limiting rates. When tested with yeast hexokinase, the plot indicated phosphorylation of glucose and fructose at the same site. Conversely, with a mixture of yeast hexokinase and galactokinase it indicated phosphorylation of glucose and galactose at different sites. In both cases the observed behaviour agreed with the known properties of the enzymes. A slight modification to the definition of this plot allows it to be applied also to enzymes that deviate from Michaelis-Menten kinetics.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cornish-Bowden A., Koshland D. E., Jr Diagnostic uses of the Hill (Logit and Nernst) plots. J Mol Biol. 1975 Jun 25;95(2):201–212. doi: 10.1016/0022-2836(75)90390-3. [DOI] [PubMed] [Google Scholar]
- DelaFuente G., Lagunas R., Sols A. Induced fit in yeast hexokinase. Eur J Biochem. 1970 Oct;16(2):226–233. doi: 10.1111/j.1432-1033.1970.tb01075.x. [DOI] [PubMed] [Google Scholar]
- Dumas R., Job D., Ortholand J. Y., Emeric G., Greiner A., Douce R. Isolation and kinetic properties of acetohydroxy acid isomeroreductase from spinach (Spinacia oleracea) chloroplasts overexpressed in Escherichia coli. Biochem J. 1992 Dec 15;288(Pt 3):865–874. doi: 10.1042/bj2880865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletterick R. J., Bates D. J., Steitz T. A. The structure of a yeast hexokinase monomer and its complexes with substrates at 2.7-A resolution. Proc Natl Acad Sci U S A. 1975 Jan;72(1):38–42. doi: 10.1073/pnas.72.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEINRICH M. R. THE PURIFICATION AND PROPERTIES OF YEAST GALACTOKINASE. J Biol Chem. 1964 Jan;239:50–53. [PubMed] [Google Scholar]
- KORNBERG A., PRICER W. E., Jr Enzymatic phosphorylation of adenosine and 2,6-diaminopurine riboside. J Biol Chem. 1951 Dec;193(2):481–495. [PubMed] [Google Scholar]
- Mastrantonio S., Nucci R., Vaccaro C., Rossi M., Whitehead E. P. Analysis of competition for substrate sites in an allosteric enzyme with co-operative kinetics. Effects of dAMP and dUMP on donkey spleen deoxycytidylate aminohydrolase. Eur J Biochem. 1983 Dec 15;137(3):421–427. doi: 10.1111/j.1432-1033.1983.tb07845.x. [DOI] [PubMed] [Google Scholar]
- Ricard J., Noat G., Got C., Borel M. The theory of alternative substrates in enzyme kinetics and its application to yeast hexokinase. Eur J Biochem. 1972 Nov 21;31(1):14–24. doi: 10.1111/j.1432-1033.1972.tb02494.x. [DOI] [PubMed] [Google Scholar]
- WEBB E. C., MORROW P. F. The activation of an arysulphatase from ox liver by chloride and other anions. Biochem J. 1959 Sep;73:7–15. doi: 10.1042/bj0730007. [DOI] [PMC free article] [PubMed] [Google Scholar]


