Abstract
Sequence analysis of the Xestia c-nigrum granulovirus (XcGV) genome identified an open reading frame encoding a 469-amino-acid (54-kDa) protein with over 30% amino acid sequence identity to a region of about 150 amino acids that includes the catalytic domains of human stromelysin 1 (Str1)/matrix metalloproteinase 3 (MMP-3) (EC 3.4.24.17) and sea urchin hatching enzyme (HE). Stromelysin homologs have not been reported from baculoviruses or other viruses. Unlike human Str1 and sea urchin HE, the putative XcGV-MMP does not have a signal peptide and lacks the peptide motif involved in the cysteine switch that maintains other MMPs in an inactive form. The putative XcGV-MMP, however, possesses a conserved zinc-binding motif in its putative catalytic domain. The XcGV-MMP homolog was cloned, and a recombinant Bombyx mori nucleopolyhedrovirus (BmNPV) that expresses XcGV-MMP under the polyhedrin promoter was constructed. A distinct pattern of melanization was observed in B. mori larvae infected with MMP-expressing BmNPV. Fat body extracts from larvae overexpressing the 54-kDa recombinant MMP digested dye-impregnated collagen (Azocoll). The enzymatic activity was inhibited by two metalloproteinase inhibitors, EDTA and 1,10-phenanthroline. These results suggest that the XcGV MMP-3 gene homolog encodes a functional metalloproteinase.
The family Baculoviridae is composed of two genera, nucleopolyhedroviruses (NPV) and granuloviruses (GV). Xestia c-nigrum granulovirus (XcGV) has a wide host range and is highly infectious in at least six agriculturally important noctuid species (5). Recently, the entire 178,733-bp sequence of the XcGV circular DNA genome was determined and 181 putative open reading frames (ORFs) were identified (10). Using computer-assisted homology searches, one of the XcGV ORFs, ORF40, showed significant sequence identity to human stromelysin 1 (Str1) (10). A stromelysin homolog has not been identified in the genomes of previously sequenced baculoviruses (1, 2, 4, 10, 11, 15) or in other viruses. Stromelysins are one group of matrix metalloproteinases (MMPs). The MMPs are neutral proteinases that require Zn2+ and Ca2+ for their enzymatic activity. They are classified into at least four superfamilies based on their substrate specificity, primary structure, and cellular localization: the collagenases, gelatinases, stromelysins, and membrane-type MMPs (33). Human Str1 is a member of the stromelysin superfamily, which includes the well-studied sea urchin hatching enzyme (HE) from Paracentrotus lividus (17). Str1 is a stromal proteinase, which can degrade a variety of extracellular matrix substrates. It also promotes mammary carcinogenesis (19, 30). The P. lividus HE is secreted by the blastula stage of the embryo to digest the extracellular egg coat (13, 25). This process is necessary for embryo development.
Two types of proteinases have been reported in baculoviruses. Trichoplusia ni GV-enhancing factor (enhancin) possesses a zinc-binding motif and requires zinc or calcium ions for its proteinase activity (18). Enhancin is associated with occlusion bodies, and evidence suggests that it digests the peritrophic membrane of insect midgut tissue to facilitate virus infection. In contrast, cysteine proteinases have also been identified in many baculoviruses, and they presumably play a role in the breakdown of infected host tissues to facilitate horizontal transmission of the virus (12, 26). In addition to cysteine proteinases, baculoviruses possess chitinases to digest chitin for degradation of host insect larvae (8, 9, 31).
Most studies of baculovirus proteases have been done with NPVs, since cell lines are not available for efficient propagation and purification of GVs. In this report, we describe the primary structure and proteinase activity of XcGV-MMP expressed by recombinant Bombyx mori nucleopolyhedrovirus (BmNPV) in B. mori larvae.
MATERIALS AND METHODS
Viruses, cell line, and insects.
The XcGV clone (alpha-4) was isolated and provided by C. Goto. XcGV was propagated in early-third-instar Pseudaletia separata larvae as described previously (6). XcGV granules were collected from XcGV-infected fat body tissues as described previously (6). Wild-type (WT) BmNPV (T3 strain) (21), BmNPV-abb (a transfer virus) (34), BmCysPD (26), BmMMP, and BmMMPCysPD were propagated in BmN (BmN-4) cells. The BmN cell line was maintained at 28°C in TC-100 medium supplemented with 10% fetal bovine serum (20). Larvae of B. mori and P. separata were reared on an artificial diet at 28°C.
Preparation of recombinant BmNPV.
The recombinant viruses were constructed by standard methods described previously (20). The mmp gene coding region was amplified by PCR as described by Zhou et al. (34) using primers MMP1 (5′-GGAGATCTATGAACGATACGTACGAA-3′) (GG-BglII site-XcGV genome, bp 32935 to 32918) and MMP2 (5′-GGTCTAGATTAACAGTGATCTAGTAATC-3′ (GG-XbaI site-XcGV genome, bp 31526 to 31545) and XcGV genomic DNA as a template. The amplified DNA fragment was digested with restriction endonucleases and inserted at the linker sites (BglII and XbaI) of the BmNPV transfer vector pBm31, a derivative of pBm030 (34). The transfer vector consisted of pUC-derived Escherichia coli vector and a 2,845-bp HpaI fragment corresponding to BmNPV genome positions bp 127831 to 2994, in which the ORF region of the polyhedrin gene is replaced by pBm030 multiple cloning sites (4, 34). XcGV-MMP was expressed under the BmNPV polyhedrin promoter in occlusion-negative virus. Cotransfection of pBm31-MMP with BmNPV-abb was performed using Lipofectin as specified by the manufacturer (GIBCO-BRL). The BmNPV cysteine proteinase gene was deleted from BmMMP by cotransfection with a Drosophila hsp70 promoter and β-galactosidase gene cassette as described by Ohkawa et al. (26), generating BmMMPCysPD. Disruption of the cysteine proteinase gene was confirmed by PCR with genomic DNA extracted from XcGV granules using the primers Cyspro1 (5′-GTCTTAATTTTAAGATGTAA-3′) and Cyspro2 (5′-TAATAAATGACTGCAGTAG-3′). Cyspro1 and Cyspro2 hybridize 20 to 40 bp upstream and at the 3′ end, respectively, of the endogenous cysteine proteinase ORF of BmNPV.
Protein expression of XcGV-MMP in E. coli and antibody preparation.
An N-terminal His6-tagged fusion construct of the XcGV-MMP was expressed in E. coli BL21(DE3) (Novagen, Madison, Wis.) and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Atto) and purified using an electric eluter (Bio-Rad, Hercules, Calif.). Rabbit polyclonal XcGV-MMP antiserum was prepared as described by Okano et al. (27). The coding region of XcGV-MMP (10) was PCR amplified from XcGV genomic DNA using primers 5′-GTGCTAGCATGAACGATACGTACGAA-3′ and 5′-GAGAATTCACAGTGATCTAGTAATCG-3′, which generate sites suitable for cloning. The PCR-amplified DNA fragment was digested with NheI and EcoRI and subcloned into the NheI and EcoRI sites of the expression vector pET-28a(+) (Novagen). The resulting plasmid was transformed in E. coli BL21(DE3). Transformed E. coli cells were harvested 3 h after induction with 1 mM isopropyl-d-thiogalactopyranoside (IPTG). The overexpressed recombinant MMP (His-tagged rMMP) was purified using a Mini Whole Gel Eluter (Bio-Rad) as specified by the manufacturer. The purified His-tagged rMMP was digested with thrombin, and its amino acid sequence was confirmed using a 477A peptide sequencer (Applied Biosystems, Foster City, Calif.). The purified His-tagged rMMP was subcutaneously injected into a rabbit with complete Freund's adjuvant for the initial injection and incomplete Freund's adjuvant for subsequent injections (with 2 to 3 weeks between injections). Rabbit antiserum was collected 1 week after the third injection and tested by Western blot analysis (27).
SDS-PAGE and Western blotting.
SDS-PAGE with 12% polyacrylamide gels was performed as described by Laemmli (16). The gels were either fixed or stained with Coomassie brilliant blue or electrophoretically transferred to a polyvinylidene difluoride membrane using a semidry-blot apparatus (Atto) as specified in the manufacturer's guidelines. Western blots were probed with rabbit polyclonal antiserum diluted 1:3,000, washed, incubated with a 1:2,000 dilution of goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase (Zymed Laboratories, Inc., South San Francisco, Calif.), and developed using a 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium detection system (Vector Laboratories, Inc., Burlingame, Calif.) as specified by the manufacturer.
Infection of B. mori larvae with recombinant BmNPVs.
Fifth-instar B. mori (1 day postecdysis) were injected with approximately 2 × 105 PFU of wild-type or recombinant BmNPVs as described by Ohkawa et al. (26). The effects of XcGV-MMP expression were observed and photographed at 12-h intervals until 1 day after death.
Cysteine proteinase assay.
XcGV-MMP activity was assayed under standard conditions for azo dye-impregnated collagen (Azocoll) (Sigma Chemical Co., St. Louis, Mo.) hydrolysis as described by Chavira et al. (3). Fat bodies (FB) dissected from BmMMPCysPD-infected B. mori larvae were collected, washed three times with phosphate-buffered saline (PBS), resuspended in 50 mM Tris-HCl (pH 7.8)–1 mM CaCl2, and homogenized using a Dounce homogenizer. The homogenate was centrifuged at 9,000 × g, and the supernatant was collected. A volume of supernatant of FB extract containing 1 mg of protein was assayed in 1 ml of Azocoll solution (5 mg/ml in 50 mM Tris-HCl [pH 7.8]–1 mM CaCl2). A 1-ml volume of the mixture was placed in a 1.5-ml microcentrifuge tube and centrifuged at 10,000 × g for 5 min. The absorbance at 520 nm (A520) of the supernatant was measured and subtracted from the A520 of a simultaneously incubated blank (Azocoll supernatant without the added protein). The proteinase activity was assayed in triplicate at least. To calculate collagen digestion activity, collagenase type IV (EC 3.4.24.3) from Clostridium histolyticum (Sigma) was used as a standard. One collagen digestion unit liberates peptides from collagen equivalent in ninhydrin color to 1.0 μmol of leucine in 5 h at pH 7.4 and 37°C in the presence of calcium ions (Sigma).
Fractionation of FB extracts using ammonium sulfate precipitation.
Aliquots of FB extracts were fractionated with the addition of saturated ammonium sulfate solution in a volume equal to 20% of the total volume (7). The precipitated FB extracts were collected and dissolved in 50 mM Tris-HCl buffer (pH 7.8) containing 1 mM CaCl2. Fractions precipitated in 40, 60, and 80% saturated ammonium sulfate solution were subsequently collected and were dissolved in 50 mM Tris-HCl buffer (pH 7.8) containing 1 mM CaCl2. Aliquots of fractions containing approximately 100 μg of protein were separated by SDS-PAGE, and MMP was detected by using antibody against His-tagged rMMP expressed in E. coli. The relative intensities of the Western blot signals were measured using NIH-Image version 1.62.
Immunohistochemistry and confocal microscopy.
BmNPV- or mock-infected BmN cells were immunostained with the XcGV-MMP antibody as previously described by Okano et al. (27). The BmN cells were fixed in 2% formalin, incubated with rabbit anti-XcGV rMMP serum (1:100 dilution in PBS containing 1% fetal bovine serum) washed four times with PBS, and treated with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (1:200 dilution; Cappel, Aurora, Ohio). The cells were mounted with a Slow Fade light antifade kit (Molecular Probes, Eugene, Oreg.) and analyzed with a laser confocal microscope (a TCS NT instrument equipped with an Ar-Kr laser; Leica, Heidelberg, Germany).
RESULTS
A putative XcGV metalloproteinase.
XcGV-MMP (ORF40) is 1,407 nucleotides long and encodes a protein of 469 amino acids (aa) with a predicted molecular mass of 54 kDa. In contrast to all other MMPs except human MMP-23 (33), no N-terminal signal sequence was identified in XcGV-MMP by computer analysis using the algorithm developed by McGeoch (23). Further analysis of the deduced amino acid sequence revealed significant similarity to various MMPs, with the highest identities to human Str1/MMP-3 (P = 5.1 × 10−23) and sea urchin P. lividus HE (P = 2.8 × 10−23). The identities were highest in the putative catalytic domain of the sequence (Fig. 1). The putative catalytic domain, including the zinc-binding signature HEXGHXXGXXHS of XcGV-MMP, was 35 and 32% identical to the catalytic domains of human Str1 (aa 100 to 264) (29) and sea urchin HE (aa 171 to 337) (17), respectively. Human Str1 and sea urchin HE possess P-R-C-G-(V/N)-P-D, a sequence involved in cysteine switching. A complex consisting of a cysteine residue in the switching domain and the essential zinc atom in the catalytic domain of the proenzyme blocks the active site of Str1. The blocked or latent Str1 can be activated by multiple means, which involve the dissociation of a cysteine residue from the complex (32). This switching phenomenon is thought to convert Str1 from an inactive latent form to an active form. Although the P-R-C-G-(V/N)-P-D sequence is conserved in the other known MMPs (32), this switching domain was not found in the XcGV-MMP. Most MMPs, except matrilysin (MMP-7), human MMP-23 (24), and MMPs from Arabidopsis thaliana (22), possess hemopexin-like repeats in the carboxyl terminal that are connected by a proline-rich linker. The gelatinases (MMP-2 and MMP-9) also have a fibronectin type II domain inserted in their catalytic domain. The hemopexin-like repeats and fibronectin type II domain are inferred to play a role in MMP substrate binding. Neither hemopexin-like repeats nor a fibronectin type II domain was identified in XcGV-MMP. Instead of a proline-rich region, we observed a threonine- and arginine-rich region in XcGV-MMP and sea urchin HE.
FIG. 1.
Amino acid sequence alignment of XcGV-MMP, human Str1, and P. lividus HE using the program Clustal W. The box encloses the consensus zinc-binding motif. The P-R-C-G-(V/N)-P-D sequence, presumably involved in the cysteine switch, is underlined. A hemopexin-like motif and a proline-rich region of Str1 are underlined with double and dotted lines, respectively. White letters within black boxes indicate identical amino acid residues, and shaded boxes denote conserved substitutions. Dashes indicate gaps in the sequence. Amino acid numbers are indicated on the left. The human Str1/MMP-3 sequence and sea urchin HE accession numbers are U78045 and S12805, respectively. The protein motifs were analyzed using the PROSITE motif database.
Effects of metalloproteinase expressed in B. mori larvae.
A recombinant BmNPV BmMMP, carrying XcGV mmp under the polyhedrin promoter, was constructed to purify and characterize the putative metalloproteinase. The endogenous cysteine proteinase gene of BmMMP was disrupted by insertion of a Drosophila hsp70 promoter-lacZ gene cassette generating BmMMPCysPD in order to avoid any effect of the cysteine proteinase of BmNPV (26). BmMMPCysPD was purified by a plaque assay (identified by blue plaques), and the hsp70-lacZ insertion was confirmed by PCR using virus genomic DNA as a template. PCR amplification of WT BmNPV DNA using primers Cyspro1 and Cyspro2 produced a 1-kb product. The PCR product amplified from the corresponding regions of BmMMPCysPD and BmCysPD containing the 3.5-kb hsp-lacZ cassette was about 4.5 kb, which indicates that the cysteine proteinase was disrupted (data not shown).
Fifth-instar B. mori larvae injected with BmNPV, BmCysPD, or BmMMPCysPD typically died between days 5 and 6 postinfection (p.i.). As reported by Ohkawa et al. (26), the epidermis of larvae injected with WT virus turned black and limp within 1 day after death whereas that of larvae injected with BmCysPD continued to be firm and white. Larvae infected with BmMMPCysPD also turned black 1 day after death; however, the epidermis was as firm as in larvae infected with BmCysPD. In addition, melanization was observed on the dorsal and ventral sides of BmMMPCysPD-infected larvae (Fig. 2). Significant differences in 50% lethal dose and length of time from infection to death were not observed between BmNPV, BmCysPD, and BmMMPCysPD in B. mori larvae (data not shown).
FIG. 2.
B. mori larvae infected with WT or recombinant BmNPVs 24 h after death. The larvae were injected with approximately 2 × 105 PFU of a viral suspension containing 0.6 mg of kanamycin per ml. Mock, mock-infected larvae; WT, BmNPV-infected larvae; 1, BmCysPD-infected larvae; 2, BmMMPCysPD-infected larvae. (A) Dorsal side. (B) Ventral side.
Western blot analysis of XcGV-MMP expressed by BmNPV.
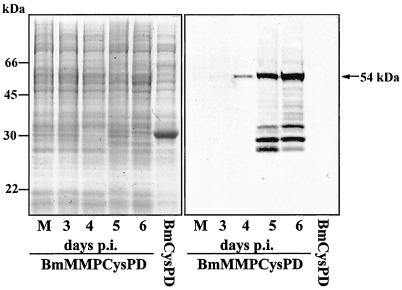
The polyclonal antibody raised against His-tagged XcGV rMMP does not cross-react with proteins from mock-infected BmN cells or FB extracted from the WT-infected larvae (Fig. 3, right panel). However, a major band of 54 kDa, the predicted size of MMP, was observed from days 4 to 6 p.i. (Fig. 3, right panel). Several bands smaller than 54 kDa were observed in FB collected 5 and 6 days p.i. The 54-kDa immunoreactive band was not detected in the hemolymph of the BmMMPCysPD-infected larvae (data not shown). According to the Western blot analysis, there was approximately 100 μg of rMMP per 20 mg of FB protein in BmMMPCysPD-infected B. mori larva on day 5 p.i. In contrast, less than 10 μg of rMMP was detected from 20 mg (3 × 107 cells) of BmMMPCysPD-infected BmN cells at 72 h p.i. (data not shown). Therefore, rMMP was produced in B. mori FB at a rate more than 10 times higher than that in BmN cells.
FIG. 3.
Coomassie brilliant blue staining (left panel) and Western blot analysis of the XcGV-MMP expressed by BmNPV using antibody against His-tagged rMMP (right panel). Aliquots of FB tissue containing approximately 100 μg of protein were collected from mock- or recombinant BmNPV-infected B. mori larvae and separated by SDS-PAGE. BmMMPCysPD, FB of BmMMPCysPD-infected B. mori larvae 3, 4, 5, and 6 days p.i.; BmCysPD, FB of BmCysPD-infected B. mori larvae collected 6 days p.i.
Proteinase activity of XcGV-MMP.
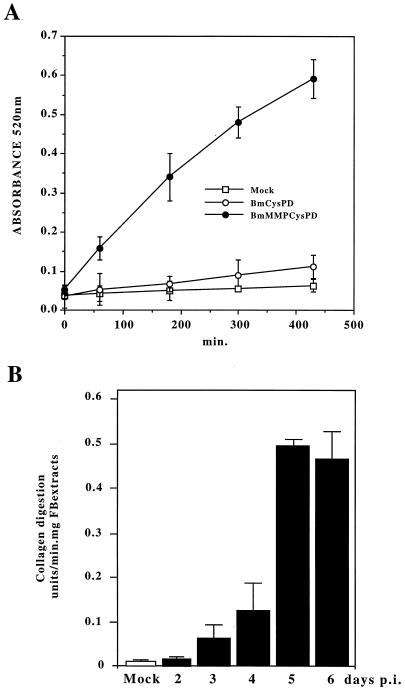
Proteinase activity was assayed with dye-impregnated collagen (Azocoll), a general proteinase substrate. FB protein extracts (1 mg) collected from mock-, BmCysPD-, or BmMMPCysPD-infected larvae were each added to 1 ml of Azocoll solution (5 mg/ml) for the proteinase assay. The A520 increases when Azocoll is digested by proteinases. As shown in Fig. 4A, mock-infected B. mori larvae showed negligible activity. A significant increase of A520 was observed when FB extracts from BmMMPCysPD-infected larvae were assayed. About 8.8-fold less proteinase activity was observed in FB extracts from BmCysPD-infected larvae. The MMP activity decreased after 180 min of incubation at 37°C. Therefore, absorbance was measured at least three times from 0 to 180 min after the FB extract was added, in order to calculate the absolute units of collagen digestion. The Azocoll digestion activity of collagenase from Clostridium histolyticum was used to generate a standard. The presence of 10 μg of collagenase type IV (429 U/mg) increased the A520 by approximately 0.8 unit in 1 h. Thus, 1 U = 0.0031 A520 unit/min was employed for calculating collagen digestion units.
FIG. 4.
Proteinase activity of recombinant XcGV-MMP expressed by BmNPV. (A) Increased A520 due to Azocoll digestion with the FB extracts from mock-, BmCysPD-, or BmMMPCysPD-infected B. mori larvae. FB extracts were assayed for proteinase activity in Azocoll solution. (B) MMP activity during infection. FB dissected from mock-infected, BmCysPD-infected (5 days p.i.), or BmMMPCysPD-infected (2, 3, 4, 5, or 6 days p.i.) larvae were assayed for proteinase activity. A 1-mg portion of FB protein was added to the Azocoll solution for the proteinase assay. Error bars indicate the range of triplicate values.
Proteinase activity was assayed with the extracts of FB dissected from BmMMPCysPD-infected larvae at 2, 3, 4, 5, and 6 days p.i. The proteinase activity of FB extracts increased gradually until it reached a maximum on day 5 p.i. (Fig. 4B). To determine whether this was metalloproteinase activity, EDTA, a chelating agent, and 10 mM 1,10-phenanthroline were added to the enzyme reaction mixture. In the presence of 10 mM or more EDTA (the experiment was not done with less than 10 mM), the proteinase activity was approximately 7% without addition of the inhibitor. Approximately 90% of the proteinase activity was inhibited by 1,10-phenanthroline. In contrast, there was a less than 5% decrease after the addition of 2.8 × 10−4 M E-64, a cysteine proteinase inhibitor, or 5 mM phenylmethylsulfonyl fluoride (PMSF), a serine proteinase inhibitor. The same level of inhibition was observed with either 10 mM EGTA or 10 mM 1,10-phenanthroline (Table 1). This suggested that XcGV-MMP requires a metal(s) for enzymatic activity, like other MMPs (24).
TABLE 1.
Effects of proteinase inhibitors on XcGV-MMP activity
| Virus | Inhibitor(s) | Collagen digestion units/min/mg of FB extracta |
|---|---|---|
| None (mock) | —b | 0.013 ± 0.001 |
| BmCysPD | — | 0.048 ± 0.003 |
| BmMMPCysPD | — | 0.475 ± 0.018 |
| EDTA (10 mM) | 0.032 ± 0.007 | |
| 1,10-Phenanthroline (10 mM) | 0.048 ± 0.005 | |
| E-64 (2.8 × 10−4 M) | 0.460 ± 0.012 | |
| PMSF (5 mM) | 0.453 ± 0.010 |
FB dissected from mock-, BmCysPD-, or BmMMPCysPD-infected larvae 5 days p.i. were assayed for MMP activity. Results are given as mean ± standard deviation.
—, no inhibitor was applied to the Azocoll proteinase assay solution.
Proteinase activities of the ammonium sulfate-precipitated FB fractions.
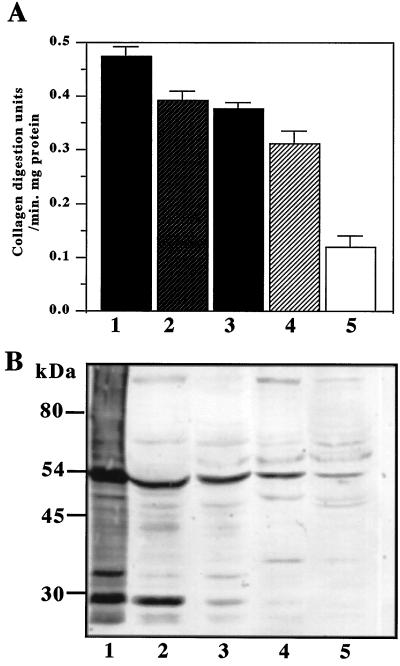
The proteinase activity of each fraction collected by ammonium sulfate precipitation was assayed by Azocoll digestion (Fig. 5A). Western blot analysis revealed that approximately equal amounts of 54- and 30-kDa proteins reacting with anti-His rMMP were present in the 20% ammonium sulfate precipitate fraction (Fig. 5B). The enzyme activity of the 20 to 40% ammonium sulfate fraction was almost the same as that of the 20% precipitate, whereas the amount of 30-kDa protein was less than one-third that of the 20% precipitate. In the 40 to 60% ammonium sulfate fraction, the proteinase activity was 75% that of the 20% ammonium sulfate fraction and no 30-kDa protein was detected.
FIG. 5.
Ammonium sulfate fractionation of FB extract containing recombinant XcGV-MMP expressed by BmMMPCysPD. (A) Proteinase activity of the FB fractions collected from ammonium sulfate precipitation. 1, FB extract; 2, 0 to 20% saturated ammonium sulfate fraction; 3, 20 to 40% saturated ammonium sulfate fraction; 4, 40 to 60% saturated ammonium sulfate fraction; 5, 60 to 80% saturated ammonium sulfate fraction. (B) Western blot analysis of the ammonium sulfate fractions. The sample numbers in panel A correspond to the sample numbers in panel B. The relative intensities of the bands reacting with MMP antibody were as follows: 1, 185.77; 2, 92.93; 3, 64.97; 4, 47.67; and 5, 19.48 (for the 54-kDa band); and 1, 153.57; 2, 90.11; 3, 13.02; 4, 2.97; and 5, 0 (for the 30-kDa band). FB extract was prepared from FB collected from BmMMPCysPD-infected B. mori larvae 5 days p.i.
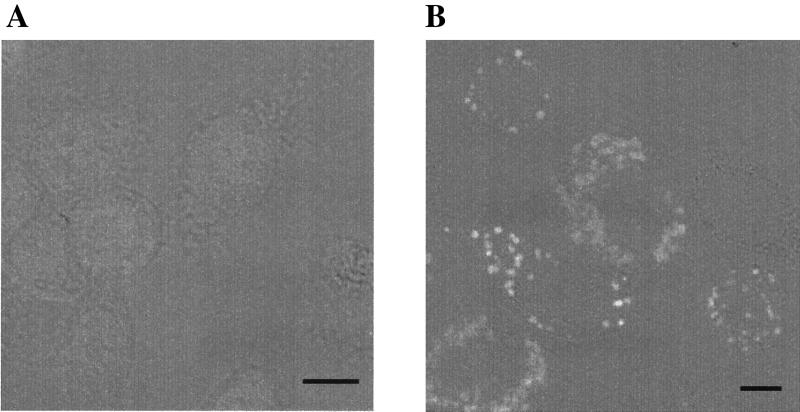
Localization of XcGV-MMP in BmN cells by immunofluorescence analysis.
Mock-infected BmN cells were not stained by the XcGV-MMP-specific antibody (Fig. 6A). Until 24 h p.i., no XcGV-MMP expression was observed in BmMMPCysPD-infected BmN cells (data not shown). By 36 h p.i., however, specific globular foci of staining were evident in the cytoplasm (Fig. 6B). The same staining patterns were observed at 48 h p.i. (data not shown). Localization of XcGV-MMP to either the nuclear or cytoplasmic membranes was not observed in infected BmN cells. In addition, XcGV-MMP was not detected in the concentrated cell medium in the Western blot analysis (data not shown).
FIG. 6.
Immunofluorescent staining of BmN cells. The cells were infected with BmMMPCysPD (multiplicity of infection, 10) or mock infected with medium. The cells were immunostained as described in Materials and Methods and examined under a confocal laser-scanning microscope. (A) Transmitted light and immunofluorescence image of mock-infected cells stained with fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G polyclonal antiserum (1:200). (B) Image of BmMMPCysPD-infected BmN cells at 36 h p.i., stained as described for panel A. Bar, 10 μm.
DISCUSSION
XcGV-MMP is a unique baculovirus proteinase.
Sequence analysis of the entire XcGV genome identified a gene encoding a protein with significant homology to MMPs. The highest homology was to human Str1/MMP-3 and a P. lividus HE. There was about 35% identity localized to the putative catalytic domain (aa 66 to 226), whereas the identity at both the N- and C-terminal regions was less than 10% (Fig. 1). XcGV-MMP lacks the recognizable signal sequence present at the N-terminal ends of all previously characterized MMPs except MMP-23 (33), suggesting that this novel enzyme is not exported and may function in an intracellular compartment. In addition, a conserved, unique P-R-C-G-(V/N)-P-D sequence found in the propeptide domain of other MMPs is not present in XcGV-MMP (Fig. 1). MMPs can be present in an inactive (latent) conformation, which is the result of formation of an intramolecular complex between the single cysteine residue in the propeptide domain and an essential zinc atom in the catalytic domain. This complex blocks the active site (32). Disruption of the Cys-zinc bond by limited proteolysis or conformational perturbations opens or unblocks the catalytic site, which leads to subsequent autocatalytic cleavage, eventually resulting in the generation of a catalytically competent enzyme (32). The absence of the conserved P-R-C-G-(V/N)-P-D sequence in XcGV-MMP, in contrast to other MMPs (Fig. 1), suggests two hypotheses: (i) these consensus residues [P-R-C-G-(V/N)-P-D] are not essential for functioning of the Cys-switch mechanism in all MMPs, and (ii) unlike other MMPs, XcGV-MMP does not require the activation process. The Western blot analysis and assay of proteinase activity in the fractions of FB extracts separated by ammonium sulfate precipitation indicated that the enzyme activity was proportional to the amount of 54-kDa protein. The 54-kDa MMP had proteinase activity in the absence of the 30-kDa protein, which was detected by anti-His-tagged XcGV rMMP as a major band on Western blots, and other smaller proteins. Therefore, we concluded that the 54-kDa protein is an active form and that processing of XcGV-MMP is not required for enzyme activation. This suggests that XcGV-MMP is regulated in a novel manner compared to other MMPs that use cysteine switching to regulate their activity.
Since a cell line is not available to efficiently propagate and purify XcGV, it is very difficult to produce a gene deletion mutant of XcGV. To analyze the function of XcGV-MMP, a recombinant BmNPV carrying the MMP gene under the BmNPV polyhedrin promoter was constructed. Furthermore, the endogenous cysteine proteinase of BmNPV was deleted to avoid the influence of the BmNPV cysteine proteinase during the in vivo observations and activity assays with Azocoll. FB extracts of BmCysPD-infected larvae showed essentially no proteinase activity in the Azocoll assay system. Similar results were previously observed with the extracts of BmCysPD-infected BmN cells. This indicated that deletion of cysteine proteinase from BmNPV eliminated proteinase activity (26).
Melanization due to hemocyte aggregation was observed on the surface of Drosophila melanogaster tissues in which the basement membrane was damaged (28). Similar melanization due to XcGV-MMP expression is observed in B. mori larvae after death. Hence, it is possible that XcGV-MMP damages the basement membranes of the host B. mori and triggers widespread hemocyte aggregation followed by melanization. It is also possible that XcGV-MMP activates cuticular phenoloxidase directly or indirectly. The Western blot analysis did not detect XcGV-MMP in the hemolymph of larvae during infection. We speculate that the MMP is released from FB cells after death and digestion of the basement membranes triggers melanization of the larvae.
The proteinase activity of FB extracts was assayed with Azocoll, which is commonly used for insect proteinase assays (14, 26). FB from BmCysPD-infected larvae showed slightly higher enzyme activity than did those from mock-infected larvae, which may be due to an FB proteinase activated by BmNPV infection. The proteinase activity of BmMMPCysPD was dramatically decreased with the chelating agents 10 mM EDTA and 10 mM 1,10-phenanthroline, indicating that XcGV-MMP requires a metal(s) cofactor for enzymatic activity.
Function of XcGV-MMP in insects.
Unlike enhancin, a metalloproteinase found in GV occlusion bodies that may be involved in enhancing NPV infection (18), XcGV-MMP is not found in occlusion bodies extracted from XcGV-infected P. separata larvae (data not shown), implying that the role of the MMP is not the enhancement of viral infection. The results obtained from expression of the MMP in B. mori larvae implied that XcGV-MMP has a function different from that of cysteine proteinase. In contrast to cysteine proteinase, which digests the larval epidermis, XcGV-MMP may destroy the basement membranes that help hold the tissue together. The pattern of XcGV-MMP localization in BmN cells was similar to the localization of Autographa californica nucleopolyhedrovirus (AcNPV) chitinase in Sf9 cells previously reported by Thomas et al. (31). They reported that with AcNPV chitinase, which possesses a signal peptide, an endoplasmic reticulum retention signal (KDEL) was localized to the endoplasmic reticulum membrane and vacuoles. Unlike AcNPV chitinase, XcGV-MMP seems to have neither a signal peptide nor an ER retention signal. Thus, it is likely that XcGV-MMP may pool in a specific organelle in insect cells and be released after cell lysis. It may be advantageous for the virus to delay release of the proteinase to digest the extracellular matrix of the host insects until maximal virus proliferation has occurred. Cysteine proteinase and chitinase proteins are thought to degrade larval tissues in order to facilitate horizontal virus transmission. Although XcGV also possesses homologs of these proteins (10), the additional metalloproteinase may destroy the basement membranes that help hold tissues together to assist in dissemination of the virus in a greater range of hosts and under various environmental conditions.
ACKNOWLEDGMENTS
We thank Masaaki Kurihara for help with the culture of BmN cells and purification of recombinant viruses. We thank Shogo Matsumoto for sequencing the recombinant MMP peptide expressed in E. coli. We thank George F. Rohrmann and Shizuo G. Kamita for critical reading of the manuscript.
This research was partially supported by grants from the USDA (9802852), the Core Research for Evolutional Science and Technology (CREST) Project, JST, Japan, and the Special Postdoctoral Researchers Program, RIKEN.
Footnotes
Dedicated to the memory of Susumu Maeda.
REFERENCES
- 1.Ahrens C H, Russell R L, Funk C J, Evans J T, Harwood S H, Rohrmann G F. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. doi: 10.1006/viro.1997.8448. [DOI] [PubMed] [Google Scholar]
- 2.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 3.Chavira R, Jr, Burnett T J, Hageman J H. Assaying proteinases with azocoll. Anal Biochem. 1984;136:446–450. doi: 10.1016/0003-2697(84)90242-2. [DOI] [PubMed] [Google Scholar]
- 4.Gomi S, Majima K, Maeda S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol. 1999;80:1323–1337. doi: 10.1099/0022-1317-80-5-1323. [DOI] [PubMed] [Google Scholar]
- 5.Goto C, Hayakawa T, Maeda S. Genome organization of Xestia c-nigrum granulovirus. Virus Genes. 1998;16:199–210. doi: 10.1023/a:1007972108026. [DOI] [PubMed] [Google Scholar]
- 6.Goto C, Minobe Y, Iizuka T. Restriction endonuclease analysis and mapping of the genomes of granulosis viruses isolated from Xestia c-nigrum and five other noctuid species. J Gen Virol. 1992;73:1491–1497. doi: 10.1099/0022-1317-73-6-1491. [DOI] [PubMed] [Google Scholar]
- 7.Green A A, Hughes W L. Protein fractionation on the basis of solubility in aqueous solutions of salts and organic solvents. Methods Enzymol. 1955;1:68–96. [Google Scholar]
- 8.Hawtin R E, Arnold K, Ayres M D, Zanotto P M, Howard S C, Gooday G W, Chappell L H, Kitts P A, King L A, Possee R D. Identification and preliminary characterization of a chitinase gene in the Autographa californica nuclear polyhedrosis virus genome. Virology. 1995;212:673–685. doi: 10.1006/viro.1995.1525. [DOI] [PubMed] [Google Scholar]
- 9.Hawtin R E, Zarkowska T, Arnold K, Thomas C J, Gooday G W, King L A, Kuzio J A, Possee R D. Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology. 1997;238:243–253. doi: 10.1006/viro.1997.8816. [DOI] [PubMed] [Google Scholar]
- 10.Hayakawa T, Ko R, Okano K, Seong S I, Goto C, Maeda S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology. 1999;262:277–297. doi: 10.1006/viro.1999.9894. [DOI] [PubMed] [Google Scholar]
- 11.Ijkel W F, van Strien E A, Heldens J G, Broer R, Zuidema D, Goldbach R W, Vlak J M. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J Gen Virol. 1999;80:3289–3304. doi: 10.1099/0022-1317-80-12-3289. [DOI] [PubMed] [Google Scholar]
- 12.Kang W, Tristem M, Maeda S, Crook N E, O'Reilly D R. Identification and characterization of the Cydia pomonella granulovirus cathepsin and chitinase genes. J Gen Virol. 1998;79:2283–2292. doi: 10.1099/0022-1317-79-9-2283. [DOI] [PubMed] [Google Scholar]
- 13.Kato K H, Takemoto K, Kato E, Miyazaki K, Kobayashi H, Ikegami S. Inhibition of sea urchin fertilization by jaspisin, a specific inhibitor of matrix metalloendoproteinase. Dev Growth Differ. 1998;40:221–230. doi: 10.1046/j.1440-169x.1998.00011.x. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi M, Mori H, Yaginuma T. Stimulation of acid protease activity in the isolated pupal abdomens of the silkworm, Bombyx mori, infected with nuclear polyhedrosis virus. J Invertebr Pathol. 1985;46:202–204. doi: 10.1016/0022-2011(90)90032-2. [DOI] [PubMed] [Google Scholar]
- 15.Kuzio J, Pearson M N, Harwood S H, Funk C J, Evans J T, Slavicek J M, Rohrmann G F. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lepage T, Gache C. Purification and characterization of the sea urchin embryo hatching enzyme. J Biol Chem. 1989;264:4787–4793. [PubMed] [Google Scholar]
- 18.Lepore L S, Roelvink P R, Granados R R. Enhancin, the granulosis virus protein that facilitates nucleopolyhedrovirus (NPV) infections, is a metalloprotease. J Invertebr Pathol. 1996;68:131–140. doi: 10.1006/jipa.1996.0070. [DOI] [PubMed] [Google Scholar]
- 19.Linder C, Bystrom P, Engel G, Auer G, Aspenblad U, Strander H, Linder S. Correlation between basic fibroblast growth factor immunostaining of stromal cells and stromelysin-3 mRNA expression in human breast carcinoma. Br J Cancer. 1998;77:941–945. doi: 10.1038/bjc.1998.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda S. Expression of foreign genes in insects using baculovirus vectors. Annu Rev Entomol. 1989;34:351–372. doi: 10.1146/annurev.en.34.010189.002031. [DOI] [PubMed] [Google Scholar]
- 21.Maeda S, Kawai T, Obinata M, Fujiwara H, Horiuchi T, Saeki Y, Sato Y, Furusawa M. Production of human alpha-interferon in silkworm using a baculovirus vector. Nature. 1985;315:592–594. doi: 10.1038/315592a0. [DOI] [PubMed] [Google Scholar]
- 22.Maidment J M, Moore D, Murphy G P, Murphy G, Clark I M. Matrix metalloproteinase homologues from Arabidopsis thaliana. Expression and activity. J Biol Chem. 1999;274:34706–34710. doi: 10.1074/jbc.274.49.34706. [DOI] [PubMed] [Google Scholar]
- 23.McGeoch D J. On the predictive recognition of signal peptide sequences. Virus Res. 1985;3:271–286. doi: 10.1016/0168-1702(85)90051-6. [DOI] [PubMed] [Google Scholar]
- 24.Nagase H, Woessner J F., Jr Matrix metalloproteinases. J Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 25.Nomura K, Shimizu T, Kinoh H, Sendai Y, Inomata M, Suzuki N. Sea urchin hatching enzyme (envelysin): cDNA cloning and deprivation of protein substrate specificity by autolytic degradation. Biochemistry. 1997;36:7225–7238. doi: 10.1021/bi9629790. [DOI] [PubMed] [Google Scholar]
- 26.Ohkawa T, Majima K, Maeda S. A cysteine protease encoded by the baculovirus Bombyx mori nuclear polyhedrosis virus. J Virol. 1994;68:6619–6625. doi: 10.1128/jvi.68.10.6619-6625.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okano K, Mikhailov V S, Maeda S. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J Virol. 1999;73:110–119. doi: 10.1128/jvi.73.1.110-119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizki R M, Rizki T M. Hemocyte responses to implanted tissues in Drosophila melanogaster larvae. Roux's Arch Dev Biol. 1980;189:207–213. doi: 10.1007/BF00868679. [DOI] [PubMed] [Google Scholar]
- 29.Saus J, Quinones S, Otani Y, Nagase H, Harris E D, Jr, Kurkinen M. The complete primary structure of human matrix metalloproteinase-3. Identity with stromelysin. J Biol Chem. 1988;263:6742–6745. [PubMed] [Google Scholar]
- 30.Sternlicht M D, Lochter A, Sympson C J, Huey B, Rougier J P, Gray J W, Pinkel D, Bissell M J, Werb Z. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–146. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas C J, Brown H L, Hawes C R, Lee B Y, Min M K, King L A, Possee R D. Localization of a baculovirus-induced chitinase in the insect cell endoplasmic reticulum. J Virol. 1998;72:10207–10212. doi: 10.1128/jvi.72.12.10207-10212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Wart H E, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc Natl Acad Sci USA. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velasco G, Pendas A M, Fueyo A, Knauper V, Murphy G, Lopez-Otin C. Cloning and characterization of human MMP-23, a new matrix metalloproteinase predominantly expressed in reproductive tissues and lacking conserved domains in other family members. J Biol Chem. 1999;274:4570–4576. doi: 10.1074/jbc.274.8.4570. [DOI] [PubMed] [Google Scholar]
- 34.Zhou C E, Ko R, Maeda S. Polyhedron-like inclusion body formation by a mutant nucleopolyhedrovirus expressing the granulin gene from a granulovirus. Virology. 1998;240:282–294. doi: 10.1006/viro.1997.8927. [DOI] [PubMed] [Google Scholar]