Abstract
Although γδ T cells are known to participate in immune dysregulation in solid tumors, their relevance to human microsatellite-stable (MSS) colorectal cancer (CRC) is still undefined. Here, using integrated gene expression analysis and T cell receptor sequencing, we characterized γδ T cells in MSS CRC, with a focus on Vδ1 + T cells. We identified Vδ1+ T cells with shared motifs in the third complementarity-determining region of the δ-chain, reflective of antigen recognition. Changes in gene and protein expression levels suggested a dysfunctional effector state of Vδ1+ T cells in MSS CRC, distinct from Vδ1+ T cells in microsatellite-instable (MSI). Interaction analysis highlighted an immunosuppressive role of fibroblasts in the dysregulation of Vδ1+ T cells in MSS CRC via the TIGIT-NECTIN2 axis. Blocking this pathway with a TIGIT antibody partially restored cytotoxicity of the dysfunctional Vδ1 phenotype. These results define an operative pathway in γδ T cells in MSS CRC.
Subject terms: Innate immune cells, Tumour immunology
Although γδ T cells are known to participate in immune dysregulation in solid tumors, their relevance to human microsatellite-stable (MSS) colorectal cancer (CRC) is less well-studied. Here, using single-cell RNA-sequencing, the authors identify a Vδ1 + T cell subset, which are functionally impaired in MSS CRC via a TIGIT-NECTIN2 interaction.
Introduction
Immune checkpoint blockade has revolutionized cancer treatment1. Colorectal cancer (CRC) can be classified into microsatellite instable (MSI) and microsatellite stable (MSS) cancers that differ in their clinical course and response to treatment. Only MSI CRC benefit from targeting the PD-1/PDL1 or CTLA-4 axis2. Recently, Vδ1 + T cells were found to be highly relevant in advancing anti-tumor effector functions after checkpoint therapy in HLA class I-negative MSI CRC, underpinning the potential of γδ T cells in CRC immunotherapy3. Since MSS CRC comprises the majority of all CRC, the discovery of next-generation immunotherapies for MSS CRC is of critical importance to a large patient group4.
In an attempt to decipher immunologic mechanisms, a large computational study analyzed a broad range of distinct carcinomas including CRC and summarized that the best correlate of overall survival was a γδ T cell gene signature5. As first-line effector cells, γδ T cells recognize infected and transformed cells in infection and cancer rapidly in a major histocompatibility complex-unrestricted manner. On their surface, γδ T cells are positive for the γ and δ chains of the T cell receptor (TCR) in contrast to most other human T cells, which express αβ TCR chains6,7. Different subsets of γδ T cells are defined by varying usage of the TCR δ chain, of which Vδ1 and Vδ3 chains are considered to be expressed by “tissue-resident” γδ T cells, whereas the Vδ2 chain is mainly found on blood T cells8. Their arsenal of cytotoxic functions is still under investigations but γδ T cells have been shown to lyse tumor cells using the perforin-granzyme pathway9. Furthermore, γδ T cells can directly induce apoptosis in cancer cells when engaging Fas ligand (FasL) and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)10. The capacity of γδ T cells to secrete cytokines, such as interferon-γ (IFN-γ) and tumor necrosis factor alpha (TNF-α), and to present antigens11,12 strongly argues for their potential as direct therapeutic targets. Reis et al. performed single-cell RNA- and γδ-TCR sequencing of γδ T cells in human CRC specimens and deciphered in mouse models for CRC that γδ T cells responses depend on differences in their TCR usage13. A recent publication identified Vδ1+ and Vδ3+ cells which expressed markers of conventional T cell exhaustion but maintained effector functions in renal cancer14. In light of the heterogeneity of CRC, it is vital to comprehensively characterize the full spectrum of γδ T cells applicable to the majority of patients with MSS CRC as a prerequisite to target them with immunotherapeutics.
Here, we combined single-cell RNA-sequencing (scRNA-seq), αβ-/γδ-TCR sequencing (scTCRαβ-seq; scTCRγδ-seq) and flow cytometry approaches with functional in vitro assays to identify targetable molecular features of γδ T cells in MSS CRC in comparison to corresponding distant healthy colon (HC) of the same individuals. We observed signs for clonal expansion in γδ T cells of MSS CRC as shared identity patterns of the complementarity-determining region 3 (CDR3) suggested the presence of γδ T cell clones recognizing the same antigens in CRC. Through integrated gene expression analysis, we found Vδ1+ cells from MSS CRC to contribute to tumor immune escape by upregulation of exhaustion-associated and downregulation of effector genes. However, Vδ1+ cells from MSS CRC did not meet all criteria for conventional T cell exhaustion and exhibit clear distinctions in their gene expression compared to exhausted Vδ1+ cells from MSI CRC. Our findings highlighted alterations of Vδ1 + T cells to be – at least partly – orchestrated by distinct immunoregulatory cell subsets of fibroblasts via the TIGIT-NECTIN axis. Finally, by phenotypical and functional profiling, we demonstrated that Vδ1+ cells of MSS CRC displayed restricted capabilities to lyse CRC cell lines which could in part be rescued by activation and blocking of TIGIT.
Results
Favored Vδ1+ usage and expanded CDR3 identity patterns in γδ T cells of MSS CRC
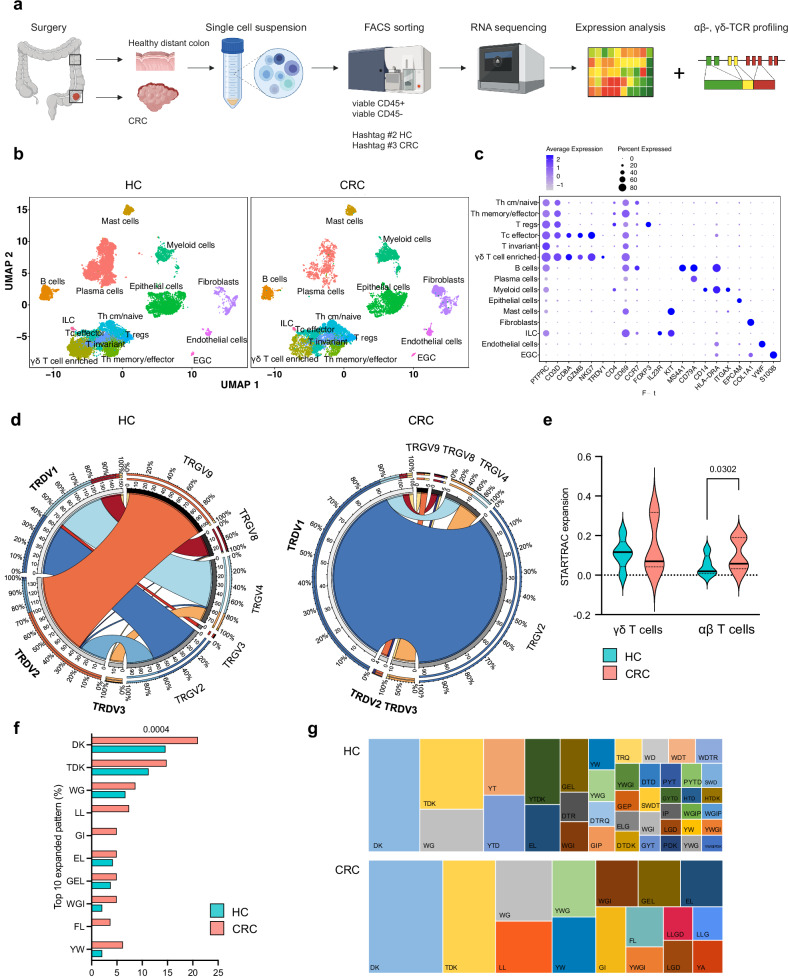
We employed droplet-based 5′ scRNA-seq of tumor samples and distant healthy colon (HC) of 17 patients with treatment-naïve MSS CRC (patient characteristics Supplementary Table S1). Additional αβ-/γδ-TCR sequencing was performed in 7 samples (Fig. 1a). After quality control, we obtained a total of 46,491 cells. Visualization of cells using uniform manifold approximation and projection for dimension reduction (UMAP)15 revealed 30 distinct clusters (Fig. 1b, S1a). Each cluster was represented by 13.9 samples, with a minimum of 7 and a maximum of 17 samples (Fig. S1b). Cluster-specific gene expression profiles and known marker gene expression supported the assignment of clusters, including various conventional and non-conventional T cell subsets, a B cell population (MS4A1 + , CD79a + , CD3D − ), plasma cell subpopulations (all CD79a + , CD3D − ), myeloid cells (CD14 + , HLA-DRA + , ITGAX + ), innate lymphoid cells (ILC; IL23R + , KIT + , CD3D − ) and mast cells (KIT + ). Non-immune cell clusters included EPCAM+ epithelial cells, COL1A1+ fibroblasts, VWF+ endothelial cells and S100B+ enteric glial cells (EGC; Fig. 1c). Gene expression separated subtypes of T cells characterized by CD3D+ expression subdivided into Th cm/naïve cells (CD4 + , CD69 + , CCR7 + ), Th memory/effector cells (CD4 + , CD69 + , CCR7 − ), regulatory T cells (Tregs; CD4 + , FOXP3 + ), cytotoxic effector T cells (Tc effector; CD8A + , GZMB + , NKG7 + , CD69 + ) and invariant T cells (CD3D + , CD69 + , CD8A − , CD4 − ). Within the T cell population, we observed a γδ T cell enriched cell cluster with expression of the Vδ1 chain (TRDV1 + ) which was additionally positive for CD8A, GZMB, NKG7 and CD69. CellTypist16 confirmed curated cluster and cell type assignment and corresponded well with the immune cell clusters (Fig. S1c). We further applied a TCR-based selection strategy, as the TCR provides a reliable molecular tag to track γδ T cells. In order to identify γδ T cells, TCRα/TCRβ and TCRγ/TCRδ clones were overlaid onto UMAP plots (Fig. S1d, Supplementary Table S2), which we further used to perform downstream analyses of the γδ TCR. We next assessed the clonality and subtypes of γδ T cells based on single-cell TCRγδ sequencing (samples 24-30). The number of shared γδ T cell clonotypes calculated between all samples showed 11 overlapping clonotypes within HC and CRC (Fig. S1e). The analysis of γδ T cell clones suggests that γδ T cell subsets in CRC are predominantly characterized by the pairing of TRDV1 with various TRGV (TRGV2, TRGV4, TRGV8, TRGV9). This indicates an altered TCR chain usage pattern within the tumor microenvironment, with a notable skewing towards TRDV1, compared to HC (Fig. 1d). A major fraction of γδ T cells in HC was characterized by pairing of TRDV2 and TRGV9, which may reflect influx of peripheral γδ T cell in HC that may be interrupted in CRC (32.69% HC vs. 15% CRC). We further found a preferred pairing between TRGV2 and TRDV1 as well as TRGV4 and TRDV1 in both HC and CRC (20.06% HC vs. 90% CRC; 18.77% HC vs. 6.67% CRC). This finding is in line with Reis et al. who demonstrated Vδ1 and Vγ4 gene usage by tumor-infiltrating γδ T cells13. We further observed a recurrent usage of TRDV1 and TRGV8 among different individuals in HC and CRC (8.09% HC vs. 2.5% CRC). To address clonal expansion as a sign of antigen recognition of γδ T cells in CRC in comparison to conventional αβ T cells we employed TCR tracking (STARTRAC) indices17. We used γδ T cells and αβ T cells based on their TCR-γδ and -αβ expression and calculated STARTRAC expansion indices (expa). In comparison to αβ T cells which demonstrated a significant increasing index and therefore exhibited signs for clonal expansion, the γδ T cells displayed a smaller index. We were wondering if the observed lack of clonality of γδ T cells in comparison to αβ T cells might be explained by a concept called TCR clustering where distinct clones can recognize the same epitopes based on similar patterns of identity in the CDR3 region18. We used GLIPH2 to assess these shared identity patterns of the δ-chain19. The identity pattern DK was observed in 21% of all CDR3 regions in all γδ T cells clones in CRC (vs. 14.6% in HC), followed by TDK (14.8% vs. 11.3% in HC), WG (8.6% vs. 6.7% in HC), LL (7.4% vs. 0% in HC) and GI (4.9% vs. 0% in HC; Fig. 1f). The patterns of identity (indicated by different colors) and their frequencies are visualized in tree maps in HC and CRC (Fig. 1g). In summary, our results demonstrate the presence of a highly diverse γδ T cell infiltrate in HC with reshaped TCRγδ chain pairing in CRC. Distinct patterns of identity in the CDR3 arise in CRC, which indicate that γδ T cells potentially recognize antigens present in the tumor, which is in line with previous published work13.
Fig. 1. Single-cell RNA- and TCR sequencing reveal favored Vδ1+ usage with expanded CDR3 identity patterns in γδ T cells of MSS CRC.
CD45 + /CD45– cells were sorted from human MSS CRC and distant HC of resection tissue and processed for 10X genomics RNA and αβ-, γδ-TCR sequencing. a Graphical abstract of the work flow. CRC tissue and healthy distant colon was obtained from 17 and 16 patients, respectively. Single-cell RNA-sequencing and αβ-/γδ-TCR sequencing were performed from seven patients. b UMAP of cells in HC and CRC displayed according to the similarity of their transcriptome. c Marker genes of depicted cell subsets. d Circos plots displaying chain pairing of TRDV and TRGV in HC compared to CRC. e STARTRAC expansion index of γδ T cells (left) and αβ T cells (right). αβ T cells but not γδ T cells showed an increasing index indicative of antigen-specific clonal expansion. f Percentage of top 10 expanded patterns of identity in the CDR3 region of γδ T cells in CRC and corresponding HC using GLIPH219. g Visual comparison of tree map graphs showing the identity pattern in the CDR3 region of γδ T cells in HC and CRC. In tree maps, each rectangle reflects a unique pattern, as indicated, where the size of a spot equals the percentage. Violin plots display median and quartiles (e); two-tailed paired t-test (e, f). EGC, enteric glial cells. ILC, innate lymphoid cells. p values are shown on the graphs.
TRDV1+ cells demonstrate profound dysregulation but not exhaustion in MSS CRC
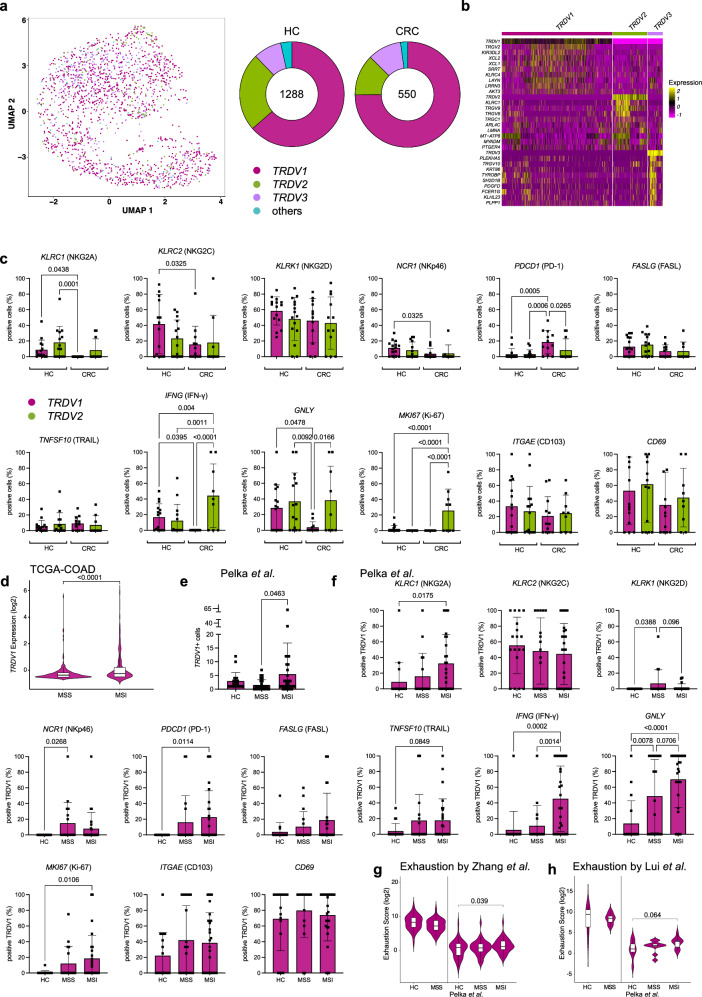
In samples where scTCRγδ-seq was not available, we identified γδ T cells based on the marker gene expressions of TRDV1, TRDV2 or TRDV3. We added all TRDV1 + /TRDV2 + /TRDV3+ cells to the previous data set and performed analysis on 1838 γδ T cells. TRDV1 comprised the majority of γδ T cells in HC (64.4%) with an increased percentage in CRC (74.4%), followed by TRDV2 (HC 23.1% vs. CRC 13.0%) and TRDV3 (HC 8.8% vs. CRC 10.5%; Fig. 2a). We summarized double or triple expression of δ chains, which accounted for 3.7% and 2.1% in HC and CRC, respectively (Fig. S2a). Doublets and triplets were excluded from further analysis. We next wanted to identify possible distinctions on the mRNA level within γδ T cell subsets in HC and MSS CRC. We identified the top differentially upregulated genes in TRDV1, TRDV2 and TRDV3+ cells (Fig. 2b). Among those were genes previously associated with NK cell cytotoxicity (KIRDL2, KLRC4 in TRDV1 + , KLRC1 in TRDV2 + , TYROBP in TRDV3 + ) and T cell exhaustion (LAYN in TRDV1 + ). We assessed the two major subsets (TRDV1 + , TRDV2 + ) for their expression of γδ T cell-associated genes (Fig. 2c). We observed significant alterations of inhibitory (KLRC1, KLRC2) and activating receptors (NCR1) in γδ T cell subsets in CRC, thus suggesting perturbances in the innate immune defense of these cells. Interestingly, we saw significantly increased numbers of PDCD1 (PD-1) positive TRDV1+ cells in CRC suggesting an activated state that could have implications for an immune regulatory reaction in response to the tumor. Interestingly, genes responsible for encoding a part of the cytotoxic machinery (IFNG, GNLY) exhibited a significantly decreased expression in TRDV1+ cells isolated from CRC. TRDV2+ cells demonstrated a significant capability to proliferate in CRC according to their MKI67 expression. ITGAE (CD103) was expressed by approximately 20−30% of cells in both subsets and the majority of cells were positive for CD69 in HC, pointing to cells with dedicated tissue retention. In the light of recent observations demonstrating responsiveness of γδ T cells in MSI CRC to immune checkpoint blockade3, we became interested in comparing TRDV1+ cells at the gene expression level from MSI and MSS CRC which has not been done so far. Analysis of bulk RNA-sequencing data from TCGA-COAD showed significant upregulation of TRDV1 exclusively expressed by Vδ1 + T cells in MSI compared to MSS CRC (Fig. 2d). We further investigated the gene expression of TRDV1 in a data set by Pelka et al.20. who performed single-cell sequencing on 36 HC, 28 MSS and 34 MSI CRC with adjacent tissue (HC). In this data set, TRDV1+ cells were significantly elevated in MSI CRC confirming our results of the TCGA-COAD analysis (Fig. 2e). When examining the relevant activating and inhibitory receptors, along with tissue retention markers of TRDV1+ cells as done previously (Fig. 2c), we found that TRDV1+ cells from MSI significantly increased the expression of several genes, such as KLRC1 (NKG2A), PDCD1 (PD-1), IFNG, GNLY and MKI67 in CRC compared to HC that were not altered in TRDV1+ cells from MSS (Fig. 2f). The expression of IFNG was significantly upregulated in TRDV1+ cells from MSI CRC compared to MSS and HC indicating functionally relevant differences of TRDV1+ cells between these two CRC cohorts. In addition, the expression of GNLY as another proinflammatory gene was significantly elevated in MSI CRC compared to HC and demonstrated a tendency towards upregulation compared to MSS CRC. Overall, TRDV1+ cells from MSI tumors seemed to maintain and even enhance crucial effector functions, whereas TRDV1+ cells from MSS CRC showed reduced expression of effector genes. Of note, in the Pelka et al. data set individuals of HC were not matched to MSS and MSI tumors which could potentially be relevant for the observed differences between the two data sets. As TRDV1+ cells dominate γδ T cell subsets in HC and CRC, we were interested in CRC-associated differences and determined the top significantly overexpressed genes in TRDV1+ cells in CRC (Fig. S2b). Genes associated with TCR signaling and downstream T cell activation were altered in TRDV1+ cells (LAT, CD48, LCP1, LCP2, TRDV1, TRGV2, JUN, CD3D, TRAF3IP3, NFATC2, SKAP1), arguing for enhanced T cell activation and potentially antigen-dependent TCR triggering in TRDV1+ cells in CRC. Consistent with this finding, genes associated with antigen processing were significantly increased (HLA-DPA1, HLA-DRB1, CD2, PSMB9). RNA profiles displayed overexpressed cytokine signaling (IL32, LTB, CKLF, FYB1), upregulated cell motility (RHOB, MYL12A, TMSB10, ACTB) and increased cell crosstalk (CCL4, CCL4L2, SEMA4D, EMP3) in TRDV1+ cells of CRC, featuring the adaption of these cells to the tumor microenvironment. We found cell apoptosis-regulating molecules upregulated in TRDV1+ cells of CRC compared to HC (BCL2, TNFAIP8, STK17A). Moreover, several transcription factors mediating differentiation and immune responses were upregulated in TRDV1+ cells of CRC (IRF1, IRF2, JUN, PURA). To further address if the observed alterations in TRDV1+ cells with significant upregulation of PD-1 were due to exhaustion, we used a recently reported exhaustion gene marker set of conventional αβ T cells21 to compare HC and CRC (Fig. 2g). In our data, we did not observe significant upregulation of exhaustion scores between HC and CRC. We verified this finding by comparing the exhaustion scores of the HC and MSS CRC of Pelka et al., which resembles our patient cohort. However, exhaustion of TRDV1+ was significantly upregulated in MSI CRC confirming published observation in MSI CRC3. Application of another reported exhaustion marker gene set22 to our data and the Pelka et al. data set confirmed these findings (Fig. 2h).
Fig. 2. TRDV1+ cells demonstrate profound dysregulation but not exhaustion in MSS CRC.
γδ T cells subsets were analyzed according to the expression of marker genes TRDV1, TRDV2 and TRDV3. (a) UMAP based on the expression of TRDV1, TRDV2 and TRDV3 in HC and CRC. On the right side, donut charts illustrating the quantities of cells, indicating the counts of TRDV1, TRDV2 and TRDV3 expressing cells in both HC and CRC. b Top 10 differentially overexpressed genes for each subset TRDV1, TRDV2 and TRDV3. c Expression of individual genes displayed as percentage of positive cells for TRDV1 and TRDV2 in HC and CRC reveal distinct effector states of both subsets (n = 16 for HC, n = 17 for CRC). d Expression of TRDV1 in MSI and MSS of TCGA-COAD (n = 217 for MSS, n = 121 for MSI). e TRDV1+ cells per sample in HC, MSS CRC and MSI CRC in the Pelka et al. data set20 (n = 36 for HC, n = 34 for MSI, n = 28 for MSS). f Expression of individual genes displayed as percentage of positive cells for TRDV1 in HC, MSS CRC and MSI CRC using Pelka et al. (n = 36 for HC, n = 28 for MSS, n = 34 for MSI). g, h Depiction of exhaustion scores using genes sets by Zhang et al.21. and Lui et al.22. as previously published of TRDV1+ cells in HC and CRC (left: n = 16 for HC, n = 17 for CRC, right: n = 36 for HC, n = 28 for MSS, n = 34 for MSI). Data points and error bars represent the mean ± SD (c, e, f), Violin plots display median, quartiles ±SD (d, g, h); one-way ANOVA with Fisher’s LSD (c, f), two-tailed unpaired t-test (d, e, g, h). p values are shown on the graphs.
Summarizing, both γδ T cell subsets express genes with distinctive variations as a prerequisite for functional differences in HC and CRC. TRDV1+ cells in MSS CRC showed no elevation in effector genes to an extend of TRDV1+ cells in MSI CRC despite significantly overexpressed genes associated with downstream TCR signaling in the overall RNA profile. This suggests a dampened immune response. Furthermore, although markers associated with exhaustion are significantly upregulated, TRDV1+ cells of MSS CRC do not display elevated exhaustion scores as previously reported for conventional T cells and γδ T cells in CRC of mice23 and human renal cancer14.
γδ T cell are reduced in MSS CRC compared to distant healthy colon
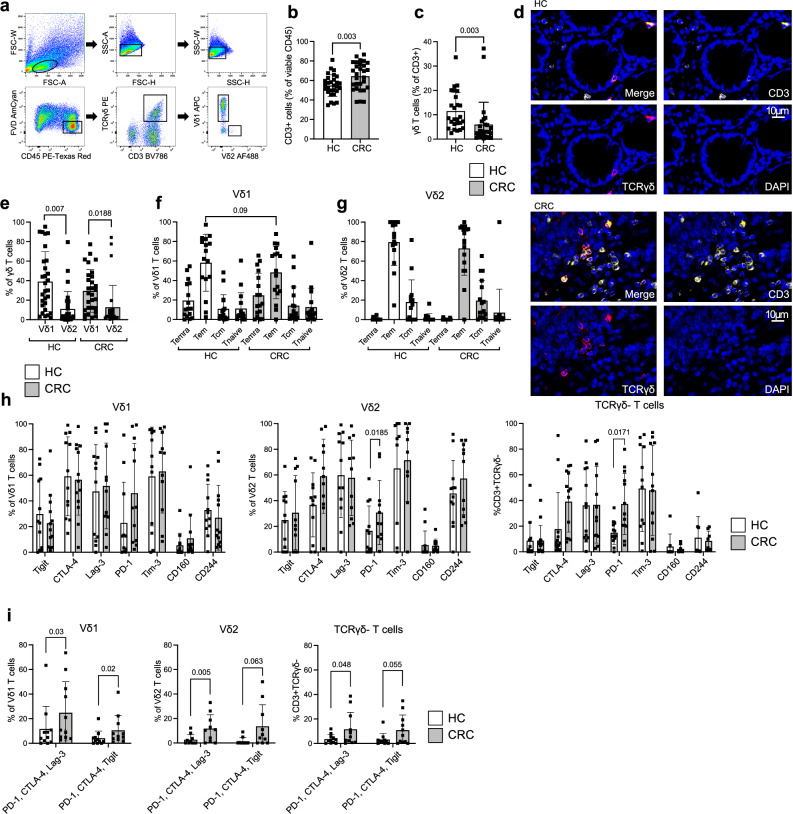
To gain further insight into the relevance of γδ T cells in CRC and validate our bioinformatic analysis of single-cell sequencing data, we performed immunophenotyping in human treatment-naïve MSS CRC and matching HC from the same patients for γδ + CD3 + T cells using flow cytometry (Fig. 3a, patient characteristics Supplementary Table S1). In both HC and CRC, CD3+ cells comprised the majority of viable CD45+ cells with a significant increase in CRC (54.8% ± 11.1 in HC vs. 64.4% ± 14.3 in CRC, Fig. 3b). Of note, γδ T cells showed high inter-individual variation, ranging from 2.4% to 33.6% in HC and 0.2% to 37.2% of viable CD3+ cells in CRC. They represented 11.5% ± 7.7 in HC with a significant decrease in CRC lesions to 5.9% ± 9.2 (of all CD3 + T cells; Fig. 3c). We performed fluorescent immunohistochemistry stainings of TCRγδ and CD3 to verify our flow cytometry-based findings. Increased density of CD3+ cells in the CRC potentially reflects the previously observed enhanced infiltration of CD3+ cells in cancerous lesions with intermingled TCRγδ + T cells (Fig. 3d). We next assessed the expression of the earlier identified two main γδ T cells subsets, Vδ1 and Vδ2 (Fig. 3e). In HC, 39% ± 30.6 of γδ T cells were positive for Vδ1 and 10.9% ± 17.8 for Vδ2. These values partly corroborate our findings in samples on the gene expression level (Fig. 2a). The observed differences may be due to individual variation of these two cohorts and comparison between two different methods. Four subsets of human γδ T cells have been identified as based upon the expression of CD45RA and CD27: naïve (CD45RA + , CD27 + ), Tcm (CD45RA − , CD27 + ), Tem (CD45RA − , CD27 − ), and Temra (CD45RA + , CD27 − )24. On the one hand, when investigating γδ T cell subsets in HC and CRC, Vδ1 + T cells were mostly comprised of Tem and Temra with a tendency towards less Tem in the CRC (p = 0.09; Fig. 3f). On the other hand, Tem dominated the Vδ2+ subsets in both tissues (HC: 79.5% ± 24.2 vs. CRC: 73% ± 27.5 of Vδ2 + ; Fig. 3g). In summary, despite similar Vδ1+ and Vδ2+ subset distributions in HC and CRC, γδ T cells as a fraction of all CD3+ cells were significantly diminished in MSS CRC which is in stark contrast to MSI CRC25. It has been previously shown that the majority of γδ T cells exhibit an effector memory phenotype26. However, the observed differences in effector cell states of Vδ1+ and Vδ2+ cells imply that both γδ T cells subsets potentially execute distinct functions.
Fig. 3. γδ T cells are reduced in MSS CRC compared to HC and upregulate co-expression of inhibitory receptors.
Flow cytometry analysis of γδ T cell abundance and subsets in human MSS CRC compared to HC. a Gating strategy for γδ T cells from human mononuclear cells isolated from CRC. b Significant increase in CD3+ cells depicted as mean percentage of viable CD45+ cells in CRC compared to HC (n = 29 per group). c Significant decrease of TCRγδ + cells in CRC depicted as mean percentage of viable CD3+ cells compared to HC (n = 29 per group). d Representative image of multicolor fluorescent immunohistochemistry staining of TCRγδ + cells (TCRγδ PE/ CD3 A647) in HC and CRC. e Significantly more Vδ1 + populate HC and CRC compared to Vδ2 + , assessed by flow cytometry and depicted as percentage of positive cells of viable TCRγδ + CD3 + cells (n = 29 per group). Vδ1 + (f) and Vδ2 + (g) display different T cell effector states according to their CD27 and CD45RA expression (n = 16 per group). Temra: CD45RA + CD27–, Tem: CD45RA–CD27–, Tcm: CD45RA–CD27 + , Tnaive: CD45RA + CD27 + . h Expression of inhibitory checkpoint receptors on Vδ1 + (n = 13), Vδ2 + (n = 11) and TCRγδ negative T cells (n = 13) in HC (white) and CRC (gray). i Co-expression of depicted receptors on Vδ1 + , Vδ2 + and TCRγδ negative T cells in HC (white) and CRC (gray; n = 11 per group). Co-expression was assessed by first gating on PD-1+ cells and then calculating the percentage of double-positive cells. Data points and error bars represent the mean ± SD (b, c, e−i); two-tailed paired t-test (b, c, e, i), one-way ANOVA with Fisher’s LSD (f−h); Temra, effector memory RA; Tem, effector memory; Tcm, central memory. p values are shown on the graphs.
Co-expression of inhibitory receptors is upregulated on γδ T cell subsets and conventional αβ T cells in MSS CRC
We next sought to explore whether observed alterations regarding elevated signs of exhaustion in the transcriptome of tumor-infiltrating γδ T cells are relevant on the protein level and how this compares to conventional αβ T cells. It has been previously described that the acquisition of an exhaustion phenotype is not a unique feature of conventional αβ T cells. For γδ T cells, upregulation of inhibiting checkpoint receptors has been demonstrated in breast cancer27, ovarian cancer28 and recently renal cancer14. Detailed immunophenotyping and – more importantly – implications for functional consequences have been lacking especially in MSS CRC. Vδ1 + T cells in CRC did not demonstrate significant altered expression of the tested checkpoint receptors compared to HC (Fig. 3h). Significantly more Vδ2 + T cells showed elevated PD-1 expression and a tendency towards increased CTLA-4 expression in CRC compared to HC, which was similar to the expression pattern of tumor-infiltrating conventional TCRγδ negative T cells. Since the simultaneous co-expression of several checkpoint molecules on a single cell is a well-established indicator of exhaustion29, we assessed triple-checkpoint receptor-positive γδ T cells and TCRγδ negative T cells as controls (Fig. 3i). Indeed, the percentage of tumor-infiltrating Vδ1 + , Vδ2 + , and TCRγδ negative T cells co-expressing PD-1/CTLA-4/Lag-3 or PD-1/CTLA-4/TIGIT was significantly elevated. The representative examples for each investigated subset demonstrated double-positive cells for CTLA-4 and TIGIT on previously gated PD-1+ cells (Fig. S3). To summarize, we found that all investigated T cells, conventional and non-conventional T cells, show a significant increase in the co-expression of checkpoint receptors in MSS CRC.
Dysfunctional cytotoxic potential of Vδ1 + T cells from MSS CRC can be restored in vitro
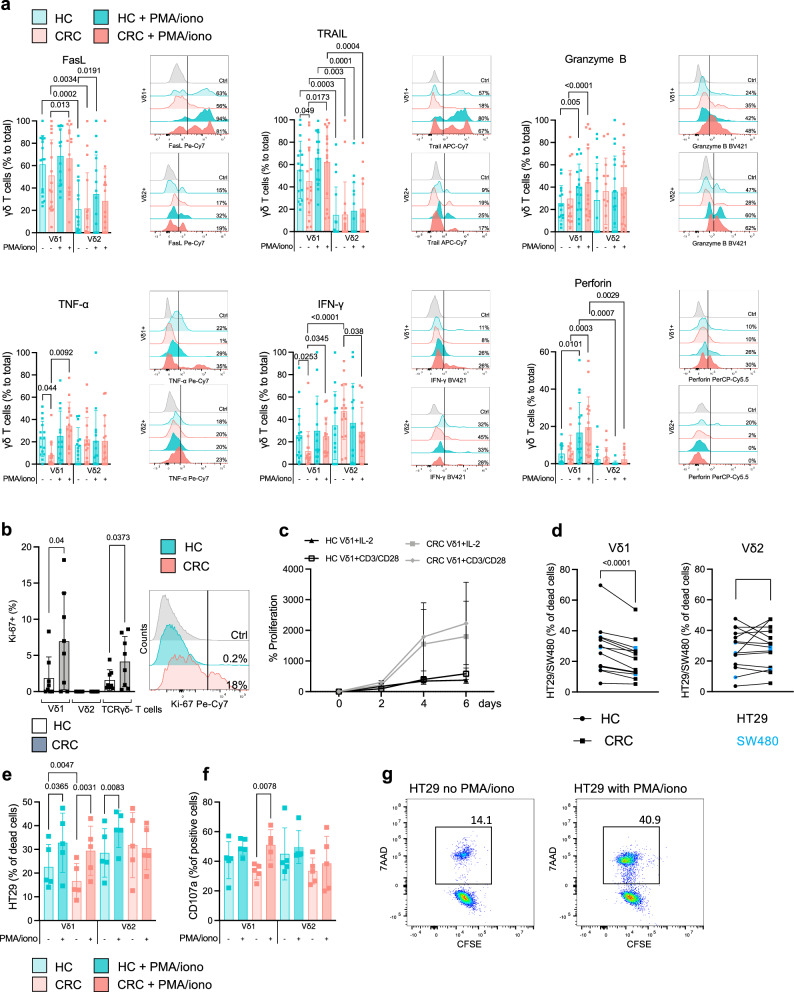
We next sought to determine whether the observed alterations in cytokine expression and effector cell differentiation on the transcriptome level in Vδ1 + T cells from CRC lesions translate to a reduced anti-tumor activity in vitro. To investigate distinct functional characteristics of Vδ1+ and Vδ2 + T cells as effector cells, we performed flow cytometry for cytokines and molecules associated with cytotoxicity in HC and CRC. We found that the majority of Vδ1 + T cells expressed FasL (61.2% ± 25.77) and TRAIL (55.2% ± 25.6) in HC (Fig. 4a). In HC, 25.5% ± 15.76 of the Vδ1 + T cells expressed granzyme B, 22.7% ± 15.66 were positive for TNF-α and 25.9% ± 24.01 for IFN-γ. 5.4% ± 5.35 of Vδ1 + T cells stained positive for perforin in HC. In contrast, we observed a significant decrease in TNF-α- (7.9% ± 11.36), IFN-γ- (11.6% ± 17.41) and TRAIL- (45% ± 30.61) positive Vδ1 + T cells in CRC, indicating reduced killing capacity of these cells in the tumor microenvironment. We also noticed a trend towards decreased expression of FasL on Vδ1 + T cells in CRC (51.2% ± 31.91), while other tested molecules were not affected. We next aimed to address whether the reduction of cytotoxic effector molecules of γδ T cells from the tumor microenvironment is reversible and therefore stimulated tissue-derived cells with phorbol 12-myristate-13-acetate/ionomycin (PMA/iono) for 5 h. In CRC, we observed a significantly increased expression of FasL, TRAIL, TNF-α and IFN-γ in stimulated Vδ1 + T cells to values comparable to HC. This evidenced that upon appropriate stimulation the CRC-associated dysfunctional phenotype of Vδ1 + T cells is potentially reversible. We performed the same experiments with Vδ2 + T cells. Compared to the Vδ1+ counterpart, Vδ2 + T cells expressed significantly less FasL (21.1% ± 25.9) and TRAIL (14.8% ± 25.1) in both HC and CRC. Unlike the Vδ1+ subset, Vδ2 + T cells displayed no significant differences in either HC or CRC under stimulated and unstimulated conditions, suggesting that both subsets display a discrete phenotype with distinct characteristics. While Vδ2+ cells have so far been the main target of γδ T cell-centered immunotherapeutic strategies30–32, our data indicate that Vδ1+ cells are present in higher numbers (Fig. 3e) and significantly exceed Vδ2+ in regard of cytotoxic potential (Fig. 4a), which is in line with previously published reports33,34. In HC, 1.9% of Vδ1 + T cells expressed Ki-67, compared to 6.9% in CRC, indicating a significantly increased proliferative capacity in the tumor (Fig. 4b). This pattern was also observed in TCRγδ negative T cells. It is noteworthy that the Vδ2+ subset showed no expression of Ki-67 in either HC or CRC, suggesting their migration from peripheral blood rather than local proliferation. The difference between MKI67-expressing TRDV2+ cells (Fig. 2c) and Vδ2 + T cells lacking Ki67 on the protein level in CRC could be explained by differences in the sensitivity of the methods and onset of MKI67 translation that is not (yet) detectable by flow cytometry. Next, we tested the proliferation rate of isolated Vδ1 + T cells in vitro for up to 6 days. Higher numbers of Ki-67 + Vδ1 + T cells from CRC compared to HC were reflected in the proliferation rate, as Vδ1 + T cells isolated from CRC demonstrated an increased proliferation compared to HC under steady state conditions (IL-2 only) as well as upon CD3/CD28 stimulation (Fig. 4c). We further sought to determine how well tumor-infiltrating γδ T cells can recognize and ultimately kill CRC cells. Since γδ T cells do not lyse tumor cells in an MHC-restricted fashion, their cytotoxic anti-tumor response can be assessed with non-autologous tumor lines. To this end, we used the MSS CRC cell lines HT29 and SW480 to mirror the patient cohort of interest. We found that Vδ1 + T cells isolated from CRC were significantly impaired in their ability to kill cancer cells when compared to Vδ1 + T cells isolated from matched HC (Fig. 4d). We did not observe this cytotoxicity pattern with the Vδ2 + T cell subset. To further assess the responsiveness of γδ T cells to cell stimulation, we exposed the cells to PMA/iono before adding HT29 (Fig. 4e). Again, Vδ1 + T cells from CRC killed significantly less HT29 compared to Vδ1 + T cells from HC. However, the killing capacity of both Vδ1 + T cells and partially Vδ2 + T cells significantly increased following treatment with PMA/iono (Fig. 4g). This was in case of Vδ1 + T cells from CRC, mirrored by the expression of CD107a after the killing assay (Fig. 4f). Specifically, Vδ1 + T cells from CRC exhibited significantly higher CD107a expression, indicating augmented degranulation upon stimulation. To sum up, we demonstrate that Vδ1 + T cells display a reduced expression of main cytotoxic molecules in CRC which can be restored upon stimulation. The observed shift from a subdued effector state of Vδ1 + T cells in CRC to an upsurge upon in vitro activation opens up the possibility of clinical translation.
Fig. 4. Dysfunctional cytotoxic potential of Vδ1 + T cells from MSS CRC can be restored in vitro.
Flow cytometry analysis of intracellular cytokine staining of Vδ1 + and Vδ2 + T cells in HC or CRC. a Overlay of representative histograms and corresponding bar plots of tested cytokines by flow cytometric analysis in Vδ1 + and Vδ2 + T cells in HC (light cyan) or CRC (light pink) and stimulated (PMA/iono) HC (cyan) or CRC (pink, n = 16 per group). In CRC, Vδ1 + display reduced expression of TRAIL, TNF-α and IFN-γ which is reversible upon stimulation. b Percentage of Ki-67 + Vδ1 + , Vδ2 + and TCRγδ negative T cells in HC (white) compared to CRC (gray; n = 8 per group). Increased expression of Ki-67 in Vδ1 + and TCRγδ negative T cells in CRC indicate in tissue proliferation in contrast to the Ki-67 negative Vδ2 + T cells which potentially influx from the periphery. Overlay of representative histograms of Ki-67 expression in HC (cyan) and CRC (pink). c Increased Ki-67+ expression of Vδ1 + T cells isolated of CRC was mirrored in elevated proliferation (gray) during IL-2 alone or CD3/CD28 stimulation as compared to HC (white; n = 3 per group). d Pairwise comparison of killing assay with Vδ1 + and Vδ2 + T cells with two different CRC lines (HT29 in black, SW480 in blue; n = 14 per group). e Percentage of dead HT29 after exclusion of baseline HT29 cell death in a killing assay with Vδ1 + and Vδ2 + T cells isolated from HC (cyan) and CRC (pink). Two conditions were tested: Vδ1 + and Vδ2 + T cells with (cyan, pink) and without PMA/iono stimulation (light cyan, light pink; n = 5 per group). f CD107a expression of Vδ1 + and Vδ2 + T cells from HC (cyan) and CRC (pink) after killing of HT29 as in (e). g Representative FACS plot of dead HT29 in a killing assay with Vδ1 + T cells with no stimulation and stimulated with PMA/ionomycin. Data points and error bars represent the mean ± SD (a−f); one-way ANOVA with Fisher’s LSD (a−f); Ctrl; Control; p values are shown on the graphs.
Interaction of fibroblasts with an inflammatory phenotype with Vδ1 + T cells via TIGIT in MSS CRC
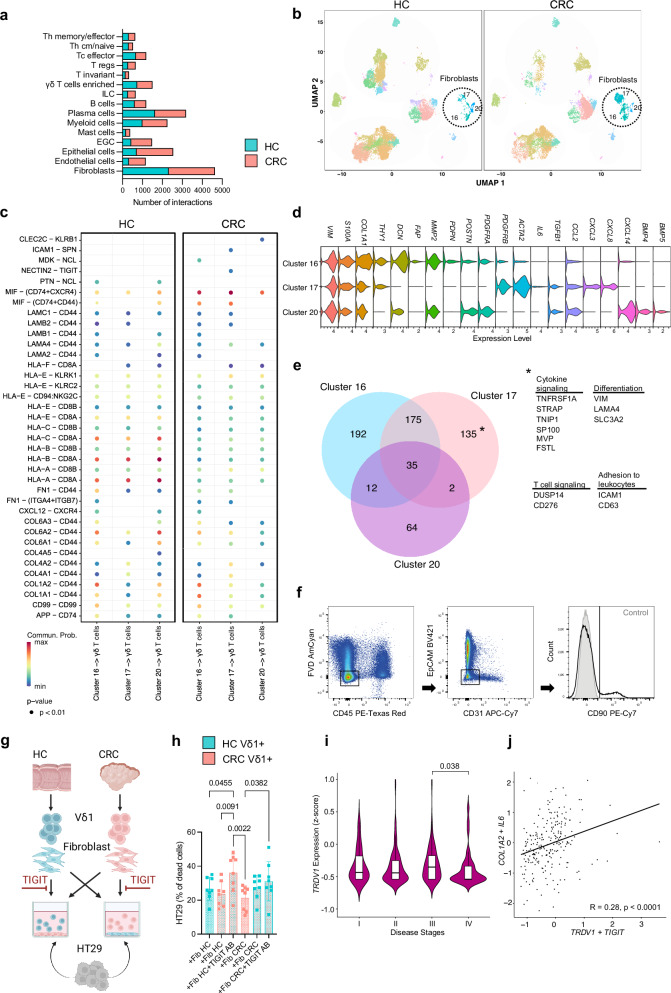
To address possible causes of the altered state of Vδ1 + T cells within the tumor microenvironment, we employed CellChat, a tool which predicts the probability of cell-cell communication and receptor-ligand interactions by using gene expression profiles35. Compared to other cell subsets, fibroblasts demonstrated the highest number of interactions in HC and CRC (Fig. 5a). Since fibroblasts can limit proliferation and induce the expression of exhaustion marker in conventional αß T cell subsets36,37, we addressed the influence of fibroblasts on γδ T cells in CRC. Therefore, we analyzed significant receptor-ligand interactions as predicted by CellChat between the three fibroblasts subsets (Fig. 5b) and γδ T cells in HC and CRC (Fig. 5c). Fibroblasts in HC maintained a variety of interactions with γδ T cells through activating (KLRK1) and inhibiting (KLRC2) receptors potentially underlining the regulatory role of fibroblasts. When focusing on therapeutically targetable immunomodulatory pathways only present in CRC, we found the NECTIN2-TIGIT interaction in fibroblasts from cluster 17, thus potentially reflecting an immunosuppressive influence of these fibroblast subset on γδ T cells in MSS CRC (Fig. 5c). NECTIN2 was found to be overexpressed in fibroblasts of cluster 17 (Fig. S4a). TIGIT, as an inhibitory receptor, has been previously reported to negatively impact conventional αß T cell cytotoxicity by facilitating inhibitory signals38. However, immunosuppressive properties of fibroblasts towards γδ T cells in CRC have not been studied so far. Next, we investigated predicted cellular communication using CellChat in another dataset20 with MSS and MSI CRC, where we found similar interaction patterns between fibroblasts and γδ T cells via NECTIN2/TIGIT (Fig. S4b). Fibroblasts subsets of both MSS and MSI significantly overexpressed NECTIN2 compared to HC (Fig. S4c). Since we detected differences between the predicted interaction of distinct fibroblast clusters and γδ T cells, we further analyzed the gene expression profile of the UMAP clusters 16, 17 and 20 (Fig. 5d). All three clusters expressed known marker genes for fibroblasts such as VIM, S100A4, COL1A2, confirming their identity. Similar to the myofibroblastic fibroblasts described in previous studies36,39,40, cluster 16 in particular displayed genes associated with contractile process, featuring the presence of THY1 (CD90), DCN, FAP, MMP2, PDPN, POSTN, PDGFRA, PDGFRAB and ACTA2. Cluster 17 exhibited gene expression linked to inflammatory fibroblasts identified earlier, including IL6, TGFB1, CCL2, CXCL3 and CXCL8. Cells within cluster 20 exhibited significant elevation of CXCL14 paired with BMP4 and BMP5 expression, features previously associated with a subset of fibroblasts in colorectal cancer20,41. The role of this subpopulation remains incompletely understood and reports on CXCL14 in CRC are controversial. Patients of the TCGA-COAD with high CXCL14 expression were found to have a better survival probability and longer progress free interval42. Another study reported a correlation of elevated CXCL14 expression from stage III/IV patients with worse overall survival43. Since the NECTIN2-TIGIT pathway was absent in fibroblasts of cluster 16 and 20, we investigated the gene signature of these cluster compared to fibroblast cluster 17. We analyzed the Top overexpressed genes of the three clusters in CRC (Fig. 5e). Cluster 17 exhibited a distinct gene expression profile, characterized by the presence of various genes encoding regulators of cytokine signaling pathways such as TNF-α (TNFRSF1A, FSTL), interferon (CD276, HNRNPA2B1), Fas (SP100), TGF-β (STRAP) and IL-23 (TNIP1). Additionally, genes associated with leukocyte adhesion (ICAM1, CD63), T cell regulation (CD276, DUSP14) and signaling pathways including ERK and mTOR (LAMA4, SLC3A2) were significantly upregulated in CRC within this cluster. These findings suggest an immunoregulatory phenotype within cluster 17 with potential interaction sites with leukocytes. In conclusion, fibroblasts were predicted to exert immunoregulatory effects on γδ T cells in CRC. In particular, the NECTIN2/TIGIT axis is active in fibroblasts displaying an inflammatory phenotype in CRC.
Fig. 5. Impaired killing capacity by Vδ1 + T cells can be reinstated after TIGIT blockade.
a Number of predicted overall interactions of depicted cell clusters in HC and CRC using CellChat. b UMAP of CD45+ and CD45– cells of HC and CRC displayed according to the similarity of their transcriptome. Fibroblasts clusters are highlighted. c Significant receptor-ligand interactions of fibroblasts cell clusters to γδ T cells as predicted by CellChat in HC and CRC. d Marker gene expression of fibroblast clusters. e Co-expression analysis of differentially overexpressed genes of each fibroblast cluster in CRC. f Gating strategy of fibroblast isolation for experiment as performed in (g). g Viable Vδ1 + T cells and fibroblasts were isolated from HC and CRC, fibroblasts were seeded, with or without antibody-blocking of TIGIT, Vδ1 + T cells were added and killing assay with HT29 was performed. h Percentage of dead HT29 after exclusion of baseline HT29 cell death in a killing assay as stated in (g); n = 8 per group. i TRDV1 expression of patients with disease stages I, II, III, IV in the MSS CRC cohort of the TCGA-COAD (n = 29 for I, n = 64 for II, n = 77 for III, n = 38 for IV). j Correlation between COL1A2, IL6 with TRDV1, TIGIT expressing cells in the MSS cohort of the TCGA-COAD. Data points and error bars represent the mean ± SD (h), Violin plots display median, quartiles ±SD (i); Wilcoxon Rank Sum test (c). one-way ANOVA with Fisher’s LSD (h), two-tailed paired t-test (i), Pearson Correlation (j). p values are shown on the graphs.
Impaired killing capacity by Vδ1 + T cells in CRC can be reinstated after TIGIT blockade
Next, we aimed to investigate the functional relevance of the predicted interaction of fibroblasts with Vδ1 + T cells via the TIGIT axis. For this, we isolated fibroblasts and Vδ1 + T cells from HC and CRC (Fig. 5f) to assess the cytotoxic activity of Vδ1 + T cells against the CRC cell line HT29 (Fig. 5g). We co-cultured fibroblasts with Vδ1 + T cells with and without antibody-blocking TIGIT before adding the cancer cells. Fibroblasts from HC and CRC did not influence the killing capacity of Vδ1 + T cells in our co-culture assay (Fig. 5h). However, blocking TIGIT significantly increased the cytotoxic activity of Vδ1 + T cells from CRC against HT29 (Fig. 5h). In a control trial, introducing the TIGIT blocking antibody to Vδ1 + T cells from HC and CRC alone, without fibroblasts, led to no significant increase in killed HT29 cells (Fig. S4d). This suggests that the TIGIT blocker likely does not directly activate the cells. These functional characteristics of isolated Vδ1 + T cells from CRC patients corroborate our findings of decreased expression of cytotoxic molecules and altered effector functions on the RNA level and suggest fibroblasts to interact with Vδ1 + T cells via TIGIT. Analysis of bulk RNA-sequencing data from MSS patients in the TCGA-COAD showed a significant decrease in TRDV1 expression from stage III to stage IV of the disease (Fig. 5i). This suggests a reduced infiltration of TRDV1+ cells in the most advanced stages of MSS CRC, highlighting a correlation with clinical disease progression. Furthermore, we identified a significant correlation between the expression levels of COL1A2 and IL6 with TRDV1 and TIGIT in MSS patients of the TCGA-COAD dataset (Fig. 5j). Analysis of our single-cell RNA-sequencing data indicated that COL1A2 was predominantly expressed by cells within fibroblast clusters 16 and 17, while TRDV1 and TIGIT were upregulated in cluster 2, primarily comprising γδ T cells (Fig. S4e).
In essence, we identified distinct fibroblast subsets exhibiting unique characteristics within MSS CRC. Specifically, one subset of fibroblasts showing elevated expression of markers linked to an inflammatory phenotype was predicted to interact specifically with γδ T cells in CRC through the TIGIT-NECTIN2 axis. At the gene level, this subset displayed particular capabilities to modulate cytokine signaling. Blocking the TIGIT pathway resulted in heightened killing abilities of Vδ1 T cells isolated from CRC. Additionally, reduced TRDV1 expression was associated with an advanced disease stage in MSS CRC. Moreover, the expression of markers linked to inflammatory fibroblasts positively correlated with TIGIT-positive Vδ1 T cells.
Discussion
Our study provides a comprehensive single-cell analysis of cancer-specific changes in γδ T cells, with a particular focus on γδ T cells in primary human MSS CRC. Using scTCRγδ-seq, we show an expansion of similar identity patterns in the CDR3 regions indicative of shared antigen recognition via TCRγδ. Notably, distinct gene expression profiles among subsets of γδ T cells pointed towards the downregulation of effector molecules within TRDV1+ cells in MSS CRC. Linking phenotypic characterization to scRNA-seq, we found increased co-expression of checkpoint receptors, impaired cytotoxic potential in in vitro experiments and killing of CRC cancer cell lines which could be partly restored by stimulation. We compared our findings with the immunologically interesting phenotype of MSI CRC. γδ T cells in MSI tumors tended to be skewed towards cytotoxicity whereas the same genes remain unaffected or downregulated in MSS CRC. Moreover, employing conventional T cell exhaustion scores, we observed a significant exhaustion phenotype in TRDV1+ cells in MSI, contrasting starkly with their MSS counterparts, which did not exhibit elevated signs of exhaustion on the transcriptional level. Further investigation into receptor-ligand interactions underscored an augmented influence of inflammatory fibroblasts on TRDV1+ cells in CRC, mediated specifically through TIGIT. Notably, blocking TIGIT reinstated the killing capacity of these cells. In conclusion, while Vδ1 + T cells in MSS exhibit certain markers associated with T cell exhaustion, they fail to meet all criteria. Contrasting Vδ1 + T cells in MSI, Vδ1 + T cells in MSS display diminished effector functions. However, these functions are potentially reversible through activation, possibly influenced by the modulatory impact of inflammatory fibroblasts on the fate of Vδ1 + T cells in MSS CRC.
Other studies previously hinted to the clinical relevance of Vδ1 + T cells in cancer. One investigation found that NKp46 on tissue-resident Vδ1 + T cells in the gut was associated with high cytolytic potential against the K562 cell line and lower risk of developing metastatic disease stages33. In a study on triple-negative breast cancer, increased Vδ1 + T cell levels were observed to be associated with remission and better overall survival44. Additionally, in non-small cell lung cancer, a tissue-resident Vδ1 + T cell population was associated with survival45. Distinct phenotypes, especially regulatory46 and IL-17-producing pro-tumorgenic γδ T cell subsets47, have been described. There are discrepancies regarding the significance of IL-17-producing γδ T cells in human CRC13,48. This scepticism is primarily due to the substantial biological differences between human and murine γδ T cells, making it difficult to extrapolate findings from mouse models to humans49. However, a large study analyzing the immune infiltrate of 18,000 cancer cases in 25 malignancies clearly identified a dominant γδ T cell gene signature as the best predictor of favorable prognosis5.
The majority of Vδ1 + T cells in HC and CRC displayed a Tem phenotype, which is in line with other studies that observed dominance of Tem Vδ1+ cells in tumor lesions26,28. We further confirmed that the percentage of γδ T cells in correlation with TCRγδ negative CD3+ cells was decreased in MSS CRC50. So far, γδ TCRs have been studied mostly in infections51, development52,53 and celiac disease54,55. In human colonic γδ T cells, Di Marco et al. demonstrated that γδ T cells are dominated by the usage of TRGV4 and TRDV156, which is consistent with our data. Equivalent studies in cancer are sparse. One investigation found a distinct γδ TCR repertoire in the blood of lung carcinoma patients57. Reis et al. reported clonally expanded γδ T cell in CRC with enrichment for Vδ1 and Vγ4 in CRC13 which is in line with our findings. Although we did not detect clonal expansion, we observed an increase in similar pattern within the CDR3 region, which governs antigen recognition. However, it’s important to exercise caution due to the limited data available on γδ T cells with TCR sequencing.
Exhaustion describes the status of immune cell dysfunction, usually in the setting of chronic infection and cancer in response to continuous stimulation58. In general, the features of exhausted T cells include altered effector function such as decreased cytokine production, high expression of multiple inhibitory checkpoints, reduced proliferative capacity, altered transcriptional program involving TOX and metabolic reprogramming59. Targeting exhausted T cells with checkpoint blockade is a major mechanism in immunotherapeutic strategies29. In this study, we demonstrate that γδ T cells in MSS CRC exhibit certain features typically linked to exhaustion in conventional αβ T cells. Nevertheless, compared to MSI CRC, Vδ1 T cells in MSS do not exhibit the typical transcriptomic profile of exhaustion. Consequently, we refer to them as dysfunctional.
Some characteristics of cancer-associated exhausted γδ T cells have been described previously and are in line with the findings of this study. In ovarian cancer, for example, Vδ1 + T cells were found to display high levels of co-expression of the checkpoint receptors TIGIT and PD-1 comparable to Vδ1 + T cells in CRC in our analyses28. PD-1+ γδ T cells were found to be enriched in MSI CRC25, and diminished effector function of γδ T cells has been observed in acute myeloid leukemia60. In renal cancer, Vδ2- γδ T cells were reported to express an exhausted transcriptional program similar to conventional T cells and a Vδ2- γδ T cell signature correlated with favorable clinical response after PD-1 blockade14. PD-1 + Vδ1 and Vδ3 were seen to be the dominating effector cells in a study investigating anti-tumor responses in HLA class I-negative MSI CRC after checkpoint therapy3. In contrast to γδ T cells in MSS CRC, Vδ1 and Vδ3 isolated from MSI tumors displayed elevated features of cytotoxic activity and were found to highly infiltrate these tumors. We noted a significant rise in PDCD1 (PD-1) expression at the single-cell level and increased co-expression of checkpoint receptors in Vδ1 cells isolated from MSS CRC. However, the count of CD107a+ degranulated cells surpassed these numbers. Thus, we cannot definitively state that tumor reactivity is exclusively limited to PD-1-positive cells among γδ T cells in MSS CRC.
Fibroblasts regulate migration and activation in the tumor microenvironment through secretion of a variety of soluble factors, which contribute to remodeling of the extracellular matrix and shaping of immune cells61,62. In CRC, tumors with a dominant myofibroblasts signature were found to be enriched with T cells and dendritic cells. On the other hand, the number of anti-tumor effector cells such as NK cells and T cells was significantly reduced in CRC with high inflammatory fibroblasts infiltrate63. T cells in the tumor may be inhibited by fibroblasts expressing checkpoint molecule ligands or by the upregulation of these molecules on other cells. For example, fibroblasts in cell cultures of lung cancer can suppress T cell function via PD-L1/PD-L2, which was reduced by blockade of the ligands64. The influence of a direct NECTIN2-TIGIT axis of fibroblasts to γδ T cells has not been proposed so far. Recently, a study underscored the role of CRC-associated fibroblasts in inhibiting effector T cells via the Nectin2 pathway, suggesting that this interaction is active not only in γδ T cells, as we have demonstrated, but also in conventional T cells65. As γδ T cells interact with numerous factors within the tumor microenvironment and further research is needed to substantiate this finding, it is crucial to cautiously interpret findings for clinical application. Further investigations, utilizing mouse models or clinical trials, are warranted to clarify the precise factors influencing the phenotype and function of γδ T cells in CRC. While recognizing that numerous aspects of γδ T cell biology remain elusive, our study endeavors to shed light on their role in MSS CRC.
Contrasting to other papers with γδ T cells single-cell sequencing, we sorted for viable CD45+ and CD45- cells. Therefore, the distribution of γδ T cells in our data reflects their overall numerically contribution in the colon and the tumor microenvironment in MSS CRC. This approach allows to study γδ T cell interactions with other cell subsets. However, owing to reduced γδ T cell numbers in MSS CRC, the quantity of sequenced γδ T cells in our dataset is comparatively limited. We did not consider the localization of the tumor within the colon, which may introduce potential bias. The anatomical site is known to be of relevance for the microbiome, mutational status and metastatic spread66, factors that could possibly influence the γδ T cells phenotype.
In summary, our findings indicate that Vδ1+ cells play a significant role in MSS CRC, characterized by a dysfunctional state rather than typical exhaustion. Consequently, γδ T cells in MSS CRC do not exhibit a response to checkpoint inhibitors targeting PD-1, which are effective in MSI CRC therapy. The trajectory of γδ T cell fate likely parallels that of conventional T cells in both tumor types, with responsiveness to PD-1 inhibition seen in MSI but not in MSS CRC. The underlying factors contributing to this difference in conventional T cells of MSS CRC may also affect γδ T cells, including lower mutational burden, reduced neoantigen recognition, decreased overall immune cell numbers, an immunosuppressive microenvironment, and alternative immune evasion mechanisms. Our findings support these hypotheses at the γδ T cell level, emphasizing the absence of clear signs of clonal expansion, reduced γδ T cell numbers, the presence of immunoregulatory fibroblast subsets and the responsiveness of γδ T cells to TIGIT blockade as an alternative immune evasion mechanism.
Methods
Patients and tissue material
Tissue samples from patients with histologically verified MSS CRC with no history of irradiation therapy or cytoablative treatment were obtained after surgical resection at the Division of Visceral Surgery, Department of General Surgery, Medical University of Vienna. Colon tissue from the most distal part of the tumor obtained from the surgical resection specimen without apparent pathology from the same patients was referred to as “distant healthy colon” (HC) throughout the manuscript. HC were located at the distal part (≥10 cm in length) of the removed tumor core and were considered “tumor-free/healthy” after a conventional macroscopic examination by pathologists. The patients were enrolled into the study after submitting written informed consent. Studies were performed according to the Declaration of Helsinki and approved by the local Ethics Committee of the Medical University of Vienna (1374/2014). Patient and tumor characteristics are listed in Supplementary Table S1. For the cytotoxicity assay, the HT29 (HTB-38) and SW480 (CCL-228) cell lines were purchased from the American Type Culture Collection (ATCC). All sample sizes (n) in this study represent biological replicates.
Cell isolation and sorting
The weight of the obtained tissue samples was assessed. Human colon tissue, HC and CRC, was cut into small pieces using a sterile scalpel. Phosphate buffered saline (PBS, Gibco) with 10% fetal bovine serum (FBS, Gibco) was used to rinse the tissue. Human colon samples were digested with medium, consisting of plain RPMI 1640 medium (Gibco) supplemented with collagenase IV (10 U/ml, Sigma-Aldrich) and deoxyribonuclease I (10 mg/ml, DNAse, Sigma-Aldrich). Digestion was performed using a digestion machine (gentleMACS Octo dissociator with heaters, Miltenyi) with the preset 37 °C_h_TDK1 protocol. The cell suspension was passed through a 70 µm cell strainer using the backside of a syringe. RPMI 1640 medium supplemented with 10% FBS was used to rinse the strainers.
For flow cytometry analysis and cell sorting, the cells were stained with fluorescent-labeled antibodies, as listed in Supplementary Table S3. Appropriate isotype controls were used as negative controls. Sample acquisition and cell sorting was performed on a FACSAria III (BD Biosciences) with FACSDiva Software. The acquired samples were analyzed with FlowJo software (v10.8). The investigators conducted data collection and analysis in a blinded manner regarding group allocation. After duplet exclusion, we gated on viable CD45+ leukocytes. The Vδ1 + T cells (CD45 + CD3 + TCRγδ + Vδ1 + ) and the Vδ2 subset of the γδ T cells (CD45 + CD3 + TCRγδ + Vδ2 + ) were sorted from the single-cell suspensions for further experiments. Cell counting and viability assessment for further experiments was done via Trypan Blue Exclusion Assay (Sigma Aldrich).
Cell stimulation
Expression of cell surface markers, cytokines and chemokines as well as cytotoxicity marker expression were assessed upon stimulation of the single-cell suspensions with PMA/iono (Cell activation cocktail, BioLegend) for 4 h at 37 °C. The cells were then fixed, permeabilized and labeled using fluorescence-labeled antibodies. Stained samples were acquired with FACSAria III and analyzed with FlowJo software (v10.8). Cell proliferation of sorted Vδ1 + T cells and Vδ2 + T cells upon stimulation with plate bound anti-CD3 antibody/anti-CD28 antibody (5 μg/ml; 2 μg/ml) was assessed as the percentage of increase in cell numbers every 2 days over 6 days at 37 °C.
Killing assays
For the killing assays, viable Vδ1 + T cells (viable CD45 + CD3 + TCRγδ + Vδ1 + ) and Vδ2 + (viable CD45 + CD3 + TCRγδ + Vδ2 + ) were sorted from human CRC samples and corresponding HC tissue as mentioned above. The target cells were washed and labeled with carboxyfluorescein succinimidyl ester (CFSE) solution, having a concentration of 5 µM. The labeled cells were counted using Trypan blue solution and the Neubauer counting chamber. Isolated and sorted γδ T cells were co-cultured with CFSE-labeled HT29 cells or SW480 in an effector-to-target cell ratio of 1:1 in a flat-bottom 96-well plate. The plate was incubated for 4 h at 37 °C, after which the samples were transferred into FACS tubes and stained again. 7-Aminoactinomycin D (7AAD) viability dye (BioLegend) was added 3 min before analysis with a flow cytometer.
In case of prior stimulation, cells were sorted as described above. Cells were split and transferred into a flat-bottom 96-well plate. Cell media was supplemented with PMA/iono (Cell activation cocktail without Brefeldin, BioLegend) in a concentration stated in the manufacturer’s instructions. The plate was incubated for 4 h at 37 °C. Media was supplemented with Monensin (BioLegend) and PE anti-human CD107a antibody (BioLegend) in a concentration stated in the manufacturer’s instructions. CFSE-labeled HT29 cells were added and experiment performed as described above.
For the co-culture experiments with fibroblasts viable Vδ1 + T cells (viable CD45 + CD3 + TCRγδ + Vδ1 + ) and viable fibroblasts (viable CD45-, EpCAM-, CD31-, CD90 + ) were FACS-sorted. Cells were transferred and rested for 2 h at 37 °C in a flat-bottom 96-well plate. Vδ1 + T cells were cultured in RPMI 1640 medium supplemented with 1% P/S and Fibroblasts in DMEM supplemented with 10% FCS and 1% penicillin/streptomycin (P/S). Fibroblasts medium was removed and replaced with RPMI 1640 medium supplemented with 1% P/S. TIGIT antibody (BioLegend) was added in a blocking concentration as stated in the manufacturer’s manual and Vδ1 + T cells were added. Cells were co-cultured for additional 2 h at 37 °C. Killing assay way performed as described prior.
Fluorescent immunohistochemistry staining
Fluorescent immunohistochemistry staining for γδ T cells was performed on 5 µm colon cryosections. Labeling of cells on acetone-fixed cryosections was performed with directly and indirectly labeled monoclonal antibodies (listed in Supplementary Table S3) and 4′,6-diamidino-2-phenylindole (DAPI) as a nuclear marker. After incubation with primary antibodies for 2 h at room temperature, the sections were stained with corresponding fluorescence-labeled secondary antibodies for 30 min at room temperature and subsequently stained with DAPI. Matched isotype controls were used as negative control. The stained tissue sections were obtained with a Z1 Axio Observer microscope using a LD Plan-Neofluar 20 × 0.4 objective (Zeiss).
Tissue processing for single-cell RNA-sequencing
Frozen colon samples (HC & CRC) were thawed using RPMI 1640 medium with 10% FBS. The cells were stained for viability and with the leukocyte marker CD45 in a 100 µl staining mix. Hashtag antibodies (4 μl, TotalSeq™-C0252 anti-human Hashtag 2; TotalSeq™-C0253 anti-human Hashtag 3 Antibody; BioLegend) − #2 for HC tissue and #3 for CRC tissue − were added and the samples were incubated for 30 minutes at 4 °C. After staining, the cells were washed using cold PBS. 10.000 viable CD45+ and CD45− cells from the HC sample and the CRC sample were sorted in 0.08% bovine serum albumin (BSA) in PBS. HC and CRC samples from the same patient were combined.
Single-cell RNA-sequencing, αβ-, γδ- TCR sequencing and bioinformatic analysis
Droplet-based single-cell RNA-seq was performed using the 10x Genomics Chromium Single Cell Controller with the Chromium Single Cell 5’ V3 Kit (10x Genomics) following the manufacturer’s instructions. After quality control, libraries were sequenced on the Illumina HiSeq 4000 platform in 2 × 75bp paired-end mode. Raw sequencing data were processed with the CellRanger v3.0.2 software (10x Genomics) for demultiplexing and alignment to the GRCh38 human reference transcriptome. Seurat (v5.0.1) was used for downstream 10x scRNA-seq data analysis67. Our dataset comprised 67,759 cells collected from 17 patients. Following rigorous filtration, cells exhibiting a mitochondria gene content surpassing 15%—indicative of cellular deterioration—were systematically eliminated, resulting in a refined cohort of 46,491 viable cells for subsequent analysis. Cells with <200 genes and >3000 numbers of detected genes were excluded, as were genes detected in <3 cells. We implemented the single-cell variational inference (scVI) method for batch correction and integration of data68. Raw unique molecular identifier counts were scaled and normalized through log transformation. The top 4000 variable genes were selected for principle-component analysis. The top 30 principal components were selected for Uniform Manifold Approximation and Projection (UMAP) and clustering analysis. Seurat function FindAllMarkers (logFC threshold = 0.25) was used to find cluster- and sample-specific marker genes. Further, UMAP clusters were annotated with known marker genes and marker genes detected by function FindAllMarkers. FindMarkers identified the differentially expressed genes between CRC and HC. The functions FeaturePlot, VlnPlot, DoHeatmap and DotPlot were used for feature plots, violins plots, heatmaps and dot plots, respectively, with default parameters.
KEGG pathway enrichment and GO enrichment analyses were done by using the KEGGA and GOANA methods respectively from limma package69. Enriched pathways were manually selected at corrected p value (p-adj < 0.05). We used scRepertoire (v1.4.0) to visualize the γδ T cell clones70 and calculate STARTRAC expa indices17. ScTCRγδ-seq and scTCRαβ-seq was used as input data. We employed GLIPH version 2 via the web portal to identify shared CDR3 motifs (http://50.255.35.37:8080)19. We combined raw count matrix for the integration of different data sets (Fig. 2). We further performed batch correction by using Harmony with Seurat. If available, scTCRγδ-seq was used to delineate γδ T cells. In cases where scTCRγδ-seq was not performed, we identified γδ T cell subsets based on the expression of TRDV1, TRDV2 or TRDV3. Double and triple positives were excluded from further analysis. We applied the CellChat package35 to perform cell-cell communication analyses among cell populations. We utilized the CellChatDB.human database for information on ligand-receptor interactions. Interactions were filtered with minimum cell <10. We used netVisual_circle and netVisual_bubble functions from CellChat to visualize the strength or weakness of cell-cell communication networks from the γδ T cells cluster to other cell clusters in the single-cell dataset. We have calculated the exhaustion marker scores and plotted by using the ggpubr function from ggscatter (v0.6.0) R package. We used TCGAbiolinks package to retrieve the TCGA clinical and molecular data for correlation analysis for MSS and MSI patients71. We identified MSS CRC samples as previously published72. For Fig. 5j, samples were included if both TIGIT and TRDV1 expression levels were above zero.
Treemaps were generated with Excel (version 16.16.27). Circos plots were generated using the online circos tool (http://circos.ca/circos_online).
Statistical analysis
The statistical analyses were performed with GraphPad PRISM (version 9.4.1, GraphPad Software). To assess the level of significance, paired t-tests were used when comparing two groups, and the two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was applied when comparing three or more groups. The level of significance is indicated for each experiment in the figures. Due to limited data availability matching patients based on relevant covariates to create comparable groups was not possible.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We are grateful to the Biomedical Sequencing Facility (BSF) at the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences for assistance with next-generation sequencing. The authors acknowledge the Core Facilities of the Medical University of Vienna, a member of VLSI. We thank the team of the Department of Pathology, Vienna Biobank KIP, Medical University of Vienna for clinical expertise. We thank Karl Thomanek for the editorial language review. Graphical abstracts were designed with BioRender. This work was supported by the Ingrid-Shaker-Nessmann Foundation granted to V.S. M.B. and M.F.F. have been funded by the Vienna Science and Technology Fund (WWTF) [10.47379/LS20042].
Author contributions
V.S., M.B., H.S., and N.P. designed the study. V.S. recruited patients, performed experiments, drafted the manuscript, analyzed flow cytometric and functional data. R.V.P. analyzed bioinformatic data. F.D. contributed crucial feedback to the bioinformatic analysis. J. List performed flow cytometry, cell sorting, and stimulation assays. L.K. prepared human tissues. M.F.F. oversaw library preparation for single-cell sequencing. J.K., J. Laengle, V.G., R.O., A.B.V., S.L., and G.S. provided critical input to the conceptualization of the study. L.W. provided clinical expertise on sample selection. M.B., H.S., and N.P. supervised the experiments. All authors contributed to writing the paper and provided critical feedback on the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Processed single-cell RNA-seq data have been deposited in Gene Expression Omnibus (GEO) under accession code GSE236384 Pelka et al.‘s dataset was accessed under the accession code GSE178341 Source data are provided with this paper.
Competing interests
The authors declare no competing interests regarding the published data. The research work of M.B. was supported by Boehringer-Ingelheim (BI), Bristol Myers Squibb (BMS) and a FFG spin off fellow-ship (CRC-OC-OV). M.B. received consultant fees and travel support from BI, BMS and Intuitive.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Michael Bergmann, Nina Pilat.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51025-1.
References
- 1.Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med.363, 711–723 (2010). 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overman, M. J. et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol.18, 1182–1191 (2017). 10.1016/S1470-2045(17)30422-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Vries, N. L. et al. gammadelta T cells are effectors of immunotherapy in cancers with HLA class I defects. Nature613, 743–750 (2023). 10.1038/s41586-022-05593-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soreide, K., Janssen, E. A., Soiland, H., Korner, H. & Baak, J. P. Microsatellite instability in colorectal cancer. Br. J. Surg.93, 395–406 (2006). 10.1002/bjs.5328 [DOI] [PubMed] [Google Scholar]
- 5.Gentles, A. J. et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med.21, 938–945 (2015). 10.1038/nm.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Born, W. et al. Peptide sequences of T-cell receptor delta and gamma chains are identical to predicted X and gamma proteins. Nature330, 572–574 (1987). 10.1038/330572a0 [DOI] [PubMed] [Google Scholar]
- 7.Hayday, A. C. et al. Structure, organization, and somatic rearrangement of T cell gamma genes. Cell40, 259–269 (1985). 10.1016/0092-8674(85)90140-0 [DOI] [PubMed] [Google Scholar]
- 8.Parker, C. M. et al. Evidence for extrathymic changes in the T cell receptor gamma/delta repertoire. J. Exp. Med.171, 1597–1612 (1990). 10.1084/jem.171.5.1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandoz, P. A. et al. Modulation of lytic molecules restrain serial killing in gammadelta T lymphocytes. Nat. Commun.14, 6035 (2023). 10.1038/s41467-023-41634-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todaro, M. et al. Efficient killing of human colon cancer stem cells by gammadelta T lymphocytes. J. Immunol.182, 7287–7296 (2009). 10.4049/jimmunol.0804288 [DOI] [PubMed] [Google Scholar]
- 11.Gao, Y. et al. Gamma delta T cells provide an early source of interferon gamma in tumor immunity. J. Exp. Med.198, 433–442 (2003). 10.1084/jem.20030584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramstead, A. G. & Jutila, M. A. Complex role of gammadelta T-cell-derived cytokines and growth factors in cancer. J. Interferon Cytokine Res.32, 563–569 (2012). 10.1089/jir.2012.0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reis, B. S. et al. TCR-Vgammadelta usage distinguishes protumor from antitumor intestinal gammadelta T cell subsets. Science377, 276–284 (2022). 10.1126/science.abj8695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rancan, C. et al. Exhausted intratumoral Vdelta2(-) gammadelta T cells in human kidney cancer retain effector function. Nat. Immunol.24, 612–624 (2023). [DOI] [PMC free article] [PubMed]
- 15.Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol.37, 38–44 (2019). [DOI] [PubMed]
- 16.Dominguez Conde, C. et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science376, eabl5197 (2022). 10.1126/science.abl5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature564, 268–272 (2018). 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- 18.Glanville, J. et al. Identifying specificity groups in the T cell receptor repertoire. Nature547, 94–98 (2017). 10.1038/nature22976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, H., Wang, C., Rubelt, F., Scriba, T. J. & Davis, M. M. Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat. Biotechnol.38, 1194–1202 (2020). 10.1038/s41587-020-0505-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelka, K. et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell184, 4734–4752.e4720 (2021). 10.1016/j.cell.2021.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, C. et al. Prioritizing exhausted T cell marker genes highlights immune subtypes in pan-cancer. iScience26, 106484 (2023). 10.1016/j.isci.2023.106484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, B. et al. Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat. Cancer3, 108–121 (2022). 10.1038/s43018-021-00292-8 [DOI] [PubMed] [Google Scholar]
- 23.Yu, L. et al. Tumor-infiltrating gamma delta T-cells reveal exhausted subsets with remarkable heterogeneity in colorectal cancer. Int. J. Cancer153, 1684–1697 (2023). 10.1002/ijc.34669 [DOI] [PubMed] [Google Scholar]
- 24.Odaira, K. et al. CD27(-)CD45(+) gammadelta T cells can be divided into two populations, CD27(-)CD45(int) and CD27(-)CD45(hi) with little proliferation potential. Biochem. Biophys. Res. Commun.478, 1298–1303 (2016). 10.1016/j.bbrc.2016.08.115 [DOI] [PubMed] [Google Scholar]
- 25.de Vries, N. L. et al. High-dimensional cytometric analysis of colorectal cancer reveals novel mediators of antitumour immunity. Gut69, 691–703 (2020). 10.1136/gutjnl-2019-318672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meraviglia, S. et al. Distinctive features of tumor-infiltrating gammadelta T lymphocytes in human colorectal cancer. Oncoimmunology6, e1347742 (2017). 10.1080/2162402X.2017.1347742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fattori, S. et al. Quantification of immune variables from liquid biopsy in breast cancer patients links Vdelta2(+) gammadelta T cell alterations with lymph node invasion. Cancers13, 441 (2021). [DOI] [PMC free article] [PubMed]
- 28.Weimer, P. et al. Tissue-specific expression of TIGIT, PD-1, TIM-3, and CD39 by gammadelta T cells in ovarian cancer. Cells11, 964 (2022). [DOI] [PMC free article] [PubMed]
- 29.Wherry, E. J. & Kurachi, M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol.15, 486–499 (2015). 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, Y. et al. Allogeneic Vgamma9Vdelta2 T-cell immunotherapy exhibits promising clinical safety and prolongs the survival of patients with late-stage lung or liver cancer. Cell Mol. Immunol.18, 427–439 (2021). 10.1038/s41423-020-0515-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Gassart, A. et al. Development of ICT01, a first-in-class, anti-BTN3A antibody for activating Vgamma9Vdelta2 T cell-mediated antitumor immune response. Sci. Transl. Med.13, eabj0835 (2021). 10.1126/scitranslmed.abj0835 [DOI] [PubMed] [Google Scholar]
- 32.Lin, M. et al. Irreversible electroporation plus allogenic Vgamma9Vdelta2 T cells enhances antitumor effect for locally advanced pancreatic cancer patients. Signal Transduct. Target Ther.5, 215 (2020). 10.1038/s41392-020-00260-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikulak, J. et al. NKp46-expressing human gut-resident intraepithelial Vdelta1 T cell subpopulation exhibits high antitumor activity against colorectal cancer. JCI Insight4, 24 (2019). [DOI] [PMC free article] [PubMed]
- 34.Maeurer, M. J. et al. Human intestinal Vdelta1+ lymphocytes recognize tumor cells of epithelial origin. J. Exp. Med.183, 1681–1696 (1996). 10.1084/jem.183.4.1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun.12, 1088 (2021). 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koncina, E. et al. IL1R1(+) cancer-associated fibroblasts drive tumor development and immunosuppression in colorectal cancer. Nat. Commun.14, 4251 (2023). 10.1038/s41467-023-39953-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinchuk, I. V. et al. PD-1 ligand expression by human colonic myofibroblasts/fibroblasts regulates CD4+ T-cell activity. Gastroenterology135, 1228–1237 (2008). 10.1053/j.gastro.2008.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, X. et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol.10, 48–57 (2009). 10.1038/ni.1674 [DOI] [PubMed] [Google Scholar]
- 39.Elyada, E. et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov.9, 1102–1123 (2019). 10.1158/2159-8290.CD-19-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, Z. et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat. Commun.11, 5077 (2020). 10.1038/s41467-020-18916-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, S. et al. An integrated map of fibroblastic populations in human colon mucosa and cancer tissues. Commun. Biol.5, 1326 (2022). 10.1038/s42003-022-04298-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, L. et al. Comprehensive analysis of CXCL14 uncovers its role during liver metastasis in colon cancer. BMC Gastroenterol.23, 273 (2023). 10.1186/s12876-023-02896-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng, J. et al. Chemokine CXCL14 is associated with prognosis in patients with colorectal carcinoma after curative resection. J. Transl. Med.11, 6 (2013). 10.1186/1479-5876-11-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, Y. et al. An innate-like Vdelta1(+) gammadelta T cell compartment in the human breast is associated with remission in triple-negative breast cancer. Sci. Transl. Med.11, eaax9364 (2019). [DOI] [PMC free article] [PubMed]
- 45.Wu, Y. et al. A local human Vdelta1 T cell population is associated with survival in nonsmall-cell lung cancer. Nat. Cancer3, 696–709 (2022). 10.1038/s43018-022-00376-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chabab, G. et al. Identification of a regulatory Vdelta1 gamma delta T cell subpopulation expressing CD73 in human breast cancer. J. Leukoc. Biol.107, 1057–1067 (2020). 10.1002/JLB.3MA0420-278RR [DOI] [PubMed] [Google Scholar]
- 47.Wu, P. et al. gammadeltaT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity40, 785–800 (2014). 10.1016/j.immuni.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mensurado, S. & Silva-Santos, B. Battle of the γδ T cell subsets in the gut. Trends Cancer8, 881–883 (2022). 10.1016/j.trecan.2022.08.006 [DOI] [PubMed] [Google Scholar]
- 49.Silva-Santos, B., Serre, K. & Norell, H. gammadelta T cells in cancer. Nat. Rev. Immunol.15, 683–691 (2015). 10.1038/nri3904 [DOI] [PubMed] [Google Scholar]
- 50.Noble, A. et al. Altered immunity to microbiota, B cell activation and depleted gammadelta/resident memory T cells in colorectal cancer. Cancer Immunol. Immunother.71, 2619–2629 (2022). 10.1007/s00262-021-03135-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davey, M. S. et al. Clonal selection in the human Vdelta1 T cell repertoire indicates gammadelta TCR-dependent adaptive immune surveillance. Nat. Commun.8, 14760 (2017). 10.1038/ncomms14760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papadopoulou, M. et al. TCR sequencing reveals the distinct development of fetal and adult human Vgamma9Vdelta2 T Cells. J. Immunol.203, 1468–1479 (2019). 10.4049/jimmunol.1900592 [DOI] [PubMed] [Google Scholar]
- 53.Tieppo, P. et al. The human fetal thymus generates invariant effector gammadelta T cells. J. Exp. Med.217, e20190580 (2020). [DOI] [PMC free article] [PubMed]
- 54.Eggesbo, L. M. et al. Single-cell TCR sequencing of gut intraepithelial gammadelta T cells reveals a vast and diverse repertoire in celiac disease. Mucosal Immunol.13, 313–321 (2020). 10.1038/s41385-019-0222-9 [DOI] [PubMed] [Google Scholar]
- 55.Mayassi, T. et al. Chronic inflammation permanently reshapes tissue-resident immunity in celiac disease. Cell176, 967–981.e919 (2019). 10.1016/j.cell.2018.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Di Marco Barros, R. et al. Epithelia use Butyrophilin-like molecules to shape organ-specific gammadelta T cell compartments. Cell167, 203–218.e217 (2016). 10.1016/j.cell.2016.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen, H. et al. Profiling the pattern of the human T-cell receptor gammadelta complementary determinant region 3 repertoire in patients with lung carcinoma via high-throughput sequencing analysis. Cell Mol. Immunol.16, 250–259 (2019). 10.1038/cmi.2017.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bengsch, B. et al. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity48, 1029–1045.e1025 (2018). 10.1016/j.immuni.2018.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott, A. C. et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature571, 270–274 (2019). 10.1038/s41586-019-1324-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, K. et al. Vdelta2 T cell subsets, defined by PD-1 and TIM-3 expression, present varied cytokine responses in acute myeloid leukemia patients. Int. Immunopharmacol.80, 106122 (2020). 10.1016/j.intimp.2019.106122 [DOI] [PubMed] [Google Scholar]
- 61.Ji, Q. et al. Primary tumors release ITGBL1-rich extracellular vesicles to promote distal metastatic tumor growth through fibroblast-niche formation. Nat. Commun.11, 1211 (2020). 10.1038/s41467-020-14869-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi, J. et al. Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat. Commun.13, 1742 (2022). 10.1038/s41467-022-29366-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng, Z., Ye, M., Ding, H., Feng, Z. & Hu, K. Spatial transcriptomics atlas reveals the crosstalk between cancer-associated fibroblasts and tumor microenvironment components in colorectal cancer. J. Transl. Med.20, 302 (2022). 10.1186/s12967-022-03510-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nazareth, M. R. et al. Characterization of human lung tumor-associated fibroblasts and their ability to modulate the activation of tumor-associated T cells. J. Immunol.178, 5552–5562 (2007). 10.4049/jimmunol.178.9.5552 [DOI] [PubMed] [Google Scholar]
- 65.Agorku, D. J., Bosio, A., Alves, F., Ströbel, P. & Hardt, O. Colorectal cancer-associated fibroblasts inhibit effector T cells via NECTIN2 signaling. Cancer Lett.595, 216985 (2024). 10.1016/j.canlet.2024.216985 [DOI] [PubMed] [Google Scholar]
- 66.Baran, B. et al. Difference between left-sided and right-sided colorectal cancer: a focused review of literature. Gastroenterol. Res.11, 264–273 (2018). 10.14740/gr1062w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol.36, 411–420 (2018). 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods15, 1053–1058 (2018). 10.1038/s41592-018-0229-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res.43, e47 (2015). 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borcherding, N., Bormann, N. L. & Kraus, G. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res9, 47 (2020). 10.12688/f1000research.22139.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Colaprico, A. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res.44, e71 (2016). 10.1093/nar/gkv1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu, Y. et al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell33, 721–735.e728 (2018). 10.1016/j.ccell.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed single-cell RNA-seq data have been deposited in Gene Expression Omnibus (GEO) under accession code GSE236384 Pelka et al.‘s dataset was accessed under the accession code GSE178341 Source data are provided with this paper.