Abstract
Artificial neural networks (ANNs) are biologically inspired algorithms designed to simulate the way in which the human brain processes information. In sample preparation for bioanalysis, liquid–liquid extraction (LLE) represents an important step with the extraction solvent selection is the key laborious step. In the current work, a robust and reliable ANNs model for LLE solvent prediction was generated which could predict the suitable solvent for analyte extraction. The developed ANNs model takes a set of chosen descriptors for the cited analyte as an input and predicts the corresponding Hansen solubility parameters of the suitable extraction solvent as a model output. Then, from the solvent combination’s appendix, the analyst can identify the proposed extraction solvents' combination for the cited analyte easily and efficiently. For the experimental validation of the model prediction capabilities, twenty structurally diverse drugs belonging to different pharmacological classes were extracted from human plasma. The extraction process was performed using the predicted extraction solvent combination for each drug and quantitively estimated by HPLC/UV methods to assess their extraction recovery. The developed LLE solvent prediction model is in- line with the global trend towards green chemistry since it limits the consumption of organic solvents.
Keywords: Liquid–liquid extraction, Artificial Neural Network model, Hansen solubility parameters, Extraction solvent prediction, Model validation
Subject terms: Analytical chemistry, Bioanalytical chemistry
Introduction
Identification and quantitation of analytes in biological fluids, such as whole blood, blood plasma, serum, urine, and saliva represent the most common definition of bioanalysis of pharmaceuticals. Bioanalysis has multidisciplinary applications, for example, in hospitals, it is essential to ensure that patients are properly medicated and compliant. In addition, bioanalysis plays an important role during the drug development and clinical trial stages for pharmacokinetics, bioequivalence, bioavailability and toxicokinetic investigation as well as for ADME studies performed for the newly developed drugs1,2. Many factors influence the development of a robust bioanalytical method including the matrices of interest, the range over which analytes need to be measured, the physicochemical properties of the analyte as well as the analyte(s) extraction process which is a very crucial step in most analytical procedures, especially bioanalytical ones3–5. To ensure the robustness of the developed analytical method, simplification of the complex biological sample should be carried out while keeping the analytes that are present in extremely low levels6. Thus, sample preparation is one of the key steps in bioanalytical procedures which is considered the most challenging one as it consumes time and effort for selecting the best extraction solvent system that efficiently extract the target analyte with high recovery and purity7.
A key technique in sample preparation is liquid–liquid extraction (LLE) which can provide extracts with low levels of the co-extracted matrix material. LLE involves the distribution of sample components between two immiscible liquid phases where analytes must be soluble in the extraction solvent and have high partition coefficients in it, for that, several solvent combinations are to be tested to achieve the best analyte extraction recovery8,9. LLE process offers several advantages such as simplicity of the technique, high throughput, elimination of environmental hazards, and high selectivity of separation10. Despite the obvious advantages of LLE, some challenges are associated with the traditional LLE such as multistage time-consuming procedures and the consumption of large amounts of organic solvents11,12.
Hansen Solubility Parameters (HSPs) were developed by Charles M. Hansen13,14 to predict if one substance will dissolve in a solvent forming a solution. Each solvent is given three HSPs which measure the interaction energies between its molecules and the solute, viz, dD (the dispersion interaction energy), dP (the dipolar intermolecular forces energy) and dH (the hydrogen bond energy). HSPs are powerful descriptors for evaluating interactions of molecules and their solubility in different liquids, thus, they have been used for several purposes such as understanding the solubility and dispersion properties of carbon nanotubes and buckyballs. Furthermore, they are used for the fast selection of safer and cheaper solvent combinations where an undesirable solvent can be rationally replaced by a combination of more desirable solvents whose combined HSPs equal those of the original solvent15–19.
Artificial neural networks are a commonly used machine learning algorithm for data modeling which adapt to complex relations between input and output data on the basis of their supervised learning20. In any modeling study, model validation is a crucial step as it evaluates the predictive ability of the generated model and ensures the model's significance and that the model results are not merely due to a statistical chance21. Model validation is carried out using internal validation (e.g., cross-validation) as well as external validation which uses unseen test set to validate that the obtained model is not merely the result of a descriptor-target property chance correlation22.
The primary aim of the current study is to develop a robust and reliable LLE solvent prediction model based on the analyte’s descriptors to predict the suitable solvent, or solvent combination, for the extraction of this specific analyte from aqueous-based matrices e.g., plasma. The developed model should save time and effort facilitating the extraction process and reducing the number of trials and the volume of consumed organic solvents making liquid–liquid extraction easier, straightforward and more eco-friendly in line with green chemistry aspects. For the experimental validation of the model prediction capabilities, twenty structurally diverse drugs from different pharmacological classes were extracted from human plasma using the model predicted solvent combination for each drug and quantitively estimated by HPLC/UV methods to study their extraction recovery.
Experimental
Materials and reagents
The used drugs were supplied by different pharmaceutical companies. HPLC-grade acetonitrile and methanol were purchased from Sigma-Aldrich (Germany). Ortho-phosphoric acid, acetic acid and potassium hydroxide were supplied by EL-Nasr Pharmaceutical Chemicals Co., Egypt. Potassium dihydrogen phosphate and ammonium acetate were supplied by Sigma-Aldrich (Germany). Bi-distilled water was produced in-house (Aquatron Water Still, A4000D, UK). Membrane filters of size 0.22 μm were purchased from ChromTech (UK). Human blank plasma was obtained from the Holding Company for Biological Products and Vaccines (VACSERA, Egypt) and stored at – 70 °C.
Instrumentation
The HPLC instrument (Agilent1100 series) was composed of an Agilent isocratic pump G1310A, Agilent UV–visible detector G1314A, an Agilent manual injector G1328B with (20 mL) injector loop and Inertsil ODS-3 column (5 µm, 150 mm × 4.6 mm). An Agilent syringe, (50 mL, USA) and a Powersonic 405 ultrasonic processor (Human Lab INC- Hwaseong city, Korea) were employed. The pH was adjusted by the addition of ortho-phosphoric acid or potassium hydroxide by means of a pH meter equipped with a glass electrode (Jenway, 3505, Essex, UK).
LLE modeling
Dataset construction
The extraction data of sixty-three structurally diverse drug molecules belonging to different pharmacological classes covering a wide range of physicochemical properties were self-collected from literature. The selected extraction solvents were ethyl acetate, diethyl ether, tert-butyl methyl ether, and dichloromethane, whereas drugs extracted with toxic solvents (e.g., chloroform) were excluded. The values of HSPs of the solvents were obtained from Dr Manuel Díaz de los Ríos, Director of Derivatives Division, ICIDCA23.
Drawing structures and molecular descriptors calculation
Molecular Operating Environment (MOE, 2020.0901) software was used for all the molecular modeling studies. Canonical SMILES of the sixty-three drugs were imported from PubChem24 into the MOE which were then converted into 3D structures. Energy minimization was performed for the built compounds until a RMS gradient of 0.05 kcal mol−1 Å−2 with MMFF94x force field and the partial charges were automatically calculated. MOE molecular mechanics descriptors were calculated for each compound and RapidMiner 7.1.000 Basic Edition25 was used to remove low variance descriptors using Remove Useless Attributes operator as they add no additional information to the model ''redundant descriptors'', this left a pool of 301 descriptors. Based on the relation of the different descriptors to the target parameters ''Hansen solubility parameters'', we found that dipole moment (Dipole), Van der Waals volume Å3 (Vdw_vol), Van der Waals energy (E_vdw), and log octanol/water partition coefficient (logP(o/w)) are the most important descriptors.
Training set and test set generation
The selected 63 drugs were split manually in a random manner into a training set of 48 molecules and an external test set of 15 molecules such that the test set maintains the same distribution of Hansen solubility parameters (HSPs) in the original dataset by keeping the ratio of the different solvents in the training and test sets equals to the original dataset (Supplementary Table S126–73 and Supplementary Table S274–88 in Supplementary File).
LLE model generation
MATLAB (version: 7.12.0.635) (R2011a) was used for generating the ANN models. Mean Absolute Error (MAE) is the model evaluation metric used to describe the average model performance. Linear Layer (design) was used in the ANN model generation.
Model validation
To assess the prediction ability and the robustness of the generated models, the developed model was validated using:
Internal validation: this was carried out using leave-20%-out cross-validation (CVL20%O) in which the training set was split into five subsets and training and test subsets were chosen such that each point appears in the test subset once. Five ANN models were generated using linear layer design network.
External validation: This was carried out by using the generated model to predict Hansen solubility parameters for the independent test set. This should be a direct simulation of the real case scenario which requires the prediction of new compounds (unseen by the model).
Experimental validation
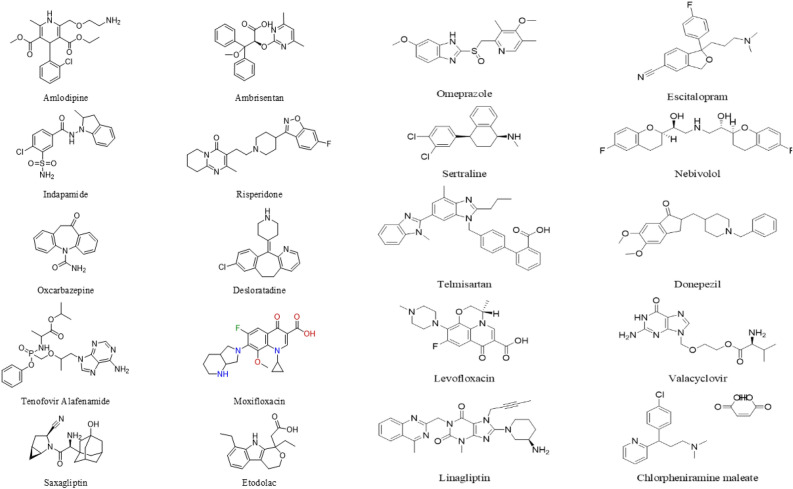
To test the generated model in a real case scenario, experimental validation of the model prediction was carried out. The developed ANN model was applied on twenty structurally diverse drugs from different pharmacological classes (Fig. 1) to predict their suitable extraction solvent combinations.
Figure 1.
Chemical structures of the investigated drugs.
Prediction of the HSPs of the extraction solvent combination
First, the model’s four descriptors of the 20 drugs were calculated using MOE (Supplementary Table S3 in Supplementary File) and then ANN linear layer design model was applied on them to predict the HSPs of the solvent combinations to be used to extract each drug and using the solvent combination appendix (Supplementary Table S4 in Supplementary File) the corresponding solvent mixture for each drug was determined based on its predicted HSPs.
Determination of the solute recovery from spiked plasma using the predicted solvent combinations
The twenty drugs were extracted from spiked human plasma using the predicted solvent combinations. Various mobile phases and chromatographic conditions were used for the separation and quantitation of those drugs using HPLC/UV methods (Supplementary Table S5 and Supplementary Fig. S1 in Supplementary File). Selectivity of the developed chromatographic methods was confirmed by the absence of any interfering peaks from plasma samples at the retention times of the investigated drugs (Supplementary Fig. S2 in Supplementary File).
Preparation of standard solutions
Stock solutions (1 mg/ml) were prepared by dissolving each drug in the appropriate HPLC-grade solvent (water, methanol or acetonitrile) and stored at 4 °C nominal. These stock solutions were diluted with a mixture of methanol and water (50: 50, v/v) to attain the required working solutions (100, 200 and 300 μg/ml).
Preparation of human plasma samples and analyte extraction
Plasma samples (0.5 ml) containing the analyte were vortexed for 30 s. The extraction solvent mixture was added to the spiked plasma and blank samples. Samples were vortexed for 1.5 min, centrifuged at 4500 rpm for 10 min. The clear supernatant was transferred into a clean Wassermann tube then evaporated to dryness at 45 °C under the stream of Nitrogen then dried extract was reconstituted with 100 μl of the mobile phase.
Procedure for extraction recovery calculations
The recovery following the sample preparation using the LLE model was evaluated by comparing the mean peak area of three extracted samples of low, medium, and high concentrations to the mean peak area of three plain standards of equivalent concentrations. Six replicates for each concentration were performed with the established extraction procedure.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Results and discussion
A correlation between some of the molecular mechanical descriptors of the drugs, dipole moment, Van der Waals volume, and log octanol/water partition coefficient and the target property (HSPs) of the extraction solvents using ANN was performed. The selection of the descriptors was based on showing high mutual solubility intercorrelation. The target property (HSPs) are physicochemical parameters that are commonly used to estimate the form of interactive forces that cause material compatibility. The HSP assumes that cohesive energy (E) can be divided into three parts: atomic dispersion (Ed), molecular dipolar interactions (Ep), and hydrogen-bonding interactions (Eh). ANNs are a type of computer programs that can be taught to mimic relationships in data sets. After the ANN has been ‘trained,' it can be used to predict the outcome of a new set of input data, such as a different composite system. Linear Layer (design) was used in the ANN model generation (Fig. 2). The generation was done using a custom script written on MATLAB (version: 7.12.0.635) (R2011a).
Figure 2.
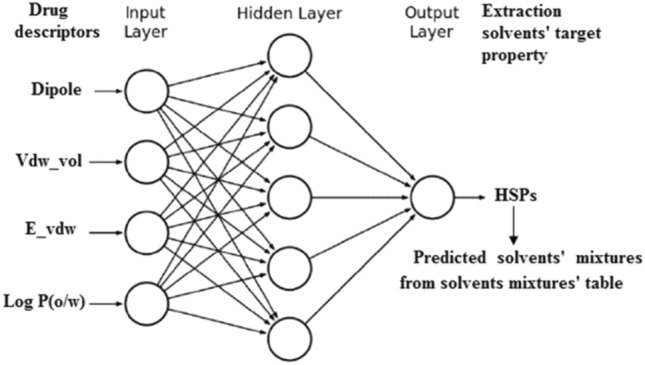
The developed ANN model structure.
The mean absolute error of a model represents the mean of the absolute values of the individual prediction errors on the overall instances in the dataset. Each prediction error is the difference between the predicted value and the true value for the instance.
where ŷi is the predicted value, yi is the true value, and n is the sample size.
MAE of internal validation
MAE of Hansen solubility parameters was found to be: 0.77 ± 0.48 for Hansen D, 1.19 ± 0.87 for Hansen P, and 1.12 ± 0.46 for Hansen H. The MAE of CV was calculated by absolute subtracting the predicted values from reported ones then divided by the number of the training set (Supplementary Table S6 in Supplementary File).
MAE of external validation
MAE of Hansen solubility parameters was found to be: 0.79 ± 0.56 for Hansen D, 1.14 ± 0.97 for Hansen P, and 1.23 ± 0.35 for Hansen H. the MAE of external validation was calculated by absolute subtracting the predicted values from reported ones then divided by the number of the test set (Supplementary Table S7 in Supplementary File).
Predicted HSPs of the investigated drugs and the predicted solvents' combinations
The predicted Hansen solubility parameters of the twenty drugs were obtained from the application of the developed ANN model on those drugs. For better extraction recovery results, the use of extraction solvents’ combination is recommended than the use of a single extraction solvent. Supplementary Table S5 in Supplementary File shows the Hansen solubility parameters of different combinations of the four extraction solvents used in the developed model with different ratios. The fraction of ratio of each solvent has been multiplied to its Hansen solubility parameters then HSP values for both solvents have been summed giving the HSPs for the solvents' combination. By visual inspection of the solvents' combinations' table and the predicted HSP values obtained from the model, one or two solvent combinations could be selected that have HSP values close to the predicted values obtained from the prediction model (Table 1).
Table 1.
Predicted Hansen solubility parameters and extraction solvents' mixtures of the investigated drugs using linear layer design model with their extraction recovery.
| Drug | Hansen D | Hansen P | Hansen H | Predicted solvents' mixture (1) | Predicted solvents' mixture (2) | Extraction recovery* |
|---|---|---|---|---|---|---|
| Amlodipine | 15.398 | 4.814 | 5.879 | Ethyl acetate: TBME (60:40)* | Diethyl ether: Dichloromethane (60:40) | 92.73 ± 0.34% |
| Ambrisentan | 15.904 | 5.533 | 6.250 | Dichloromethane: TBME (50:50) | Ethyl acetate: Dichloromethane (90:10)* | 55.93 ± 0.45% |
| Risperidone | 15.453 | 4.901 | 5.981 | Ethyl acetate: TBME (60:40) | Dichloromethane: TBME (30:70)* | 85.29 ± 0.35% |
| Indapamide | 15.557 | 5.024 | 6.208 | Ethyl acetate: TBME (70:30) | Ethyl acetate: Diethyl ether (80:20)* | 93.29 ± 0.48% |
| Tenofovir alfenamide | 15.045 | 4.336 | 5.807 | Ethyl acetate: TBME (30:70) | Ethyl acetate: Diethyl ether (50:50)* | 71.95 ± 0.40% |
| Oxcarbazepine | 16.129 | 5.812 | 6.553 | Ethyl acetate: Dichloromethane (80:20)* | Dichloromethane: TBME (60:40) | 88.78 ± 0.67% |
| Desloratadine | 15.776 | 5.330 | 6.118 | Dichloromethane: TBME (40:60)* | Diethyl ether: Dichloromethane (50:50) | 98.63 ± 0.41% |
| Saxagliptin | 15.554 | 5.021 | 6.322 | Ethyl acetate: Diethyl ether (70:30)* | Ethyl acetate: TBME (70:30) | 69.45 ± 0.60% |
| Moxifloxacin | 15.845 | 5.450 | 6.389 | Ethyl acetate: Diethyl ether (90:10)* | Dichloromethane: TBME (50:50) | 86.22 ± 0.69% |
| Etodolac | 15.262 | 4.595 | 5.764 | Ethyl acetate: TBME (40:60)* | Ethyl acetate: Diethyl ether (50:50) | 83.02 ± 0.31% |
| Omeprazole | 15.626 | 5.120 | 6.177 | Ethyl acetate: Diethyl ether (90:10) | Ethyl acetate: TBME (80:20)* | 98% ± 0.45% |
| Escitalopram | 15.324 | 4.696 | 5.723 | Ethyl acetate: TBME (50:50) | Diethyl ether: Dichloromethane (60:40)* | 87% ± 0.41% |
| Sertraline | 15.598 | 5.075 | 5.917 | Diethylether:Dichloromethane (50:50) | Dichloromethane: TBME (30:70)* | 91% ± 0.48% |
| Nebivolol | 15.708 | 5.257 | 6.141 | Ethyl acetate: Diethyl ether (90:10)* | Dichloromethane: TBME (40:60) | 87% ± 0.30% |
| Telmisartan | 14.902 | 4.158 | 5.035 | Diethyl ether: TBME (10:90) | Ethyl acetate: Diethyl ether (30:70)* | 53% ± 0.48% |
| Donepezil | 15.423 | 4.857 | 5.824 | Ethyl acetate: TBME (50:50) | Diethyl ether: Dichloromethane(60:40)* | 66% ± 0.56% |
| Levofloxacin | 16.215 | 5.962 | 6.752 | Ethyl acetate: Diethyl ether (90:10) | Diethyl ether: Dichloromethane (50:50)* | 61% ± 0.24% |
| Valacyclovir | 15.569 | 5.033 | 6.414 | Ethyl acetate: TBME (70:30)* | Ethyl acetate: Diethyl ether (80:20) | 92% ± 0.49% |
| Linagliptin | 15.611 | 5.140 | 6.098 | Ethyl acetate: Diethyl ether (90:10)* | Ethyl acetate: TBME (80:20) | 90% ± 0.36% |
| Chlorpheniramine maleate | 15.122 | 4.433 | 5.633 | Ethyl acetate: TBME (30:70)* | Ethyl acetate: Diethyl ether (40:60) | 94% ± 0.32% |
*Extraction recovery is the average recovery of three concentrations for each drug where each concentration was repeated six times.
*TBME tertiary-Butyl methyl ether.
Recovery of the investigated drugs
Recovery of each drug was performed by comparing the results obtained from the analysis of plasma spiked with three different concentrations to non-extracted samples of equivalent concentrations (Table 1).
Conclusion
A robust and validated LLE solvent prediction model which helps in predicting the organic extraction solvents' combinations for different drugs from aqueous-based matrices was built and validated. This was performed by making a correlation between some of the molecular mechanical descriptors of the drugs and the target property (HSPs) of the extraction solvents using ANN. Assessment of the prediction ability and the robustness of the generated model has been performed by internal and external validation. The generated ANN model has been applied on twenty drugs from different pharmacological classes. The extraction process of the investigated drugs was performed using the predicted extraction solvents' combination for each drug and quantitively estimated by HPLC/UV methods to study their extraction recovery. Good extraction recoveries were achieved. Therefore, bioanalysis could be much easier and more eco-friendly with the aid of the developed LLE solvent prediction model. The generated ANN model can be continuously improved by adding more input data to get more prediction capabilities.
Supplementary Information
Author contributions
Study conception and design was performed by E.F. and M.S. Material preparation, data collection and analysis were performed by F.H., E.A. and E.M. Molecular descriptors calculation and validation of the model was performed by A.M.. The first draft of the manuscript was written by F.H. and revised by M.S., A.A., E.F., M.A., A.M. and E.M. and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). The research was conducted without any funding sources.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files. Should any raw data files be needed in another format they are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-68981-9.
References
- 1.Hansen, F., Øiestad, E. L. & Pedersen-Bjergaard, S. Bioanalysis of pharmaceuticals using liquid-phase microextraction combined with liquid chromatography–mass spectrometry. J. Pharm. Biomed. Anal.189, 113446 (2020). 10.1016/j.jpba.2020.113446 [DOI] [PubMed] [Google Scholar]
- 2.Prabu, S. L., Suriyaprakash, T. N. K. Extraction of Drug from the Biological Matrix: A Review (IntechOpen, 2012).
- 3.Shah, V. P. The history of bioanalytical method validation and regulation: Evolution of a guidance document on bioanalytical methods validation. AAPS J.9, E43 (2007). 10.1208/aapsj0901005 [DOI] [Google Scholar]
- 4.Chan, C. C., Lee, Y. C., Lam, H., Zhang, X.-M. Analytical Method Validation and Instrument Performance Verification (Wiley, 2004).
- 5.Murugan, S., Pravallika, N., Sirisha, P. & Chandrakala, K. A review on bioanalytical method development and validation by using LC-MS/MS. J. Chem. Pharm. Sci.6, 41–45 (2013). [Google Scholar]
- 6.Sangster, T. & Oliver, M. Interview: Challenges faced by the modern bioanalytical laboratory. Bioanalysis.4, 2329–2333 (2012). 10.4155/bio.12.244 [DOI] [PubMed] [Google Scholar]
- 7.Pawliszyn, J. Sample preparation: quo vadis?. Anal. Chem.75, 2543–2558 (2003). 10.1021/ac034094h [DOI] [PubMed] [Google Scholar]
- 8.Wells, D. A. High throughput bioanalytical sample preparation-methods and automation strategies. Prog. Pharm. Biomed. Anal.2003, (2003).
- 9.Kyle, P. B. Toxicology: GCMS. In Mass Spectrom. Clin. Lab. 131–163 (Elsevier, 2017).
- 10.Kumar, A., Kishore, L., Kaur, N. & Nair, A. Method development and validation: Skills and tricks. Chronicles Young Sci.3, 3 (2012). 10.4103/2229-5186.94303 [DOI] [Google Scholar]
- 11.Rawa-Adkonis, M., Wolska, L., Przyjazny, A. & Namieśnik, J. Sources of errors associated with the determination of PAH and PCB analytes in water samples. Anal. Lett.39, 2317–2331 (2006). 10.1080/00032710600755793 [DOI] [Google Scholar]
- 12.Temerdashev, Z. A., Musorina, T. N., Chervonnaya, T. A. & Arutyunyan, Z. V. Possibilities and limitations of solid-phase and liquid extraction for the determination of polycyclic aromatic hydrocarbons in environmental samples. J. Anal. Chem.76, 1357–1370 (2021). 10.1134/S1061934821120133 [DOI] [Google Scholar]
- 13.Hansen, C. M. Hansen Solubility Parameters: A User’s Handbook (CRC Press, 2007).
- 14.Hansen, C. M. Polymer science applied to biological problems: Prediction of cytotoxic drug interactions with DNA. Eur. Polym. J.44, 2741–2748 (2008). 10.1016/j.eurpolymj.2008.07.005 [DOI] [Google Scholar]
- 15.Tait, J. G. et al. Determination of solvent systems for blade coating thin film photovoltaics. Adv. Funct. Mater.25, 3393–3398 (2015). 10.1002/adfm.201501039 [DOI] [Google Scholar]
- 16.Wang, S.-H. et al. Hansen solubility parameter analysis on the dispersion of zirconia nanocrystals. J. Colloid Interface Sci.407, 140–147 (2013). 10.1016/j.jcis.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 17.Wieneke, J. U., Kommoß, B., Gaer, O., Prykhodko, I. & Ulbricht, M. Systematic investigation of dispersions of unmodified inorganic nanoparticles in organic solvents with focus on the Hansen solubility parameters. Ind. Eng. Chem. Res.51, 327–334 (2012). 10.1021/ie201973u [DOI] [Google Scholar]
- 18.Süß, S., Sobisch, T., Peukert, W., Lerche, D. & Segets, D. Determination of Hansen parameters for particles: A standardized routine based on analytical centrifugation. Adv. Powder Technol.29, 1550–1561 (2018). 10.1016/j.apt.2018.03.018 [DOI] [Google Scholar]
- 19.Zuaznabar-Gardona, J. C. & Fragoso, A. Determination of the Hansen solubility parameters of carbon nano-onions and prediction of their dispersibility in organic solvents. J. Mol. Liq.294, 111646 (2019). 10.1016/j.molliq.2019.111646 [DOI] [Google Scholar]
- 20.Dondeti, S., Kannan, K. & Manavalan, R. Principal component artificial neural network calibration models for simultaneous spectrophotometric estimation of phenobarbitone and phenytoin sodium in tablets. Acta Chim. Slov.52, 138–144 (2005). [Google Scholar]
- 21.Fouad, M. A., Tolba, E. H., El-Shal, M. A., El Kerdawy, A. M. QSRR modeling for the chromatographic retention behavior of some β-lactam antibiotics using forward and firefly variable selection algorithms coupled with multiple linear regression. J. Chromatogr. A (2018). [DOI] [PubMed]
- 22.Vaghela, A., Patel, A., Patel, A., Vyas, A. & Patel, N. Sample preparation in bioanalysis: A review. Int. J. Sci. Technol. Res.5, 6–10 (2016). [Google Scholar]
- 23.Díaz de los Ríos, M., Hernández Ramos, E. Determination of the Hansen solubility parameters and the Hansen sphere radius with the aid of the solver add-in of Microsoft Excel. SN Appl. Sci.2, 1–7 (2020).
- 24.- The PubChem Project. https://pubchem.ncbi.nlm.nih.gov/.
- 25.- Data Science Platform|RapidMiner. https://rapidminer.com/.
- 26.Harahap, Y., Azizah, N. & Andalusia, R. Simultaneous analytical method development of 6-mercaptopurine and 6-methylmercaptopurine in plasma by high performance liquid chromatography-photodiode array. J. Young Pharm.9, S29–S34. 10.5530/jyp.2017.1s.8 (2017). 10.5530/jyp.2017.1s.8 [DOI] [Google Scholar]
- 27.Chandramowli, B. & Rajkamal, B. B. A validated LC-MS/MS method for the estimation of boceprevir and boceprevir D6 (IS) in human plasma employing liquid-liquid extraction. Int. J. Pharm. Pharm. Sci.8, 133–137 (2016). [Google Scholar]
- 28.Wang, Y. Y. et al. A sensitive, simple and rapid HPLC-MS/MS method for simultaneous quantification of buprenorpine and its N-dealkylated metabolite norbuprenorphine in human plasma. J. Pharm. Anal.3, 221–228. 10.1016/j.jpha.2012.12.002 (2013). 10.1016/j.jpha.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mürdter, T. E. et al. Sensitive and rapid quantification of busulfan in small plasma volumes by liquid chromatography-electrospray mass spectrometry. Clin. Chem.47, 1437–1442. 10.1093/clinchem/47.8.1437 (2001). 10.1093/clinchem/47.8.1437 [DOI] [PubMed] [Google Scholar]
- 30.Park, M. S., Shim, W. S., Yim, S. V. & Lee, K. T. Development of simple and rapid LC-MS/MS method for determination of celecoxib in human plasma and its application to bioequivalence study. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.902, 137–141. 10.1016/j.jchromb.2012.06.016 (2012). 10.1016/j.jchromb.2012.06.016 [DOI] [PubMed] [Google Scholar]
- 31.Oswald, S., Peters, J., Venner, M. & Siegmund, W. LC-MS/MS method for the simultaneous determination of clarithromycin, rifampicin and their main metabolites in horse plasma, epithelial lining fluid and broncho-alveolar cells. J. Pharm. Biomed. Anal.55, 194–201. 10.1016/j.jpba.2011.01.019 (2011). 10.1016/j.jpba.2011.01.019 [DOI] [PubMed] [Google Scholar]
- 32.Xie, Z. et al. Development and full validation of a sensitive quantitative assay for the determination of clemastine in human plasma by liquid chromatography-tandem mass spectrometry. J. Pharm. Biomed. Anal.44, 924–930. 10.1016/j.jpba.2007.03.019 (2007). 10.1016/j.jpba.2007.03.019 [DOI] [PubMed] [Google Scholar]
- 33.Rechberger, G. N. et al. Quantitative analysis of clindamycin in human plasma by liquid chromatography/electrospray ionisation tandem mass spectrometry using d1-N-ethylclindamycin as internal standard. Rapid Commun. Mass Spectrom.17, 135–139. 10.1002/rcm.887 (2003). 10.1002/rcm.887 [DOI] [PubMed] [Google Scholar]
- 34.Mohammadi, A., Kanfer, I., Sewram, V. & Walker, R. B. An LC-MS-MS method for the determination of cyclizine in human serum, J. Chromatogr. B Anal. Technol. Biomed. Life Sci.824, 148–152. 10.1016/j.jchromb.2005.07.015 (2005). 10.1016/j.jchromb.2005.07.015 [DOI] [PubMed] [Google Scholar]
- 35.Jiang, H. et al. A sensitive and accurate liquid chromatography–tandem mass spectrometry method for quantitative determination of the novel hepatitis C NS5A inhibitor BMS-790052 (daclastasvir) in human plasma and urine. J. Chromatogr. A.1245, 117–121. 10.1016/j.chroma.2012.05.028 (2012). 10.1016/j.chroma.2012.05.028 [DOI] [PubMed] [Google Scholar]
- 36.Hotha, K. K., Bharathi, D. V. & Jagadeesh, B. Development and validation of a highly sensitive LC-MS/MS method for quantitation of dexlansoprazole in human plasma: Application to a human pharmacokinetic study. Biomed. Chromatogr.26, 192–198. 10.1002/bmc.1645 (2012). 10.1002/bmc.1645 [DOI] [PubMed] [Google Scholar]
- 37.Arisoy, G. G. et al. Development and validation of HPLC-UV method for the determination of diclofenac in human plasma with application to a pharmacokinetic study. Turk. J. Pharm. Sci.13, 292–299. 10.4274/tjps.2016.02 (2016). 10.4274/tjps.2016.02 [DOI] [Google Scholar]
- 38.Liu, Y. et al. Determination of diethylstilbestrol in human plasma with measurement uncertainty estimation by liquid chromatography-tandem mass spectrometry. J. Liq. Chromatogr. Relat. Technol.37, 353–366. 10.1080/10826076.2012.745140 (2014). 10.1080/10826076.2012.745140 [DOI] [Google Scholar]
- 39.Liu, F., Luo, Y., Feng, J. L. & Hu, X. Y. Determination of dihydroetorphine in biological fluids by gas chromatography-mass spectrometry using selected-ion monitoring. J. Chromatogr. B Biomed. Appl.679, 113–118. 10.1016/0378-4347(96)00044-8 (1996). 10.1016/0378-4347(96)00044-8 [DOI] [PubMed] [Google Scholar]
- 40.Dasandi, B., Shah, S. & Shivprakash,. Development and validation of a high throughput and robust LC-MS/MS with electrospray ionization method for simultaneous quantitation of diltiazem and its two metabolites in human plasma: Application to a bioequivalence study. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.877, 791–798. 10.1016/j.jchromb.2009.02.016 (2009). 10.1016/j.jchromb.2009.02.016 [DOI] [PubMed] [Google Scholar]
- 41.Nichol, H., Vine, J. & Thomas, J. Quantification of doxapram in blood, plasma and urine. J. Chromatogr. B Biomed. Appl.182, 191–200 (1980). 10.1016/S0378-4347(00)81623-0 [DOI] [PubMed] [Google Scholar]
- 42.Kohlhof, K. J., Stump, D. & Zizzamia, J. A. Analysis of doxylamine in plasma by high-performance liquid chromatography. J. Pharm. Sci.72, 961–962. 10.1002/jps.2600720834 (1983). 10.1002/jps.2600720834 [DOI] [PubMed] [Google Scholar]
- 43.Ramachandran, G. et al. Simple and rapid liquid chromatography method for determination of efavirenz in plasma. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.835, 131–135. 10.1016/j.jchromb.2006.03.014 (2006). 10.1016/j.jchromb.2006.03.014 [DOI] [PubMed] [Google Scholar]
- 44.Sythana, S., Lavanya, A. S. K., Sankar, P. & Shanmugasundaram, V. Determination of entecavir in human plasma by LC-MS/MS and method validtion. Int. J. PharmTech Res.4, 1721–1729 (2012). [Google Scholar]
- 45.Bonnaire, Y., Plou, P., Pages, N., Boudene, C. & Jouany, J. M. GC/MS confirmatory method for etorphine in horse urine. J. Anal. Toxicol.13, 193–196. 10.1093/jat/13.4.193 (1989). 10.1093/jat/13.4.193 [DOI] [PubMed] [Google Scholar]
- 46.Chen, X., Gardner, E. R., Price, D. K. & Figg, W. D. Development and validation of an LC-MS assay for finasteride and its application to prostate cancer prevention trial sample analysis. J. Chromatogr. Sci.46, 356–361. 10.1093/chromsci/46.4.356 (2008). 10.1093/chromsci/46.4.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liapatas, G., Kousoulos, C. & Koupparis, M. A. LC-Ion Trap-MS method for the determination of fluconazole in plasma for bioequivalence studies of pharmaceutical formulations using semi-automated sample handling. J. Liq. Chromatogr. Relat. Technol.38, 1808–1814. 10.1080/10826076.2015.1113545 (2015). 10.1080/10826076.2015.1113545 [DOI] [Google Scholar]
- 48.Bae, J. W. et al. HPLC analysis of plasma glipizide and its application to pharmacokinetic study. J. Liq. Chromatogr. Relat. Technol.32, 1969–1977. 10.1080/10826070903091712 (2009). 10.1080/10826070903091712 [DOI] [Google Scholar]
- 49.Gonçalves, T. M. et al. Determination of indinavir in human plasma and its use in pharmacokinetic study. Rev. Bras. Ciencias Farm. J. Pharm. Sci.43, 639–647. 10.1590/S1516-93322007000400018 (2007). 10.1590/S1516-93322007000400018 [DOI] [Google Scholar]
- 50.Dwivedi, A., Singh, B., Sharma, S., Lokhandae, R. S. & Dubey, N. Ultra-performance liquid chromatography electrospray ionization-tandem mass spectrometry method for the simultaneous determination of itraconazole and hydroxy itraconazole in human plasma. J. Pharm. Anal.4, 316–324. 10.1016/j.jpha.2013.09.005 (2014). 10.1016/j.jpha.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu, M. L., Xu, M. & Ye, Q. Quantitative determination of ketoconazole by UPLC-MS/MS in human plasma and its application to pharmacokinetic study. Drug Res. (Stuttg)64, 548–552. 10.1055/s-0033-1363966 (2014). 10.1055/s-0033-1363966 [DOI] [PubMed] [Google Scholar]
- 52.Tang, J., Zhu, R., Zhao, R., Cheng, G. & Peng, W. Ultra-performance liquid chromatography-tandem mass spectrometry for the determination of lacidipine in human plasma and its application in a pharmacokinetic study. J. Pharm. Biomed. Anal.47, 923–928. 10.1016/j.jpba.2008.04.018 (2008). 10.1016/j.jpba.2008.04.018 [DOI] [PubMed] [Google Scholar]
- 53.Wichitnithad, W., Jithavech, P., Sanphanya, K., Vicheantawatchai, P. & Rojsitthisak, P. Determination of levocetirizine in human plasma by LC-MS-MS: Validation and application in a pharmacokinetic study. J. Chromatogr. Sci.53, 1663–1672. 10.1093/chromsci/bmv069 (2015). 10.1093/chromsci/bmv069 [DOI] [PubMed] [Google Scholar]
- 54.Ahmed, R. M., Hadad, G. M., El-Gendy, A. E. & Ibrahim, A. Development of HPLC method for determination of sitagliptin in human plasma using fluorescence detector by experimental design approach. Anal. Chem. Lett.8, 813–828. 10.1080/22297928.2018.1545603 (2018). 10.1080/22297928.2018.1545603 [DOI] [Google Scholar]
- 55.Kazemifard, A. G., Gholami, K. & Dabirsiaghi, A. Optimized determination of lorazepam in human serum by extraction and high-performance liquid chromatographic analysis. Acta Pharm.56, 481–488 (2006). [PubMed] [Google Scholar]
- 56.Young, H. K., Hye, Y. J., Park, E. S., Chae, S. W. & Hye, S. L. Liquid chromatography-electrospray ionization tandem mass spectrometric determination of lornoxicam in human plasma. Arch. Pharm. Res.30, 905–910. 10.1007/bf02978844 (2007). 10.1007/bf02978844 [DOI] [PubMed] [Google Scholar]
- 57.Siddiraju, S., Lal Prasanth, M. L. & Sirisha, T. A novel LC-MS/MS assay for methylprednisolone in human plasma and its pharmacokinetic application. Asian J. Pharm. Sci.11, 459–468. 10.1016/j.ajps.2015.06.005 (2016). 10.1016/j.ajps.2015.06.005 [DOI] [Google Scholar]
- 58.Lee, H. W. et al. Determination of metoclopramide in human plasma using hydrophilic interaction chromatography with tandem mass spectrometry. J Chromatogr. B Anal. Technol. Biomed. Life Sci.877, 1716–1720. 10.1016/j.jchromb.2009.04.027 (2009). 10.1016/j.jchromb.2009.04.027 [DOI] [PubMed] [Google Scholar]
- 59.Chen, Y. A. & Hsu, K. Y. Development of a LC-MS/MS-based method for determining metolazone concentrations in human plasma: Application to a pharmacokinetic study. J. Food Drug Anal.21, 154–159. 10.1016/j.jfda.2013.05.004 (2013). 10.1016/j.jfda.2013.05.004 [DOI] [Google Scholar]
- 60.Dalton, J. T., Geuns, E. R. & Lai-Sim Au, J. High-performance liquid chromatographic determination of mitomycin C in rat and human plasma and urine. J. Chromatogr. B Biomed. Sci. Appl.495, 330–337. 10.1016/S0378-4347(00)82641-9 (1989). 10.1016/S0378-4347(00)82641-9 [DOI] [PubMed] [Google Scholar]
- 61.Janchawee, B. et al. A high-performance liquid chromatographic method for determination of mitragynine in serum and its application to a pharmacokinetic study in rats. Biomed. Chromatogr.21, 176–183. 10.1002/bmc (2007). 10.1002/bmc [DOI] [PubMed] [Google Scholar]
- 62.Sahoo, N. K., Sahu, M., Rao, P. S. & Ghosh, G. Development and validation of liquid chromatography-mass spectroscopy/mass spectroscopy method for quantitative analysis of naproxen in human plasma after liquid-liquid extraction. Trop. J. Pharm. Res.13, 1503–1510. 10.4314/tjpr.v13i9.17 (2014). 10.4314/tjpr.v13i9.17 [DOI] [Google Scholar]
- 63.Chen, H. et al. Development and validation of a rapid andsensitive UHPLC-MS/MS method for thedetermination of paliperidone in beagle dog plasma, Asian. J. Pharm. Sci.9, 286–292. 10.1016/j.ajps.2014.07.008 (2014). 10.1016/j.ajps.2014.07.008 [DOI] [Google Scholar]
- 64.Kaddoumi, A., Mori, T., Nakashima, M. N., Wada, M. & Nakashima, K. High performance liquid chromatography with fluorescence detection for the determination of phenylpropanolamine in human plasma and rat’s blood and brain microdialysates using DIB-Cl as a label. J. Pharm. Biomed. Anal.34, 643–650. 10.1016/S0731-7085(03)00633-2 (2004). 10.1016/S0731-7085(03)00633-2 [DOI] [PubMed] [Google Scholar]
- 65.Souri, E., Jalalizadeh, H. & Saremi, S. Development and validation of a simple and rapid Hplc method for determination of pioglitazone in human plasma and its application to a pharmacokinetic study. J. Chromatogr. Sci.46, 809–812. 10.1093/chromsci/46.9.809 (2008). 10.1093/chromsci/46.9.809 [DOI] [PubMed] [Google Scholar]
- 66.Matabosch, X. et al. Detection and characterization of prednisolone metabolites in human urine by LC-MS/MS. J. Mass Spectrom.50, 633–642. 10.1002/jms.3571 (2015). 10.1002/jms.3571 [DOI] [PubMed] [Google Scholar]
- 67.Nirogi, R., Kandikere, V. & Mudigonda, K. Sensitive liquid chromatography positive electrospray tandem mass spectrometry method for the quantitation of tegaserod in human plasma using liquid-liquid extraction. J. Chromatogr. Sci.47, 164–169. 10.1093/chromsci/47.2.164 (2009). 10.1093/chromsci/47.2.164 [DOI] [PubMed] [Google Scholar]
- 68.Gilant, E., Kaza, M., Szlagowska, A., Serafin-Byczak, K. & Rudzki, P. J. Validated HPLC method for determination of temozolomide in human plasma. Acta Pol. Pharm. Drug Res.69, 1347–1355 (2012). [PubMed] [Google Scholar]
- 69.Langmann, P. et al. High performance liquid chromatographic method for the determination of HIV-1 protease inhibitor tipranavir in plasma of patients during highly active antiretroviral therapy. Eur. J. Med. Res.13, 52–58 (2008). [PubMed] [Google Scholar]
- 70.Singhal, P. et al. Enantiomeric separation of verapamil and its active metabolite, norverapamil, and simultaneous quantification in human Plasma by LC-ESI-MS-MS. J. Chromatogr. Sci.50, 839–848. 10.1093/chromsci/bms080 (2012). 10.1093/chromsci/bms080 [DOI] [PubMed] [Google Scholar]
- 71.Yang, F., Wang, H., Hu, P. & Jiang, J. Validation of an UPLC-MS-MS method for quantitative analysis of vincristine in human urine after intravenous administration of vincristine sulfate liposome injection. J. Chromatogr. Sci.53, 974–978. 10.1093/chromsci/bmu164 (2015). 10.1093/chromsci/bmu164 [DOI] [PubMed] [Google Scholar]
- 72.Qian, J. et al. Rapid and sensitive determination of vinorelbine in human plasma by liquid chromatography-tandem mass spectrometry and its pharmacokinetic application. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.879, 662–668. 10.1016/j.jchromb.2011.01.039 (2011). 10.1016/j.jchromb.2011.01.039 [DOI] [PubMed] [Google Scholar]
- 73.Jiang, Y. et al. Development and validation of a liquid chromatography-tandem mass spectrometry method for the determination of zofenopril and its active metabolite zofenoprilat in human plasma. J. Pharm. Biomed. Anal.55, 527–532. 10.1016/j.jpba.2011.02.010 (2011). 10.1016/j.jpba.2011.02.010 [DOI] [PubMed] [Google Scholar]
- 74.Patil, J. S., Suresh, S., Sureshbabu, A. & Rajesh, M. Development and validation of liquid chromatography-Mass spectrometry method for the estimation of rifampicin in plasma. Indian J. Pharm. Sci.73, 558–563. 10.4103/0250-474X.99014 (2011). 10.4103/0250-474X.99014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu, C. et al. Simultaneous determination of ivabradine and N-desmethylivabradine in human plasma and urine using a LC-MS/MS method: Application to a pharmacokinetic study. Acta Pharm. Sin. B.2, 205–212. 10.1016/j.apsb.2012.01.004 (2012). 10.1016/j.apsb.2012.01.004 [DOI] [Google Scholar]
- 76.Zhou, N. et al. Development and validation of LC-MS method for the determination of hydroxyzine hydrochloride in human plasma and subsequent application in a bioequivalence study. Chromatographia.66, 481–486. 10.1365/s10337-007-0372-x (2007). 10.1365/s10337-007-0372-x [DOI] [Google Scholar]
- 77.Yuan, G., Rong, L., Duanyun, S. & Changxiao, L. Determination of 5-fluorouracil in human plasma by highperformance liquid chromatography (HPLC). Trans. Tianjin Univ.16, 167–173. 10.1007/s12209 (2010). 10.1007/s12209 [DOI] [Google Scholar]
- 78.Shah, I., Baker, J., Batron, S. J. & Naughton, D. P. A novel method for determination of fenofibric acid in human plasma using HPLC-UV: Application to a pharmacokinetic study of new formulations. J. Anal. Bioanal. Tech. S12, 5–8. 10.4172/2155-9872.s12-009 (2014). 10.4172/2155-9872.s12-009 [DOI] [Google Scholar]
- 79.Dalmora, S. L. et al. Determination of phenobarbital in human plasma by a specific liquid chromatography method: Application to a bioequivalence study. Quim. Nova.33, 124–129. 10.1590/S0100-40422010000100023 (2010). 10.1590/S0100-40422010000100023 [DOI] [Google Scholar]
- 80.Satyadev, T. N., Ch, B. & Sundar, B. S. Development and validation of high performance liquid chromatographic method for the determination of Dolutegravir in human plasma. Der. Pharm. Sin.6(4), 65–72 (2015). [Google Scholar]
- 81.Kim, M. S. et al. Quantification of nimesulide in human plasma by high-performance liquid chromatography with ultraviolet detector (HPLC-UV): Application to pharmacokinetic studies in 28 healthy korean subjects. J. Chromatogr. Sci.50, 396–400. 10.1093/chromsci/bms014 (2012). 10.1093/chromsci/bms014 [DOI] [PubMed] [Google Scholar]
- 82.Park, J. H. et al. Quantification of isradipine in human plasma using LC-MS/MS for pharmacokinetic and bioequivalence study. J Chromatogr. B Anal. Technol. Biomed. Life Sci.877, 59–64. 10.1016/j.jchromb.2008.11.021 (2009). 10.1016/j.jchromb.2008.11.021 [DOI] [PubMed] [Google Scholar]
- 83.Manimala, M. & Karpagam, S. LC-MS-MS method for the determination of digoxin in human plasma. Int. J. Pharm. Pharm. Sci.5, 131–132 (2013). [Google Scholar]
- 84.Zhao, Y. et al. Determination of nimodipine in human plasma by HPLC-ESI-MS and its application to a bioequivalence study. J. Chromatogr. Sci.48, 81–85. 10.1093/chromsci/48.2.81 (2010). 10.1093/chromsci/48.2.81 [DOI] [PubMed] [Google Scholar]
- 85.Salem, I. I., Alkhatib, M. & Najib, N. LC-MS/MS determination of betamethasone and its phosphate and acetate esters in human plasma after sample stabilization. J. Pharm. Biomed. Anal.56, 983–991. 10.1016/j.jpba.2011.07.020 (2011). 10.1016/j.jpba.2011.07.020 [DOI] [PubMed] [Google Scholar]
- 86.Jaussaud, P. et al. Pharmacokinetics of tolfenamic acid in the horse. Equine Vet. J. Suppl.10.1111/j.2042-3306.1992.tb04778.x (1992). 10.1111/j.2042-3306.1992.tb04778.x [DOI] [PubMed] [Google Scholar]
- 87.Yan, M. et al. Quantification of prochlorperazine maleate in human plasma by liquid chromatography-mass spectrometry: Application to a bioequivalence study. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.877, 3243–3247. 10.1016/j.jchromb.2009.07.038 (2009). 10.1016/j.jchromb.2009.07.038 [DOI] [PubMed] [Google Scholar]
- 88.Kang, W., Liu, K. H., Ryu, J. Y. & Shin, J. G. Simultaneous determination of ebastine and its three metabolites in plasma using liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.813, 75–80 (2004). 10.1016/j.jchromb.2004.09.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files. Should any raw data files be needed in another format they are available from the corresponding author upon reasonable request. Source data are provided with this paper.