Abstract
The human cytomegalovirus US3, an endoplasmic reticulum (ER)-resident transmembrane glycoprotein, forms a complex with major histocompatibility complex (MHC) class I molecules and retains them in the ER, thereby preventing cytolysis by cytotoxic T lymphocytes. To identify which parts of US3 confine the protein to the ER and which parts are responsible for the association with MHC class I molecules, we constructed truncated mutant and chimeric forms in which US3 domains were exchanged with corresponding domains of CD4 and analyzed them for their intracellular localization and the ability to associate with MHC class I molecules. All of the truncated mutant and chimeric proteins containing the luminal domain of US3 were retained in the ER, while replacement of the US3 luminal domain with that of CD4 led to cell surface expression of the chimera. Thus, the luminal domain of US3 was sufficient for ER retention. Immunolocalization of the US3 glycoprotein after nocodazole treatment and the observation that the carbohydrate moiety of the US3 glycoprotein was not modified by Golgi enzymes indicated that the ER localization of US3 involved true retention, without recycling through the Golgi. Unlike the ER retention signal, the ability to associate with MHC class I molecules required the transmembrane domain in addition to the luminal domain of US3. Direct interaction between US3 and MHC class I molecules could be demonstrated after in vitro translation by coimmunoprecipitation. Together, the present data indicate that the properties that allow US3 to be localized in the ER and bind MHC class I molecules are located in different parts of the molecule.
The importance of cytotoxic T-lymphocyte (CTL)-mediated immune responses in limiting and clearing viral infections has been well documented for a number of viral systems (11). Human cytomegalovirus (HCMV) causes benign but persistent infections in immunocompetent individuals. This implies a balance between immune control of the virus and immune escape by the virus (40). A number of viruses encode proteins that can inhibit or abolish the surface expression of major histocompatibility complex (MHC) class I molecules on infected cells. HCMV encodes an endoplasmic reticulum (ER)-resident glycoprotein, US3, that prevents intracellular transport of MHC class I molecules (1, 22). HCMV US3 binds physically to MHC class I heterodimers and sequesters them in the ER. Therefore, the downregulation of MHC class I molecules by US3 very likely serves to protect HCMV-infected cells from CTL recognition. The primary structure of the US3 protein (1) consists of a signal sequence of 15 amino acids followed by a luminal domain of 146 amino acids. This portion of the US3 protein is separated from a short cytoplasmic tail of 5 amino acids by 20 membrane-spanning residues. The protein contains an N-glycosylation site in the luminal domain.
At least two separate properties of the US3 protein make it particularly interesting. First, a 7-kb region of the US part of the HCMV genome encodes a family of eight type I glycoproteins of 20 to 30 kDa (21) (US2, US3, US6, and US7 to US11), all of which share some degree of sequence homology (1, 6) and are dispensable for viral replication (21). Despite their structural relatedness, some members (US2, US3, US6, and US11) of this family are independently capable of preventing MHC class I surface expression while the others (US7, US8, US9, and US10) do not affect the intracellular transport of MHC class I molecules (2). More interestingly, the molecular mechanisms by which US2, US3, US6, and US11 downregulate the cell surface expression of MHC class I molecules are quite different. US2 and US11 induce the rapid export of MHC class I molecules out of the ER into the cytosol, where they are degraded by proteasomes (54, 55). US6 inhibits transporter associated with antigen processing (TAP)-mediated peptide translocation (2). It was therefore of interest to find out in what properties US3 differs from the other proteins of the US family with regard to its unique action on MHC class I molecules.
A second interesting property of the US3 protein is its cellular localization. In general, ER proteins can reach their specific localization either by direct retention or by retrieval from distal compartments in the secretory pathway. The mammalian KDEL and yeast HDEL sequence at the carboxyl-terminal end has been shown to function as an ER retention signal for ER luminal proteins (32). The carboxyl-terminal dilysine motif (KKXX or KXKXX) of type I transmembrane proteins has also been characterized as an ER retention signal (20, 33). In a manner analogous to the action of US3, the E19 protein of adenovirus type 2 binds to MHC class I molecules, thereby interfering with their cell surface expression. Its cytosolic tail contains a dilysine motif which is both necessary and sufficient for ER localization (19). It is widely believed that ER proteins containing the K(H)DEL or dilysine motif are recognized by a receptor in the Golgi and shuttled back to the ER (27, 38). Some ER membrane proteins do not contain KKXX-like signals but seem to be restricted to the ER without undergoing retrieval (15, 51). Since US3 does not contain any known ER retention signals such as the carboxyl-terminal dilysine consensus motif, the mechanism by which US3 becomes an ER resident is not yet clear. In this study, we examined whether the ability of US3 to bind MHC class I molecules and to be retained in the ER is vested in one or indifferent parts of the molecule. We found that the luminal domain of the US3 protein is sufficient for retention in the ER and that the ER localization of US3 involves true retention without recycling through the Golgi. On the other hand, the transmembrane domain, in addition to the luminal domain, was required for the interaction of US3 with MHC class I molecules. Our results also showed that US3 directly interacts with MHC class I molecules in vitro.
MATERIALS AND METHODS
Cell lines and cell culture.
HeLa cells were cultured in minimum essential medium (Life Technologies, Rockville, Md.) supplemented with 10% fetal bovine serum (HyClone, Logan, Utah), penicillin (50 U/ml), and streptomycin (50 μg/ml). HLA-A, -B, and -C-negative LCL 721.221 and tapasin-negative LCL 721.220 cells were cultured in RPMI 1640 medium (Life Technologies) (10, 16). TAP1/TAP2-negative T2 (44) and calnexin-negative CEM-NKR (46) cells were cultured in Iscove modified Dulbecco medium (Life Technologies).
Transfection and viral infection.
The mammalian expression vector was transfected into the cells by the calcium phosphate precipitation method (7). Recombinant vaccinia viruses expressing US3 were generated by homologous recombination essentially as previously described (9) and plaque purified three times on thymidine kinase-deficient 143B cells under bromodeoxyuridine (50 μg/ml) selection. Cells were infected with recombinant vaccinia viruses at a multiplicity of infection of 25 PFU/cell for 1 h in 500 μl of RPMI 1640 medium supplemented with 10% bovine serum albumin (Sigma, St. Louis, Mo.) at 37°C.
Constructs.
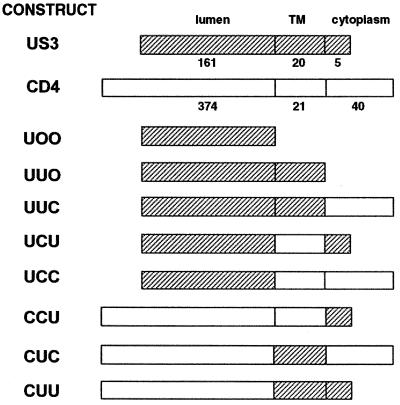
Plasmids expressing chimeric proteins were constructed as shown in Fig. 1. Respective DNA fragments were obtained by either restriction digestion or PCR amplification. Chimeric proteins are designated by three letters, which refer to the luminal (extracellular), transmembrane, and cytoplasmic domains. U, C, and O refer to US3, human CD4, and no domain, respectively. For enzymatic manipulation, the unique restriction sites BglII and ClaI were introduced at the junctions between the luminal and transmembrane domains and between the transmembrane and cytoplasmic domains of US3, respectively. These caused the addition of three amino acids, YRL and ADI, at the junctions, respectively. Chimera UCC is a cDNA that encodes the luminal domain of US3 (amino acids 1 to 161) attached to the transmembrane and cytoplasmic domains of CD4 (amino acids 375 to 435). CUU is a cDNA that encodes the luminal domain of CD4 (amino acids 1 to 374) fused to the transmembrane and cytoplasmic domains of US3 (amino acids 162 to 186). UUC contains the luminal and transmembrane domains of US3 (amino acids 1 to 181), followed by the cytoplasmic domain of CD4 (amino acids 396 to 435). CCU contains the luminal and transmembrane domains of CD4 (to amino acid 395), followed by the cytoplasmic domain of US3 (amino acid sequence RLRFI at positions 182 to 186). UCU contains the luminal domain of US3 (amino acids 1 to 161), followed by the transmembrane domain of CD4 (amino acids 375 to 395) and the cytoplasmic domain of US3 (amino acids 182 to 186). CUC contains the luminal domain of CD4 (to amino acid 374), followed by the transmembrane domain of US3 (amino acids 162 to 181) and the cytoplasmic domain of CD4 (amino acids 396 to 435). UOO and UUO were constructed by introducing a stop codon at amino acid positions 161 and 182, respectively. All constructs were confirmed by DNA sequencing, and the constructs were subsequently subcloned into mammalian cell expression vector pcDNA3.1 (Invitrogen, San Diego, Calif.).
FIG. 1.
Schematic representation of chimeras consisting of US3 and CD4. The three letters represent the luminal, transmembrane (TM), and cytoplasmic domains, respectively, and U refers to US3, C refers to CD4, and O indicates lack of any domain. Each domain is shown as either a filled box (US3 origin) or an open box (CD4 origin). Details of the constructions are described in Materials and Methods.
Antibodies.
MHC class I-specific antisera K455 and K355 were raised against purified human class I heterodimers with human β2-microglobulin (β2m) or human β2m, respectively (3). K455 recognizes the MHC class I heavy chain (HC) and β2m in both assembled and nonassembled forms. K355 recognizes both free and complexed β2m. Monoclonal antibody (MAb) W6/32 recognizes only the complex of HC and β2m, and MAb OKT4 specifically reacts with human CD4 (24). Polyclonal antiserum detecting US3 was raised against the synthetic peptides corresponding to the luminal NH2-terminal portion of the proteins (1). Rabbit polyclonal antibody to PDI (SPA-890) was purchased from Stress Gen (Victoria, British Columbia, Canada). Rabbit polyclonal antibody to p58 (25) and mannosidase II (30) were kindly provided by Ralf F. Pettersson (Ludwig Institute for Cancer Research) and K. Moremen (University of Georgia), respectively. Fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit and anti-mouse immunoglobulin G (IgG) were purchased from Sigma.
Metabolic labeling and immunoprecipitation.
Cells were methionine starved for 30 min in a methionine-free medium prior to pulse-labeling for 30 min using [35S]methionine (TranS-label; Amersham, Arlington Heights, Ill.) at 0.1 mCi/ml. The label was chased at various time points with minimum essential medium containing 10% fetal bovine serum. After one wash with cold phosphate-buffered saline (PBS), cells were lysed using 1% Nonidet P-40 (NP-40; Sigma) in PBS or 1% digitonin (Calbiochem) in PBS for 30 min at 4°C. After incubation with primary antibody, the lysates were incubated with protein A-Sepharose beads (Pharmacia, Uppsala, Sweden). The beads were washed four times with 0.1% NP-40 or 0.1% digitonin, and the immunoprecipitates were eluted by boiling in sodium dodecyl sulfate (SDS) sample buffer and separated by SDS-polyacrylamide gel electrophoresis (PAGE). The gel was dried, exposed to BAS film, and analyzed by Phosphor Imaging System BAS-2500 (Fuji Film Company). For endo-N-acetylglucosaminidase H (endo H) treatment, immunoprecipitates were digested with 3 mU of endo H (Boehringer, Mannheim, Germany) for 16 h at 37°C in 50 mM sodium acetate (NaOAc; pH 5.6)–0.3% SDS–150 mM β-mercaptoethanol. For endo-N-acetylglucosaminidase D (endo D) treatment, immunoprecipitates were washed and then boiled in 10 μl of 50 mM NaOAc (pH 5.6)–0.5% SDS. Then, 10 μl of 50 mM NaOAc (pH 5.6)–40 mM EDTA (pH 7.5)–3% Triton X-100–2 mU of endo D (Boehringer) was added and the mixture was incubated overnight at 37°C.
In vitro transcription and translation.
HLA-A2.1, β2m, and US3 proteins were in vitro transcribed and translated by using a T7-coupled rabbit reticulocyte lysate system (Promega) in accordance with the manufacturer's instructions. Each cDNA was subcloned into plasmid pcDNA3.1 (Invitrogen). The reaction was carried out in the presence of [35S]methionine and canine pancreatic microsomes. After the reaction, the microsomes were sedimented (10 min, 100,000 × g) and lysed in 1% digitonin lysis buffer. Immunoprecipitation was done as described above. For reprecipitation, immunoprecipitated material was denatured in 1% SDS at 100°C for 10 min and, after dilution to 0.05% SDS with 1% NP-40 in PBS, again immunoprecipitated with the respective antibodies.
Flow cytometric analysis and immunofluorescence.
Expression of MHC class I glycoproteins on the membrane was determined by flow cytometry (FACScalibur; Becton Dickinson, Mountain View, Calif.) after indirect immunofluorescence using anti-MHC class I MAb W6/32 and an FITC-conjugated goat anti-mouse antibody. For immunofluorescent staining of permeabilized cells, HeLa cells were fixed in 4% formaldehyde and permeabilized with 0.1% Triton X-100, followed by incubation with the appropriate primary antibody for 1 h. Bound antibody was visualized with an FITC-conjugated secondary antibody. Cell surface staining of human CD4 was obtained with MAb OKT4, followed by secondary-antibody incubation. For treatment with nocodazole, cells on coverslips were incubated with medium containing 20 μM nocodazole (5-mg/ml stock in dimethyl sulfoxide) at 37°C in a CO2 incubator.
RESULTS
The luminal domain of US3 is sufficient for its retention in the ER.
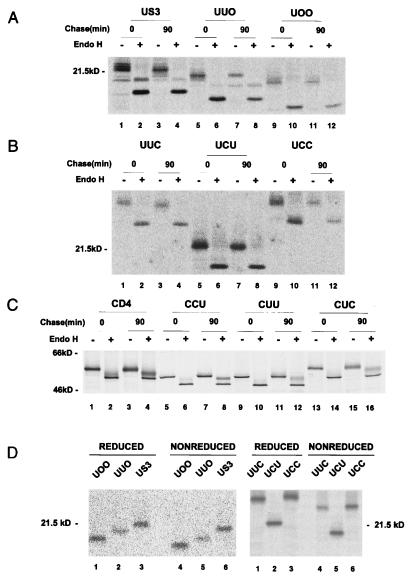
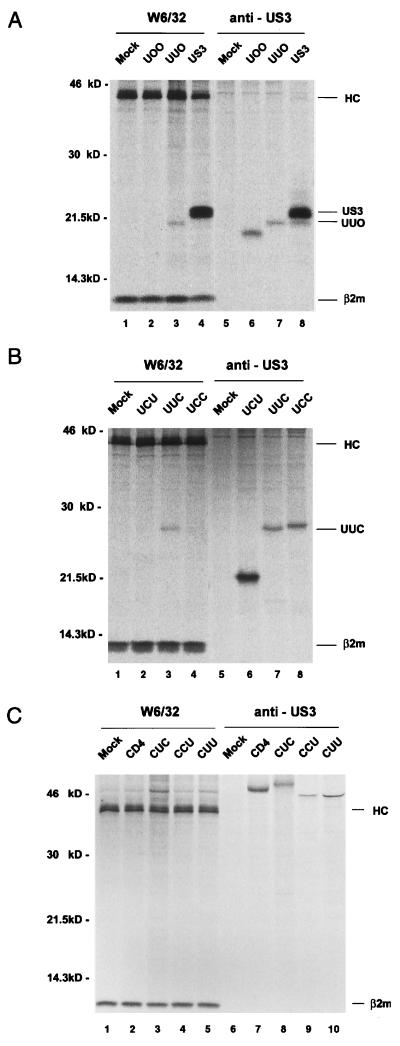
In order to identify the regions that are responsible for the retention of US3 in the ER, we constructed truncated mutant forms of US3 and a series of chimeras in which structural domains of US3 and human CD4, a plasma membrane protein, were reciprocally exchanged (Fig. 1). Intracellular transport of the chimeric glycoproteins was monitored by assaying the sensitivity of their glycans to endoglycosidase treatment after pulse-chase labeling. Endo H removes high-mannose but not complex forms of N-linked glycans (52). Sensitivity to endo H indicates that a protein did not reach at least the medial Golgi compartment. Transfected HeLa cells were pulse-labeled with [35S]methionine and then chased for 90 min. The soluble truncation mutant forms UOO and UUO, in which the transmembrane and cytoplasmic domains and the cytoplasmic tail of US3 were deleted, respectively, remained sensitive to endo H digestion (Fig. 2A). These results suggested that the luminal domain of US3 is sufficient for retention in a premedial Golgi compartment, probably the ER. This notion was further supported by the observation that UCC, in which the luminal domain of CD4 was replaced with that of US3, was retained in the ER, as indicated by endo H sensitivity after a 90-min chase (Fig. 2B, lane 12). In line with these results, chimeras having the luminal domain of US3 in common, UUC and UCU, were sensitive to endo H digestion after the chase (Fig. 2B, lanes 4 and 8). In contrast, all of the chimeras containing the luminal domain of CD4 (CCU, CUU, and CUC) were resistant to endo H (Fig. 2C). The luminal domain of human CD4 contains two N-linked glycans, and only one of them becomes endo H resistant (49). In accordance with this, after endo H digestion of chased material, wild-type CD4 and the chimeras revealed both endo H-resistant and -sensitive forms. These results indicate that the luminal domain of US3 is sufficient for protein retention in the ER and that both the transmembrane and cytoplasmic domains of US3 are not required for its retention in the ER.
FIG. 2.
Endo H sensitivity of glycoproteins synthesized in HeLa cells. Cells transfected with plasmids encoding truncation mutants or chimeras were labeled with [35S]methionine for 30 min and chased for 90 min with unlabeled methionine. The expressed proteins were immunoprecipitated with the appropriate antibodies. Immunoprecipitated proteins were then left untreated (−) or treated with endo H (+) before analysis by SDS-PAGE and autoradiography. (A) Endo H sensitivity of truncated mutants. (B) Endo H sensitivity of chimeras containing the US3-derived luminal domain. (C) Endo H sensitivity of chimeras containing the CD4-derived luminal domain. (D) Disulfide formation of chimeric molecules. Labeled cells were lysed in the presence of 10 mM iodoacetamide. Immunoprecipitates were divided into two aliquots and either reduced with 200 mM dithiothreitol (lanes 1 to 3 in each panel) or run under nonreducing conditions (lanes 4 to 6 in each panel).
Since misfolded proteins are usually retained in the ER independently of the presence of a specific retention signal (17), it was important to establish that the luminal domain of US3 in the various chimeric constructs was able to maintain its proper conformation. To test for proper conformation, we performed different experiments. First, we examined the mobility of chimeras by SDS-PAGE under nonreducing conditions. As under reducing conditions (Fig. 2D, lanes 1 to 3 in each panel), wild-type US3 and all of the chimeras also ran as monomers under nonreducing conditions (lanes 4 to 6 in each panel), suggesting that the chimeras did not form disulfide-bridged aggregates indicative of misfolded proteins. It is possible that the chimeric proteins form large aggregates, which may not enter the gel. However, as we did not detect any aggregates during our pulse-chase protocol under nonreducing conditions (data not shown), we believe that the chimeric proteins do not aggregate. Interestingly, when nonreduced, the chimeras migrated with slightly faster mobility than when reduced (compare lanes 1 to 3 with lanes 4 to 6, respectively). This implies the existence of an intramolecular disulfide bond(s) which could maintain a more compact structure of the protein under nonreducing conditions. Second, since the ER chaperones calnexin and calreticulin are known to be involved in the quality control of several glycoproteins (17), we examined if the chimeric proteins were bound to ER chaperones. None of the chimeric proteins, which were pulse-chased for 90 min, was coimmunoprecipitated by either an anticalnexin or an anticalreticulin antibody (data not shown). Taken together, these results, although not conclusive, support the idea that retention of these chimeras in the ER was not due to misfolding.
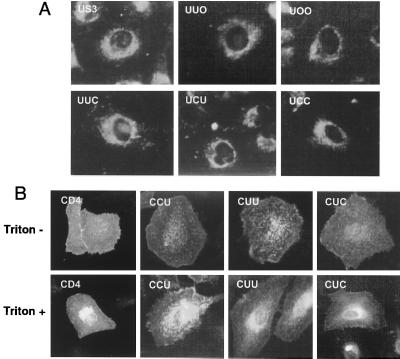
To further ascertain whether the luminal domain of US3 has ER retention properties, the subcellular localization of chimeras was examined by indirect immunofluorescence microscopy. In agreement with our previous observation (1), wild-type US3 expressed in HeLa cells exhibited strong perinuclear staining along with staining of the reticular network extending throughout the cytoplasm, characteristic of the ER (Fig. 3A). Similar ER fluorescence patterns were observed for cells expressing chimeras UUO, UOO, UUC, UCU, and UCC (Fig. 3A), supporting the above-described biochemical finding that the sugar chains of the chimeric proteins were sensitive to endo H digestion. In contrast, in nonpermeabilized cells, fluorescent staining of CD4 with MAb OKT4 revealed typical surface labeling (Fig. 3B). Fluorescence could be detected on cells transfected with CUC, CUU, or CCU cDNA with or without permeabilization, indicating that the expressed chimeric proteins were transported to the cell surface. Taken together, these findings demonstrate that the luminal domain of US3 functions as a retention signal in the ER.
FIG. 3.
Intracellular localization of chimeric proteins. (A) HeLa cells expressing chimeric proteins were permeabilized and immunostained with anti-US3 antibodies, followed by FITC-conjugated goat anti-rabbit IgG. (B) Cells were fixed, permeabilized with Triton X-100 (lower panel) or not permeabilized (upper panel), and immunostained with anti-CD4 MAb OKT4, followed by FITC-conjugated anti-mouse IgG. Shown are representative fields from multiple independent transfections. Transfection efficiencies were generally 30 to 50%.
ER localization of US3 arises from true retention without recycling through the Golgi or the ER-Golgi intermediate compartment.
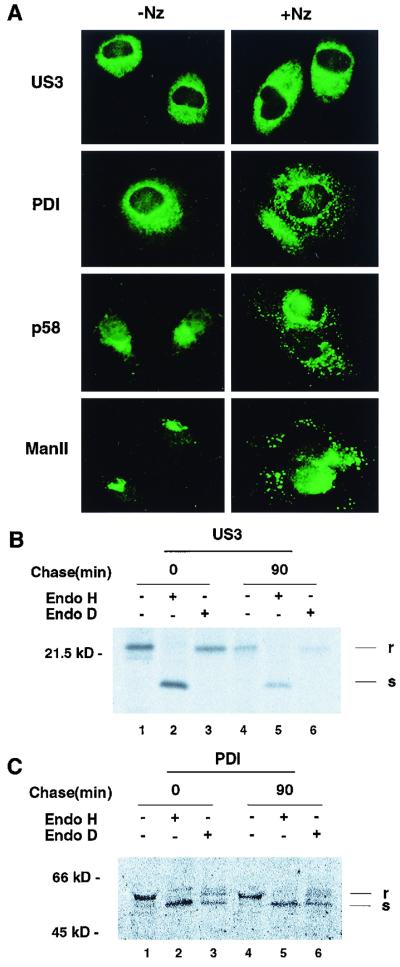
The above data still do not discern between US3's being strictly retained in the ER and its being transported beyond the ER and then returned to the ER, as occurs with many ER-resident proteins (39). To address this question, cells expressing US3 glycoproteins were treated with nocodazole. Nocodazole disrupts microtubules, leading to disintegration of the Golgi and interruption of traffic among the Golgi, the ERGIC (the ER-Golgi intermediate compartment), and the ER (28). The intracellular distribution of US3 was compared with that of PDI, an ER-resident marker that contains a KDEL signal for retrieval. After treatment with 20 μM nocodazole for 5 h, the staining pattern of PDI disappeared from the ER whereas much of the immunoreactive PDI was concentrated in large spots (Fig. 4A). Under the same conditions, the US3 staining pattern (Fig. 4A) remained unchanged in the ER. As expected, the ERGIC marker p58 (25) exhibited perinuclear staining without treatment, which changed to more punctate staining after treatment with nocodazole (Fig. 4A). In the presence of nocodazole, the distribution of Man II, a Golgi marker, also changed from a compact juxtanuclear to a punctate perinuclear pattern (Fig. 4A). Therefore, we believe that the US3 glycoproteins do not cycle between the ER and the Golgi complex.
FIG. 4.
Static retention of US3 in the ER. (A) Effect of nocodazole treatment on the intracellular distribution of US3. Subconfluent HeLa cells grown on coverslips were infected with vaccinia virus recombinant US3 at a multiplicity of infection of 10 PFU/cell. At 2 h postinfection, the cells were incubated for an additional 5 h in the presence (right) or absence (left) of 20 μM nocodazole. Cells were then fixed, permeabilized with Triton X-100, and labeled with anti-US3, anti-PDI (ER), anti-p58 (ERGIC), or anti-Man II (Golgi) antibody. Note the changes in the staining pattern of PDI after treatment of the cells with nocodazole, while the ER-like staining pattern of US3 remains unchanged. (B and C) Insensitivity of US3 to endo D digestion. HeLa cells infected with vaccinia virus recombinant US3 were pulse-labeled with [35S]methionine for 30 min and chased for 90 min. Cell lysates were immunoprecipitated either with anti-US3 antibody (B) or with anti-PDI antibody (C); this was followed in the indicated cases by treatment with endo D or endo H and analysis by SDS-PAGE. r, endo D or H resistant; s, endo D or H sensitive.
To further support this notion, we looked for the modifications that their glycans had potentially acquired in the compartment into which they had transited. In pulse-chase experiments (Fig. 2A), we showed that the US3 glycoprotein remained sensitive to endo H, suggesting that it did not reach the medial Golgi compartment, where the modification of glycoproteins to endo H-resistant forms occurs. To rule out the possibility that the US3 glycoprotein reached the cis Golgi compartment and then was recycled back to the ER, immunoprecipitates were digested with endo D. As shown in Fig. 4B, glycosylated US3 was resistant to endo D (lanes 3 and 6) while it was again susceptible to endo H digestion (lanes 2 and 5). In contrast, PDI, a positive control, was susceptible to both endo D (lanes 3 and 6) and endo H digestion (Fig. 4C, lanes 2 and 5). Since glycoproteins become sensitive to endo D after being processed by α-mannosidase 1A, which is located in the cis Golgi (4), these results suggested that the US3 glycoproteins did not reach the cis Golgi compartment. Taken together, these results led us to conclude that the US3 glycoproteins are strictly retained in the ER and do not cycle through the Golgi or the ERGIC.
Both the luminal and transmembrane domains of US3 are required for binding to MHC class I molecules.
We had previously demonstrated that US3 physically associates with MHC class I heterodimers (1). This prompted us to determine which parts of the US3 protein are crucial for its association with MHC class I molecules. HeLa cells were transfected either with different US3/CD4 hybrid gene constructs or with various truncated US3 mutants, labeled with [35S]methionine, and solubilized with digitonin. Possible complex formation between mutant proteins and MHC class I molecules was monitored by coimmunoprecipitation using appropriate antibodies in comparison to the wild-type protein. As can be seen in cells expressing wild-type US3 (Fig. 5A, lane 4), an additional band of 22 kDa which was not observed in mock-transfected cells (lane 1) coprecipitated with MHC class I molecules. Interestingly, the truncated mutants and chimeras exhibited different capacities to form complexes with MHC class I molecules. Using anti-HC antibody, coprecipitation with MHC class I molecules was observed only for chimeras containing both the luminal and transmembrane domains of US3 (UUO and UUC) (Fig. 5A, lane 3, and 5B, lane 3, respectively), suggesting that the cytoplasmic tail of US3 is not directly involved in the interaction with MHC class I molecules. Neither the luminal domain (UOO and UCC) nor the transmembrane domain (CUC) of US3 alone could independently mediate the association with MHC class I molecules (Fig. 5A, lane 2, 5B, lane 4, and 5C, lane 3, respectively). In accordance with this result, replacement of either the luminal or the transmembrane domain of US3 with the corresponding domain of CD4 abolished the association between the two molecules (Fig. 5C, lane 5, CUU, and 5B, lane 2, UCU, respectively). These results demonstrate that both the luminal and transmembrane domains of US3 are required for its interaction with MHC class I molecules. In the reciprocal experiment using anti-US3 antibody, no materials corresponding to MHC class I molecules were coprecipitated (Fig. 5A, lanes 5 to 8, and 5B, lanes 5 to 8). We assume that binding of the anti-US3 antibody could be prevented by the binding of MHC class I molecules to the respective epitope. Considering that the anti-US3 antibody was raised against the peptide sequences corresponding to the luminal segment of US3 (residues 78 to 97) (1), it is conceivable that this region could play an important role in the US3 binding of MHC class I molecules. Alternatively, the antibody could displace MHC class I molecules during the immunoprecipitation procedure.
FIG. 5.
Coimmunoprecipitation of chimeric proteins with MHC class I molecules. Cells transfected with the indicated cDNAs were labeled with [35S]methionine for 30 min and lysed with 1% digitonin lysis buffer. The antibodies used for immunoprecipitation were MAb W6/32 (A and B, lanes 1 to 4, respectively; C, lanes 1 to 5), an anti-US3 antibody (A and B, lanes 5 to 8, respectively), and MAb OKT4 (C, lanes 6 to 10).
Lack of coprecipitation does not always correlate with lack of transport inhibition, as has been seen at least with adenovirus E19 (48), the functional homolog to US3. To examine whether the binding of the chimeras to MHC class I molecules correlated with the downregulation of the cell surface expression of MHC class I molecules, the identical sets of transfectants were subjected to fluorescence-activated cell sorter analysis. Cell surface expression of MHC class I molecules was lower only on cells transfected with cDNAs encoding either UUO or UUC (Fig. 6). In contrast, all of the mutant cell lines in which coprecipitation had been undetectable expressed normal levels of MHC class I molecules on the cell surface. Both the coimmunoprecipitation and fluorescence-activated cell sorter data are therefore in agreement. Thus, we conclude that the luminal and transmembrane domains of US3 are required for binding of MHC class I molecules and subsequently cause downregulation of MHC class I molecules on the cell surface whereas the cytoplasmic domain of US3 is dispensable.
FIG. 6.
MHC class I surface expression in HeLa cells expressing chimeric proteins. HeLa cells were transiently transfected with the individual cDNAs encoding chimeric proteins. After 48 h, the cell surface expression of MHC class I molecules was monitored by flow cytometry using MAb W6/32. Open areas represent the staining of mock-transfected cells, and filled areas represent the staining of cDNA-transfected cells.
US3 directly interacts with MHC class I molecules.
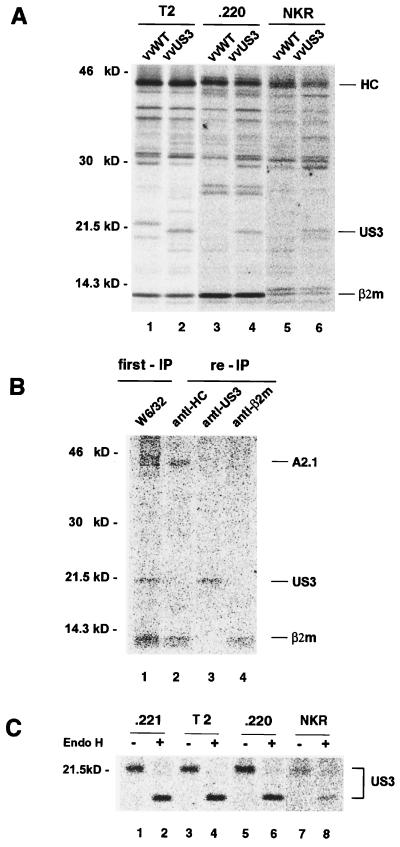
Assembly of MHC class I molecules is initiated in the ER, where unfolded MHC class I molecules associate with the ER-resident chaperone calnexin (35). Subsequent binding of the MHC class I part to β2m then causes dissociation of calnexin (43). The MHC class I-β2m heterodimer then associates with TAP (36, 50). In this process, tapasin, another ER-resident chaperone, plays an important role in bridging MHC class I to TAP (42). To explore the possibility that calnexin, tapasin, or functional TAP plays a role in the association of US3 with MHC class I molecules, we infected calnexin-negative CEM-NKR, tapasin-negative LCL 721.220, and TAP1/TAP2-negative T2 cells with recombinant vaccinia virus expressing US3. Possible association between US3 and MHC class I was assessed by coimmunoprecipitation. Because of variation in infection efficiency between the cell lines, quantitative assessment of US3 coimmunoprecipitation was difficult. Nonetheless, after performing several experiments, it became obvious that the coprecipitation of US3 protein with the MHC class I molecules was maintained in all of the mutant cell lines analyzed (Fig. 7A, lanes 2, 4, and 6). These results, therefore, indicate that at least calnexin, tapasin, or TAP1/TAP2 is not essential for the interaction of US3 with MHC class I molecules. Furthermore, a direct interaction between US3 and MHC class I molecules can also be demonstrated in vitro (Fig. 7B). HLA-A2.1 cDNA was in vitro transcribed and translated together with β2m and US3 in the presence of canine microsomes. After centrifugal sedimentation, the pellet fraction was lysed in detergent. Immunoprecipitation with MAb W6/32 recovered A2.1 associated with β2m, together with US3 (lane 1), as evidenced by reprecipitation of the relevant polypeptides with K455, anti-US3 antibody, or K355 (lanes 2, 3, and 4, respectively). These results thus support the notion that US3 directly binds to MHC class I molecules without further components being involved.
FIG. 7.
Identification of minimal requirements for the association of US3 with MHC class I molecules and retention of US3 in the ER. (A) Calnexin-, tapasin-, or TAP-deficient cells were infected with US3-expressing recombinant vaccinia virus for 1 h, incubated for 2 h, and then labeled with [35S]methionine for 30 min. The labeled cells were lysed with digitonin, and the lysate was subjected to coimmunoprecipitation using MAb W6/32. (B) Proteins were in vitro transcribed and translated in the presence of [35S]methionine using a rabbit reticulocyte lysate supplemented with canine pancreatic microsomes. Immunoprecipitation (IP) was done as described in Materials and methods. (C) The labeled cells were chased for 90 min and lysed with detergent. The lysates were then treated with anti-US3 antibody, and the immunoprecipitates were digested with endo H and analyzed by SDS-PAGE.
Another interesting finding was that the US3 glycoprotein expressed in these cell lines remained sensitive to endo H digestion upon a 90-min chase (Fig. 7C). This suggests that the ER retention of US3 is at least not mediated via interactions with calnexin, tapasin, or TAP, all representatives of ER-resident proteins, but is most likely mediated by its own signal.
DISCUSSION
We have shown earlier that US3, a glycoprotein of HCMV, specifically binds to MHC class I molecules in the ER, inhibiting their transport to the cell surface (1). Since CTLs recognize antigens associated with MHC class I molecules, US3 may allow infected cells to evade virus-specific CTLs by preventing antigen presentation of MHC class I molecules. This function may play a crucial role in the establishment of persistent and latent infections, as well as in an acute viral infection. Two properties of US3 enable it to block cell surface expression of MHC class I complexed with HCMV peptides (1, 22). First, US3 is retained in the ER, the mechanism of which is still unknown. A second key property of the US3 protein is that it can bind to MHC class I molecules. Therefore, the identification of retention signals and the elucidation of the structural requirements for US3 to be able to bind to MHC class I molecules are important for the understanding of mechanisms of viral pathogenesis and protein compartmentalization.
In our study, we identified the signal for ER retention of the US3 protein in its luminal domain. The ER localization of US3 is accomplished by static retention: no recycling through the Golgi. The luminal domain of US3 is necessary and sufficient for ER retention. Interestingly, though, the association of US3 with MHC class I molecules requires the transmembrane domain in addition to the luminal domain of US3, while lack of the cytoplasmic domain does not affect US3 binding. Our in vitro data also establish that US3 directly interacts with MHC class I molecules.
The HCMV US3 glycoprotein is a functional and structural homolog of the adenovirus E3/19K gene product (E19), although there is no amino acid sequence homology between them. The proteins are similar in size, and they are ER-resident type I transmembrane glycoproteins featuring a short cytoplasmic tail and a bulky luminal domain. Compared to E19, however, our data show that the MHC class I binding function and the ER retention function of US3 are assigned to different regions of the protein. The cytoplasmic domain of E19 mediates ER retention through a carboxyl-terminal dilysine motif (KKXX) (37), which is also present in other ER proteins (19). This motif allows retrieval of the protein from the Golgi and transfer to the ER in a coatomer-dependent manner (26). Although the cytoplasmic tail of US3 lacks a conventional dilysine motif, there is the carboxyl-terminal sequence RLRFI that might function as a retrieval motif similar to KXKXX, since diarginine motifs could play an analogous role (53). However, our data argue against this view. The transfer of the cytoplasmic tail of US3 to a plasma membrane reporter protein, CD4, did not confer ER targeting on the chimeric protein (CCU). Furthermore, unlike E19, immunolocalization of the US3 glycoproteins and our analysis of their glycans confirmed that this protein is strictly retained in the ER. Generally, the retention signals of resident ER membrane proteins have been localized within the transmembrane or the cytoplasmic domain (12, 18, 20, 31, 41). In contrast to these proteins, we identified the luminal domain as containing the retention signal of the US3 glycoprotein. To our knowledge, this is the first time that an ER localization motif of a type I transmembrane glycoprotein has been mapped to the luminal domain.
There are several possible mechanisms by which the luminal domain of US3 could mediate ER retention. First, although US3 does not contain any of the known linear sequences signaling for ER retention, it is possible that the signal consists of a “patch signal” made up of several interacting regions, as has been suggested for export signals (29). Second, another possible way to achieve protein retention in a membrane organelle is the formation of oligomers too large to be included in transport vesicles (45). Oligomerization as a mechanism for retention has been suggested for some Golgi proteins (8) and p63, a protein localized in the cis Golgi network (47). Although little is known about the structural properties of US3, the US3 protein may be able to form multimers by “kin recognition,” as is the case with N-acetylglucosaminyltransferase I, a Golgi transmembrane protein (34). The formation of homodimers may facilitate further oligomerization of US3, which then could result in the apparent immobilization of US3 in the ER. Third, proteins can also be retained in the ER indirectly by interaction with ER-resident proteins. For example, luminal chaperones, including BiP, calnexin, and calreticulin, interact with newly synthesized proteins in the ER lumen and mediate transient or stable retention of proteins that are devoid of intrinsic ER retention-retrieval sequences (17). As the bulk of the US3 protein is on the luminal side of the ER membrane, there is every opportunity for US3 to interact with these ER-resident proteins. However, our attempt to identify tapasin, calnexin, or TAP as such a partner protein was unsuccessful.
We analyzed US3-derived chimeras for the ability to interact with MHC class I molecules. In contrast to the adenovirus E19 glycoprotein, in which the luminal domain is sufficient to bind to MHC class I molecules (13, 14), our findings document that the interaction of US3 with MHC class I engages both intact luminal and transmembrane domains of US3. Neither the luminal nor the transmembrane domain of US3 alone could mediate association with MHC class I molecules. This finding is consistent with the idea that both domains represent a distinct functional and structural unit such that the absence or replacement of either domain alters the tertiary structure of this unit, thus abrogating binding to MHC class I molecules. Stated another way, the binding site in US3 for MHC class I molecules is not a simple linear sequence but rather is embodied in the tertiary structure of the luminal and transmembrane regions of US3. Another possible scenario is that the transmembrane region of US3 simply plays a structural role, allowing the luminal domain of US3 to extend out from the ER membrane and be exposed correctly to MHC class I molecules. However, this does not really provide an appropriate explanation. If the transmembrane domain functioned in this manner in the binding of MHC class I molecules, we would have found that the chimeric proteins UCU and UCC, which contain membrane regions derived from CD4, were associated with MHC class I molecules. At present, we have no experimental data favoring any one of these possibilities. In any case, it seems obvious that unique properties present in the transmembrane region of US3 but not in that of CD4 are critically involved in the binding of MHC class I molecules. In the context of the conserved structural features among the US proteins (1), it is interesting that the luminal domain of US3 contains two cysteine residues sandwiching a glycosylation site. This structural feature is also present in E19 of adenovirus, where it has been shown to be essential for the binding of MHC class I molecules (47). Deletion studies and site-directed mutagenesis experiments may reveal whether this structural feature is crucial for the binding of US3 to MHC class I molecules.
It remains unclear which structure of the MHC class I molecules is recognized by US3 and whether US3 displays differential binding preferences among different MHC class I alleles. The α1 and α2 domains of the hypervariable regions of MHC class I molecules, which form the peptide-binding cleft, do not appear to interact with US3 based on results we obtained in a previous study in which we demonstrated that US3 does not block the ability of MHC class I molecules to bind peptides in the ER (1). We recently reported that US3 is capable of binding HLA-G, a nonclassical MHC class I molecule, as well as HLA-C alleles and HLA-A and -B (23). In extending our considerations to include our observations with other MHC molecules, we propose that the US3 glycoprotein has a broad ability to bind MHC class I alleles. It is thus likely that the α3 domain of HCs, a region highly conserved between different alleles (5), is the primary site of interaction with US3.
In summary, we provide evidence that the properties of intact US3 protein to bind MHC class I molecules and to be retained in the ER are encoded in different parts of the molecule. The region of US3 conferring ER targeting was mapped to the luminal segment of the protein, while the structures responsible for the association of US3 with MHC class I molecules are embedded in the luminal and transmembrane domains of the molecule.
ACKNOWLEDGMENTS
This work was supported by a grant from the Korea Science and Engineering Foundation.
We thank K. Moremen, Ralf F. Pettersson, and Peter Cresswell for the generous gift of antibodies or cell lines and K. Fruh and Y. Yang for helpful discussions.
REFERENCES
- 1.Ahn K, Angulo A, Ghazal P, Peterson P A, Yang Y, Fruh K. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc Natl Acad Sci USA. 1996;93:10990–10995. doi: 10.1073/pnas.93.20.10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn K, Gruhler A, Galocha B, Jones T R, Wiertz E J, Ploegh H L, Peterson P A, Yang Y, Fruh K. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity. 1997;6:613–621. doi: 10.1016/s1074-7613(00)80349-0. [DOI] [PubMed] [Google Scholar]
- 3.Andersson M, Paabo S, Nilsson T, Peterson P A. Impaired intracellular transport of class I MHC antigens as a possible means for adenoviruses to evade immune surveillance. Cell. 1985;43:215–222. doi: 10.1016/0092-8674(85)90026-1. [DOI] [PubMed] [Google Scholar]
- 4.Beckers C J, Keller D S, Balch W E. Semi-intact cells permeable to macromolecules: use in reconstitution of protein transport from the endoplasmic reticulum to the Golgi complex. Cell. 1987;50:523–534. doi: 10.1016/0092-8674(87)90025-0. [DOI] [PubMed] [Google Scholar]
- 5.Bjorkman P J, Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- 6.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox J H, Bennink J R, Yewdell J W. Retention of adenovirus E19 glycoprotein in the endoplasmic reticulum is essential to its ability to block antigen presentation. J Exp Med. 1991;174:1629–1637. doi: 10.1084/jem.174.6.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox J H, Yewdell J W, Eisenlohr L C, Johnson P R, Bennink J R. Antigen presentation requires transport of MHC class I molecules from the endoplasmic reticulum. Science. 1990;247:715–718. doi: 10.1126/science.2137259. [DOI] [PubMed] [Google Scholar]
- 10.DeMars R, Rudersdorf R, Chang C, Petersen J, Strandtmann J, Korn N, Sidwell B, Orr H T. Mutations that impair a posttranscriptional step in expression of HLA-A and -B antigens. Proc Natl Acad Sci USA. 1985;82:8183–8187. doi: 10.1073/pnas.82.23.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doherty P C, Allan W, Eichelberger M, Carding S R. Roles of alpha beta and gamma delta T cell subsets in viral immunity. Annu Rev Immunol. 1992;10:123–151. doi: 10.1146/annurev.iy.10.040192.001011. [DOI] [PubMed] [Google Scholar]
- 12.Duvet S, Cocquerel L, Pillez A, Cacan R, Verbert A, Moradpour D, Wychowski C, Dubuisson J. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J Biol Chem. 1998;273:32088–32095. doi: 10.1074/jbc.273.48.32088. [DOI] [PubMed] [Google Scholar]
- 13.Flomenberg P, Szmulewicz J, Gutierrez E, Lupatkin H. Role of the adenovirus E3–19k conserved region in binding major histocompatibility complex class I molecules. J Virol. 1992;66:4778–4783. doi: 10.1128/jvi.66.8.4778-4783.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabathuler R, Lévy F, Kvist S. Requirements for the association of adenovirus type 2 E3/19K wild-type and mutant proteins with HLA antigens. J Virol. 1990;64:3679–3685. doi: 10.1128/jvi.64.8.3679-3685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaynor E C, te Heesen S, Graham T R, Aebi M, Emr S D. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994;127:653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenwood R, Shimizu Y, Sekhon G S, DeMars R. Novel allele-specific, post-translational reduction in HLA class I surface expression in a mutant human B cell line. J Immunol. 1994;153:5525–5536. [PubMed] [Google Scholar]
- 17.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 18.Honsho M, Mitoma J Y, Ito A. Retention of cytochrome b5 in the endoplasmic reticulum is transmembrane and luminal domain-dependent. J Biol Chem. 1998;273:20860–20866. doi: 10.1074/jbc.273.33.20860. [DOI] [PubMed] [Google Scholar]
- 19.Jackson M R, Nilsson T, Peterson P A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson M R, Nilsson T, Peterson P A. Retrieval of transmembrane proteins to the endoplasmic reticulum. J Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones T R, Hanson L K, Sun L, Slater J S, Stenberg R M, Campbell A E. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J Virol. 1995;69:4830–4841. doi: 10.1128/jvi.69.8.4830-4841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones T R, Wiertz E J, Sun L, Fish K N, Nelson J A, Ploegh H L. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci USA. 1996;93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jun Y, Kim E, Jin M, Sung H C, Han H, Geraghty D E, Ahn K. Human cytomegalovirus gene products US3 and US6 down-regulate trophoblast class I MHC molecules. J Immunol. 2000;164:805–811. doi: 10.4049/jimmunol.164.2.805. [DOI] [PubMed] [Google Scholar]
- 24.Kung P, Goldstein G, Reinherz E L, Schlossman S F. Monoclonal antibodies defining distinctive human T cell surface antigens. Science. 1979;206:347–349. doi: 10.1126/science.314668. [DOI] [PubMed] [Google Scholar]
- 25.Lahtinen U, Hellman U, Wernstedt C, Saraste J, Pettersson R F. Molecular cloning and expression of a 58-kDa cis-Golgi and intermediate compartment protein. J Biol Chem. 1996;271:4031–4037. doi: 10.1074/jbc.271.8.4031. [DOI] [PubMed] [Google Scholar]
- 26.Letourneur F, Gaynor E C, Hennecke S, Demolliere C, Duden R, Emr S D, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 27.Lewis M J, Pelham H R. Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell. 1992;68:353–364. doi: 10.1016/0092-8674(92)90476-s. [DOI] [PubMed] [Google Scholar]
- 28.Lippincott-Schwartz J, Donaldson J G, Schweizer A, Berger E G, Hauri H P, Yuan L C, Klausner R D. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- 29.Locker J K, Opstelten D J, Ericsson M, Horzinek M C, Rottier P J. Oligomerization of a trans-Golgi/trans-Golgi network retained protein occurs in the Golgi complex and may be part of its retention. J Biol Chem. 1995;270:8815–8821. doi: 10.1074/jbc.270.15.8815. [DOI] [PubMed] [Google Scholar]
- 30.Moremen K W, Touster O, Robbins P W. Novel purification of the catalytic domain of Golgi alpha-mannosidase. II. Characterization and comparison with the intact enzyme. J Biol Chem. 1991;266:16876–16885. [PubMed] [Google Scholar]
- 31.Munro S. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 1998;8:11–15. doi: 10.1016/S0962-8924(97)01197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munro S, Pelham H R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 33.Nilsson T, Jackson M, Peterson P A. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 1989;58:707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson T, Rabouille C, Hui N, Watson R, Warren G. The role of the membrane-spanning domain and stalk region of N-acetylglucosaminyltransferase I in retention, kin recognition and structural maintenance of the Golgi apparatus in HeLa cells. J Cell Sci. 1996;109:1975–1989. doi: 10.1242/jcs.109.7.1975. [DOI] [PubMed] [Google Scholar]
- 35.Nossner E, Parham P. Species-specific differences in chaperone interaction of human and mouse major histocompatibility complex class I molecules. J Exp Med. 1995;181:327–337. doi: 10.1084/jem.181.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortmann B, Androlewicz M J, Cresswell P. MHC class I/beta 2-microglobulin complexes associate with TAP transporters before peptide binding. Nature. 1994;368:864–867. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 37.Paabo S, Bhat B M, Wold W S, Peterson P A. A short sequence in the COOH-terminus makes an adenovirus membrane glycoprotein a resident of the endoplasmic reticulum. Cell. 1987;50:311–317. doi: 10.1016/0092-8674(87)90226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelham H R. Evidence that luminal ER proteins are sorted from secreted proteins in a post-ER compartment. EMBO J. 1988;7:913–918. doi: 10.1002/j.1460-2075.1988.tb02896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelham H R. Sorting and retrieval between the endoplasmic reticulum and Golgi apparatus. Curr Opin Cell Biol. 1995;7:530–535. doi: 10.1016/0955-0674(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 40.Rinaldo C R., Jr Immune suppression by herpesviruses. Annu Rev Med. 1990;41:331–338. doi: 10.1146/annurev.me.41.020190.001555. [DOI] [PubMed] [Google Scholar]
- 41.Rothman J E, Wieland F T. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 42.Sadasivan B, Lehner P J, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 43.Sadasivan B K, Cariappa A, Waneck G L, Cresswell P. Assembly, peptide loading, and transport of MHC class I molecules in a calnexin-negative cell line. Cold Spring Harbor Symp Quant Biol. 1995;60:267–275. doi: 10.1101/sqb.1995.060.01.031. [DOI] [PubMed] [Google Scholar]
- 44.Salter R D, Howell D N, Cresswell P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 45.Schweizer A, Rohrer J, Hauri H P, Kornfeld S. Retention of p63 in an ER-Golgi intermediate compartment depends on the presence of all three of its domains and on its ability to form oligomers. J Cell Biol. 1994;126:25–39. doi: 10.1083/jcb.126.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott J E, Dawson J R. MHC class I expression and transport in a calnexin-deficient cell line. J Immunol. 1995;155:143–148. [PubMed] [Google Scholar]
- 47.Sester M, Burgert H-G. Conserved cysteine residues within the E3/19K protein of adenovirus type 2 are essential for binding to major histocompatibility complex antigens. J Virol. 1994;68:5423–5432. doi: 10.1128/jvi.68.9.5423-5432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Severinsson L, Martens I, Peterson P A. Differential association between two human MHC class I antigens and an adenoviral glycoprotein. J Immunol. 1986;137:1003–1009. [PubMed] [Google Scholar]
- 49.Shin J, Dunbrack R L, Jr, Lee S, Strominger J L. Signals for retention of transmembrane proteins in the endoplasmic reticulum studied with CD4 truncation mutants. Proc Natl Acad Sci USA. 1991;88:1918–1922. doi: 10.1073/pnas.88.5.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suh W K, Cohen-Doyle M F, Fruh K, Wang K, Peterson P A, Williams D B. Interaction of MHC class I molecules with the transporter associated with antigen processing. Science. 1994;264:1322–1326. doi: 10.1126/science.8191286. [DOI] [PubMed] [Google Scholar]
- 51.Szczesna-Skorupa E, Kemper B. An N-terminal glycosylation signal on cytochrome P450 is restricted to the endoplasmic reticulum in a luminal orientation. J Biol Chem. 1993;268:1757–1762. [PubMed] [Google Scholar]
- 52.Tarentino A L, Maley F. Purification and properties of an endo-beta-N-acetylglucosaminidase from Streptomyces griseus. J Biol Chem. 1974;249:811–817. [PubMed] [Google Scholar]
- 53.Teasdale R D, Jackson M R. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 54.Wiertz E J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 55.Wiertz E J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]