Abstract
Directed evolution techniques allow us to genuinely mimic molecular evolution in vitro. To enhance this imitation of natural evolutionary processes on a laboratory scale in even more detail, we developed an in vitro method for the generation of random deletions and repeats. The pairwise fusion of two fragments of the same gene that are truncated by exonuclease BAL-31 either at the 3′ or 5′ side results in a deletion or a repeat at the fusion point. Although in principle the method randomly covers the whole gene, it can also be limited to a predefined area in the sequence by controlling the level of the initial truncation. To test the procedure and to illustrate its potential, we used haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 (DhlA) as a model enzyme, since the adaptation of this enzyme towards new substrates is known to occur via the generation of this type of mutation. The results show that the mutagenesis method presented here is an effective tool for accessing formerly unexplorable sequence space and can contribute to the success of future directed evolution experiments.
INTRODUCTION
Recent developments in molecular biology have made it possible to mimic the process of natural evolution on a laboratory scale, turning directed evolution into a key technology in protein engineering. Methods for generating genetic diversity include chemical mutagenesis (1,2), the use of mutator strains (3,4), error-prone PCR (5,6) or recombination of a set of homologous genes (7,8). An important breakthrough came with the development of suitable in vitro recombination methods (9–11). In particular, the introduction of DNA shuffling (9) enhanced the efficiency of the directed evolution process. By combining these techniques, protein engineers can now virtually mimic the whole Darwinian pathway of repeated rounds of mutation, recombination and selection in vitro.
An important in vivo evolutionary mutagenesis event that is still lacking a proper in vitro equivalent is the random generation of segmental mutations (12). Using PCR, it is possible to create deletions or insertions in a gene, but the need for predefined primers implies that this can only be done in a directed fashion. Eustance et al. (13) reported a semi-random method, using BAL-31 exonuclease activity on a construct containing overlapping fragments of the same gene. However, by initiating the digestion at a unique restriction site in between the two fragments the product becomes heavily biased, since both ends of the construct are exposed to exonuclease activity for the same length of time. The method we present here ensures an entirely random introduction of deletions and repeats in vitro. By combining two independently generated batches of DNA, containing either 3′ or 5′ truncated gene fragments, a library of random fusions is created, which results in the generation of deletions and repeats at the fusion point. By controlling the level of initial truncation, the sequence area subjected to segmental mutagenesis can easily be governed, limiting the size of the resulting library (see Fig. 1). Combination of this strategy with random mutagenesis and gene shuffling should lead to an in vitro evolution process that allows the exploration of a much larger and so far inaccessible area of sequence space.
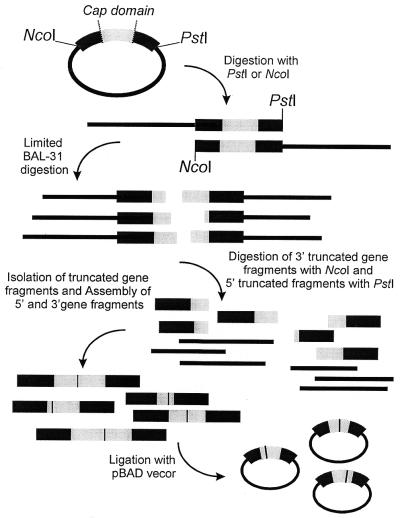
Figure 1.
Schematic illustration of the segmental mutagenesis procedure. The vector is first linearized either at the 5′ or 3′ end of the dehalogenase gene. Progressive exonuclease action and removal of the remaining vector DNA yields two batches of either 3′ or 5′ truncated gene fragments. After assembly of these two ends, the segmental mutagenesis library is ligated into a fresh vector, transformed to E.coli and can be screened for activity.
As a model system we used haloalkane dehalogenase (DhlA) from Xanthobacter autotrophicus GJ10. This hydrolytic dehalogenase is the key enzyme in the metabolic pathway of the bacterium when it is growing on 1,2-dichloroethane (DCE) (14). It can convert a broad range of small chlorinated and brominated compounds to their corresponding alcohols (15), which makes it interesting for bioremediation (16,17), since several of these substrates occur widely as environmental pollutants. The generation of repeats and deletions in its sequence is known to play a role in the adaptation to new substrates (18,19). The results illustrate the effectiveness of the method, and the remarkable diversity that was found emphasizes the potential value of this mutagenesis method for in vitro molecular evolution experiments.
MATERIALS AND METHODS
Plasmid construction
The presumed primitive dehalogenases DhlAS and DhlAT (see Fig. 2) were constructed according to Kunkel (20) using pGEF(Δ+F172L) as a template. This plasmid contains a Phe172Leu mutant of the wild-type DhlA with an additional 27-bp deletion, comprising amino acids Phe161−Ala169, which occurred spontaneously during random mutagenesis of Phe172 (21). Using 5′-ACCCAGCCTGCGGATGGCTTTAG/CCGCCTGGAAATAC-3′ as a primer resulted in a 50% mixture of dhlAS (ACC) and dhlAT (AGC). Mutant sequences were established by the Center for Biomedical Technology, University of Groningen (The Netherlands). After PCR amplification with the primers MPfwdI (5′-AGATATACCATGGTAAATGCAA-3′, NcoI site underlined) and MPrevIII (5′- ATAATACTGCAGCGCTATTCTGTCTCGGCAAA-3′, PstI site underlined) the mutant genes were cloned into the multiple cloning site of pBAD/myc-HisA (Invitrogen). This vector provides a tightly regulated protein expression system based on the araBAD promoter, which is activated in the presence of arabinose.
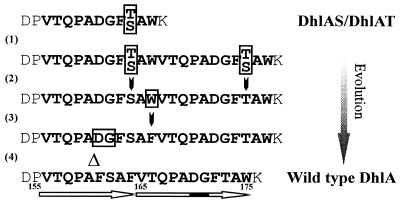
Figure 2.
Proposed route for the evolution of DhlA. The putative predecessor is indicated as DhlAS/Dh1AT, denoting both the serine- and threonine-containing species. The order of the mutations that occurred after the initial duplication of an 11-amino acid stretch (1) is not clear. Event (2) represents a substitution of either a T→S or S→T, depending on which amino acid was present in the original sequence, and (3) indicates the substitution of W→F. The last step (4) shows the deletion of 2 amino acids in the first half of the repeat.
Library construction
For the construction of the segmental mutations library (Fig. 1) a 50–50 mixture of pBAD/dhlAS&T was cut either with PstI (‘left’ or 5′ batch) or NcoI (‘right’ or 3′ batch). The linear DNA fragments were subjected to digestion with BAL-31 exonuclease (New England Biolabs). Reaction mixtures contained 2 µg DNA and 1 U BAL-31 in 1× BAL-31 exonuclease buffer and were incubated at 30°C. The time required to reach the cap domain section of the DNA sequence (removal of ∼450 bp from the 5′ side or 240 bp from the 3′ side) was estimated by analyzing periodically withdrawn samples on an agarose gel. Starting material for the library was obtained by taking samples with time intervals estimated to be required to remove 5 bp, ensuring an accurate diversity of the truncated fragments. The exonuclease was inactivated in 20 mM EGTA by heating at 65°C for 10 min. All samples of a separate batch were then pooled and purified (PCR purification kit, Qiagen). Subsequently the DNA fragments were treated with Klenow (New England Biolabs) to repair staggered ends, required for the blunt-end fusion. Finally the ‘left’ batch was digested with NcoI and the ‘right’ batch with PstI. For a final removal of undesired fragments, DNA within the proper range (450–690 bp and 240–480 bp for the ‘left’ and ‘right’ batch, respectively) was isolated from a 2% low melting point agarose gel. After treating the ‘right’ batch with alkaline phosphatase, ‘left’ and ‘right’ ends were assembled in a ligation reaction using the Rapid Ligation kit (Roche Molecular Biochemicals). This yielded long stretches of DNA since the PstI and NcoI sites will also combine. To prepare the fusion product for ligation into the expression vector pBAD/myc-HisA, one could choose to digest the product with the two restriction enzymes directly. To enhance the yield we included a few additional PCR cycles with MPfwdI and MPrevIII.
Screening
Escherichia coli TOP10 (Invitrogen) was used as a host for the mutant library. This strain can transport arabinose, but is not capable of metabolizing it, ensuring that arabinose levels will remain constant during cultivation. Expression was induced using 0.02% arabinose in solid media and 0.001% in liquid media. Initial screening on solid medium was achieved by including a mixture of 45 mg/l eosin and 7 mg/l methylene blue (22) in the LB-agar. Upon incubation with a halogenated substrate, which was carried out by exposing the plates in a sealed box to the evaporated substrate, active colonies turn blue. Positive clones were cultured overnight at 30°C in a 96-well plate. Identical aliquots of these cultures were retested using a pH assay (23) with 5 mM of the halogenated substrate. Activity was determined by following absorption at 560 nm. Specific activity in cell-free extract was measured using colorimetric detection of halide release (15). Protein concentrations were determined with Coomassie Brilliant Blue using bovine serum albumin as a standard. Activity measurements were routinely performed at pH 8.2, since this is the optimum for the wild-type enzyme.
RESULTS
Creation of the library and selection of the mutants
The X-ray structure of DhlA (24,25) shows that the enzyme is composed of an α/β-hydrolase-fold main domain (26) and an additional α-helical domain. This cap domain is extruding from the main domain and comprises roughly 80 amino acids, starting approximately halfway along the primary sequence. The cap domain is covering the active site and is important in modulating the substrate specificity. When the wild-type dhlA gene was introduced in a Pseudomonas strain that is capable of growing on hexanol, forced adaptation towards 1-chlorohexane yielded several mutants with improved Km or Vmax values for this substrate (19). Of the 12 independently isolated mutants, as many as 10 contained either a deletion or repeat in the N-terminal part of the cap domain. The wild-type enzyme itself exhibits a perfect and an imperfect repeat in the cap domain. Because DCE is a synthetic compound and has only been present in the environment during the last century, these repeats are believed to have played a role in the recent adaptation of DhlA to DCE. The repeats can easily be traced back to a putative predecessor (see Fig. 2) and when this hypothetical ancestor protein was constructed, it indeed lacked the capability of DCE conversion, while it could still convert various other halogenated substrates, such as 1,2-dibromoethane (DBE). This primitive dehalogenase (DhlAS/T) was taken as the starting point in our segmental mutagenesis experiment. We hypothesized that the introduction of the initiating repeat [Event (1) in Figure 2] would restore the capacity to hydrolyze DCE.
Starting with both forms of the primitive ancestor (DhlAS/T), we generated two batches of either 3′ or 5′ truncated gene fragments (Fig. 1). After linearizing the vector by cutting either at the 3′ (PstI) or the 5′ (NcoI) end of the dehalogenase gene, nuclease BAL-31 was used to engender a progressive shortening of the gene. In contrast to the truncational method employed by Ostermeier et al. (27,28) who used exonuclease III, which can only act on 3′ recessed ends, using this particular exonuclease circumvents the need for specific restriction sites since it degrades both 3′ and 5′ termini of duplex DNA. The truncation was allowed to proceed until it reached the part of the sequence coding for the cap domain (amino acid residue 150–230) and by sampling at appropriate intervals the entire cap domain was covered. The truncated vector fragments were removed by digesting the intact restriction site flanking opposite ends of the gene. Subsequently the 5′ and 3′ fragments were randomly ligated, causing either a repeat or a deletion at the fusion point. In this way we generated a library covering mutants ranging from the complete deletion of the cap domain, via reassembly of the wild type to mutants with a double cap domain. After ligation in pBAD the library was expressed in E.coli and screened for activity. Upon hydrolysis of a halogenated substrate active colonies turned blue.
Preparatory tests showed that visual screening for DCE activity directly on solid media would require an activity of at least 10% of the wild-type level. The wild-type DhlA sequence shows several mutations [Events (2), (3) and (4) in Fig. 2] that must have occurred after the emergence of the initial repeat. All these adaptations most likely have served to further optimize the activity towards DCE. Therefore, there would be a reasonable chance that the hypothetical predecessor, carrying only the initial repeat VTQPADGFT/SAW, would have only a very low activity that could not be detected. For most mutants of DhlA we have observed that cleavage of the carbon−halogen bond becomes rate limiting (21,29). The primitive mutants DhlAS and DhlAT are not capable of converting DCE. However, they can hydrolyze DBE, albeit at a lower rate than wild-type enzyme, probably because bromine is a better leaving group in bimolecular nucleophilic substitutions than chlorine. Assuming that improved DBE conversion would indicate a more efficient catalysis of carbon−halogen bond cleavage and thus might be accompanied by the ability to convert DCE as well, we decided to search our library for mutants with enhanced activity towards DBE.
Screening of approximately 60 000 colonies on pH-indicator medium resulted in the selection of 10 mutants with significantly enhanced DBE activity. The sequences of these mutants are shown in Figure 3. The results show a striking correspondence with former in vivo 1-chlorohexane adaptation experiments (19) and the proposed pathway for the evolutionary development of the wild-type enzyme (Fig. 2), as all isolated mutants contain a repeat in the very N-terminal part of the cap domain. The size of the repeats shows a remarkable variation, ranging from 5 amino acids (10D7) to as many as 21 (3D3). Also the exact location varies: the majority of the mutants carry a repeat of a sequence fragment upstream of the expected VTQPADGFT/SAW repeat, starting as early as Ile145. Only mutants 2C7, 3D3 and 3F9 cover the expected repeat to a significant extent.
Figure 3.
Sequence alignment of the primitive dehalogenase (DhlAS/T), the hypothetical mutant arising after the duplication of the 11-amino acid stretch, wild-type DhlA and the 10 selected mutants. The sequences are arranged according to an increasing length of the N-terminal part of the fusion. Repeats in the mutants are boxed, the part of the sequence involved in the repeat leading to the actual wild type is underlined.
Activity of the mutant enzymes
The assumption that an enhanced hydrolysis of DBE might be accompanied by a restoration of the capability to convert DCE proved to be right. Table 1 shows the results of activity measurements for DBE, DCE and 1-bromobutane (BB). Compared with their primitive DhlAS and DhlAT parents, the mutants show a significantly enhanced DBE hydrolysis. Surprisingly, most of the mutants even turn out to surpass the original wild-type DhlA. Six out of the 10 isolated mutants are also capable of hydrolyzing DCE. Strikingly, however, the mutants that most seem to resemble wild type, i.e. those carrying the major part of the expected VTQPADGFT/SAW repeat (2C7, 3D3 and 3F9), do not display any activity towards DCE. Since screening for activity in whole cells implies selection at in vivo pH, we also tested activity after lowering the pH from 8.2 (wild-type optimum) to 7, but the majority of the mutants appeared to have a wild-type-like pH dependence.
Table 1. Specific activities of the mutants compared with wild-type enzyme and the two forms of the primitive dehalogenase in cell-free extracta.
| DBEb (pH 8.2) U/mg | DBEb (pH 7.5) U/mg | DCEb (pH 8.2) U/mg | BBb (pH 8.2) U/mg | DBE/BB ratio | |
|---|---|---|---|---|---|
| Wild type |
2.41 |
2.06 |
2.92 |
0.60 |
4.0 |
| DhlAS |
1.30 |
1.13 |
−c |
0.51 |
2.6 |
| DhlAT |
1.31 |
1.06 |
− |
0.52 |
2.5 |
| 2A2 |
2.17 |
1.43 |
− |
0.50 |
4.3 |
| 10D7 |
6.88 |
6.50 |
0.21 |
1.68 |
4.1 |
| D2 |
4.35 |
4.12 |
0.11 |
1.06 |
4.1 |
| D8 |
4.78 |
4.28 |
0.16 |
0.95 |
5.0 |
| 1C12 |
4.75 |
3.80 |
0.46 |
0.95 |
5.0 |
| 3B2 |
5.63 |
5.09 |
0.32 |
1.13 |
5.0 |
| 3C11 |
3.96 |
3.49 |
0.12 |
0.58 |
6.8 |
| 2C7 |
2.00 |
2.03 |
− |
0.59 |
3.4 |
| 3D3 |
3.30 |
3.03 |
− |
0.87 |
3.8 |
| 3F9 | 1.95 | 2.23 | − | 0.86 | 2.3 |
aAnalysis with SDS−PAGE confirmed that expression levels were similar.
bSubstrate concentrations were 5 mM.
c−, Not detectable/<0.02 U/mg.
We measured activity with BB to examine the effect of a longer carbon chain. The enhanced activity of the mutants that were obtained with the in vivo adaptation of wild-type DhlA to chlorohexane (19) was attributed to the generation of more space or flexibility in the active site. Since all our mutants comprise elongated sequences, we expected them also to have a preference for longer chain substrates, such as BB. For the majority of the mutants the activity with this substrate appears to be enhanced, but the ratio between activity towards DBE and BB turns out to be equal or even higher than for the wild-type enzyme. This preference for the smaller substrate is remarkable, since the active site cavity of the wild-type enzyme was believed to have evolved optimally toward accommodating C2 compounds.
DISCUSSION
We have developed a method for the random generation of repeats and deletions in vitro. The availability of a method to create segmental mutations allows further exploitation of the sequence space. The diversity we observed among the isolated dehalogenase mutants underscores the potential of this type of mutation in directed evolution. Although the exact position and length of the repeats appear to vary considerably, the isolated DhlA mutants all have in common that their sequences are extended by a repeat in front of the (wild-type) Trp175, which is considered to have an important catalytic function. Together with Trp125, this residue stabilizes the charge developing on the halogen atom in the transition state during carbon−halogen bond cleavage and acts as a binding site for the halide ion. Pushing this tryptophan residue downstream the sequence seems to be a key determinant in the adaptation towards new substrates. The additional, mainly hydrophobic, cluster that this extension is generating seems of substantial importance for the integrity of the cap domain. The segmental mutagenesis library was created by a random combinatorial fusion, which theoretically should also produce new amino acids, arising from the in-frame combination of two incomplete codon fragments. The mutants 2A2, 10D7 and 3B2 indicate that this type of point mutation is indeed formed and tolerated.
A drawback of the fusion method we used is the large amount of unproductive constructs: only one-third of the assembled gene products will be in frame. The combination with other mutagenesis and recombination techniques causes a further expansion of the explorable sequence space and consequently libraries might become excessively large, making it impossible to perform comprehensive screening. To downsize the number of mutants to manageable proportions, it is desirable to increase the frequency of mutations that give a favorable phenotype. The segmental mutagenesis method presented here can be applied to a specific segment of a target gene that is expected to be capable of hosting beneficial mutations. In DhlA a good target for such an approach is the cap domain, which covers the active site. A strategy that confines mutations to a specific area of interest, however, will always require some prior knowledge of the structure and the active site of an enzyme, either based on a 3D structure or on homology modeling. Developments in screening methods are promising: besides end-point or time-dependent UV/Vis or fluorescence measurements, techniques such as HPLC, mass spectrometry and capillary electrophoresis can now be performed in robotics-based high-throughput systems. Bylina and co-workers recently presented a method that can tackle quantities far beyond traditional high-throughput systems (30–32). This combination of digital imaging spectroscopy with solid-phase screening of microcolonies might also be attractive for DhlA selection since a convenient solid state assay is available.
Some of the isolated mutants show an up to 3-fold enhancement of DBE hydrolysis compared with the wild-type enzyme. The activity of the mutants for DCE, however, is still 10–20 times lower than the activity of wild-type enzyme. Optimization by random mutagenesis is likely to yield further improvement, since the amino acid content of a repeat is completely determined by the original sequence. In the wild-type sequence this kind of additional optimization likely has caused the W→F mutation and the deletion of DG in the first half of the repeat. DhlA appears to be particularly susceptible to this type of evolutionary process. In view of the recent appearance of the xenobiotic compound DCE in the biosphere and the relatively poor catalytic performance of the enzyme (kcat = 3.3 s–1, Km = 0.5 mM), it seems likely that the enzyme is standing only at the beginning of its evolutionary track. However, attempts to enhance the enzyme’s performance by random mutagenesis did not result in any catalytic improvement so far. Screening 50 000 colonies of a library generated using an E.coli mutator strain only yielded a mutant with a lower pH optimum (33). Analysis of over 100 000 colonies of an error-prone PCR library prepared in our own laboratory also failed to identify mutants with enhanced catalytic properties toward DCE (M.G.Pikkemaat, unpublished results). This makes us suspect that the wild-type enzyme in its present form is situated at a local evolutionary optimum. It might very well be that by going one step back in evolution, random mutagenesis of the current set of mutants would liberate the enzyme from this evolutionary trap and lead to a much more efficient catalyst. Creating primitive enzymes therefore also holds promise in providing a useful scaffold for the adaptation towards other halogenated substrates, offering a new set of enzymes for the detoxification of xenobiotic compounds.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Peter Dubbelhuis for the construction and characterization of the primitive dehalogenase mutants. This work was supported by a grant from the Netherlands Foundation for Chemical Research (C.W.) with financial aid from the Netherlands Organization for Scientific Research (N.W.O.).
REFERENCES
- 1.Shortle D. and Nathans,D. (1978) Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc. Natl Acad. Sci. USA, 75, 2170–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Myers R.M., Lerman,L.S. and Maniatis,T. (1985) A general method for saturation mutagenesis of cloned DNA fragments. Science, 229, 242–247. [DOI] [PubMed] [Google Scholar]
- 3.Cox E.C. (1976) Bacterial mutator genes and the control of spontaneous mutation. Annu. Rev. Genet., 10, 135–156. [DOI] [PubMed] [Google Scholar]
- 4.Bornscheuer U.T., Altenbuchner,J. and Meyer,H.H. (1998) Directed evolution of an esterase for the stereoselective resolution of a key intermediate in the synthesis of epothilones. Biotechnol. Bioeng., 58, 554–559. [DOI] [PubMed] [Google Scholar]
- 5.Leung D.W., Chen,E. and Goeddel,D.V. (1989) A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique, 1, 11–15. [Google Scholar]
- 6.Cadwell R.C. and Joyce,G.F. (1992) Randomization of genes by PCR mutagenesis. PCR Methods Appl., 2, 28–33. [DOI] [PubMed] [Google Scholar]
- 7.Crameri A., Raillard,S.A., Bermudez,E. and Stemmer,W.P. (1998) DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature, 391, 288–291. [DOI] [PubMed] [Google Scholar]
- 8.Ness J.E., Welch,M., Giver,L., Bueno,M., Cherry,J.R., Borchert,T.V., Stemmer,W.P. and Minshull,J. (1999) DNA shuffling of subgenomic sequences of subtilisin. Nat. Biotechnol., 17, 893–896. [DOI] [PubMed] [Google Scholar]
- 9.Stemmer W.P. (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature, 370, 389–391. [DOI] [PubMed] [Google Scholar]
- 10.Shao Z., Zhao,H., Giver,L. and Arnold,F.H. (1998) Random-priming in vitro recombination: an effective tool for directed evolution. Nucleic Acids Res., 26, 681–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H., Giver,L., Shao,Z., Affholter,J.A. and Arnold,F.H. (1998) Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol., 16, 258–261. [DOI] [PubMed] [Google Scholar]
- 12.Ohno S. (1984) Repeats of base oligomers as the primordial coding sequences of the primeval earth and their vestiges in modern genes. J. Mol. Evol., 20, 313–321. [DOI] [PubMed] [Google Scholar]
- 13.Eustance R.J., Bustos,S.A. and Schleif,R.F. (1994) Reaching out. Locating and lengthening the interdomain linker in AraC protein. J. Mol. Biol., 242, 330–338. [DOI] [PubMed] [Google Scholar]
- 14.Janssen D.B., Scheper,A., Dijkhuizen,L. and Witholt,B. (1985) Degradation of halogenated aliphatic compounds by Xanthobacter autotrophicus GJ10. Appl. Environ. Microbiol., 49, 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keuning S., Janssen,D.B. and Witholt,B. (1985) Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J. Bacteriol., 163, 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stucki G. and Thuer,M. (1994) Increased removal capacity for 1,2-dichloroethane by biological modification of the granular activated carbon process. Appl. Microbiol. Biotechnol., 42, 167–172. [DOI] [PubMed] [Google Scholar]
- 17.Stucki G. and Thuer,M. (1995) Experiences of a large-scale application of 1,2-dichloroethane degrading microorganisms for groundwater treatment. Environ. Sci. Technol., 29, 2339–2345. [DOI] [PubMed] [Google Scholar]
- 18.Damborsky J. and Koca,J. (1999) Analysis of the reaction mechanism and substrate specificity of haloalkane dehalogenases by sequential and structural comparisons. Protein Eng., 12, 989–998. [DOI] [PubMed] [Google Scholar]
- 19.Pries F., van den Wijngaard,A.J., Bos,R., Pentenga,M. and Janssen,D.B. (1994) The role of spontaneous cap domain mutations in haloalkane dehalogenase specificity and evolution. J. Biol. Chem., 269, 17490–17494. [PubMed] [Google Scholar]
- 20.Kunkel T.A. (1985) Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA, 82, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schanstra J.P., Ridder,I.S., Heimeriks,G.J., Rink,R., Poelarends,G.J., Kalk,K.H., Dijkstra,B.W. and Janssen,D.B. (1996) Kinetic characterization and X-ray structure of a mutant of haloalkane dehalogenase with higher catalytic activity and modified substrate range. Biochemistry, 35, 13186–13195. [DOI] [PubMed] [Google Scholar]
- 22.Loos M.A. (1975) Indicator media for microorganisms degrading chlorinated pesticides. Can. J. Microbiol., 21, 104–107. [DOI] [PubMed] [Google Scholar]
- 23.Holloway P., Trevors,J.T. and Lee,H. (1998) A colorimetric assay for detecting haloalkane dehalogenase activity. J. Microbiol. Meth., 32, 31–36. [Google Scholar]
- 24.Franken S.M., Rozeboom,H.J., Kalk,K.H. and Dijkstra,B.W. (1991) Crystal structure of haloalkane dehalogenase: an enzyme to detoxify halogenated alkanes. EMBO J., 10, 1297–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridder I.S., Rozeboom,H.J. and Dijkstra,B.W. (1999) Haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 refined at 1.15 Å resolution. Acta Crystallogr. D Biol. Crystallogr., 55, 1273–1290. [DOI] [PubMed] [Google Scholar]
- 26.Ollis D.L., Cheah,E., Cygler,M., Dijkstra,B., Frolow,F., Franken,S.M., Harel,M., Remington,S.J., Silman,I., Schrag,J. et al. (1992) The alpha/beta hydrolase fold. Protein Eng., 5, 197–211. [DOI] [PubMed] [Google Scholar]
- 27.Ostermeier M., Nixon,A.E., Shim,J.H. and Benkovic,S.J. (1999) Combinatorial protein engineering by incremental truncation. Proc. Natl Acad. Sci. USA, 96, 3562–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostermeier M., Nixon,A.E. and Benkovic,S.J. (1999) Incremental truncation as a strategy in the engineering of novel biocatalysts. Bioorg. Med. Chem., 7, 2139–2144. [DOI] [PubMed] [Google Scholar]
- 29.Schanstra J.P., Ridder,A., Kingma,J. and Janssen,D.B. (1997) Influence of mutations of Val226 on the catalytic rate of haloalkane dehalogenase. Protein Eng., 10, 53–61. [DOI] [PubMed] [Google Scholar]
- 30.Bylina E.J., Coleman,W.J., Dilworth,M.R., Silva,C.M., Yang,M.M. and Youvan,D.C. (1999) Solid phase enzyme kinetics screening in microcolonies. US Patent. 5914245.
- 31.Bylina E.J., Coleman,W.J., Dilworth,M.R., Robles,S.J., Tanner,M.A., Yang,M.M. and Youvan,D.C. (2000) Solid-phase enzyme screening. ASM News, 66, 211–217. [Google Scholar]
- 32.Delagrave S., Murphy,D.J., Pruss,J.L., Maffia,A.M.,III, Marrs,B.L., Bylina,E.J., Coleman,W.J., Grek,C.L., Dilworth,M.R., Yang,M.M. and Youvan,D.C. (2001) Application of a very high-throughput digital imaging screen to evolve the enzyme galactose oxidase. Protein Eng., 14, 261–267. [DOI] [PubMed] [Google Scholar]
- 33.Chang C.H., Schindler,J.F., Unkefer,C.J., Vanderberg,L.A., Brainard,J.R. and Terwilliger,T.C. (1999) In vivo screening of haloalkane dehalogenase mutants. Bioorg. Med. Chem., 7, 2175–2181. [DOI] [PubMed] [Google Scholar]