Abstract
Human T-lymphotropic virus type 1 (HTLV-1), a complex retrovirus, causes adult T-cell lymphoma/leukemia and is linked to a variety of immune-mediated disorders. The roles of proteins encoded in the pX open reading frame (ORF) II gene region in HTLV-1 replication or in mediating virus-associated diseases remain to be defined. A nucleus-localizing 30-kDa protein, p30II, encoded within pX ORF II has limited homology with the POU family of transcription factors. Recently, we reported that selected mutations in pX ORF II diminish the ability of HTLV-1 to maintain high viral loads in infected rabbits. Herein we have tested the transcriptional ability of p30II in mammalian cells by using yeast Gal4 fusion protein vectors and transfection of luciferase reporter genes driven by CREB-responsive promoters. p30II as a Gal4 DNA-binding domain (DBD) fusion protein transactivates Gal4-driven luciferase reporter gene activity up to 25-fold in 293 and HeLa-tat cells. We confirmed nuclear localization of p30II and demonstrate dose-dependent binding of p30II-Gal4(DBD) to Gal4 DNA-binding sites. The transcriptional activity of p30II-Gal4(DBD) was independent of TATA box flanking sequences, as shown by using two different Gal4 reporter systems. Studies of selected p30II mutants indicated that domains that mediate transcription are restricted to a central core region of the protein between amino acids 62 and 220. Transfection of a p30II-expressing plasmid repressed cellular CRE-driven reporter gene activity, with or without Tax expression. In contrast, p30II at lower concentrations enhanced HTLV-1 long terminal repeat-driven reporter gene activity independent of Tax expression. These data are the first to demonstrate a transcriptional function for p30II and suggest a mechanism by which this nuclear protein may influence HTLV-1 replication or cellular gene expression in vivo.
Human T-lymphotropic virus type 1 (HTLV-1) is a complex retrovirus that encodes typical gag, pol, and env gene products as well as unique regulatory and accessory genes (11). HTLV-1 causes adult T-cell leukemia/lymphoma (ATL) and is etiologically linked to tropical spastic paraparesis/HTLV-associated myelopathy (HAM/TSP), a chronic neurodegenerative disorder (12, 21), as well as a variety of other immune-mediated diseases (10). The role of HTLV-1 in mediating these diseases is not clear but is likely related to the ability of the virus to evoke lymphocyte activation (14). The complex genome of HTLV-1 contains unique regulatory and accessory genes in four open reading frames (ORFs), I to IV, of the pX region. ORFs IV and III of HTLV-1 encode the well-characterized Tax and Rex proteins, respectively. Tax is a 40-kDa nucleus-localizing phosphoprotein which increases viral transcription from the HTLV-1 long terminal repeat (LTR) as well as many cellular genes involved in host cell proliferation (19). Rex is a 27-kDa nucleolus-localizing phosphoprotein that increases the cytoplasmic accumulation of nonspliced and singly spliced viral RNA (13).
In contrast to the extensive knowledge of Tax and Rex structure and function, little is known about the role of pX ORF I and ORF II in the replication or pathogenesis of HTLV-1. However, emerging evidence supports the expression of pX ORFs I and II both in vitro and in vivo and the importance of these conserved ORFs in the replication of HTLV-1. At least eight alternatively spliced mRNAs are expressed from the 3′ or pX region of HTLV-1 (2). Reverse transcription-PCR assays identified mRNAs from infected cell lines and freshly isolated cells from HTLV-1-infected subjects (17). Cereseto et al. (3) reported the detection of the same RNA species from patients with ATL and HAM/TSP using a semiquantitative RNase protection assay. Importantly, cytotoxic CD8+ T cells from HTLV-1-infected individuals have recently been demonstrated to recognize pX ORF I- and II-derived peptides, indicating that these viral proteins are expressed in vivo (22). Despite evidence for the expression of pX ORF I and ORF II, these viral genes do not appear to be required for viral infectivity, replication, or transformation in typical cell culture systems. In contrast, using an infectious molecular clone of HTLV-1 (7) with selective mutations that ablated the mRNA from ORF I (encoding p12I), we were the first to identify a functional role of pX ORF I in establishment of infection in an animal model (6).
ORF II is spliced to the first tax exon and encodes two proteins, a full-length p30II and an internally initiated p13II. The smaller protein, p13II, is derived from initiation at the first internal methionine codon in ORF II and represents the carboxyl-terminal 87 residues of p30II. The p30II and p13II proteins were initially found to localize to the nucleolus and nucleus, respectively (16), but p13II also localizes to mitochondrial membranes (5). The cellular segregation of ORF II gene products suggests specific roles for these proteins in the regulation of HTLV-1 expression or as mediators of virus-cell interactions. The p30II protein contains serine- and threonine-rich regions with distant homology to transcription factors Oct-1 and -2, Pit-1, and POU-M1 (4). We have recently reported that mutations in the ACH.p30II/p13II viral clone which destroy the initiator methionine of the mRNA encoding p13II and insert an artificial termination codon in the mRNA encoding p30II prevent the virus from obtaining normal levels in rabbits (1).
In this study, we have tested the transcriptional ability of p30II in mammalian cells by using a yeast Gal4 fusion protein system and transfection of luciferase reporter genes driven by CREB-responsive promoters. Our data indicate that p30II, as a Gal4 fusion protein, significantly transactivates Gal4-driven luciferase reporter gene activity in multiple cell types. Furthermore, we provide data demonstrating that the transcriptional activity of p30II-Gal4 DNA-binding domain (DBD) (DBD) is independent of TATA box flanking sequences by comparing two different Gal4 reporter systems. Mutational studies of p30II indicated that the transactivation domains reside within the central portion of the protein (between amino acids 62 and 220). Interestingly, small amounts of p30II expression transactivated HTLV-1 LTR-driven reporter gene activity, even in the presence of Tax, whereas higher concentrations repressed LTR and CRE-driven reporter gene activity. Our data provide the first evidence to support the transcriptional activity of p30II and suggest an important role for the nuclear protein in HTLV-1 replication and cellular gene expression.
MATERIALS AND METHODS
Cell lines.
All cultured cells (293 cells obtained from American Type Culture Collection, no. CRL-1573, and HeLa-tat cells were from the National Institutes of Health AIDS Research and Reference Reagent Program [catalog no. 502]) were grown in 10-cm tissue culture dishes in Dulbecco's minimal essential medium (DMEM) containing 10% fetal bovine serum and 1% streptomycin and penicillin at 37°C. Cells were split and cultured in six-well plates to 50% confluence 16 h before transfection according to the manufacturer's protocol (Lipofectamine-Plus; Gibco-BRL).
Gal4-mediated transcription assay. (i) Reporter plasmids.
Plasmid p5XGT-TATA-Luc, a kind gift of P. Quinn (The Pennsylvania State University, Hershey, Pa.), contains five tandem Gal4 DNA-binding sequences upstream of a TATA box, derived from positions −64 to +1 of the phosphoenolpyruvate carboxykinase (PEPCK) gene in a luciferase reporter gene plasmid (25). pGL2-TATA-Luc was constructed by ligating the TATA box of the adenovirus E1b gene, derived from plasmid E1b-CAT (24), into the XhoI and BglII sites of pGL2-basic, a luciferase reporter gene vector (Promega), and then subcloning tandem copies of Gal4 DNA-binding sequence using KpnI and XhoI sites.
(ii) Effector plasmids.
The p30II-Gal4(DBD) expression vector was constructed by replacing the CREB-(1-247)-encoding sequence of CREB-Gal4-pCRG4-11 (a kind gift of P. Quinn, The Pennsylvania State University) with the p30II-encoding sequence synthesized by PCR amplification with 5′ primer 5′-A [ATATGAATTCATGGCACTATGCTGTTCGCC) and 3′ primer 3′-A (TATAACTAGTTTAGAGGTTCTCGGGTG) from the HTLV-1 molecular clone ACH (15), including 5′ EcoRI and 3′ SpeI restriction sites (underlined). Similarly, all truncated mutants (MT-1 through MT-6) of the p30II expression vector were constructed by replacing CREB-(1-247)-encoding sequence of CREB-Gal4-pCRG4-11 (containing Gal4-DBD) with appropriate truncated p30II-encoding sequences synthesized by PCR from ACH using the following primers: for MT-1, p30II(1-220) 5′-A (above) and TATAACTAGTGGGCACCAGTCGCCTTGT (3′-B); for MT-2, p30II(1-179) 5′-A and TATAACTAGTGGTTAACTTTGTATCTGT (3′-C); for MT-3, p30II(1-132) 5′-A and TATAACTAGTGGAAGAGTTAAAGGACAA (3′-D); for MT-4, p30II(1-62) 5′-A and TATAACTAGTGCGGGAGAAAGAGGAGGA (3′-E); for MT-5, p30II(62-220) ATATGAATTCATGTCTTTTTTTCGCTTCCTC (5′-B) and 3′-B; for MT-6, p30II(179-241) ATATGAATTCATGCTTATTATCAGCCCA (5′-C) and 3′-A (above).
All of the inserted p30 gene sequences were in frame and had similar expression efficiencies, which was verified by immunoblot assay of transfected HeLa-tat cells. The p30II-HA expression vector (pCMV-p30II-HA) was a kind gift of G. Franchini (National Cancer Institute, Bethesda, Md.) (16).
(iii) Gal4 transcription assay.
For each trial, 0.3 μg of reporter plasmids (p5XGT-TATA-Luc and pGL2-TATA-Luc) and 0 to 1.5 μg of effector plasmids (p30II-Gal4-pCRG4-11, pCMV-p30II-HA, and Gal4-pCRG4-11) were transfected as indicated in figure legends. As an internal control for transfection efficiency, 0.1 μg of pRSV-β-gal (P. Quinn, The Pennsylvania State University) was also used in each transfection. pBlue-Script (Stratagene) was used as carrier DNA to equalize DNA concentrations for each transfection. Transfected cells were lysed with 1× lysis buffer (Promega) using 0.5 ml/well at room temperature for 15 min. Twenty microliters of each lysate was used to test luciferase reporter gene activity using an enhanced luciferase assay kit (Promega) according to the manufacturer's protocol. To normalize transfection experiments, 5 μl of each lysate was assayed for β-galactosidase activity according to the manufacturer's protocol (Lumigen, Southfield, Mich.). Results were expressed as mean fold increase ± standard deviation (SD) in arbitrary light units of luciferase activity in four independent trials for each set of experiments. Statistical comparisons of data sets were performed by a standard two-sample t test.
CRE- and TRE-mediated transcription assay.
pCRE-α-Luc, a kind gift of S. McKnight (University of Washington), has a promoter with three repeats of the CRE element within the sequences from −168 to +45 of the α-subunit of the human glycoprotein hormone gene (18). The HTLV-1 LTR-Luc plasmid is driven by the complete HTLV-1 LTR, which has three repeats of the Tax-responsive element (TRE), and was constructed by cloning the LTR sequence of HTLV-1 into pGL2-basic vector (20). The p30II-HA expression vector (pCMV-p30II-HA) is described above (Gal4 assay). The HTLV-1 Tax expression vector (pCMV-Tax) has been described previously (20). For each transfection, 0.3 μg of reporter plasmids (pCRE-α-Luc and pLTR-Luc) and 0 to 1.0 μg of effector plasmids (pCMV-p30II-HA and pCMV-Tax) as indicated in the figure legends were used for each transfection. As an internal control for transfection efficiency, 0.1 μg of pRSV-β-gal was also used in each transfection. pBlue-Script (Stratagene) was used as carrier DNA to equalize DNA concentrations for each transfection. Luciferase activity and transfection control methods were the same as in the Gal4 transcription assay above.
Preparation of nuclear extracts.
Nuclear extracts were prepared by a modification of the method previously described by Dignam et al. (9). Briefly, 106 transfected cells were washed twice with ice-cold phosphate-buffered saline (PBS), harvested in 5 ml of PBS, and centrifuged at 400 × g for 5 min at 4°C. The pellet was washed with 4 packed cell volumes of hypertonic buffer containing 10 mM Tris-HCl (pH 7.8), 1.5 mM MgCl2, and 10 mM KCl and left on ice for 10 min. The cells were then lysed by 10 strokes of a Dounce homogenizer using a type B pestle. The presence of intact cell nuclei was determined by cytospin analysis. Intact nuclei were sedimented at 4,500 × g for 5 min at 4°C, resuspended in 2 packed cell volumes of hypotonic buffer containing 420 mM KCl, 20 mM Tris-HCl (pH 7.8), 1.5 mM MgCl2, and 20% glycerol, and incubated at 4°C with gentle agitation for 1 h. Immediately before use, 0.5 mM dithiothreitol (DTT), 0.4 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg of microcystin, and 2 μg each of leupeptin and pepstatin per ml were added fresh to the nucleus preparation buffer. After complete lysis of nuclei, the nuclear extract was centrifuged at 10,000 × g for 30 min at 4°C. The supernatant was dialyzed twice against 500 ml of dialysis buffer containing 20 mM Tris-HCl (pH 7.8), 100 mM KCl, 0.2 mM EDTA, and 20% glycerol for 4 h at 4°C. The nuclear lysate then was aliquoted and frozen in liquid N2 immediately and stored at −80°C. A mouse monoclonal antibody that recognizes the 27-kDa Fas-associated death domain protein (Transduction Labs) and a rabbit polyclonal antibody that recognizes histone H1 (Upstate Biotechnology) were used to verify cytoplasmic and nuclear fractions, respectively, by Western immunoblot assay.
Expression of p30II proteins.
HeLa-tat cells at approximately 65% confluence in 10-cm tissue culture dishes were transfected using calcium phosphate with 10 μg of pCMV-p30II-HA, p30II-Gal4-pCRG4-11, or p30II-mutant-Gal4-pCRG4-11 plasmids MT-1 through MT-6. At 48 h posttransfection, the nuclei and cytosol of transfected cells were prepared as described above. Whole cell lysates were made by lysis of cells (90% confluent in 10-cm dish) using 0.4 ml of radioimmunoprecipitation buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) containing 1 mM PMSF, 2 μg of aprotinin, 2 μg of pepstatin A, and 1 μg of leupeptin per ml. Each 50 μg of nuclear, cytosolic, and whole-cell lysates was electrophoresed by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose membrane (Millipore), blocked using 5% nonfat milk in 1× Tris buffer (0.5 M Tris-HCl [pH 7.5], 1.5 M NaCl, 0.1% NP-40) for 1 h at 4°C. The blocked membrane was incubated with primary polyclonal anti-HA serum (Santa Cruz Biotechnology) or polyclonal anti-Gal(DBD) serum (Santa Cruz) (each 1:1,000 in Tris buffer containing 3% bovine serum albumin) overnight at 4°C. After extensively washing the membrane with Tris-Tween buffer solution (TTBS; 0.5 M Tris-HCl [pH 7.5], 1.5 M NaCl, 0.075% Tween 20) the membrane was blotted with anti-rabbit antibody serum (1:1,000 in TTBS containing 5% nonfat dry milk) for 1 h at room temperature. After washing the membrane three times with TTBS, the membrane was developed using enhanced chemiluminescence and exposed to film according to the manufacturer's protocols (Amersham).
To detect p30II by immunofluorescence, HeLa-tat cells were seeded in chamber slides (Fisher Scientific) at approximately 40% confluence 18 h prior to transfection. Transfection with 4 μg of pCMV-p30II-HA was performed using Lipofectamine plus (Sigma). At 48 h posttransfection, DMEM was removed and cells were washed twice in PBS. Fixation of the cells for 15 min using 4% paraformaldehyde was performed at room temperature. Cells were then incubated with a monoclonal anti-HA antiserum (Babco) overnight at 4°C, followed by incubation with indocarbocyanine-labeled anti-mouse immunoglobulin (Jackson Immunogen) for 1 h at room temperature. The expression of p30II-HA was evaluated by immunofluorescence microscopy (Zeiss Axioplan2). A digital camera (Diagnostic Instruments Inc.) was used to produce standard light microscopic and immunofluorescent photomicrographs.
EMSA.
For the electromobility shift assay (EMSA), nuclear extracts from HeLa-tat cells transfected with p30II-Gal4(DBD) expression vector (10 μg/10-cm dish) were prepared as described above. Tandem copies of Gal4 DNA-binding sequence (CGGAGGACTCGTCTCCG) were synthesized and used as the probe in the EMSA. The probe was 32P labeled using T4 kinase (Promega) at 37°C for 30 min. The 32P-labeled 2× Gal4-DNA-binding sequence oligonucleotide was separated from free [32P]ATP by a Sephedex G50 filtration column (Amersham). Binding was performed in 30 μl of binding reaction buffer containing 20 mM Tris, 1 mM MgCl, 12% glycerol, 0.1 mM DTT, 10 to 15 μg of nuclear protein, and 1 μg of poly(dI-dC) (Pharmacia). Various concentrations of unlabeled Gal4-DNA-binding sequence oligonucleotide were used as a specific competitor of probe. Specific antiserum (2 μg) for Gal4(DBD) (Santa Cruz) was added to the binding reaction mixture for the supershift assay. After 25 min of incubation at 4°C, 0.5 to 1.0 ng (20,000 to 40,000 cpm) of 32P-labeled 2× Gal4(DBD) oligonucleotide probe was added to each reaction and further incubated for 30 min at 25°C. The sample was electrophoresed in 5% nondenatured polyacrylamide gels in 0.5× Tris-borate-EDTA at a constant 180 V for 4 h at 4°C. The gel was subsequently dried, and bands were visualized by autoradiography.
RESULTS
HTLV-1 p30II localizes to the nucleus.
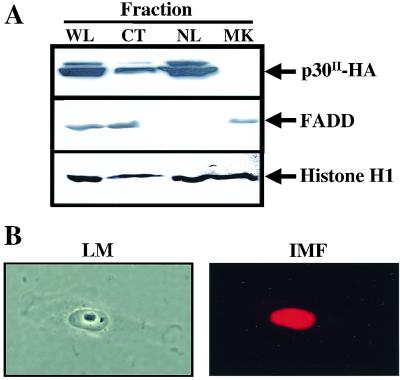
Nuclear localization or conditional translocation to the nucleus is a characteristic of most proteins that function as a transcription factor. Previous studies have demonstrated by standard immunofluorescence methods that HTLV-1 p30II was localized in the nucleolus of transfected cells and contained two nuclear localization signals (16). Consistent with previous reports, p30II was detected predominantly in the nuclear fraction of our transiently transfected HeLa-tat cells by immunoblot assay (Fig. 1A). We verified these results using an immunofluorescence assay (Fig. 1B). Our results indicate that, following transfection, p30II accumulates in the nucleus. This subcellular localization and the predicted regional homology of p30II with the POU family of transcription factors suggested to us that p30II could play a role in the regulation of transcription.
FIG. 1.
p30II accumulates in the nuclear fraction of transfected HeLa-tat cells. (A) Whole-cell (WL), nuclear lysate (NL), and cytoplasmic extracts (CT) were prepared from HeLa-tat cells transfected with pCMV-p30II-HA for 48 h as described in the text. Twenty-five micrograms of cellular extract was separated in SDS–10% PAGE gels and blotted on nitrocellulose membranes. p30II-HA was detected by immunoblot analysis using polyclonal rabbit anti-HA serum. A mouse monoclonal antibody that recognizes the 27-kDa Fas-associated death domain protein (FADD; Transduction Labs) and a rabbit polyclonal antibody that recognizes histone H1 (Upstate Biotechnology) were used to verify cytoplasmic and nuclear fractions, respectively. (B) Light microscopic (LM) and immunofluorescence assay (IMF) of HeLa-tat cells 48 h after transfection with 4 μg of pCMV-p30II-HA as described in the text. p30II-HA expression in cell nuclei was detected using a monoclonal anti-HA antiserum by indirect immunofluorescence assay (Zeiss Axioplan2) and stored from the same microscopic field as photomicrographs using a digital camera (Diagnostic Instruments Inc.).
p30II increases Gal4-driven luciferase reporter gene activity.
HTLV-1 p30II has several important structural characteristics of a transcription factor, including nuclear localization sequences, serine/threonine-rich motifs, and regional homology with the transcription factor Oct-1 in DNA-binding regions. While these observations suggest that p30II may serve a role as a transcription factor, the absence of any information about the cis-acting element(s) in viral or cellular promoters that may interact with p30II precludes standard approaches to testing DNA-binding regions as targets of the viral protein. Therefore, to test if p30II could influence transcription, we used a Gal4 system and constructed p30II as a chimeric protein with the DBD of Gal4. The Gal4(DBD) (amino acids 1 to 147) was cloned into the carboxyl-terminal region of full-length p30II (amino acids 1 to 241) to form the p30II-Gal4(DBD) expression vector (p30II-Gal4-pCRG4-11) (Fig. 2A). We initially tested a reporter construct, p5XGT-TATA-Luc, whose promoter contains five copies of the Gal4 DNA-binding site upstream of the TATA box derived from the PEPCK promoter (−61 to +1) (Fig. 2A). As expected, cotransfection of the parent Gal4-pCRG4-11 vector with p5XGT-TATA-Luc resulted in no significant luciferase reporter gene activity (Fig. 2B). In contrast, in a dose-dependent manner, p30II-Gal4-pCRG4-11 elicited up to an 18-fold mean increase in reporter gene activity (Fig. 2B). The fact that p30II in the absence of Gal4(DBD) did not significantly promote reporter gene activity indicates a requirement for localization of p30II to the Gal4 promoter (Fig. 2B).
FIG. 2.
Activation of Gal4-driven reporter gene by Gal4(DBD)-p30II fusion protein. (A) Schematic illustration of reporter and effector plasmids used in the Gal4 transcription assay. (B) HeLa-tat cells were transiently cotransfected with 0.3 μg of p5XGT-TATA-luciferase plasmid and 0 to 1.5 μg of p30II-Gal4-pCRG4-11, pCMV-p30II-HA, or Gal4-pCRG4-11 plasmid as indicated. (C) HeLa-tat cells transfected with 0.3 μg of p5XGT-TATA-Luc or pGL2-TATA-luciferase reporter plasmid and 0 to 1.5 μg of p30II-Gal4-pCRG4-11 plasmid. Results are expressed as mean fold increase ± SD in luciferase activity (normalized to β-galactosidase activity) for four independent trials. (D) 293 cells were transiently cotransfected with 0.3 μg of p5XGT-TATA-luciferase plasmid and 0 to 1.5 μg of p30II-Gal4-pCRG4-11, pCMV-p30II-HA, or Gal4-pCRG4-11 plasmid to confirm the results using HeLa-Tat cells.
To test whether the observed transactivation of reporter gene activity by p30II-Gal4-pCRG4-11 was dependent on specific flanking sequences adjacent to the TATA box region, the p5XGT-TATA-Luc containing the TATA box from the PEPCK (−61 to +1) gene promoter was compared with pGL2-TATA-Luc. pGL2-TATA-Luc contains a minimal TATA box derived from the E1b gene promoter of adenovirus. Both luciferase reporter gene activities were significantly increased (up to 18- to 25-fold) by cotransfection with the p30II-Gal4-pCRG4-11 expression vector (Fig. 2C). These data indicate that transactivation of reporter gene activity by p30II is independent of the flanking sequence of the TATA box. We confirmed the transcriptional activity of p30II in a dose-dependent manner using 293 cells (Fig. 2D).
p30II-Gal4(DBD) binds to the Gal4 promoter in a dose-dependent manner.
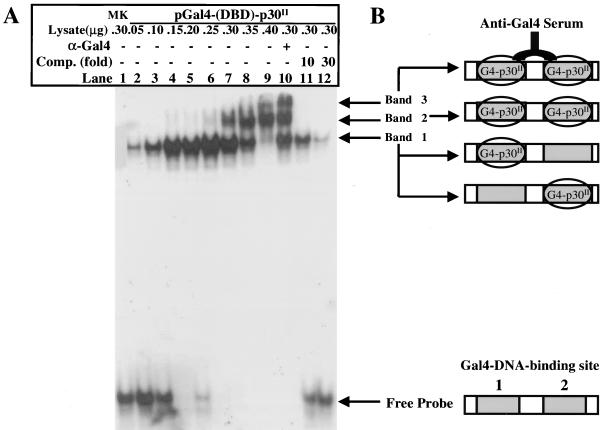
To test if p30II-Gal4(DBD) bound the Gal4 promoter directly, we quantitatively analyzed the promoter-binding activity of p30II-Gal4(DBD) fusion protein in transfected HeLa-tat cells by EMSA. p30II-Gal4(DBD) in nuclear lysates of HeLa-tat cells bound to Gal4-DNA-binding sequences efficiently and in a dose-dependent manner (Fig. 3). Nuclear proteins (50 to 250 ng) from HeLa-tat cells transfected with 10 μg of p30II-Gal4-pCRG4-11 plasmid for 48 h bound to only one site of the double Gal4-DNA-binding probe (site 1), which resulted in a single shifted band (Fig. 3A, lanes 2 to 6, band 1). Higher concentrations of nuclear lysates (300 to 350 ng) containing p30II-Gal4(DBD) elicited an additional band corresponding to binding of a second binding site by p30II-Gal4(DBD) (Fig. 3A, lanes 7 and 8, band 2). When the nuclear proteins were further increased to 400 ng, both Gal4-DNA-binding sites of the probe were occupied, which resulted in a predominant and slower migrating band 2 (Fig. 3A, lane 9). The addition of Gal4(DBD)-specific antiserum resulted in a supershifted band (Fig. 3A, lane 10, band 3). In addition, when a 10- to 30-fold excess of nonlabeled Gal4-DNA-binding oligonucleotides were added as competitors to the binding reaction, the shifted bands were efficiently attenuated (Fig. 3A, lanes 11 and 12). Figure 3B illustrates the EMSA band shifts. Collectively, these data indicated that the observed shifted probe bands were p30II-Gal4(DBD) specific and provided direct evidence to show the specific interactions between Gal4(DBD)-containing p30II and the promoter of the Gal4 reporter gene.
FIG. 3.
EMSA demonstrates that Gal4(DBD)-p30II binds Gal4 DNA element in a dose-dependent manner. (A) EMSA using nuclear extracts obtained from HeLa-tat cells 48 h posttransfection with p30II-Gal4-pCRG4-11 as described in the text. The DNA oligonucleotide probe containing tandem Gal4-DNA-binding elements was labeled with γ-32P by T4 kinase. The amounts of nuclear lysates and presence of anti-Gal4 serum (lane 10) and competitors (Comp.) (lanes 11 and 12) are indicated at the top of the gel. Free probe, probe complexes, and shifted bands are represented in panel B.
Mutation of p30II reveals a central core that mediates transcriptional activity.
In order to determine the structural motifs of p30II that mediated the transcriptional activity in our Gal4 system, a series of six truncated p30II mutants were fused to Gal4(DBD). These mutants of p30II contained progressive deletions in both the amino- and carboxyl-terminal regions of the protein (Fig. 4A). Each of the p30II mutant proteins was expressed at the expected molecular weight, as indicated by immunoblot analysis (Fig. 4B). Equal amounts of wild-type p30II-Gal4(DBD) and each of the mutant proteins were evaluated by cotransfection with our p5XGT-TATA-Luc reporter gene plasmid in HeLa-tat cells. The luciferase activity elicited by p30II-Gal4(DBD) was compared to luciferase activity elicited by each of the six p30II-Gal4(DBD) mutants (Fig. 4C). Luciferase activity elicited by the serially deleted mutants MT-1 through MT-4 was progressively reduced compared to wild-type p30II-Gal4(DBD) (from 75% to less than 10% of wild-type levels). These data indicate that the amino acid sequence from 62 to 220 of p30II is essential for the transcriptional activity observed in our assays. This observation was further confirmed by mutant MT-5, which represents the central amino acid sequence from 62 to 220 of p30II, which retained 85% of the transactivation of wild-type p30II. Mutant MT-6 includes sequences encoding p13II and elicited only 35% of the transcriptional activity of wild-type p30II, suggesting that p13II by itself does not effectively mediate the transcriptional activity. However, p13II sequences, when deleted from the full-length p30II (MT-1), lost approximately 25% of the transcriptional activity of the wild-type protein. These data are consistent with the mitochondrial localization of p13II, implying that the protein does not effect nuclear transcription events (5).
FIG. 4.
Effects of p30II deletion mutants on Gal4-driven luciferase reporter gene activity. (A) Diagram of reporter plasmid p5XGT-TATA-Luc and structures of wild-type (WT) and deletion mutants of Gal4(DBD)-p30II effector plasmids. (B) Expression of wild-type (WT) and deletion mutants of Gal4(DBD)-p30II effector plasmids by immunoblot assay following transfection in HeLa-Tat cells. (C) Reporter gene activity from HeLa-tat cells cotransfected with 0.3 μg of p5XGT-TATA-luciferase reporter plasmids with 1 μg of each indicated Gal4(DBD)-p30II expression plasmid (MT-1 to MT-6). The reporter gene activity by individual Gal4(DBD)-p30II mutants is represented as a percentage of the reporter gene activity of wild-type (WT) Gal4(DBD)-p30II normalized for β-galactosidase activity. Results are expressed as mean percent change in arbitrary light units (ALU) ± SD in luciferase activity (normalized to β-galactosidase activity) for four independent trials.
p30II differentially influences CRE- and TRE-mediated reporter gene activity.
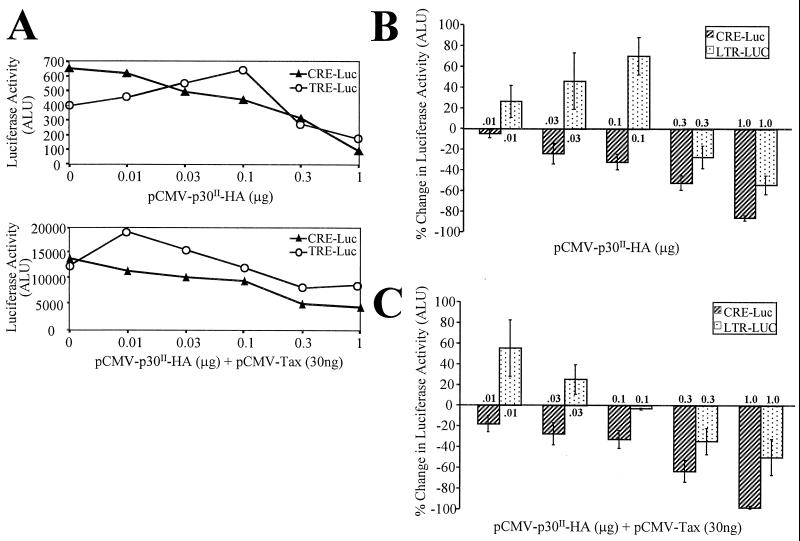
To test p30II transcriptional activity in the context of the HTLV-1 promoter, we compared the ability of p30II expressed from a pCMV-p30II-HA expression plasmid to mediate CRE- and TRE-mediated reporter gene activity. Because of the known importance of HTLV-1 Tax in the regulation of these promoter elements, we tested each of our reporter gene systems with simultaneous Tax expression in a dose-dependent manner. Cotransfection of pCMV-Tax consistently increased the basal luciferase activity of both reporter gene constructs from 15- to 25-fold (Fig. 5A). The pCMV-p30II-HA plasmid was cotransfected into 293 cells with the CRE- and TRE-luciferase reporter plasmids in the absence and in the presence of Tax (pCMV-Tax). p30II repressed the cellular CRE-driven reporter gene activity in a dose-dependent manner (Fig. 5B) and also reduced the positive transcriptional effects of Tax (Fig. 5C). Interestingly, lower concentrations of the p30II plasmid (<0.1 μg) consistently activated HTLV-1 LTR reporter gene activity, but increased amounts (>0.1 μg) of the plasmid repressed LTR reporter gene activity (Fig. 5B). Tax expression only modestly influenced this differential pattern of p30II effects on LTR-mediated transcription, and the positive effect of lower concentrations of p30II in LTR-mediated reporter gene activity were additive to typical Tax effects (Fig. 5C). Cotransfection of pCMV-p30II-HA had no effect on expression of pRSV-β-gal (data not shown). These data suggest that low concentrations of p30II have the potential to differentially interfere with the transcription of CRE-driven gene activity while promoting LTR-mediated transcription.
FIG. 5.
p30II differentially modulates CRE- and TRE-mediated transcription. (A) Luciferase activity of 0.3 μg of α-CRE-luciferase reporter plasmid (solid triangles) or 0.3 μg of pLTR-luciferase reporter plasmid (open circles) and 0 to 1.0 μg of pCMV-p30II HA. The lower panel demonstrates parallel transfections but with 30 ng of pCMV-Tax as well. Results are expressed as arbitrary light units (ALU) to indicate basal activity (without pCMV-p30II HA) and activity following transfection of plasmids. (B) 293 cells transiently cotransfected with 0.3 μg of α-CRE-luciferase reporter plasmid (hatched bars) or 0.3 μg of pLTR-luciferase reporter plasmid (dotted bars) and 0 to 1.0 μg of pCMV-p30II HA. (C) Same transfections as in panel A but also concurrently transfected with 30 ng of pCMV-Tax. Cotransfection of pCMV-p30II-HA had no effect on expression of pRSV-β-gal, used as a transfection control. Results are expressed as mean percent change in arbitrary light units (ALU) ± SD in luciferase activity for four independent trials.
DISCUSSION
Our data are the first to demonstrate the functional role of p30II in modulating transcription. Using a yeast Gal4 fusion protein system and transfection of luciferase reporter genes driven by CREB-responsive promoters, we provide evidence to support the ability of p30II to serve as a transcription factor in mammalian cells. Transcriptional activity of the nucleus-localizing p30II-Gal4(DBD) was independent of TATA box flanking sequences, and selected mutant proteins indicate that the transactivation domains of p30II are localized between amino acids 62 and 220. Furthermore, we demonstrated that p30II repressed cellular CRE-driven reporter gene activity, with or without Tax expression, while small amounts of p30II enhanced HTLV-1 LTR-driven reporter gene activity, even in the presence of Tax. Higher concentrations of the viral protein repressed LTR-driven reporter gene activity. Collectively, our data have important implications for the role of p30II in the replication of HTLV-1 and suggest a mechanism by which this nuclear protein may differentially influence HTLV-1 replication or cellular gene expression in vivo.
A growing body of evidence indicates the importance of HTLV-1 pX ORFs I and II in the replication of the virus in vivo. The exact function of the proteins encoded by HTLV-1 ORF II, p30II and p13II, remains elusive. Selective mutations of the infectious ACH clone designed to eliminate p30II and p13II expression do not affect in vitro viral infectivity of HTLV-1 in human peripheral blood mononuclear cells or alter the Gag and Env composition of virus particles or influence Tax function in transfected cell lines (26). However, pX ORF II is highly conserved by the virus. We have recently reported that selected mutations which prevent the expression of full-length p30II and eliminate the start codon for p13II dramatically influence the ability of the proviral clone ACH to maintain proviral loads in infected rabbits (1). Furthermore, Pique et al. (22) have recently reported that cytotoxic CD8+ T cells from HTLV-1-infected individuals recognize pX ORF I- and II-derived peptides, indicating that these viral proteins are expressed in vivo.
HTLV-1 p30II (also referred to as Tof) contains several important features of many transcription factors. The protein contains a region rich in serine and threonine residues, which are conserved in the transcriptionally important domains of several octamer-binding transcription factors such as Oct-1, Oct-2, and Pit-1. In addition, p30II has nuclear localization signal sequences. These features imply that p30II functions as a transcription factor in HTLV-1-infected cells. Our data suggest that the enhancement of Gal4 reporter gene activity by p30II-Gal4(DBD) specifically results from the interactions between p30II and the transcription complex bound to or associated with the transcription start site. To further define the molecular mechanism of p30II-mediated transactivation, it will be important to identify binding proteins for p30II among the transcription machinery complex.
The results from our p30II mutant studies indicated that the transactivation motif of p30II was localized in the middle region of the protein (residues 62 to 220). This core region includes the defined nuclear localization signal and serine/threonine-rich regions (4). We found that p30II-Gal4(DBD) mutants with progressive deletions in the C-terminal sequences of p30II (from MT-1 to MT-4) correspondingly lost their ability to mediate transcription. MT-4, representing only the first 62 amino acids of the N terminus, lost almost all of its ability to mediate transcription. Our conclusions are further supported by data from the evaluation of MT-5 (62 to 220), which retained the ability to promote reporter gene activity nearly as efficiently as wild-type p30II despite the fact it lacks N-terminal (1 to 62) and C-terminal (220 to 241) sequences. MT-6 (retaining sequences corresponding to p13II) lost as much as 75% of the transactivation of reporter gene activity, suggesting that p13II serves a different role in the viral life cycle (5). Further studies using site-directed mutations are needed to define the particular amino acid residues of p30II that serve as the transactivation domain of the protein.
Our data showing that p30II differentially influences CRE- and TRE-driven reporter gene activity are not without precedence. Similarly, the regulation of the immediate-early (IE) gene promoter of herpes simplex virus type 1 (HSV-1) is dependent on the interplay between cellular and viral transcription factors. VP16, a potent transcription factor from HSV-1, binds the host cell protein HCF, which allows the viral protein to form a stable complex with Oct-1 (29). The IE gene promoter contains an Oct-1-like motif (TAATGARAT) that is critical for IE gene expression. Cellular octamer-binding proteins can mediate the inhibition of IE promoters. The TAATGARAT motif (where R is a purine) has been demonstrated to cause both positive and negative effects, depending on the context of these cellular transcription factors and VP16 (27). As a result, these motifs have been postulated to mediate active transcription of HSV-1 during lytic cycles of replication but silence the IE genes during HSV-1 latency by serving as a target for inhibitory octamer-binding proteins. HTLV-1 p30II may modulate transcription by similar mechanisms. Further studies are in progress to identify the DNA-binding sites or cellular proteins that interact with p30II.
Our data also demonstrated that small amounts of p30II could transactivate HTLV-1 LTR-driven reporter gene activity, whereas increasing concentrations of p30II repressed LTR reporter gene activity. The positive effect of lower concentrations of p30II in LTR-mediated reporter gene activity was additive to the influence of Tax, which only modestly altered the differential pattern of p30II effects on LTR-mediated transcription. A previous report suggested that expression of p30II had no positive influence on LTR-Tax reporter gene activity (4). However, this study did not report the concentrations of expression plasmids used to monitor LTR-mediated reporter gene activity or whether p30II was tested in a dose-dependent manner. Our data indicate that the transcriptional effects of p30II on LTR reporter gene activity are concentration dependent. In context to the infected cell, in which small amounts of p30II are likely to be expressed, this viral protein may successfully promote viral transcription while suppressing basal CRE-mediated gene expression.
In summary, this report provides the first evidence that p30II mediates transcriptional activity. We believe that p30II functions in infected cells as either a transcriptional activator or repressor, depending on the cis-acting sequence of the promoter and p30II expression levels. Further studies are required to identify DNA or protein targets that form functional partners with p30II before the role of the viral protein is delineated in context to the replication of HTLV-1 or in mediating the pathogenesis of virus-associated diseases.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants RR-14324 from the National Center for Research Resources and CA-70259 from the National Cancer Institute, awarded through the Ohio State University Comprehensive Cancer Center. W. Zhang is supported by a David White Fellowship award. M. Lairmore is supported by an Independent Scientist Career Award from the National Institutes of Health (K02 AI01474).
We thank Tim Vojt for preparation of figures. We also thank P. Quinn, G. Franchini, S. McKnight, and L. Ratner for valuable reagents and B. Albrecht, P. Green, and K. Boris-Lawrie for critical review of the manuscript.
REFERENCES
- 1.Bartoe J T, Albrecht B, Collins N D, Robek M D, Ratner L, Green P L, Lairmore M D. Functional role of pX open reading frame II of human T-lymphotropic virus type 1 in maintenance of viral loads in vivo. J Virol. 2000;74:1094–1100. doi: 10.1128/jvi.74.3.1094-1100.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berneman Z N, Gartenhaus R B, Reitz M S, Blattner W A, Manns A, Hanchard B, Ikehara O, Gallo R C, Klotman M E. Expression of alternatively spliced human T-lymphotropic virus type 1 pX mRNA in infected cell lines and in primary uncultured cells from patients with adult T-cell leukemia/lymphoma and healthy carriers. Proc Natl Acad Sci USA. 1992;89:3005–3009. doi: 10.1073/pnas.89.7.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cereseto A, Berneman Z, Koralnik I, Vaughn J, Franchini G, Klotman M E. Differential expression of alternatively spliced pX mRNAs in HTLV-I-infected cell lines. Leukemia. 1997;11:866–870. doi: 10.1038/sj.leu.2400665. [DOI] [PubMed] [Google Scholar]
- 4.Ciminale V, Pavlakis G N, Derse D, Cunningham C P, Felber B K. Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV type I. J Virol. 1992;66:1737–1745. doi: 10.1128/jvi.66.3.1737-1745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciminale V, Zotti L, Dagostino D M, Ferro T, Casareto L, Franchini G, Bernardi P, Chiecobianchi L. Mitochondrial targeting of the p13(II) protein coded by the x-II ORF of human T-cell leukemia/lymphotropic virus type I (HTLV-I) Oncogene. 1999;18:4505–4514. doi: 10.1038/sj.onc.1203047. [DOI] [PubMed] [Google Scholar]
- 6.Collins N D, Newbound G C, Albrecht B, Beard J L, Ratner L, Lairmore M D. Selective ablation of human T-cell lymphotropic virus type 1 p12(I) reduces viral infectivity in vivo. Blood. 1998;91:4701–4707. [PubMed] [Google Scholar]
- 7.Collins N D, Newbound G C, Ratner L, Lairmore M D. In vitro CD4+ lymphocyte transformation and infection in a rabbit model with a molecular clone of human T-cell lymphotropic virus type 1. J Virol. 1996;70:7241–7246. doi: 10.1128/jvi.70.10.7241-7246.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derse D, Mikovits J, Ruscetti F. X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology. 1997;237:123–128. doi: 10.1006/viro.1997.8781. [DOI] [PubMed] [Google Scholar]
- 9.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase in soluble extract from isolated mammalian cell nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira O C, Planelles V, Rosenblatt J D. Human T-cell leukemia viruses: epidemiology, biology, and pathogenesis. Blood Rev. 1997;11:91–104. doi: 10.1016/s0268-960x(97)90015-1. [DOI] [PubMed] [Google Scholar]
- 11.Franchini G. Molecular mechanisms of human T-cell leukemia/lymphotropic virus type I infection. Blood. 1995;86:3619–3639. [PubMed] [Google Scholar]
- 12.Gessain A, Barin F, Vernant J, Gout O, Maurs L, Calender A, Dethe G. Antibodies to human T lymphotropic virus type 1 in patients with tropical spastic paresis. Lancet. 1985;ii:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 13.Green P L, Chen I S Y. Molecular features of the human T-cell leukemia virus: mechanisms of transformation and leukemogenicity. In: Levy J A, editor. The retroviridae. New York, N.Y: Plenum Press; 1994. pp. 277–311. [Google Scholar]
- 14.Hollsberg P. Mechanisms of T-cell activation by human T-cell lymphotropic virus type I. Microbiol Mol Biol Rev. 1999;63:308–333. doi: 10.1128/mmbr.63.2.308-333.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimata J T, Wong F, Wang J, Ratner L. Construction and characterization of infectious human T-cell leukemia virus type 1 molecular clones. Virology. 1994;204:656–664. doi: 10.1006/viro.1994.1581. [DOI] [PubMed] [Google Scholar]
- 16.Koralnik I J, Fullen J, Franchini G. The p12, p13, and p30 proteins encoded by human T-cell leukemia/lymphotropic virus type-1 open reading frames I and II are localized in three different cellular compartments. J Virol. 1993;67:2360–2366. doi: 10.1128/jvi.67.4.2360-2366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koralnik I J, Gessain A, Klotman M E, Lo Monico A, Berneman Z N, Franchini G. Protein isoforms encoded by the pX region of human T-cell leukemia/lymphotropic virus type 1. Proc Natl Acad Sci USA. 1992;89:8813–8817. doi: 10.1073/pnas.89.18.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mellon P, Clegg C, Correll L, McKnight G. Regulation of the transcription of cyclic AMP-dependent protein kinase. Proc Natl Acad Sci USA. 1989;86:4887–4891. doi: 10.1073/pnas.86.13.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mesnard J M, Devaux C. Multiple control levels of cell proliferation by human T-cell leukemia virus type 1 tax protein. Virology. 1999;257:277–284. doi: 10.1006/viro.1999.9685. [DOI] [PubMed] [Google Scholar]
- 20.Newbound G C, Andrews J M, Orourke J, Brady J N, Lairmore M D. Human T-cell lymphotropic virus type 1 Tax mediates enhanced transcription in CD4+ T lymphocytes. J Virol. 1996;70:2101–2106. doi: 10.1128/jvi.70.4.2101-2106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. HTLV-I associated myelopathy, a new clinical entity. Lancet. 1986;i:1031–1032. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 22.Pique C, Uretavidal A, Gessain A, Chancerel B, Gout O, Tamouza R, Agis F, Dokhelar M C. Evidence for the chronic in vivo production of human T cell leukemia virus type I Rof and Tof proteins from cytotoxic T lymphocytes directed against viral peptides. J Exp Med. 2000;191:567–572. doi: 10.1084/jem.191.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potter J, Cheneval D, Dang C, Ressar L, Mezey E, Yang X. The upstream stimulatory factor binds to and activates the promoter of the rat class I alcohol dehydrogenase gene. J Biol Chem. 1991;266:15457–15463. [PubMed] [Google Scholar]
- 25.Quinn P G. Distinct activation domains within cAMP response element-binding. J Biol Chem. 1993;268:16999–17009. [PubMed] [Google Scholar]
- 26.Robek M D, Wong F H, Ratner L. Human T-cell leukemia virus type 1 pX-I and pX-II open reading frames are dispensable for the immortalization of primary lymphocytes. J Virol. 1998;72:4458–4462. doi: 10.1128/jvi.72.5.4458-4462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas S, Coffin R, Watts P, Gough G, Latchman D S. The TAATGARAT motif in the herpes simplex virus immediate-early gene promoter can confer both positive and negative responses to cellular octomer-binding proteins when it is located within the viral genome. J Virol. 1998;72:3495–3500. doi: 10.1128/jvi.72.4.3495-3500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verrijzer C P, Van der Vliet P C. POU domain transcription factors. Biochim Biophys Acta. 1993;1173:1–21. doi: 10.1016/0167-4781(93)90237-8. [DOI] [PubMed] [Google Scholar]
- 29.Wilson A C, Freeman R W, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human T-cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]