Abstract
The prevalence of chronic, non-communicable diseases has risen sharply in recent decades, especially in industrialized countries. While several studies implicate the microbiome in this trend, few have examined the evolutionary history of industrialized microbiomes. Here we sampled 235 ancient dental calculus samples from individuals living in Great Britain (~2200 BCE to 1853 CE), including 127 well-contextualized London adults. We reconstructed their microbial history spanning the transition to industrialization. After controlling for oral geography and technical biases, we identified multiple oral microbial communities that coexisted in Britain for millennia, including a community associated with Methanobrevibacter, an anaerobic Archaea not commonly prevalent in the oral microbiome of modern industrialized societies. Calculus analysis suggests that oral hygiene contributed to oral microbiome composition, while microbial functions reflected past differences in diet, specifically in dairy and carbohydrate consumption. In London samples, Methanobrevibacter-associated microbial communities are linked with skeletal markers of systemic diseases (for example, periostitis and joint pathologies), and their disappearance is consistent with temporal shifts, including the arrival of the Second Plague Pandemic. This suggests pre-industrialized microbiomes were more diverse than previously recognized, enhancing our understanding of chronic, non-communicable disease origins in industrialized populations.
Modern, industrialized microbiomes are linked to a wide range of non-communicable, chronic diseases, including obesity, cardiovascular disease, allergies and poor mental health1,2, which are increasing rapidly in industrialized countries and are predicted to rise in low- and middle-income countries in the future3,4. As such, determining the evolutionary background of these microbial communities is critical to understanding the origins and aetiologies of these diseases. To date, the origins and evolution of industrialized microbiomes are primarily investigated by examining ‘pre-industrialized’ microbiomes of other primates or extant Indigenous peoples who practise traditional subsistence lifeways (such as hunting and foraging)5–9. Such research has suggested that shifts in diet (for example, reductions in dietary fibre8) and the loss of microorganisms (for example, Helicobacter pylori) have shaped industrialized gut microbiomes5, alongside changes in environmental and social factors10. Studies tracking gut microbiomes of immigrants to industrialized countries, such as the United States of America, have similarly shown a decrease in diversity and a loss of certain species upon the adoption of ‘Western’ lifestyles11,12, confirming that industrialization has substantial impacts on the human gut microbiome. As a response, scientists have called for the biobanking of Indigenous people’s microorganisms before those microorganisms become extinct13.
Despite these findings, the extent and rate of microbial extirpations in industrialized societies remain poorly understood, as the approaches used to describe pre-industrialized microbiota are problematic. First, each population has a unique evolutionary history with distinct genetics, environments, diets and selection pressures that shape its microbiome in unique ways14. Consequently, modern non-industrialized populations or immigrants may not accurately reflect the microorganisms that existed in the ancestors of industrialized peoples today15,16. Second, this research places unnecessary responsibilities and obligations on Indigenous communities to participate in microbiome research, the benefits of which may not directly serve Indigenous peoples17. Therefore, a more direct path towards reconstructing pre-industrialized human microbiomes is needed. One such approach is to use the available bioarchaeological record of communities that predate industrialized populations through the analysis of ancestral archaeological human remains. Although this approach involves a number of complex challenges in recovering gut microbiomes18,19, reconstructing ancient oral microbiomes preserved within calcified dental plaque (called calculus) is an established way of tracing past oral microbial histories20,21.
Results
Filtering and authentication of British oral microbiomes
We performed an ancient dental calculus study (n = 235 samples assessed), reconstructing authenticated oral microbiota using shotgun metagenomics from 183 pre-industrialized individuals who were excavated across 27 archaeological sites in England and Scotland (Fig. 1 and Supplementary Table 1) from ~2200 bce to 1853 ce, to directly describe the history of a pre-industrialized population’s microbiome. These samples originate from eight geographic regions and include individuals who resided in Britain before Roman colonization and up to the ‘Industrial Revolution’ (Supplementary Section 1 and Supplementary Table 1). We used a multi-tiered assessment procedure to authenticate and control for contamination in this dataset (Supplementary Section 3, Supplementary Fig. 3 (summary) and Supplementary Tables 2–5 and 13). First, we included only high-quality samples with more than 100,000 taxonomically assigned sequences22 and more than 5 phyla20. We authenticated ancient DNA fragmentation with a novel, reference-free DNA damage program called ChangePoint23 (Supplementary Table 5) and the gold-standard, reference-based approach called MapDamage2.0 (ref. 24); damage consistent with these archaeological ages was present in known oral species (Streptococcus sanguinis, Porphyromonas gingivalis, Actinomyces oral taxon 414, Anaerolineaceae bacterium oral taxon 439 and Methanobrevibacter oralis) and less in common contaminant species (Burkholderia multivorans, Comamonas testosteroni, Escherichia coli and Flavobacteriaceae bacterium; Supplementary Table 13). We were conservative and limited the effects of potential laboratory and environmental contaminant DNA by removing samples whose microbial composition was similar to that of laboratory controls (Supplementary Fig. 2 and Supplementary Table 2) and conservatively filtering contaminant species identified in environmental and laboratory controls (Supplementary Fig. 4 and Supplementary Table 4). Lastly, we verified the presence of oral taxa from known modern and ancient oral microbiomes using SourceTracker1.0 (MALTx results) and SourceTracker2.0 (MALTn results; average 85% oral; Supplementary Section 3 and Supplementary Figs. 2 and 13) and confirmed that the highly abundant taxa were present in the Human Oral Microbiome Database (Supplementary Fig. 13c).
Fig. 1|. Geographical and temporal distribution of samples.
a, Map showing the locations of the archaeological sites examined in this study across the British Isles. Each site is represented by a circle, and the size of the circle corresponds to the number of dental calculus samples examined from that site. A total of 235 individuals and 27 sites were sampled. b, The number of dental calculus samples from each time period is shown. The broad time periods and associated dates are Pre-Roman Britain (–43 CE), Roman Britain (43–410 CE), Anglo-Saxon or Early Medieval Britain (410–1066 CE), Norman Britain and the Middle Ages (1066–1547 CE), Reformation (1547–1750 ce) and Industrial (1750–1900 CE).
Oral geography biases significantly impact ancient oral microbiome data
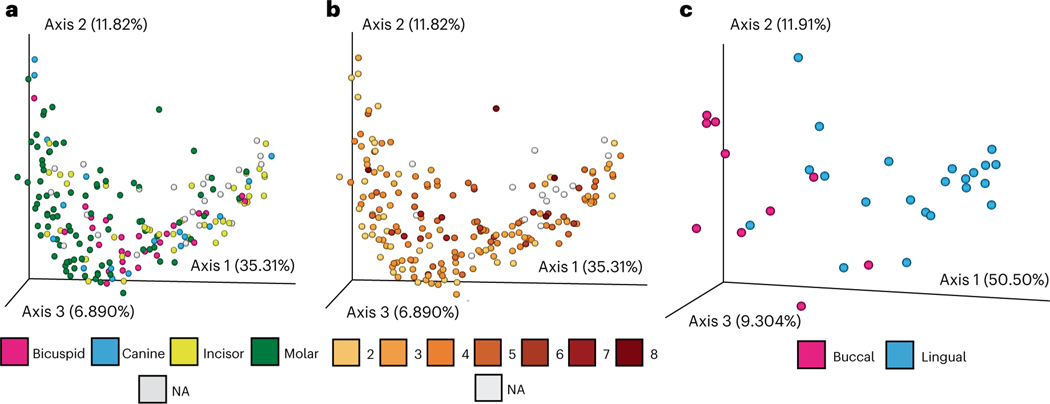
As oral geography (that is, position within the oral cavity) plays a role in oral microbiome composition25–28, we tested and identified oral geography biases in the dataset; calculus sample size and gingival location influenced taxonomic composition in molars, while tooth surface drove compositional variation in incisors (Fig. 2a–c and Supplementary Section 4). As a result, we stratified our taxonomic and functional data according to tooth type and included oral geography (that is, tooth type, surface, gingival region and calculus size) in our statistical analyses (Supplementary Tables 6–9). As these biases probably reflect biological and ecological differences in the mouth25, these differences attributed to oral geography raise questions about the interpretations of previous palaeomicrobiome studies that used calculus samples collected from a mixed dentition29–31 and suggest that future studies should control for oral geography during sampling and analysis (Supplementary Fig. 18). Overall, 954 microbial species were identified across all ancient British calculus samples, predominantly spanning the Firmicutes, Actinobacteria, Proteobacteria and Bacteroidetes phyla (Supplementary Fig. 4).
Fig. 2 |. Oral geography and microbial compositions.
a–c, The contributions of oral geography on bacterial and archaeal taxonomic compositions are shown by performing PCoA of Bray–Curtis distances for all teeth. The oral microbiota composition of each calculus sample is coloured according to the tooth that was sampled (a), the approximated size of the dental calculus sample obtained for DNA extraction as described in Supplementary Information (b) or the surface of the tooth that was sampled, shown for molar teeth only (c). When information regarding oral geography was unavailable, samples are labeled as not applicable or ‘NA’.
British oral microbiome composition is driven by two major community types
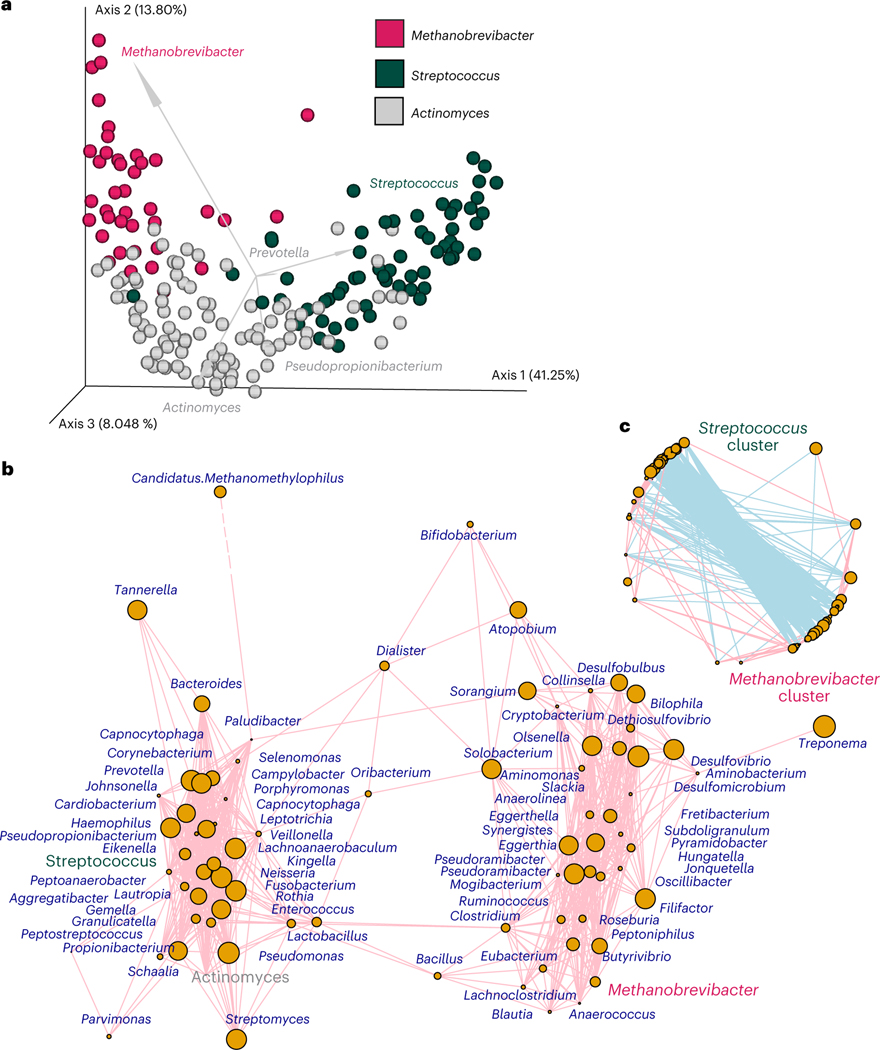
To explore drivers of variation in pre-industrialized oral microbiome composition, we performed a principal coordinate analysis (PCoA) ordination of Bray–Curtis dissimilarities with biplots (Fig. 3a and Supplementary Fig. 5a) using microbial genera identified in the samples. We observed a ‘U’-shaped curve indicative of distinct ecologies32 (axis 1 = 41.25% variation; Fig. 3a and Supplementary Table 10), and the biplots indicated that the Methanobrevibacter and Streptococcus genera were associated with the 41.2% of the variation explained by axis 1, while Actinomyces-dominated communities drove variation on axis 2 (13.8%, Fig. 3a). As such, we grouped the samples according to whether or not the Methanobrevibacter, Streptococcus and Actinomyces genera were most dominant in each sample. We then compared the two communities on the extremes of axis 1 (that is, Streptococcus and Methanobrevibacter associated) to assess major factors that drive oral microbial diversity (Fig. 3a). Streptococcus- and Methanobrevibacter-associated samples contained distinct community assemblages (dominant category; R2 ≥ 0.20, P ≤ 0.05; Supplementary Table 12). Co-occurrence analysis of the Streptococcus- or Methanobrevibacter-associated communities using CCLasso positively associated Streptococcus with the Leptotrichia, Neisseria, Gemella, Capnocytophaga, Granulicatella, Lautropia, Kingella, Aggregatibacter, Lachnoanaerobaculum and Rothia genera (Fig. 3b,c and Supplementary Table 11), as seen in modern industrialized oral microbiomes, such as those from Spain (Supplementary Fig. 6). In contrast, Methanobrevibacter positively co-occurred with genera not often described in the industrialized oral cavity, including Methanosphaera, Peptoniphilus, Anaerofustis, Syntrophomonas, Shuttleworthia, Subdoligranulum, Pseudoramibacter, Synergistes, Hungatella and Butyrivibrio taxa (Fig. 3b,c and Supplementary Table 11). Several different oral Methanobrevibacter species have now been described in ancient mouths33, and at least three of the co-occurring genera have been previously characterized in the mouth34–36 and not as contaminants20,26,30. We confirmed via a literature review that species within these co-occurring genera can cohabitate with Methanobrevibacter oral species, as all are anaerobic and can produce metabolic by-products that support methanogenesis37. However, this Methanobrevibacter-associated community has not yet been described in studies of modern dental calculus20,31,38, suggesting that a Methanobrevibacter-associated community may represent a unique oral microbial ecology not typically found in modern industrialized societies.
Fig. 3 |. Exploration of dominant taxa and communities.
a, PCoA plot showing differences in Bray–Curtis distances of microbial genera present in each sample. Biplots are also shown using arrows for the top five most significant genera, with the length of the arrow proportional to its magnitude. Samples are coloured according to which the top three genera identified via biplots (Actinomyces, Streptococcus or Methanobrevibacter) were most dominant within each sample; samples that contained either more Actinomyces, Streptococcus or Methanobrevibacter are coloured grey, green or pink, respectively. b,c, CCLasso was used to identify genera that positively (b) or negatively (c) co-occurred in all samples; the top three genera identified using biplots are coloured as in a: Actinomyces associated (grey), Streptococcus associated (green) and Methanobrevibacter associated (pink).
To explore the origins of these two microbial communities, we tested whether physiological, cultural and temporal factors previously thought to drive ancient oral microbiome composition were associated with this signal in ancient British microbiomes39. Demographic variables (that is, sex and age), broad cultural classifications (that is, religion, class, or urban and rural locations; Supplementary Table 1) or major biocultural or socio-political events (for example, civil war or plague outbreaks, such as the Second Plague Pandemic, also known as the Black Death (caused by Yersinia pestis); Supplementary Table 1) did not explain significant levels of taxonomic or functional compositional variation across Great Britain (ADONIS, P ≥ 0.05; Supplementary Table 12). A mild association between the location where individuals were buried (that is, cemetery) and microbial genera composition was observed (ADONIS, R2 = 0.158, P = 0.031; Supplementary Table 12), but this was not true when examining the data at the species level (ADONIS of contaminant species-filtered data, P ≥ 0.05; Supplementary Table 12). Unexpectedly, these findings suggest that these large-scale cultural and social factors that occurred across Britain over 2,200 years were not significant drivers of oral microbiome composition at a population scale in this dataset.
Ancient British oral microbiomes are potentially linked to oral hygiene
We next examined whether these microbial communities were linked to known signatures of oral disease, as Methanobrevibacter taxa have been linked to severe periodontitis in modern populations40. As indicated by an ADONIS test (P ≤ 0.05), oral microbiome composition in all individuals was not linked to the occurrence of periodontal disease, nor other known oral pathologies, such as caries or apical abscesses (Supplementary Table 12). Unexpectedly, species and functions linked to periodontal disease in modern populations (for example, P. gingivalis and Tannerella forsythia) were more likely to be found in Streptococcus-associated communities than those dominated by Methanobrevibacter (Supplementary Table 13), suggesting that the modern aetiology of industrial-age periodontal disease may in fact originate from the Streptococcus-associated communities. However, oral microbiome composition was directly linked with size of the calculus sample analysed (Fig. 2b and Supplementary Tables 6–9). While it would be reasonable to infer that Methanobrevibacter-associated communities may thrive within larger, potentially more mature calculus deposits, consistent with the anaerobic requirements of these taxa, we did not find this to be the case. The separation of samples into Streptococcus- or Methanobrevibacter-associated communities did not explain calculus sample size (ADONIS; P = 0.101), nor was the size of calculus samples driven by Methanobrevibacter-associated species. Rather, taxa associated with Streptococcus communities (based on the correspondence analysis in Supplementary Table 11), including Gemella and Lautropia, were associated with larger sample sizes (ALDEx2; Supplementary Fig. 17). Nevertheless, modern oral hygiene practices can reduce dental plaque and calculus formation41,42 and lead to smaller deposits, so it is possible that compositional shifts in ancient British communities are linked to dental hygiene practices.
Oral microbial functions can be used to reconstruct past diets
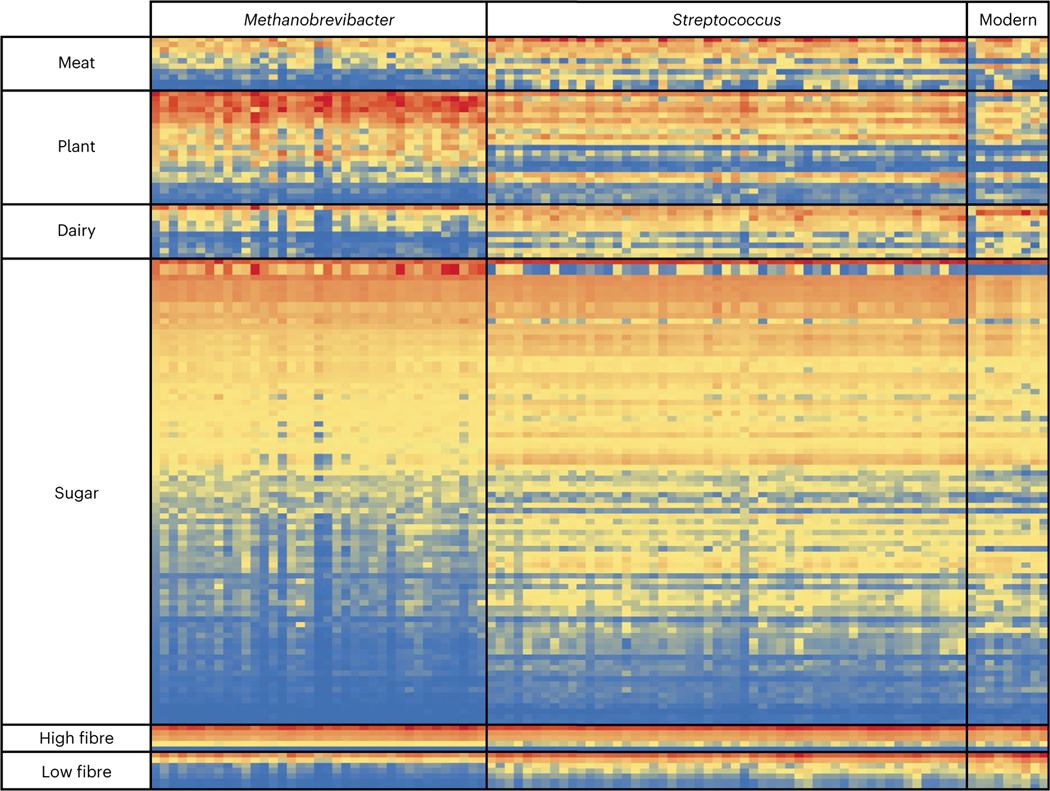
As dietary changes are proposed to be a main driver of oral microbiome evolution through time20,30,39, we further explored whether differences in diet could underpin these two distinct microbial ecologies. Direct dietary DNA signals were explored in six deeply sequenced calculus samples (that is, >100 million sequences per sample; 3 samples each from Streptococcus-associated or Methanobrevibacter-associated communities), but after careful consideration43,44, no verifiable DNA fragments could be robustly confirmed from either plants or non-host animals. As oral microorganisms in dental plaque can ferment sugars, starch molecules and amino acids in the mouth45, we then developed a novel approach to explore whether indirect dietary signals were present. We assembled a list of microbial genetic functional differences linked to dietary changes in the gut8,20,46,47, as a proxy for predicting broad dietary differences. We included 42 amino acid metabolism functions linked to either carnivorous or herbivorous diets46, 17 functions linked to high- or low-dietary-fibre digestion8, 124 carbohydrate metabolism gene families, and 30 lactose and galactose metabolism functions linked to milk consumption (Supplementary Table 14). We first validated this approach by examining the presence of these microbial functions in calculus from a modern, industrialized Spanish population (Supplementary Table 1). Our results were consistent with an omnivorous diet with high sugar (for example, lactate fermentation, fructose utilization, galactose degradation and glucose utilization) and low dietary-fibre intake (for example, glycan degradation; Fig. 4 and Supplementary Table 15). Notably, microbial functions associated with dairy consumption (that is, beta-galactosidase or lactase; EC 3.2.1.23) were highly abundant in these modern human oral microbiomes (Fig. 4 and Supplementary Table 15).
Fig. 4 |. Abundance of dietary microbial functions in ancient Britain.
Normalized relative abundances of dietary microbial functions found to be differentially abundant using a Benjamini–Hochberg-corrected P value of a two-tailed Welch’s t-test (P ≤ 0.05) in ALDEx2 are shown for Streptococcus-associated, Methanobrevibacter-associated and modern oral microbiomes. Red colouring represents high abundances, yellow is medium and blue represents low; the colouring is normalized within each dietary function category. A full list of functions tested is shown in Supplementary Table 14, and a full version of this differential abundance analysis is shown in Supplementary Table 15.
In our ancient dataset, we identified 81.0% (34 of 42) of the amino acid metabolism pathways associated with herbivory or carnivory in ancient samples (Supplementary Table 15). While 88% were significantly more abundant in one of the two microbial communities (Fig. 4 and Supplementary Fig. 8), both microbiomes possessed functions consistent with an omnivorous diet (Fig. 4 and Supplementary Fig. 8). Streptococcus-associated communities contained more microbial functions significantly linked with low-fibre (70% of the 16 identified fibre digestion functions; galactose metabolism, glycosphingolipid biosynthesis and glycan degradation; Fig. 4 and Supplementary Fig. 9) and high-carbohydrate diets (46.8% of the 124 carbohydrate-metabolism-associated genes, compared with 22.6% in Methanobrevibacter-associated communities), including pathways linked to fructose, sucrose, trehalose, mannose, beta-glucoside and maltose metabolism (Fig. 4 and Supplementary Fig. 10). By contrast, Methanobrevibacter-associated communities were enriched for functions linked to methanogenesis, gluconeogenesis and xylose utilization (Fig. 4 and Supplementary Fig. 10). Lastly, Streptococcus-associated individuals showed increased abundances of microbial functions linked to dairy consumption (26.7% of lactose metabolism genes were significantly enriched compared with 6.7% in Methanobrevibacter-associated communities; Fig. 4 and Supplementary Fig. 11), including a significant enrichment in alpha- and beta-galactosidases, similar to observations in modern individuals (Supplementary Fig. 11 and Supplementary Table 15). This observation may reflect differences in dairy consumption or access across Britain, which is consistent with the presence of milk proteins in only one-third of medieval calculus samples from Britain (800 BCE to 1895 CE)48, even though dairy products were widely available49. Together, this analysis represents a powerful instrument in a growing toolbox of biomolecular approaches to reconstruct past diets and suggests that past carbohydrate and dairy consumption may be reflected in the oral microbiome of past British populations.
Specific oral microbiome compositional shifts were observed in London
As large-scale geographic differences across Great Britain may mask factors that drive oral microbiome variation through time49, we further narrowed our analysis to 127 medieval and post-medieval (1066 to 1853 CE) individuals from London that have been extensively studied to further explore the origin of these two distinct oral communities, while again controlling for oral geography effects (Supplementary Tables 16–18). As seen in the above Britain-wide analysis, the physiological or cultural factors considered did not appear to significantly contribute to past Londoners’ oral microbiome composition (P ≥ 0.05; Table 1). However, the presence of disease, as defined by 14 detailed oral and other systemic health indicators previously recorded on the bones and teeth of the individuals included in this study in the Wellcome Osteological Research Database, did potentially explain some of the differences in oral microbiome composition. In London, periodontal disease (but not caries, abscesses or dental developmental defects) was significantly linked to oral microbiome composition (R2 > 0.05, P < 0.05; Table 1). Again, microbial species positively associated with periodontal disease were, rather unexpectedly, more commonly found in Streptococcus-associated communities, rather than in Methanobrevibacter-associated ones (Supplementary Table 19). However, the Methanobrevibacter-associated communities were associated with the presence of several skeletal markers of systemic disease, including non-specific periostitis, joint porosity, osteophytic lipping and overall scores for joint pathologies (R2 > 0.05, P < 0.05; Table 1). While there could be multiple causes for these skeletal markers, most are thought to be related to inflammatory-associated conditions. Each of these disease markers was also linked to specific genera within the Methanobrevibacter-associated community (Supplementary Table 19), such as Methanobrevibacter, Eubacterium, Pseudoramibacter, Mogibacterium and Peptoniphilus taxa (Supplementary Table 19). While not all systemic diseases contribute to morphological changes in the skeleton, the association between oral microbiomes and systemic diseases has been clearly shown in modern individuals, often through inflammatory pathways50,51. While cause and effect remain unclear, or may be the result of indirect associations (for example, socioeconomic status), our study has linked ancient microbiomes to systemic diseases that manifest in the skeleton and, therefore, provides a new model to examine the origins of modern chronic, non-communicable diseases.
Table 1 |.
ADONIS of culture and health variables for all teeth and molars, with factors based on oral geography tables accounted into models for Museum of London samples only
| Species |
Genera |
All taxa |
||||
|---|---|---|---|---|---|---|
| R 2 | Pr(>F) | R 2 | Pr(>F) | R 2 | Pr(>F) | |
|
| ||||||
| Variable (all teeth) | ||||||
|
| ||||||
| Black Death (1346–1353) | 0.042271 | 0.013 | 0.030313 | 0.048 | 0.025943 | 0.115 |
|
| ||||||
| Civil war (before, after or during) | 0.016108 | 0.135 | 0.011964 | 0.283 | 0.01031 | 0.342 |
|
| ||||||
| Date (300 year intervals) | 0.034836 | 0.048 | 0.024156 | 0.198 | 0.022078 | 0.221 |
|
| ||||||
| Date (400 year intervals) | 0.029446 | 0.013 | 0.016392 | 0.12 | 0.013127 | 0.203 |
|
| ||||||
| Overall pathology | 0.006137 | 0.755 | 0.008607 | 0.449 | 0.004815 | 0.737 |
|
| ||||||
| Blood disorder | 0.004924 | 0.874 | 0.006369 | 0.667 | 0.003584 | 0.875 |
|
| ||||||
| Non-specific periostitis | 0.011312 | 0.376 | 0.008351 | 0.485 | 0.006533 | 0.55 |
|
| ||||||
| Overall vertebral pathology | 0.007189 | 0.668 | 0.006596 | 0.6 | 0.002856 | 0.936 |
|
| ||||||
| Vertebral pathology (facets) | 0.011992 | 0.398 | 0.011231 | 0.326 | 0.008289 | 0.482 |
|
| ||||||
| Vertebral pathology (Schmorl’s nodes) | 0.01889 | 0.163 | 0.014865 | 0.201 | 0.012520 | 0.248 |
|
| ||||||
| Vertebral anomaly | 0.003516 | 0.975 | 0.00074 | 0.999 | 0.003231 | 0.928 |
|
| ||||||
| Overall joint score | 0.014469 | 0.339 | 0.013742 | 0.278 | 0.008986 | 0.476 |
|
| ||||||
| Osteophitic lipping (joints) | 0.016424 | 0.252 | 0.015658 | 0.2 | 0.013047 | 0.293 |
|
| ||||||
| Porosity (joint) | 0.018879 | 0.2 | 0.016178 | 0.216 | 0.008034 | 0.561 |
|
| ||||||
| Dental abscess | 0.007348 | 0.802 | 0.006688 | 0.831 | 0.008883 | 0.689 |
|
| ||||||
| Caries | 0.013519 | 0.365 | 0.010129 | 0.552 | 0.006764 | 0.742 |
|
| ||||||
| Hypoplasia | 0.016002 | 0.315 | 0.014795 | 0.32 | 0.012569 | 0.413 |
|
| ||||||
| Periodontitis | 0.007565 | 0.738 | 0.006348 | 0.791 | 0.003939 | 0.936 |
|
| ||||||
| Class | 0.050463 | 0.817 | 0.047845 | 0.842 | ND | ND |
|
| ||||||
| Hospital | 0.008254 | 0.583 | 0.006867 | 0.68 | 0.003706 | 0.89 |
|
| ||||||
| Rank | 0.024601 | 0.375 | 0.022691 | 0.419 | 0.013996 | 0.698 |
|
| ||||||
| Religion | 0.069326 | 0.37 | 0.065258 | 0.523 | ND | ND |
|
| ||||||
| Empire | 0.022583 | 0.161 | 0.024645 | 0.121 | 0.009261 | 0.296 |
|
| ||||||
| Sex | 0.012235 | 0.229 | 0.010149 | 0.346 | 0.007244 | 0.506 |
|
| ||||||
| Cemetery | 0.078145 | 0.066 | 0.06099 | 0.307 | 0.056177 | 0.38 |
|
| ||||||
| Rural versus urban | 0.015152 | 0.454 | 0.013429 | 0.55 | 0.008919 | 0.73 |
|
| ||||||
| Medieval versus post-medieval | 0.013773 | 0.126 | 0.013609 | 0.107 | 0.009232 | 0.233 |
|
| ||||||
| Age | 0.0334 | 0.14 | 0.040031 | 0.053 | 0.039874 | 0.074 |
|
| ||||||
| Dominant community | 0.295798 | 0.001 | 0.278079 | 0.001 | 0.270359 | 0.001 |
|
| ||||||
| Variable (molars) | ||||||
|
| ||||||
| Black Death (1346–1353) | 0.051076 | 0.141 | 0.034373 | 0.529 | ND | ND |
|
| ||||||
| Civil war (before, after or during) | 0.030608 | 0.245 | 0.025953 | 0.287 | 0.22388 | 0.33 |
|
| ||||||
| Date (300 year intervals) | 0.045842 | 0.356 | 0.040998 | 0.438 | 0.033105 | 0.646 |
|
| ||||||
| Date (400 year intervals) | 0.033868 | 0.137 | 0.027913 | 0.238 | 0.023156 | 0.329 |
|
| ||||||
| Overall pathology | 0.036885 | 0.071 | 0.053478 | 0.119 | 0.047454 | 0.014 |
|
| ||||||
| Blood disorder | 0.029438 | 0.135 | 0.04632 | 0.157 | 0.029031 | 0.121 |
|
| ||||||
| Non-specific periostitis * | 0.073858 | 0.001 | 0.068745 | 0.04 | 0.066492 | 0.002 |
|
| ||||||
| Overall vertebral pathology | 0.032115 | 0.101 | 0.022347 | 0.595 | 0.027613 | 0.146 |
|
| ||||||
| Vertebral pathology (facets) | 0.017843 | 0.464 | 0.034455 | 0.369 | 0.011874 | 0.801 |
|
| ||||||
| Vertebral pathology (Schmorl’s nodes) | 0.020959 | 0.367 | 0.0136 | 0.908 | 0.017336 | 0.485 |
|
| ||||||
| Vertebral anomaly | 0.007946 | 0.931 | 0.005958 | 0.998 | 0.012164 | 0.737 |
|
| ||||||
| Overall joint score | 0.064592 | 0.007 | 0.100103 | 0.018 | 0.059902 | 0.012 |
|
| ||||||
| Osteophitic lipping (joints) | 0.064268 | 0.007 | 0.099268 | 0.022 | 0.060218 | 0.006 |
|
| ||||||
| Porosity (joint) | 0.057578 | 0.014 | 0.099376 | 0.016 | 0.05229 | 0.022 |
|
| ||||||
| Dental abscess | 0.034019 | 0.262 | 0.035143 | 0.531 | 0.036956 | 0.209 |
|
| ||||||
| Caries | 0.019593 | 0.573 | 0.031298 | 0.502 | 0.024434 | 0.334 |
|
| ||||||
| Hypoplasia | 0.027694 | 0.268 | 0.036616 | 0.442 | 0.026032 | 0.291 |
|
| ||||||
| Periodontitis | 0.011512 | 0.833 | 0.025481 | 0.633 | 0.015703 | 0.026032 |
|
| ||||||
| Class | ND | ND | ND | ND | ND | ND |
|
| ||||||
| Hospital | 0.023396 | 0.234 | 0.035948 | 0.274 | 0.026313 | 0.177 |
|
| ||||||
| Rank | 0.024147 | 0.82 | ND | ND | 0.022222 | 0.852 |
|
| ||||||
| Religion | 0.095904 | 0.117 | ND | ND | 0.093495 | 0.104 |
|
| ||||||
| Empire | 0.032085 | 0.089 | 0.026332 | 0.185 | 0.019894 | 0.321 |
|
| ||||||
| Sex | 0.0239 | 0.376 | 0.032519 | 0.187 | 0.023802 | 0.365 |
|
| ||||||
| Cemetery | 0.087245 | 0.159 | 0.088173 | 0.158 | 0.06858 | 0.385 |
|
| ||||||
| Medieval or post-medieval | 0.030868 | 0.117 | 0.025388 | 0.169 | 0.019233 | 0.346 |
|
| ||||||
| Rural versus urban | 0.033662 | 0.503 | 0.062878 | 0.296 | 0.037808 | 0.393 |
|
| ||||||
| Age | 0.049095 | 0.619 | 0.058474 | 0.37 | 0.047467 | 0.573 |
|
| ||||||
| Dominant community | 0.221255 | 0.001 | 0.237476 | 0.001 | 0.187197 | 0.001 |
Bolding indicates a variable with significant results (P ≤ 0.05).
Significantly associated with the dominant category. The results from the ADONIS analysis on the beta diversity (Bray–Curtis) of oral microbiota from all London individuals are shown. The fit of the test (R2) and the P value for each test are shown for all species, all genera or all taxa present in each sample, after accounting for oral geography. Significant results are in italics (P ≤ 0.05). Results significant after implementing a Bonferroni correction are demarcated with *. Taxa driving these shifts (Supplementary Fig. 17) and linkages to the dominant associated taxa (Supplementary Table 23) are presented in the Supplementary Text. ND indicates not determined (e.g. tests could not be performed due to small samples sizes).
Methanobrevibacter over time
Lastly, we examine why this distinct Methanobrevibacter-associated oral ecology is not commonly found today in industrialized populations by exploring its presence over time. In all of Britain, the Methanobrevibacter-associated community was first observed in individuals ~2,200 years ago and was still present in London until at least ~1853 (Supplementary Fig. 12). We then examined compositional shifts over time in just London. Oral microbiome composition significantly shifted across 300 and 400 year intervals (P ≤ 0.05; Table 1) and significantly shifted after the arrival of the Second Plague Pandemic in London in 1348. The arrival of Y. pestis was verified in plague cemeteries (for example, East Smithfield)52 and resulted in the deaths of over 30–50% of Londoners between 1348 and 1351 alone53, changing the population structure and ways of life in the city substantially. As temporal differences can be confounded by taphonomy, we examined these compositional shifts in the context of the Second Plague Pandemic more closely by fitting a Bayesian multinomial logistic-normal linear model to the overall oral microbiome composition from historic London, which included the arrival of the Second Plague Pandemic as a covariate and minimized the impacts of oral geography and cemetery location. Our results show that 10.88% of the total variation in microbiome composition can be explained by temporal changes, including the arrival of the Second Plague Pandemic (95% credible interval: 4.98% to 19.47%), while only 65.41% of that signal could be equally explained by other factors (Supplementary Section 6). In addition, only 1.5% of the variation explained by burial location was attributed to DNA damage patterns (deltaD) a detected within oral taxa Anaerolineaceae, M. oralis, P. gingivalis and S. sanguinis (Supplementary Table 13 and Supplementary Section 6), suggesting this observation is not driven by taphonomy. While this finding needs further examination, temporal shifts in oral microbiome composition coinciding with the Second Plague Pandemic in London could be the result of disease selection and susceptibility during the pandemic54,55, subsequent advancements in public health and hygiene56, dietary shifts and/or cultural shifts that were a consequence of this devastating pandemic on the citizens of London.
Discussion
Our study reveals the existence of a now rare or potentially extinct oral microbial ecosystem that was present in British populations over at least 2,200 years alongside other oral microbiome communities. This oral microbial community persisted through major biocultural transitions and historically important socio-political events, only to diminish in recent history—a phenomenon associated with the rise of industrialization. Why this Methanobrevibacter-associated community disappeared in Britain or has not yet been described in healthy modern, industrialized societies remains unknown, but reports of broad-spread Methanobrevibacter species in the ancient calculus literature suggest that this community may have once been widespread33. Our findings suggest that advancements in modern dentistry (for example, the routine removal of large calculus deposits and the use of modern oral hygiene products), shifts in dairy and carbohydrate consumption, and medical care may have additionally contributed to its perceived absence today, although further work should explore additional lifestyle changes post-1900s, including migration and nutrition. This finding establishes a new paradigm to explore the foundations and origins of chronic, non-communicable disease in living populations and opens the door to identify unknown (now extinct) microbial diversity in past, pre-industrialized human populations.
Methods
Sample information and collection
For calculus samples obtained from individuals who lived in Britain, access was provided by the Natural History Museum, Royal College of Surgeons of England, Oxford Archaeology East and Aberdeen Museum (Supplementary Table 1). For samples from historic London, 160 archaeological samples were collected from individuals buried at eight different cemeteries in a 16 km2 section of London, which formed a continuous temporal sequence from ~1000 to 1853 CE, from the curated archaeological skeletal remains collection stored at the Museum of London. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to and generally exceed those reported in previous publications20,49. Detailed information on each sample is provided in Supplementary Table 1. Samples were handled using sterile procedures as previously outlined20. Intact calculus samples were stored in labelled, sterile plastic bags and transported to the ancient DNA facility at the Australian Centre for Ancient DNA, University of Adelaide, Australia.
Decontamination, DNA extraction and library preparation
Careful consideration was given to the risk of laboratory and environmental contamination, as endogenous signals can be easily obscured or misinterpreted owing to contaminating microbial DNA26,27. To minimize contamination, samples were processed in an ultra-sterile, specialized ancient DNA laboratory and underwent a decontamination protocol, as previously published39. Samples underwent an in-house, silica-based DNA extraction, as previously described20,57,58. Extraction blank controls were incorporated throughout to monitor laboratory and reagent contamination at a ratio of two extraction blank controls to ten samples, as well as no template controls during the amplification process59. Shotgun libraries were generated using a previous protocol20. Samples were sequenced on an Illumina NextSeq using a high-output 2 × 150 bp kit. Further details about the methods are available in the Supplementary Text.
Bioinformatic and statistical analysis
DNA sequences were demultiplexed, trimmed and merged using AdapterRemoval2.0 with a 5 bp overlap60. Taxonomic and functional information was derived from analysis-ready reads using MALTx20 against the 2014nr database. Taxonomic comparisons were also completed using MALTn against the 2017 RefSeq GCS database (Supplementary Section 3). Only collapsed reads were used because fragments greater than 300 bp were considered more likely to be modern DNA contamination19. A total of 14 samples with a Bray–Curtis dissimilarity value >0.72 (the similarity of negative controls to themselves) compared with the negative controls were excluded. Next, all species identified within the negative control samples were conservatively removed from all samples; genera and higher classifications of data were not filtered. Four samples with <100,000 DNA sequences were also removed, as these are unlikely to capture overall microbial community structures22. We also authenticated the oral signal in our samples using a range of ancient calculus, modern calculus and plaque, soil, and laboratory controls using SourceTracker2.0 (ref. 61), and we verified DNA damage consistent with known ages of samples using both a reference-free approach using ChangePoint analysis23 and the reference-based gold-standard approach MapDamage2.0 (ref. 24) against oral and contaminant species (Supplementary Section 3).
To identify correlations with metadata and taxonomic data, taxonomic information from modern and ancient samples was exported from MEGAN6 CE and imported into QIIME2 (V2020.2.0)62, and singletons were removed. We rarefied the dataset to the maximum number of sequences available (100,000 for all taxa, 60,000 for genera and 30,000 for species per sample). Bray–Curtis dissimilarity was calculated with biplots, and the ADONIS test was applied (9,999 permutations) to identify factors that shaped beta diversity in the dataset. As each ADONIS test incorporates multiple variables, a correction for multiple tests may not be necessary. Nevertheless, we report P ≤ 0.05 as significant, and we highlight which results are also significant when applying a Bonferroni correction to account for multiple ADONIS tests (that is, P ≤ 0.002; Table 1). LefSe analysis was conducted to identify specific species that increased in abundance with select metadata fields63. To explore the functional potential of the microbiomes, functional tables were exported from MEGAN6 CE into QIIME2. Amino acid functions matching the Enzyme Commission numbers identified as distinguishing of carnivores and herbivores46 were exported. For carbohydrate and dairy metabolism, all level 4 functional groups were exported. Fibre metabolism functions64 were exported from the KEGG database within MEGAN5. ADONIS tests were run on functional data as done with taxonomic data, and LefSE analysis was conducted to identify specific functions that increased in abundance with select metadata fields.
Project outreach
To promote this project and provide ways for non-scientists to engage with this work, we engaged visitors to the Museum of London in conversations during the sampling period of this study. A stall was set up in the public galleries of the museum for one afternoon within the five day sampling visit. The stall consisted of a single table, skulls and models from the museum’s Centre for Human Bioarchaeology teaching collection, and a conference poster. The teaching collection and poster served to attract attention and trigger conversation. Conversational engagement allowed the public to ask their own questions about the topics that interested them. Discussions included the active project, background to the field, broader anthropological questions and discussions of our team members’ career path. In doing this, we offered an opportunity to engage with the public and support diverse and active learning in museums, enhancing science capital65, while promoting the active research and ongoing partnerships of these institutions.
Ethics statement
Ancient human samples are not subject to institutional review board approvals; however, this study was reviewed by the University of Adelaide Human Research Ethics Committee and received approval (H-2012–108).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank C. Stringer and R. Kruszynski of the Natural History Museum, London; S. Schiffels; D. Sayer; Oxford Archaeology East; M. Farrell of the Royal College of Surgeons of England; J. Pearson of the Inverness Museum; and all of the museums for access to samples. We also thank the Museum of London for allowing us to collect and destructively analyse archaeological dental calculus samples from their collections from London, particularly J. Bekvalac and R. Redfern. We would also like to acknowledge J. VanderBerg at EnDev Geographic for producing the map used in Fig. 1. A.C., C.A. and L.W. thank the Australian Research Council for research funding (DP110105038) and Laureate (FL140100260). The work was also supported by an Australian Research Council Future Fellowship Award to L.S.W. (FT180100407). This material is also based on work supported by the National Science Foundation Graduate Research Fellowship Program awarded to A.S.G. under Grant No. DGE1255832. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Code availability
The analysis pipelines are available in the microARCH GitHub page (@microARCHlab/BritishDentalCalculus_2021), as well as in https://github.com/michellepistner/ancientDNA.
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41564-023-01527-3.
Data availability
All trimmed and merged DNA sequences (fastq) are available in the SRA database (BioProject PRJNA780005) of NCBI. The 2017 NCBI nr database and the 2017 NCBI RefSeq GCS database were used in this study. Unmerged reads can be made available upon request, as only merged sequences were assessed in full for this publication.
References
- 1.Sonnenburg ED & Sonnenburg JL The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 17, 383–390 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Broussard JL & Devkota S. The changing microbial landscape of Western society: diet, dwellings and discordance. Mol. Metab. 5, 737–742 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abegunde DO, Mathers CD, Adam T, Ortegon M. & Strong K. The burden and costs of chronic diseases in low-income and middle-income countries. Lancet 370, 1929–1938 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Mathers CD & Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 3, e442 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnorr SL et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5, 3654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemente JC et al. The microbiome of uncontacted Amerindians. Sci. Adv. 1, e1500183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martínez I. et al. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep. 11, 527–538 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Clayton JB et al. Captivity humanizes the primate microbiome. Proc. Natl Acad. Sci. USA 113, 10376–10381 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obregon-Tito AJ et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun. 6, 6505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lokmer A. et al. Response of the human gut and saliva microbiome to urbanization in Cameroon. Sci. Rep. 10, 2856 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jha AR et al. Gut microbiome transition across a lifestyle gradient in Himalaya. PLoS Biol. 16, e2005396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vangay P. et al. U.S. immigration westernizes the human gut microbiome. Cell 175, 962–972.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bello MGD, Knight R, Gilbert JA & Blaser MJ Preserving microbial diversity. Science 362, 33–34 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Skelly E, Kapellas K, Cooper A. & Weyrich LS Consequences of colonialism: a microbial perspective to contemporary Indigenous health. Am. J. Biol. Anthropol. 167, 423–437 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Schnorr SL Meanings, measurements, and musings on the significance of patterns in human microbiome variation. Curr. Opin. Genet. Dev. 53, 43–52 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Carmody RN, Sarkar A. & Reese AT Gut microbiota through an evolutionary lens. Science 372, 462–463 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Tsosie KS, Fox K. & Yracheta JM Genomics data: the broken promise is to Indigenous people. Nature 591, 529–529 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Wibowo MC et al. Reconstruction of ancient microbial genomes from the human gut. Nature 594, 234–239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weyrich LS, Dobney K. & Cooper A. Ancient DNA analysis of dental calculus. J. Hum. Evol. 79, 119–124 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Weyrich LS et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature 544, 357–361 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Warinner C, Speller C. & Collins MJ A new era in palaeomicrobiology: prospects for ancient dental calculus as a long-term record of the human oral microbiome. Philos. Trans. R. Soc. Lond. B 370, 20130376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hillmann B. et al. Evaluating the information content of shallow shotgun metagenomics. mSystems 3, e00069–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y. The Role of Epigenetic Modications and Microbiome Evolution in Bovid Adaptation to Environmental Changes. PhD thesis, University of Adelaide (2019). [Google Scholar]
- 24.Jónsson H, Ginolhac A, Schubert M, Johnson PLF & Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simón-Soro A. et al. Microbial geography of the oral cavity. J. Dent. Res. 92, 616–621 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Salter SJ et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisenhofer R, Anderson A, Dobney K, Cooper A. & Weyrich LS Ancient microbial DNA in dental calculus: a new method for studying rapid human migration events. J. Isl. Coast. Archaeol. 14, 149–162 (2019). [Google Scholar]
- 28.Thompson LR et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velsko IM et al. Ancient dental calculus preserves signatures of biofilm succession and interindividual variation independent of dental pathology. PNAS Nexus 1, pgac 148 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yates JAF et al. The evolution and changing ecology of the African hominid oral microbiome. Proc. Natl Acad. Sci. 118, e2021655118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velsko IM, Gallois S, Stahl R, Henry AG & Warinner C. High conservation of the dental plaque microbiome across populations with differing subsistence strategies and levels of market integration. Mol. Ecol. 10.1111/mec.16988 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Morton JT et al. Uncovering the horseshoe effect in microbial analyses. mSystems 2, e00166–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Granehäll L. et al. Metagenomic analysis of ancient dental calculus reveals unexplored diversity of oral archaeal Methanobrevibacter. Microbiome 9, 197 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Downes J, Munson MA, Radford DR, Spratt DA & Wade WG Shuttleworthia satelles gen. nov., sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 52, 1469–1475 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Siqueira JF & Rôças IN Pseudoramibacter alactolyticus in primary endodontic infections. J. Endod. 29, 735–738 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Huang S. et al. Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC Oral Health 11, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaci N, Borrel G, Tottey W, O’Toole PW & Brugère J-F Archaea and the human gut: new beginning of an old story. World J. Gastroenterol. 20, 16062–16078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Handsley-Davis M. et al. Heritage-specific oral microbiota in Indigenous Australian dental calculus. Evol. Med. Public Health 10, 352–362 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adler CJ et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat. Genet. 45, 450–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lepp PW et al. Methanogenic Archaea and human periodontal disease. Proc. Natl Acad. Sci. USA 101, 6176–6181 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Creeth JE et al. The effect of brushing time and dentifrice on dental plaque removal in vivo. J. Dent. Hyg. 83, 111–116 (2009). [PubMed] [Google Scholar]
- 42.Zanatta FB, Bergoli AD, Werle SB & Antoniazzi RP Biofilm removal and gingival abrasion with medium and soft toothbrushes. Oral Health Prev. Dent. 9, 177–183 (2011). [PubMed] [Google Scholar]
- 43.Mann AE et al. Do I have something in my teeth? The trouble with genetic analyses of diet from archaeological dental calculus. Quat. Int. 653–654, 33–46 (2023). [Google Scholar]
- 44.Ozga AT & Ottoni C. Dental calculus as a proxy for animal microbiomes. Quat. Int. 653–654, 47–52 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hillson S. Archaeology and the study of teeth. Endeavour 10, 145–149 (1986). [DOI] [PubMed] [Google Scholar]
- 46.Muegge BD et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ley RE et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warinner C. et al. Direct evidence of milk consumption from ancient human dental calculus. Sci. Rep. 4, 7104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ottoni C. et al. Tracking the transition to agriculture in southern Europe through ancient DNA analysis of dental calculus. Proc. Natl Acad. Sci. USA 118, e2102116118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y-H et al. Progress in oral microbiome related to oral and systemic diseases: an update. Diagnostics 11, 1283 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng X. et al. Oral microbiota in human systematic diseases. Int. J. Oral. Sci. 14, 1–11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuenemann VJ et al. Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death. Proc. Natl Acad. Sci. USA 108, E746–E752 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shrewsbury JFD A History of Bubonic Plague in the British Isles (Cambridge Univ. Press, 2005). [Google Scholar]
- 54.DeWitte SN & Wood JW Selectivity of Black Death mortality with respect to preexisting health. Proc. Natl Acad. Sci. USA 105, 1436–1441 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yaussy SL & DeWitte SN Calculus and survivorship in medieval London: the association between dental disease and a demographic measure of general health. Am. J. Phys. Anthropol. 168, 552–565 (2019). [DOI] [PubMed] [Google Scholar]
- 56.DeWitte SN Health in post-Black Death London (1350–1538): age patterns of periosteal new bone formation in a post-epidemic population. Am. J. Phys. Anthropol. 155, 260–267 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Moore NE & Weyrich LSA in The Oral Microbiome: Methods and Protocols (ed. Adami GR) 93–118 (Springer, 2021); 10.1007/978-1-0716-1518-8_7 [DOI] [Google Scholar]
- 58.Brotherton P. et al. Neolithic mitochondrial haplogroup H genomes and the genetic origins of Europeans. Nat. Commun. 4, 1764 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eisenhofer R. et al. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 27, 105–117 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Lindgreen S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes 5, 337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knights D. et al. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolyen E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Segata N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitra S. et al. Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinform. 12, S21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Archer L, Dawson E, DeWitt J, Seakins A. & Wong B. “Science capital”: a conceptual, methodological, and empirical argument for extending bourdieusian notions of capital beyond the arts. J. Res. Sci. Teach. 52, 922–948 (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All trimmed and merged DNA sequences (fastq) are available in the SRA database (BioProject PRJNA780005) of NCBI. The 2017 NCBI nr database and the 2017 NCBI RefSeq GCS database were used in this study. Unmerged reads can be made available upon request, as only merged sequences were assessed in full for this publication.