Abstract
How DNA is repaired after retrovirus integration is not well understood. DNA-dependent protein kinase (DNA-PK) is known to play a central role in the repair of double-stranded DNA breaks. Recently, a role for DNA-PK in retroviral DNA integration has been proposed (R. Daniel, R. A. Katz, and A. M. Skalka, Science 284:644–647, 1999). Reduced transduction efficiency and increased cell death by apoptosis were observed upon retrovirus infection of cultured scid cells. We have used a human immunodeficiency virus (HIV) type 1 (HIV-1)-derived lentivirus vector system to further investigate the role of DNA-PK during integration. We measured lentivirus transduction of scid mouse embryonic fibroblasts (MEF) and xrs-5 or xrs-6 cells. These cells are deficient in the catalytic subunit of DNA-PK and in Ku, the DNA-binding subunit of DNA-PK, respectively. At low vector titers, efficient and stable lentivirus transduction was obtained, excluding an essential role for DNA-PK in lentivirus integration. Likewise, the efficiency of transduction of HIV-derived vectors in scid mouse brain was as efficient as that in control mice, without evidence of apoptosis. We observed increased cell death in scid MEF and xrs-5 or xrs-6 cells, but only after transduction with high vector titers (multiplicity of infection [MOI], >1 transducing unit [TU]/cell) and subsequent passage of the transduced cells. At an MOI of <1 TU/cell, however, transduction efficiency was even higher in DNA-PK-deficient cells than in control cells. Taken together, the data suggest a protective role of DNA-PK against cellular toxicity induced by high levels of retrovirus integrase or integration. Another candidate cellular enzyme that has been claimed to play an important role during retrovirus integration is poly(ADP-ribose) polymerase (PARP). However, no inhibition of lentivirus vector-mediated transduction or HIV-1 replication by 3-methoxybenzamide, a known PARP inhibitor, was observed. In conclusion, DNA-PK and PARP are not essential for lentivirus integration.
Integration is an essential step in the retrovirus replication cycle (8). The viral integrase catalyzes both the 3′ processing of the viral DNA ends and the insertion of the viral DNA into the host chromosome. This insertion is mediated by a coordinated nucleophilic attack by the hydroxyl groups of both processed ends on both strands of the phosphodiester backbone of the host DNA, followed by the ligation of the viral 3′ ends to the cellular DNA. The result is a gapped intermediate in which the viral 5′ ends are not joined to the host DNA. Resolution of the integration intermediate leads to the chromosomal insertion of the proviral DNA trimmed of both terminal dinucleotides and flanked by a duplicated host DNA fragment. The size of the DNA duplication is virus specific. For human immunodeficiency virus (HIV), a 5-bp duplication is formed. Cellular DNA repair mechanisms are generally believed to fill in and ligate the remaining single-stranded DNA gaps, although the underlying mechanisms have not been characterized. Alternatively, viral enzymes may be involved. Reverse transcriptase could fill in the gaps and, after removal of the two overhanging nucleotides at the 5′ end of the viral DNA, the DNA splicing activity of integrase could ligate the viral DNA to the target DNA (11, 35). A report that integrase would also have the required DNA polymerase activity awaits independent confirmation (2).
In eukaryotic cells, nonhomologous end joining represents the major mechanism for the repair of double-stranded DNA (dsDNA) breaks (26, 30). In eukaryotes, dsDNA breaks occur during V(D)J recombination and during meiotic recombination and are also generated by ionizing radiation. Nonhomologous end joining is mediated by DNA-dependent protein kinase (DNA-PK), a kinase activated by dsDNA ends (19, 24). DNA-PK is composed of a 450-kDa catalytic subunit (DNA-PKCS) and the heterodimeric protein Ku, composed of 70- and 86-kDa subunits. Ku is the DNA-binding component of DNA-PK required for the activation of the catalytic subunit. Ku binds strongly to dsDNA ends and, at least in vitro, to gapped and nicked DNA molecules as well (5, 20). Mice with severe combined immunodeficiency (scid) have a DNA-PKCS truncation mutation in both alleles (6, 14). This scid mutation affects V(D)J rearrangement and double-strand break repair, resulting in the lack of mature B and T lymphocytes in scid mice (7). Primary cells derived from scid mice are deficient in DNA-PK activity (27, 30). Chinese hamster ovary (CHO) cell lines deficient in Ku86 (xrs-5 and xrs-6) are also available (15, 25). Like scid cells, these mutant cell lines are highly sensitive to irradiation.
A role for nonhomologous end joining in general and DNA-PK in particular in repairing both DNA breaks generated by retrovirus integration is certainly conceivable. Recently, DNA-PK was claimed to be required for retrovirus integration (13). It was shown that integration efficiency was reduced in DNA-PK-deficient murine scid cells and that high-titered virus stocks induced apoptosis in these cells.
Another candidate cellular enzyme that could play an important role during retrovirus integration is poly(ADP-ribose) polymerase (PARP) (22). This nuclear enzyme (EC 2.4.2.30) is a zinc finger protein of 113 kDa that can bind to both single-stranded DNA and dsDNA breaks and that is believed to participate in a number of cellular processes involving DNA break formation and rejoining (9, 16). PARP activity is stimulated by a variety of DNA-damaging agents, including ionizing irradiation, alkylating agents, and oxygen radicals. PARP catalyzes the attachment of polymers of the ADP-ribose moiety of its substrate, NAD, to various proteins, including itself. After automodification, the protein dissociates from the DNA, providing access to other components of the DNA repair system (36). PARP is believed to protect the integrity of the genome by preventing accidental homologous DNA recombination (9). Excessive stimulation of PARP may deplete NAD and lead to cell death (33). Selective PARP inhibitors, such as 3-methoxybenzamide (3-MB) are known (34). Infection of mammalian cells by recombinant retrovirus vectors is inhibited by inhibition of PARP activity (22), suggesting a role during retrovirus integration.
In the context of the establishment of a cellular integration system (10), we have investigated the possible requirements of DNA-PK and PARP for efficient lentivirus integration. To our surprise and in contrast to published reports, we have found no evidence that these enzymes are essential for integration at multiplicities of infection (MOI) below 1 transducing unit (TU)/cell.
MATERIALS AND METHODS
Cell cultures.
HeLa, CHO-K1, xrs-5, and xrs-6 cells were obtained from the American Type Culture Collection. By limiting dilution, individual clones of the Ku-deficient CHO-K1-derived cell lines xrs-5 and xrs-6 (xrs-5/2 and xrs-6/1) in which the absence of the Ku protein was verified by Western blotting were selected (data not shown). Human embryonic kidney 293T cells expressing simian virus 40 (SV40) large T antigen, were obtained from O. Danos (Evry, France). Wild-type (WT) mouse embryonary fibroblasts (MEF) were obtained from embryos of C57Black/6J and 129/OLA parents; scid MEF were obtained from CB-17/scid × scid mice. Cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing Glutamax-I and supplemented with 10% fetal calf serum (FCS) (Harlan Sera-Lab LTD) and 20 μg of gentamicin (Gibco BRL) per ml at 37°C in a 5% CO2 humidified atmosphere. 293T cells were transfected using polyethyleneimine (PEI) (average molecular weight, 25,000; Sigma-Aldrich, Bornem, Belgium) (1); MEF cells were transfected using Lipofectamine (Gibco BRL). The continuous cell lines WT MEF-T and scid MEF-T were obtained by transfecting WT and scid MEF at 50 to 70% confluence with pMSSVLT, encoding SV40 large T antigen. At 48 h after transfection, 500 μg of Geneticin (G418) per ml was added to the culture medium to select stable transfectants. The number of viable cells was determined by trypan blue exclusion. Anti-HIV activity and cytotoxicity measurements for MT-4 cells were based on viability measurements using a tetrazolium-based colorimetric method (32).
Lentivirus vector production.
HIV type 1 (HIV-1)-derived vector particles, pseudotyped with the envelope of vesicular stomatitis virus, were produced by transfecting 293T cells with a packaging plasmid encoding viral Gag, Pol, and accessory proteins (pCMVΔR8.2), a plasmid encoding the envelope of vesicular stomatitis virus (pMDG), and a plasmid carrying a reporter gene flanked by two long terminal repeats (pHR′-CMVLacZ) (31). A second-generation attenuated packaging plasmid, pCMVΔR8.91, which lacks the vif, vpr, vpu, and nef genes (39), was used as well. All plasmids were kindly provided by O. Danos and D. Trono (Geneva, Switzerland).
For transfection of a 10-cm dish of 293T cells, 20 μg of vector plasmid, 10 μg of packaging construct, and 5 μg of envelope plasmid were mixed in 700 μl of 150 mM NaCl. PEI solution (1.57 mM PEI in 150 mM NaCl) (700 μl) was added slowly. The mixture was incubated at room temperature for 15 min and then added dropwise to 293T cells in DMEM containing 1% FCS. On the next morning, the medium was replaced with DMEM containing 10% FCS. Supernatants were collected from day 2 until day 5 posttransfection. The vector particles in the supernatants were sedimented by ultracentrifugation in a swinging-bucket rotor (SW27; Beckman, Palo Alto, Calif.) at 25,000 rpm for 2 h at 4°C. Pellets were redissolved in 300 μl of phosphate-buffered saline (PBS), resulting in a 100-fold concentration. p24 antigen content was determined using an HIV-1 p24 core profile enzyme-linked immunosorbent assay (DuPont, Dreieich, Germany), and viral titers were determined by end-point dilution and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining of transduced cells. p24 content was used to normalize the different vector batches.
For transduction experiments, cells were seeded at 10,000/well in 96-well plates. After 24 h, the medium was replaced with DMEM containing 1% FCS, and the cells were infected with different amounts of vector supplemented with 2 μg of Polybrene per ml. On the next morning, the supernatant was replaced with fresh medium containing 10% FCS. β-Galactosidase reporter gene activity was measured 3 days after transduction. Cells were washed with PBS, fixed with 2% formaldehyde–0.2% glutaraldehyde in PBS, and stained with freshly prepared X-Gal substrate (5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM MgCl2, and 400 μg of X-gal per ml [Biotech Trade & Service GmbH, St. Leon-Rot, Germany] in PBS) at 37°C overnight. Individual blue cells were counted microscopically. Alternatively, total β-galactosidase activity in the cell lysate was determined. Medium was removed by gentle aspiration, and the monolayers were washed with PBS. Cells were lysed in 25 μl of 0.5% Nonidet P-40. Five microliters of the extract was used to determine total protein content according to the Bradford method (Bio-Rad, Hercules, Calif.). The remaining volume was used to measure the conversion of o-nitrophenyl-β-d-galactopyranoside (Sigma-Aldrich) in a colorimetric assay as described previously (3). Results were normalized to the total protein concentration. In experiments evaluating titer-dependent cellular toxicity, this normalization of reporter gene activity was not done.
In vivo analysis of lentivirus vector transduction.
Adult CB-17/scid × scid and C57Black/6 mice of both sexes were used. All surgical procedures were performed under chloral hydrate anesthesia (400 mg/kg of body weight intraperitoneally) using aseptic procedures. The mice were placed in a stereotactic head frame (Narishige); after midline incision of the skin, one or two small holes were drilled in the skull in the appropriate location (AP 0.5, LAT 2.0, and DV 3.0, using bregma as a reference point [21]). Two microliters of highly concentrated vector (108 pg of p24/ml) supplemented with 4 μg of Polybrene per ml was injected at a rate of 0.5 μl/min with a 30-gauge needle connected by microdialysis tubing to a 25-μl Hamilton syringe in a microinjection pump (CMA/microdialysis AB, Stockholm, Sweden). The needle was raised slowly 0.25 mm in the dorsal direction every 60 s during the 4-min injection. After the injection, the needle was left in place for an additional 4 min before being withdrawn slowly from the brain (protocol adapted from that in reference 18).
To assess lentivirus transduction, the mice were deeply anesthetized with pentobarbital and perfused transcardially with saline followed by ice-cold 4% paraformaldehyde in PBS for 15 min. The brain was removed from the skull and postfixed overnight in the same fixing solution. Coronal brain sections (50 μm thick) were cut with a vibratome and stored at 4°C in PBS containing 0.1% sodium azide. The sections were first screened for green fluorescent protein (GFP) expression with an inverted fluorescence microscope. More sensitive detection was obtained by immunohistochemical analysis with a polyclonal antibody against GFP (Clontech, Palo Alto, Calif.). The sections were treated with 3% hydrogen peroxide and preincubated in 5% normal swine serum–0.1% Triton X-100 in PBS. The anti-GFP primary antibody (1:1,000) in 5% normal swine serum–0.1% Triton X-100 was added and incubated overnight at room temperature. The sections were then incubated with biotinylated swine anti-rabbit secondary antibody, followed by Strept ABC-HRP complex (Dako, Glostrup, Denmark). Detection was accomplished with diaminobenzidine and H2O2 as a substrate.
Astrocytes and neurons were identified using a polyclonal antibody against glial fibrillary acidic protein (GFAP; Dako) and a monoclonal antibody against NeuN (Chemicon, Temecula, Calif.), respectively. TUNEL (terminal deoxynucleotidyltransferase [TdT]-mediated dUTP-biotin nick end labeling) staining was performed according to a protocol adapted from that of Young et al. (38). Briefly, sections were pretreated with 1% H2O2 in methanol and then incubated in 0.1% sodium citrate buffer containing 0.1% Triton X-100 on ice for 2 min. The labeling reaction with TdT in the presence of biotin–16-dUTP was performed for 4 h at 37°C. Next, the sections were incubated with Strept ABC-HRP complex and stained with diaminobenzidine and H2O2. Sections treated with DNase I for 30 min at 37°C prior to TdT incubation were used as a positive control.
Quantitative analysis of GFP expression was carried out by means of a Bioquant image analyzing system (R&M Biometrics, Nashville, Tenn.). For each animal, serial sections (minimum of six) were analyzed over a distance of 2 mm centered around the injection site. For each section, the relative density and the number of pixels above a threshold of intensity were measured in the area of the striatum that was positive for GFP expression.
RESULTS
Efficient transduction of scid cells by lentivirus vectors.
The role of DNA-PK during lentivirus integration was investigated using amphotropic HIV-1-derived lentivirus vectors (31, 39) that encode β-galactosidase as a marker protein. The vectors retrotranscribe and integrate their genomes into the host chromosome much like the parental virus, but they cannot replicate due to the lack of viral coding sequences. This model system can thus be used to study the requirements of lentivirus integration. It was possible to transduce MEF using lentivirus vectors, although the efficiency of transduction in these cells was lower than that in established cell lines, such as 293T, HeLa, or CHO. To verify whether DNA-PK was required for integration, scid and WT MEF cells were transduced at low MOI (<0.1 TU/cell) to prevent infection by more than one particle per cell. MEF derived from scid mice were as efficiently transduced as MEF derived from WT mice (Fig. 1). Both with WT and with attenuated vectors (the latter lacking the four HIV accessory genes), scid MEF were transduced at least as efficiently as WT MEF. Similar results were obtained for continuous cell lines (WT MEF-T and scid MEF-T) derived from MEF after transformation with SV40 T antigen. Since the transduction efficiency in these cell lines was reproducibly higher than that in primary cells, MEF-T were used for further experiments. Lentivirus transduction of cells at the low titers used in these experiments did not induce cell death. To exclude the possibility that the need for DNA-PK-mediated DNA repair is apparent only in cells undergoing mitosis, we passaged the transduced cells prior to staining. After passage, the reporter gene activity remained equally high in WT MEF-T and scid MEF-T (data not shown).
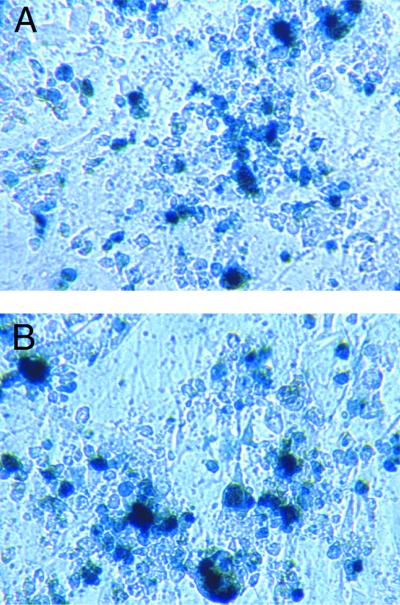
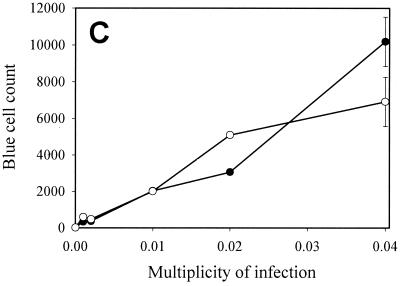
FIG. 1.
Lentivirus transduction efficiency in scid cells. Different amounts of a second-generation HIV-1-derived lentivirus vector (devoid of Vpr, Nef, Vif, and Vpu) encoding β-galactosidase were used to transduce subconfluent WT and scid MEF. At confluence, WT (A) and scid (B) MEF were stained with X-Gal, and positive WT (●) or scid (○) MEF were counted microscopically (C). Means and standard deviations of duplicate experiments are shown.
Efficient integration of lentivirus vectors in Ku-deficient cells.
In a second set of experiments, we verified the role of Ku, the DNA-binding component of DNA-PK. The CHO-K1-derived cell lines xrs-5 and xrs-6 are deficient in Ku86 protein. The absence of Ku protein was verified by Western blotting (data not shown). The relative efficiency of lentivirus vector transduction in these cell lines was compared with that in the parental CHO-K1 cell line (Fig. 2). The average transduction efficiencies were 70% in xrs-5 cells and 152% in xrs-6 cells relative to the transduction efficiency in CHO-K1 cells (Fig. 2A). Transduction was not accompanied by cell death (Fig. 2B). After passage of the transduced cells, the β-galactosidase-positive colonies were counted. Results similar to those obtained in transient transduction experiments were obtained (Fig. 2C). In mammalian cells, there is evidence that Ku plays a role in transcriptional regulation (23, 28, 29). This idea raised the possibility that in the absence of Ku, reduced transcription of integrated provirus could lead to an underestimation of apparent transduction efficiency when total reporter gene activity is measured. We quantified the total β-galactosidase activity of the cell lysate photometrically. Again, at MOI of 0.04 TU/cell, reporter gene activity was about 2-fold lower in xrs-5 cells but 1.5-fold higher in xrs-6 cells relative to the activity in the maternal cell line (data not shown). No significant reduction in reporter gene activity could thus be demonstrated in the absence of Ku.
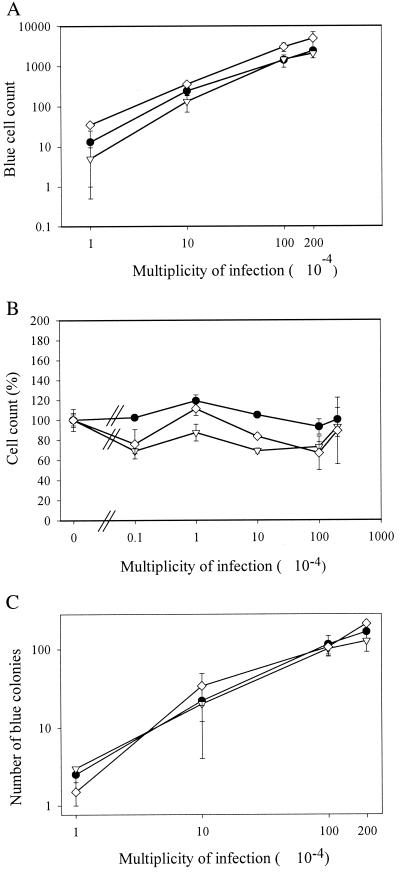
FIG. 2.
Lentivirus transduction efficiency in Ku-deficient cell lines. Subconfluent CHO-K1 (●), xrs-5/2 (▿), and xrs-6/1 (◊) cells were transduced with different amounts of a first-generation lentivirus vector and stained after 3 days. The number of β-galactosidase-positive cells (A) and the cell count (B) were determined microscopically. After passage (1/5) of the transduced CHO-K1 (●), xrs-5/2 (▿), and xrs-6/1 (□) cells, β-galactosidase-positive colonies (≥10 blue cells) were counted at confluence (C). Means and standard deviations of duplicate experiments are shown.
Efficient lentivirus transduction of scid mouse brain.
We have been studying the potential of HIV-1-derived lentivirus vectors for gene transfer into the murine brain as a step toward future gene therapy of neurodegenerative diseases (4, 31). In this context, we used scid mice in parallel with immunocompetent C57Black/6 (C57BL) mice to investigate potential immunological reactions to vector or transgene. Highly concentrated vector (2 × 105 pg of p24) encoding GFP was injected stereotactically into the striatum of adult mice, and the expression of the transgene was evaluated by immunohistochemical analysis. The levels of GFP 2 and 11 weeks after injection were as high in scid mice as in control C57BL mice (Fig. 3). GFP expression was evident in both neurons and glial cells (data not shown). Thus, although deficient for DNA-PK, brain cells of scid mice were efficiently and stably transduced by HIV-1-derived lentivirus vectors. Daniel et al. attributed the lower retrovirus integration efficiency in scid cells to increased cell death by apoptosis (13). Nissl staining of brain sections of scid and C57BL mice did not reveal extensive cell loss in the transduced area. We also performed TUNEL staining to visualize apoptotic cells after lentivirus vector-mediated gene transfer (Fig. 4). Since reactive apoptosis is likely to occur as an early event after transduction, we included a time point 2 days after transduction. GFP expression was indeed evident after 2 days, but TUNEL staining was negative for brain slices from both scid and C57BL mice at all time points analyzed (Fig. 4). In conclusion, the results obtained with our in vivo model argue against an essential role for DNA-PK during lentivirus integration.
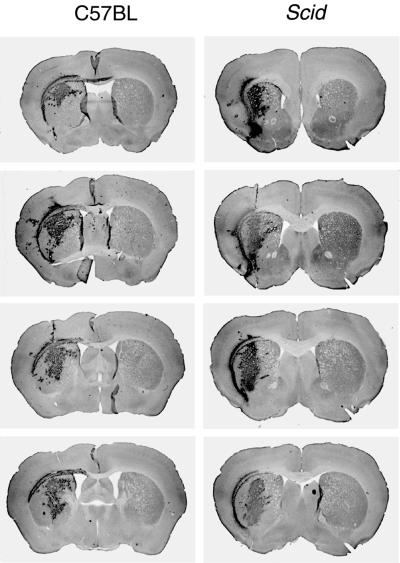
FIG. 3.
Efficient lentivirus transduction in scid mouse brain. (Top) Expression of the GFP transgene in the striatum of C57BL and scid mice 2 weeks after transduction with the same batch of lentivirus vectors. Serial sections with an interval of 250 μm showed equal transduction efficiency in both mouse strains. (Bottom) Quantification of GFP expression in serial sections over a distance of 2 mm around the injection site in the striatum of scid and C57BL mice 11 weeks after lentivirus vector transduction. The y axis indicates the area of positive GFP expression. Although there was some interindividual variation in transgene expression, no significant difference between scid and C57BL mice was observed.
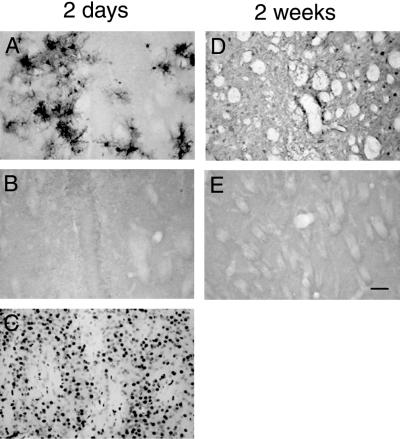
FIG. 4.
No evidence for increased apoptosis after lentivirus transduction in scid mouse brain. (A and D) GFP expression around the needle track in the striatum of scid mice 2 days (A) and 2 weeks (D) after lentivirus transduction. (B and E) TUNEL staining of adjacent sections shows no apoptotic cells in the area of the striatum positive for GFP expression 2 days (B) and 2 weeks (E) after lentivirus transduction. (C) DNase treatment of a section like that in the positive control results in numerous labeled nuclei. Scale bar, 100 μm.
No inhibition of lentivirus integration by inhibitors of PARP activity.
We extended our study to a second candidate repair enzyme, PARP. This enzyme has been thought to play an essential role during retrovirus integration, based on the inhibition of retrovirus transduction by competitive inhibitors of PARP (22). 3-MB is an inhibitor of PARP with a reported 50% inhibitory concentration (IC50) for lentivirus transduction or viral replication of approximately 0.8 mM, whereas 4-methoxybenzamide (4-MB) is an inactive analogue. We performed lentivirus transduction experiments with CHO-K1 and xrs-5 or xrs-6 cells in the presence of various concentrations of 3-MB or 4-MB. The cells were preincubated with the inhibitors for 24 h prior to infection. Lentivirus transduction was assessed by measuring β-galactosidase activity in fixed cells or in a cell lysate. At nontoxic concentrations, 3-MB did not interfere with lentivirus transduction (Table 1). We also evaluated the effects of 3-MB and 4-MB against HIV-1 (strain IIIB) replication in MT-4 cell cultures. Again, no inhibition was seen at the concentrations tested (up to 5 mM).
TABLE 1.
Lack of specific inhibition of lentivirus transduction and replication by inhibitors of PARP
| Virus and cell line | 3-MBa
|
4-MBa
|
||
|---|---|---|---|---|
| IC50 | CC50 | IC50 | CC50 | |
| HIV-1 vectorb | ||||
| CHO-K1 | >4 | 10 ± 1 | >4 | 20 ± 5 |
| xrs-5 | >4 | 8 ± 2 | >4 | 8 ± 4 |
| xrs-6 | >4 | 6 ± 2 | 2 ± 0.5 | 6 ± 2 |
| HIV-1 (strain IIIB)c, MT-4 cells | >3 | 3 ± 1 | >4 | 6 ± 1 |
The IC50 and the 50% cytotoxic concentration (CC50) are given as millimolar. Results reported as mean and standard deviation are based on two separate experiments.
CHO-derived cell lines were transduced with an HIV-1 vector (first generation) encoding β-galactosidase in the absence or presence of different concentrations of the known PARP inhibitor 3-MB or an inactive analogue, 4-MB. Two days later, cells were fixed and stained with X-Gal, and blue cells were counted. Total protein content was used to determine cellular toxicity.
Inhibition of HIV-1 replication and cellular toxicity in MT-4 cells were determined by the MTT method (32).
Cellular toxicity induced in DNA-PK-deficient cells by high titers of lentivirus vectors.
In all of our experiments so far, low MOI were used to investigate the requirement of DNA-PK for lentivirus integration. Recently, Daniel et al. proposed an essential role for DNA-PK during retrovirus integration based on results obtained with high viral titers (13). To examine the effect of high lentivirus vector titers, additional cell culture experiments were performed. We compared the lentivirus transduction of WT versus scid MEF-T (Fig. 5) and of CHO-K1 versus xrs-5 or xrs-6 cells (Fig. 6). Transduction of subconfluent cells was performed at different MOI. β-galactosidase activity was measured 3 days after transduction and at confluence after passage of the transduced cells.
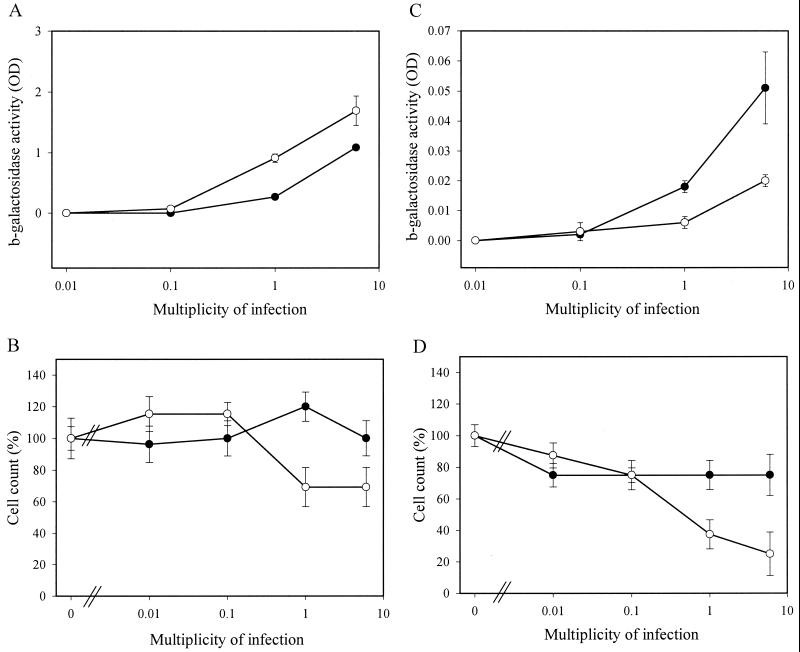
FIG. 5.
Titer- and passage-dependent cellular toxicity after lentivirus transduction of scid cells. Subconfluent WT (●) or scid (○) MEF-T were transduced in duplicate with different amounts of a second-generation lentivirus vector devoid of Vpr, Nef, Vif, and Vpu. Half of the cells were analyzed at confluence (A and B), and half of the cells were passaged (1/5) and analyzed at confluence (C and D). β-Galactosidase activity (OD, optical density) was measured in the cell lysate (A and C), and the cell count was determined by trypan blue dye exclusion (B and D). Reporter gene activity was not normalized for differential cell growth. Means and standard deviations of duplicate experiments are shown.
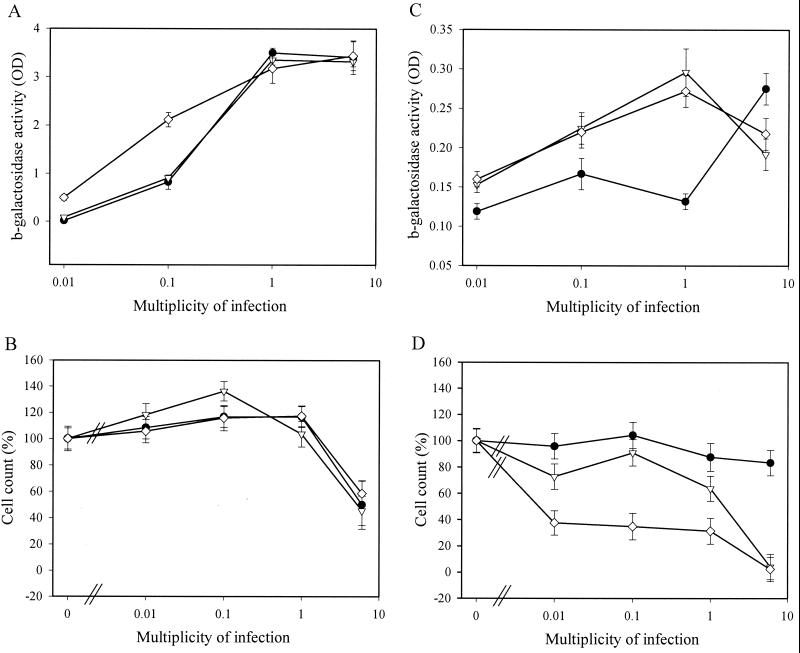
FIG. 6.
Titer- and passage-dependent cellular toxicity after lentivirus transduction of Ku-deficient cells. Subconfluent CHO-K1 (●), xrs-5/2 (▿), and xrs-6/1 (◊) cells were transduced in duplicate with different amounts of a second-generation lentivirus vector. Half of the cells were analyzed at confluence (A and B), and half of the cells were passaged (1/5) and analyzed at confluence (C and D). β-Galactosidase activity (OD, optical density) was measured in the cell lysate (A and C), and the cell count was determined by trypan blue dye exclusion (B and D). Reporter gene activity was not normalized for differential cell growth. Means and standard deviations of duplicate experiments are shown.
The reporter gene activity was always higher in scid than in WT MEF-T, even when higher MOI were used (Fig. 5A). At MOI of 6 TU/cell, mild toxicity in scid but not in WT MEF-T was evidenced by small reductions in cell count (Fig. 5B) and in total protein content (data not shown). Remarkably, scid MEF-T transduced with the highest titers of vector displayed massive cell death after passage (Fig. 5D), resulting in reduced reporter gene activity (Fig. 5C). At low MOI, this toxicity was not observed. Analogous results were obtained with the Ku-deficient cell lines (Fig. 6). Lentivirus vector-mediated transduction efficiency for xrs-5 or xrs-6 cells was not below that of the parental CHO-K1 cells (Fig. 6A). At high vector titers, some cellular toxicity was observed for Ku-deficient cell lines (Fig. 6B). However, after subcultivation, cell death was pronounced in the Ku-deficient cells transduced with high titers but not in the CHO-K1 cells (Fig. 6D). The cell death resulted in a reduction in total β-galactosidase activity in xrs-5 or xrs-6 cells (Fig. 6C).
DISCUSSION
It remains unresolved to what extent the retroviral enzymes reverse transcriptase (gap filling) and integrase (DNA splicing [11]) or the eukaryotic host proteins, such as DNA-PK and PARP, contribute to the DNA repair of the single-stranded DNA gaps that remain after retrovirus integration. A role for PARP as a nick sensor during retrovirus integration has been postulated (22). In our experimental design, the PARP inhibitor 3-MB was not capable of interfering with lentivirus vector transduction or HIV-1 replication, arguing against an important role for this enzyme during lentivirus integration.
To investigate the role of DNA-PK during retrovirus integration, we analyzed the transduction efficiency of HIV-1-derived lentivirus vectors in scid and xrs-5 or xrs-6 cells, which are deficient in the DNA-PK pathway. We previously showed that a vector carrying the inactivating D64V mutation in the integrase gene gave rise to only 5% the transduced cells obtained with the WT vector (10). After passage of transduced cells, this pseudotransduction level even fell below 1%. Since transduction by HIV vectors is thus largely dependent on the integration step, the efficiency of transduction can be used to evaluate cofactors essential for integration. We reasoned that the use of low vector titers would increase the likelihood that the requirement for a potential host factor would become apparent. Since lentivirus transduction was as efficient in scid, xrs-5, or xrs-6 cells as in WT cells, an essential role for DNA-PK in integration was ruled out. These results were confirmed with an in vivo model. We have been investigating the potential of lentivirus vectors for in vivo gene transfer after stereotactic injection into the brain (4). scid mice were used to control for the immunological response to the vector or transgene. In this in vivo model, the transduction of neurons and glial cells was at least as efficient in scid mice as in C57BL mice.
Our results with cell cultures and with the in vivo mouse model do exclude an essential role for DNA-PK during lentivirus integration. During the course of our experimental work, however, a study by Daniel et al. was published, wherein an essential role for DNA-PK during retrovirus integration was claimed (13). The claim was based on the observation that retrovirus transduction was reduced in scid cells and accompanied by cell death due to apoptosis. Intriguingly, this effect was reported to occur at viral MOI of ≥1 TU/cell. Our cell culture data were obtained at low MOI. In the in vivo model, the actual MOI is difficult to estimate. It is determined by the limited amount of virus (at maximal titer) that one can inject into the brain and the area of diffusion. One can assume that a concentration gradient exists, with high MOI around the injection site, where mechanical damage around the needle track may mask apoptosis.
Therefore, we repeated lentivirus transduction experiments at high MOI and using a colorimetric detection method. We analyzed the efficiency of both transient transduction of subconfluent cells and stable transduction after subcultivation. Daniel et al. selected for neomycin-resistant colonies after retrovirus transduction (13). In our experiments, different plating efficiencies for DNA-PK-deficient cell lines compromised this approach. Moreover, since Ku is known to be involved in transcriptional regulation, expression levels rather than transduction levels can influence test results. At an MOI of 6 TU/cell, toxicity was observed in scid and xrs-5 or xrs-6 cells 3 days after transduction. Even at this MOI, the reporter gene activity remained higher in the DNA-PK-deficient cells, ruling out interference with the integration step. Passage of the transduced cells, however, was associated with pronounced cell death in the DNA-PK-deficient cells. Here, overall reporter gene activity dropped, more by reduction of cell number than by reduction of gene expression per cell. In agreement with the published data, extensive retrovirus transduction in DNA-PK-deficient cells is apparently associated with cell death, especially when cells are forced to divide. The absence of apoptosis after in vivo transduction of neuronal cells might be due to the fact that they are postmitotic cells.
The results of Daniel et al. were based on transduction with both retrovirus and HIV-derived lentivirus vectors (13). Since our experiments were carried out only with HIV-derived lentivirus vectors, our conclusions are confined to lentivirus integration. We used an attenuated HIV-derived vector for experiments at high MOI to avoid potential cellular toxicity due to the presence of Vpr in the virus particle, as has been described elsewhere (37).
The DNA-binding component of DNA-PK was recently shown to potentiate retrotransposition of the Ty element of Saccharomyces cerevisiae in a galactose-inducible Ty1 system (17). However, the same authors reported that the level of endogenous Ty1 retrotransposition was almost twofold higher in the absence of Ku, an effect that they attributed to the transcriptional induction of genes responsive to DNA damage, such as Ty1 genes, in the abence of Ku. However, on the basis of our results with lentivirus transduction, it may be argued that the requirement of Ku for retrotransposition is linked to the overexpression of the Ty1 element. Since Ty1 retrotransposition was scored by selection of transduced colonies, the distinction between lack of integration or integration-induced cell death in the absence of Ku was not made. We hypothesize that high levels of retrotransposition induce cell death in the absence of Ku.
Our data do not exclude the possibility that an alternative cellular pathway can substitute for DNA-PK during lentivirus integration. Nevertheless, our data argue against an essential role for this enzyme and question its suitability as an antiviral target. Scepticism toward a universal role for DNA-PK in retrovirus integration was already expressed by Coffin and Rosenberg (12). It was pointed out that primary B cells from scid mice can be immortalized efficiently by infection with a retrovirus. Coffin and Rosenberg (12) suggested that, instead, DNA-PK might protect the cell from a side effect of integration. Although we were able to obtain data analogous to those of Daniel et al. (13), namely, cell death induced by lentivirus transduction of DNA-PK-deficient cells, we observed this effect only after transduction with excess vector and especially when the transduced cells were forced to divide. Since transduction was affected less than cell survival, the lack of DNA-PK apparently does not prevent integration as such. Moreover, at low viral titers, lentivirus transduction was apparently more efficient in most DNA-PK-deficient cells. Together, our data tend to support the notion of a protective role for DNA-PK during excessive retrovirus integration. In this context, DNA-PK may play a physiological role in protection against superinfection. More research is required to determine exactly what triggers cell death after excessive lentivirus infection of DNA-PK-deficient cells and what protective role DNA-PK plays in this process.
In conclusion, no evidence was found that DNA-PK or PARP is essential for lentivirus integration when low virus titers are used. Another cellular or a virus-mediated gap repair mechanism may be involved.
ACKNOWLEDGMENTS
Veerle Baekelandt and Anje Claeys contributed equally to this work.
We thank K. Craessaerts, K. Eggermont, and M. Michiels for excellent technical assistance and the laboratory of F. Vandesande (Katholieke Universiteit Leuven [KUL]) for use of the photographic and image analysis equipment. We are grateful to M. Witvrouw (KUL) for anti-HIV testing. The HIV-1-derived lentivirus vector system was a kind gift from O. Danos (Evry, France) and D. Trono (University of Geneva). We thank A. M. Skalka, R. Katz, and R. Daniel (Fox Chase Cancer Center, Philadelphia, Pa.) for helpful discussions.
V.B. and Z.D. are postdoctoral fellows of the Flemish Fund for Scientific Research (FWO). This work was funded by IDO grant 98/006 from the Katholieke Universiteit Leuven Research Council and the STWW Program of the Flemish Institute Supporting Scientific-Technological Research in Industry (IWT).
REFERENCES
- 1.Abdallah B, Hassan A, Benoist C, Goula D, Behr J P, Demeneix B A. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Hum Gene Ther. 1997;7:1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 2.Acel A, Udashkin B E, Wainberg M A, Faust E A. Efficient gap repair catalyzed in vitro by an intrinsic DNA polymerase activity of human immunodeficiency virus type 1 integrase. J Virol. 1998;72:2062–2071. doi: 10.1128/jvi.72.3.2062-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Baekelandt, V., B. De Strooper, B. Nuttin, and Z. Debyser. Gene therapeutic strategies for neurodegenerative diseases. Curr. Opin. Mol. Ther., in press. [PubMed]
- 5.Blier P R, Griffith A J, Craft J, Hardin J A. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- 6.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 7.Bosma G C, Custer R P, Bosma M J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 8.Brown P O. Integration. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–203. [PubMed] [Google Scholar]
- 9.Chatterjee S, Berger S, Berger N. Poly(ADP-ribose)polymerase: a guardian of the genome that facilitates DNA repair by protecting against DNA recombination. Mol Cell Biochem. 1999;193:23–30. [PubMed] [Google Scholar]
- 10.Cherepanov P, Pluymers W, Claeys A, Proost P, De Clercq E, Debyser Z. High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 2000;14:1389–1399. doi: 10.1096/fj.14.10.1389. [DOI] [PubMed] [Google Scholar]
- 11.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 12.Coffin J M, Rosenberg N. Closing the joint. Nature. 1999;399:413–414. doi: 10.1038/20810. [DOI] [PubMed] [Google Scholar]
- 13.Daniel R, Katz R A, Skalka A M. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 14.Danska J S, Holland D P, Mariathasan S, Williams K M, Guidos C J. Biochemical and genetic defects in the DNA-dependent protein kinase in murine scid lymphocytes. Mol Cell Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darroudi F, Natarajan A T. Cytogenetical characterization of Chinese hamster ovary X-ray-sensitive mutant cells, xrs 5 and xrs 6. IV. Study of chromosomal aberrations and sister-chromatid exchanges by restriction endonucleases and inhibitors of DNA topoisomerase II. Mutat Res. 1989;212:137–148. doi: 10.1016/0027-5107(89)90064-x. [DOI] [PubMed] [Google Scholar]
- 16.de Murcia G, de Murcia J M. Poly(ADP-ribose)polymerase: a molecular nick-sensor. Trends Biochem Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 17.Downs J A, Jackson S P. Involvement of DNA-end binding protein Ku in Ty element retrotransposition. Mol Cell Biol. 1999;19:6260–6268. doi: 10.1128/mcb.19.9.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dull T, Zufferey R, Kelly M, Mandel R J, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dvir A, Peterson S R, Knuth M W, Lu H, Dynan W S. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci USA. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falzon M, Fewell J W, Kuff E L. EBP-80, a transcription factor closely resembling the human auto-antigen Ku, recognizes single- to double-strand transitions in DNA. J Biol Chem. 1993;268:10546–10552. [PubMed] [Google Scholar]
- 21.Franklin K B J, Paxinos G. The mouse brain in stereotaxic coordinates. San Diego, Calif: Academic Press, Inc.; 1997. [Google Scholar]
- 22.Gäken J A, Tavassoli M, Gan S-U, Vallian S, Giddings I, Darling D C, Galea-Lauri J, Thomas M G, Abedi H, Schreiber V, Ménissier-de Murcia J, Collins M K L, Shall S, Farzaneh F. Efficient retroviral infection of mammalian cells is blocked by inhibition of poly(ADP-ribose) polymerase activity. J Virol. 1996;70:3992–4000. doi: 10.1128/jvi.70.6.3992-4000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giffin W, Torrance H, Rodda D J, Préfontaine G G, Pope L, Haché R J G. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 24.Gottlieb T M, Jackson S P. The DNA-dependent protein kinase: requirement of DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 25.Jeggo P A. Studies on mammalian mutants defective in rejoining double-strand breaks in DNA. Mutat Res. 1990;239:1–16. doi: 10.1016/0165-1110(90)90028-a. [DOI] [PubMed] [Google Scholar]
- 26.Jeggo P A. DNA breakage and repair. Adv Genet. 1998;38:185–219. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- 27.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M, Iliakis G, Mehta R, Jackson M. DNA-dependent protein kinase (p350) as a candidate gene for murine SCID defect. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn A, Gottleib T M, Jackson S P, Grummt I. DNA-dependent protein kinase—a potent inhibitor of transcription by RNA polymerase I. Genes Dev. 1995;9:193–203. doi: 10.1101/gad.9.2.193. [DOI] [PubMed] [Google Scholar]
- 29.Labhart P. DNA-dependent protein kinase specifically represses promoter-directed transcription initiation by RNA polymerase I. Proc Natl Acad Sci USA. 1995;92:2934–2938. doi: 10.1073/pnas.92.7.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day R S, Barron G M, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 31.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 32.Pauwels R, Balzarini J, Baba M, Snoeck R, Schols D, Herdewijn P, Desmyter J, De Clercq E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 33.Pieper A A, Verma A, Zhang J, Snyder S H. Poly(ADP-ribose) polymerase, nitric oxide and cell death. Trends Pharmacol Sci. 1999;20:171–181. doi: 10.1016/s0165-6147(99)01292-4. [DOI] [PubMed] [Google Scholar]
- 34.Rankin W P, Jacobson E L, Benjamin R C, Moss J, Jacobson M K. Quantitative studies of inhibitors of ADP-ribosylation in vitro and in vivo. J Biol Chem. 1989;264:4312–4317. [PubMed] [Google Scholar]
- 35.Roe T, Chowand S A, Brown P O. 3′-End processing and kinetics of 5′-end joining during retroviral integration in vivo. J Virol. 1997;71:1334–1340. doi: 10.1128/jvi.71.2.1334-1340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satoh M S, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature (London) 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 37.Stewart S A, Poon B, Song J Y, Chen I S Y. Human immunodeficiency virus type 1 vpr induces apoptosis through caspase activation. J Virol. 2000;74:3105–3111. doi: 10.1128/jvi.74.7.3105-3111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young D, Lawlor P A, Leone P, Dragunow M, During M J. Environmental enrichment inhibits spontaneous apoptosis, prevents seizures and is neuroprotective. Nat Med. 1999;5:448–453. doi: 10.1038/7449. [DOI] [PubMed] [Google Scholar]
- 39.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]