Abstract
The murine bacterial pathogen Chlamydia muridarum (Cm) has been used to study human Chlamydia infections in various mouse models. CD4+ T-cells, natural killer cells, and interferon-gamma (IFN-γ)–mediated immunity are important to control experimentally induced Cm infections. Despite its experimental use, natural infection by Cm has not been documented in laboratory mice since the 1940s. In 2022, the authors reported the discovery of natural Cm infections in numerous academic institutional laboratory mouse colonies around the globe. To evaluate the impact of Cm infection in severely immunocompromised mice, 19 NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice were cohoused with Cm shedding, naturally infected immunocompetent mice and/or their soiled bedding for 4 weeks and subsequently euthanized. Clinical disease, characterized by lethargy, dyspnea, and weight loss, was observed in 11/19 NSG mice, and 16/18 NSG mice had neutrophilia. All mice exhibited multifocal to coalescing histiocytic and neutrophilic bronchointerstitial pneumonia (17/19) or bronchiolitis (2/19) with intraepithelial chlamydial inclusions (CIs). Immunofluorescence showed CIs were often associated with bronchiolar epithelium. CIs were frequently detected by immunohistochemistry in tracheal and bronchiolar epithelium (19/19), as well as throughout the small and large intestinal epithelium without lesions (19/19). In a subset of cases, Cm colonized the surface epithelium in the nasopharynx (16/19), nasal cavity (7/19), and middle ear canal (5/19). Endometritis and salpingitis with intraepithelial CI were identified in a single mouse. These findings demonstrate that Cm infection acquired through direct contact or soiled bedding causes significant pulmonary pathology and widespread intestinal colonization in NSG mice.
Keywords: bronchointerstitial pneumonia, Chlamydia muridarum, elementary bodies, intraepithelial chlamydial inclusions (CIs), major outer membrane protein, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ, reticulate bodies
NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice are a highly immunocompromised, inbred mouse strain commonly used for oncology, stem cell biology, diabetes, and infectious disease research. The Prkdc and interleukin 2 receptor (Il2rg) mutations in these mice produce a phenotype that lacks numerous immune components including T-cells, B-cells, natural killer (NK) cells, and have deficient signaling for 6 cytokines (interleukin [IL]-2, IL-4, IL-7, IL-9, IL-15, and IL-21).21 In addition, NSG mice have defects in classical innate functions, including an absent hemolytic complement system, reduced dendritic cell function, and defective macrophage activity.13,16,21 These defects in adaptive and innate immunity result in severe immunosuppression leaving the mice susceptible to opportunistic bacterial agents including Enterococcus spp., Klebsiella pneumoniae, Klebsiella oxytoca, pks+ Escherichia coli, and others.5,22
Chlamydia spp. represent a diverse group of gram-negative obligate intracellular bacteria known for causing disease in numerous animal species and humans.4 Chlamydia have a tropism for mucosal epithelial cells and a distinctive developmental cycle that includes 2 forms – the infectious elementary body and the noninfectious intracellular reticulate body.4,17 Once introduced to a susceptible mucosa, Chlamydia spreads along adjacent epithelium by the entry and exit of elementary bodies from the apical surface of mucosal epithelial cells.17
Chlamydia muridarum (Cm) is a murine specific Chlamydia species that was discovered in the early 1940s when spontaneous infections were detected in both control and experimental mice while studying human respiratory viruses.6,7,10 Cm is frequently used to model urogenital infections resulting from Chlamydia trachomatis in humans.3 In such models, following intravaginal administration, the genital tract of mice is infiltrated by neutrophils during the early stage of Cm infection while macrophages and lymphocytes, including B-cells, CD4+ T-cells, and CD8+ T-cells, are recruited to genital tract tissues as the infection resolves.3 CD4+ T-cells are essential to clear Cm from the urogenital tract.3 Cm has also been used to study the immune response in the lungs following intranasal inoculation of the organism in immunocompetent mouse strains and Tlr2 knockout mice.9,23 In addition, this bacterium has been used to model gastrointestinal infections in mice following oral inoculation, resulting in asymptomatic long-lasting colonization in the intestine and persistent shedding of the organism in feces.29 Recent experimental studies have described Cm infection of the central nervous system following transmission via the olfactory and trigeminal nerves.15
Histological analysis of 2 immunocompetent mice at our institution revealed multifocal mononuclear pulmonary infiltrates prompting further investigation into the etiology. Polymerase chain reaction (PCR), immunohistochemistry (IHC), and in situ hybridization (ISH) revealed Cm infection of the lungs and intestines in these mice.14 Further investigation revealed a significant prevalence of Cm among global academic laboratory mouse colonies, with Cm detected in 33% of incoming mouse shipment received from 39 academic institutions, 63% of soiled bedding sentinels from 3 institutions, 14% of 120 institutions submitting microbiota samples, and in 16% of biological and environmental samples from a multi-institutional cohort.14 Because natural infection by Cm had not been described in laboratory mice since the 1940s and NSG mice are highly susceptible to opportunistic bacterial infections, we performed a pilot experiment in which 4 NSG mice were cohoused with recently imported naturally infected immunocompetent mice shedding Cm in their feces, principally to facilitate isolation of the bacterium.14 Development of severe clinical disease and pulmonary lesions reported in 2 mice prompted further investigation.14 The aim of this investigation was to characterize the pathology and determine the extent of disease associated with natural Cm infection in male and female NSG mice.
Materials and Methods
Animals
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (The Jackson Laboratory, Bar Harbor, Maine) mice were obtained from a colony of F1 progeny at the Memorial Sloan Kettering Cancer Center. NSG mice were free of mouse hepatitis virus, Sendai virus, mouse parvovirus, minute virus of mice, murine norovirus, murine astrovirus 2, pneumonia virus of mice, Theiler’s meningoencephalitis virus, epizootic diarrhea of infant mice (mouse rotavirus), Ectromelia virus, reovirus type 3, lymphocytic choriomeningitis virus, K virus, mouse adenovirus 1 and 2, polyoma virus, murine cytomegalovirus, mouse thymic virus, Hantaan virus, mouse kidney parvovirus, Mycoplasma pulmonis, Chlamydia muridarum, Citrobacter rodentium, Rodentibacter pneumotropicus, Helicobacter spp., segmented filamentous bacteria (SFB or Candidatus savagella), Salmonella spp., Streptococcus pneumoniae, beta-hemolytic Streptococcus spp., Streptobacillus moniliformis, Filobacterium rodentium, Clostridium piliforme, Corynebacterium bovis, Corynebacterium kutscheri, Staphylococcus aureus, Klebsiella pneumoniae, Klebsiella oxytoca, Pseudomonas aeruginosa, Pneumocystis spp., and endoparasites and ectoparasites. All procedures were performed at the Memorial Sloan Kettering Cancer Center (MSK) following institutional animal care and use committee approval, in accordance with the recommendations provided in the Guide for the Care and Use of Laboratory Animals (8th edition).11
Exposure of NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ Mice to Chlamydia muridarum Shedding Imported Mice
Eight to twelve-month-old NSG mice (9 females and 10 males) were exposed to Cm either by cohousing with Cm-positive immunocompetent mice (N = 11 NSG) or by housing in cages containing soiled bedding from those same Cm-positive immunocompetent mice (N = 8 NSG). Eleven NSG mice were exposed in November/December 2021 and 8 NSG mice were exposed in April/May 2022. Mice were maintained in individually ventilated, polysulfone shoebox cages with stainless-steel wire-bar lids and filter tops. The diet consisted of a natural ingredient, closed source, flash-autoclaved, γ-irradiated feed that was autoclaved reverse osmosis acidified water.14 Additional husbandry details are found in Mishkin et al.14 Following 3 to 4 weeks of initial exposure, mice were euthanized and a complete postmortem examination was performed.
Real-Time Quantitative Polymerase Chain Reaction
Total DNA from formalin-fixed, paraffin-embedded lung scrolls (20-μm-thick) from 7 NSG mice with pulmonary lesions consistent with Chlamydia infection were deparaffinized and extracted using a magnetic-based total nucleic acid isolation kit (MagMAX Total Nucleic Acid Isolation Kit, Life Technologies, Carlsbad, California) following internally optimized protocols. Quantitative (real-time) Polymerase Chain Reaction (qPCR) amplification was performed using the ThermoFisher qPCR Systems (Waltham, Massachusetts) according to the manufacturer’s instructions. A proprietary real-time fluorogenic 5’ nuclease PCR assay specifically targeting a 425 bp fragment of Cm 23S rRNA was used to determine the presence of Chlamydia DNA in samples as previously described.14 The double-stranded nucleotide sequences of selected purified PCR products were determined using Sanger dye-termination sequencing (Genomics Core, Tufts University Medical School, Boston, Massachusetts).
Microbiology
Lung and cecal samples were collected aseptically during necropsy using instruments with tips sterilized in a glass bead sterilizer (Germinator 500, CellPoint Scientific, Gaithersburg, Maryland) and transported in sterile Petri dishes for immediate processing. Tissue samples were inoculated onto solid agars. For aerobic culture of lungs, samples were plated on BBL Prepared Plated Media Trypticase Soy Agar II with 5% sheep blood (Becton Dickinson, Franklin Lakes, New Jersey), BBL Chocolate II Agar, BBL Columbia CNA Agar with 5% sheep blood, and BBL MacConkey II Agar. Cecal samples were also plated onto BBL Hektoen Enteric Agar. For anaerobic culture, samples were plated on BBL Brucella Agar with 5% sheep blood, hemin, and vitamin K1 (Becton Dickinson) and, in 1 case, Clostridium difficile selective-media plates (AnaeroGro Cycloserine–Cefoxitin Fructose Agar, Hardy Diagnostics, Santa Maria, California) and placed promptly into a anaerobic incubation sachet (BD GasPak EZ Container Systems, Becton Dickinson). Lung tissues were additionally immersed into BBL Fluid Thioglycolate Medium (Becton Dickinson). Samples were incubated aerobically and anaerobically at 37°C for 7 days. Subculture of positive broth cultures was performed on the agars described above. Observed colonial growth was identified to species level using the MALDI-TOF Biotyper Sirius using the MBT Compass Library #1829023, revision H, version 11, as well as FlexControl v3.4 and Maldi Biotyper v4.1.100 software (Bruker Daltonics, Bremen, Germany).
Necropsy and Histopathology
NSG mice submitted for postmortem examination were euthanized by CO2 asphyxiation and cardiac exsanguination. Complete necropsies were performed at the Laboratory of Comparative Pathology (MSK, the Rockefeller University, and Weill Cornell Medicine). Representative sections from all organ systems including heart, thymus, lungs, liver, gallbladder, kidneys, pancreas, stomach, duodenum, jejunum, ileum, cecum, colon, lymph nodes (mandibular, mesenteric), salivary glands, skin (trunk and head), urinary bladder, uterus, cervix, vagina, ovaries, oviducts, adrenal glands, spleen, thyroid gland, esophagus, trachea, spinal cord, vertebrae, sternum, femur, tibia, stifle joint, skeletal muscle, nerves, skull, nasal cavity, oral cavity, teeth, ears, eyes, pituitary gland, and brain were fixed in 10% neutral-buffered formalin, processed in alcohol and xylene, embedded in paraffin, sectioned (5-μm-thick) and stained with hematoxylin and eosin. The skull, spinal column, sternum, and hindlimb were decalcified in a formic acid and formaldehyde solution (Surgipath Decalcifier I, Leica Biosystems, Wetzlar, Germany) before processing.
Blood was collected from all mice by cardiac puncture immediately following euthanasia and subsequently transferred into tubes containing ethylenediaminetetraacetic acid (EDTA). Complete blood counts with differentials were performed using an automated hematology analyzer (IDEXX Procyte DX, Columbia, Missouri). Complete blood count values were compared to our in-house reference values from healthy, experimentally naïve, adult NSG mice.
Immunohistochemistry and Special Stains
All tissues evaluated histologically were screened for Chlamydia major outer membrane protein (MOMP) using a technique optimized and validated by the Laboratory of Comparative Pathology. Formalin-fixed, paraffin-embedded sections were stained using an automated staining platform (Leica Bond RX, Leica Biosystems). Following deparaffinization and heat-induced epitope retrieval in a citrate buffer at pH 6.0, the primary antibody against Chlamydia MOMP (NB100-65054, Novus Biologicals, Centennial, Colorado) was applied at a dilution of 1:2000. A rabbit antigoat secondary antibody (Cat. No. BA-5000, Vector Laboratories, Burlingame, California) and a polymer detection system (DS9800, Novocastra Bond Polymer Refine Detection, Leica Biosystems) was then applied to the tissues. The chromogen was 3,3’-diaminobenzidine tetrachloride (DAB), and the sections were counterstained with hematoxylin and examined by light microscopy. Reproductive tracts from TLR3-deficient mice experimentally infected with Cm strain Nigg were used as a positive control.
F4/80 and Ly6G IHC, as well as Masson’s trichrome stain, were performed on lungs in 4 representative cases with bronchointerstitial pneumonia. Formalin-fixed, paraffin-embedded sections were stained using an automated staining platform (Leica Bond RX, Leica Biosystems). Following deparaffinization and heat-induced epitope retrieval in citrate buffers at pH 6.0 (F4/80 IHC) or at pH 9.0 (Ly6G IHC), the primary antibody against F4/80 (14-4801-85, Invitrogen, Waltham, Massachusetts) or Ly6G (ab238132, Abcam, Cambridge, United Kingdom) was applied at dilutions of 1:100 (F4/80) or 1:2500 (Ly6G). For F4/80, a rabbit antirat secondary antibody (Cat. No. BA-4001, Vector Laboratories) and a polymer detection system (DS9800, Novocastra Bond Polymer Refine Detection, Leica Biosystems) were then applied to the tissues. For Ly6G, a rabbit antigoat secondary antibody (Leica Biosystems, DS9800 Kit, Reagent #3) and a polymer detection system (DS9800, Novocastra Bond Polymer Refine Detection, Leica Biosystems) were then applied to the tissues. The chromogen was DAB, and the sections were counterstained with hematoxylin and examined by light microscopy. Positive immunoreactivity for Ly6G and F4/80 was confirmed with internal mouse tissue array controls used to validate these immunoassays. A subset of tissues incubated with antibody diluents and secondary antibody only were used as negative controls for these assays.
In Situ Hybridization
Select IHC-positive tissues were included for ISH analysis. The target probe was designed to detect region 581–617 of Cm strain Nigg complete sequence, NCBI Reference Sequence NC_002620.2 (1039538-C1; Advanced Cell Diagnostics, Newark, California). The target probe was validated on reproductive tracts from mice experimentally inoculated with Cm strain Nigg. Slides were stained on an automated stainer (Leica Bond RX, Leica Biosystems) with RNAscope 2.5 LS Assay Reagent Kit-Red (322150, Advanced Cell Diagnostics) and Bond Polymer Refine Red Detection (DS9390, Leica Biosystems). Control probes detecting a validated positive housekeeping gene (mouse peptidylprolyl isomerase B, Ppib to confirm adequate RNA preservation and detection; 313918, Advanced Cell Diagnostics) and a negative control, Bacillus subtilis dihydrodipicolinate reductase gene (dapB to confirm absence of nonspecific labeling; 312038, Advanced Cell Diagnostics) were used. Positive RNA hybridization was identified as discrete, punctate chromogenic red dots under bright field microscopy.
Immunofluorescence
Paraffin-embedded blocks containing formalin-fixed lung sections (5-μm-thick) from 3 representative mice were used for this immunoassay. Tissue sections were deparaffinized in xylene and rehydrated in graded alcohols and water. Endogenous peroxidase was blocked with 1% hydrogen peroxide for 15 minutes. Antigen retrieval was performed in a steamer at 99°C for 30 minutes in citrate (10 mM pH 6.0) followed by cooling and rinsing with phosphate-buffered saline (PBS). Slides were initially incubated for 1 hour at room temperature or overnight at 4°C with the following primary antibodies anticytokeratin (1:100; Z0622, Dako, Santa Clara, California), anti-F4/80 (1:100; 14-4801-82, Invitrogen, Waltham, Massachusetts), and Chlamydia L2 MOMP (1:100; NB100-65054, Novus, Centennial, Colorado). Slides were then incubated for 1 hour at room temperature with an Alexa Fluor 488 conjugated antibody (1:500; A11055, Invitrogen) to detect Cm and an Alexa Fluor 594-labeled secondary antibody (1:500; A21207, or A21209, Invitrogen) to detect macrophages or epithelial cells. Each step was followed by washes with PBS. Slides were then incubated in 4’,6-diamidino-2-phenylindole (DAPI) for 4 minutes at room temperature, rinsed with PBS and water, and mounted with ProLong Gold Antifade reagent (Invitrogen). Autofluorescence quenching with the TrueVIEW kit (Vector Laboratories) was performed before cover slipping. Representative images were captured using a motorized, widefield fluorescence BX61 microscope with a DP80 camera and cellSens (version 1.18) Dimension software (Olympus America, Center Valley, Pennsylvania). Positive immunoreactivity for all IF was confirmed with internal mouse tissue array controls used to validate these assays. A subset of tissues incubated with antibody diluents and secondary antibody only were used as negative controls for these assays.
Transmission Electron Microscopy
Lungs from 2 representative mice were fixed with a solution of 2.5% glutaraldehyde, 4% paraformaldehyde, and 0.02% picric acid in 0.1 M sodium cacodylate buffer at pH 7.2. Samples were postfixed in a solution of 1% osmium tetroxide and 1.5% potassium ferricyanide, dehydrated through a graded ethanol series, and embedded in an Epon analog resin. Multiple semithin representative sections were prepared and mounted on glass slides for light microscopy to first identify and select regions of interest containing inclusions. Once a region of interest was selected, ultrathin sections of 60 to 80 nm in thickness were cut using a Diatome diamond knife (Diatome, Hatfield, Pennsylvania) on a Leica UltraCut S Ultramicrotome (Leica, Vienna, Austria). Sections were collected on copper grids, further contrasted with lead citrate, and viewed on a JEM 1400 electron microscope (JEOL Inc., Peabody, Massachusetts) operated at 100 kV. Images were recorded with a Veleta 2K × 2K digital camera (Olympus-SIS, Münster, Germany). Sample preparation was performed by the Weill Cornell Medicine Imaging Core Facility staff.
Results
Cm Infection in NSG Mice is Associated With Bronchointerstitial Pneumonia and Neutrophilia
Eleven of nineteen mice presented with clinical signs including weight loss, lethargy, hunched posture, and dyspnea. All mice were euthanized 3 to 4 weeks after initial exposure to Cm-shedding mice, soiled bedding, and/or other Cm-positive NSG mice. Gross abnormalities were limited to the lungs in which 17 of 19 mice showed discrete, focal to multifocal, pale pink to white foci of discoloration. The lungs of all NSG mice contained microscopic pulmonary pathology (19/19) and most animals had a leukocytosis characterized by a mild (3.17–6.50 K/μL) to moderate (6.51–15.5 K/ μL) neutrophilia (16/18) with or without a mild (0.27–0.80 K/μL) to moderate (0.81–1.76 K/μL) monocytosis (11/18) (Table 1). The most common microscopic finding was a histiocytic and neutrophilic bronchointerstitial pneumonia (17/19) with bronchiolar intraepithelial structures compatible with chlamydial inclusions (CIs) (Fig. 1a–d, Table 1). These structures were round to oval, well demarcated from the surrounding cytoplasm, measured 15 to 25 μm and were composed of a clear space containing numerous round basophilic organisms measuring approximately 0.5 to 1.0 μm in diameter (Fig. 1e). The remaining 2 mice had histiocytic and neutrophilic bronchiolitis with intraepithelial CI. The histiocytic and neutrophilic bronchointerstitial pneumonia was most often multifocal to coalescing with some cases involving an entire lung lobe (Fig. 1a). The severity of lesions varied among mice from focal inflammation and necrosis with sporadic CI, to inflammation and necrosis that involved extensive portions of the lung, up to encompassing entire lobes. MOMP immunohistochemistry (IHC) labeled the inclusions in bronchiolar epithelial cells and the interstitium of 19/19 mice (Fig. 1f, Table 1). Similarly, ISH for Chlamydia RNA highlighted inclusions in all 19 mice (Fig. 1g, Table 1). A subset of lung scrolls (N = 7/7) from IHC-positive lungs were positive for Cm by real-time qPCR (Table 1). The 23S rRNA gene fragment amplified from the scrolls contained 26,439 to 215,000 copies of the Cm 23S rRNA gene sequence. Aerobic and anaerobic cultures of the lung were negative in 14 of 18 mice. Four mice had mixed bacterial overgrowth including Ligilactobacillus murinus, Staphylococcus epidermidis, Staphylococcus hominis, and Actinomyces naeslundii. These bacteria are considered contaminants from sample processing as pulmonary lesions were not associated with other opportunistic or commensal bacterial agents.
Table 1.
Signalment, exposure, pathologic findings, tissues with Chlamydia inclusions, and ancillary tests performed in NSG mice infected with Chlamydia muridarum.
| Case Number | Sex | Agea | Exposure | Microscopic Diagnoses | Complete Blood Count Findings | Tissues with Chlamydia Inclusions (IHC and/or ISH) | Ancillary Testing |
|---|---|---|---|---|---|---|---|
| 1 | F | 12 | C | Bronchointerstitial pneumonia; Typhlocolitis | LC, N | L, SI, LI, T, NP | Isolation, IHC, ISH, qPCR |
| 2 | F | 12 | C | Bronchointerstitial pneumonia; Typhlocolitis | LC, N | L, SI, LI, T, NP, NC, ET | IHC, ISH |
| 3 | F | 12 | C | Bronchointerstitial pneumonia; Typhlocolitis | LC, N, Mo | L, SI, LI, T, NP, NC | IHC, ISH, qPCR, IFA |
| 4 | F | 12 | C | Bronchointerstitial pneumonia | LC, N | L, SI, LI, T, NP, NC, ME | IHC, ISH, qPCR |
| 5 | F | 12 | C | Bronchointerstitial pneumonia; Rhinitis | LC, N, Mo | L, SI, LI, T, NP, NC | IHC, ISH |
| 6 | F | 9 | C | Bronchointerstitial pneumonia; Rhinitis; Endometritis; Vaginitis; Salpingitis | LC, N, Mo | L, SI, LI, T, NP, V, U, O | IHC, ISH, IFA |
| 7 | F | 9 | C | Bronchointerstitial pneumonia | LC, N, Mo | L, SI, LI, T, NP, NC, ME | IHC |
| 8 | M | 9 | B | Bronchointerstitial pneumonia; Nasopharyngitis | N | L, SI, LI, T, NC | IHC |
| 9 | M | 9 | B | Bronchointerstitial pneumonia; Tracheitis | LC, N, Mo | L, SI, LI, T, ME | IHC |
| 10 | M | 9 | B | Bronchointerstitial pneumonia; Nasopharyngitis; Tracheitis | LC, N, Mo | L, SI, LI, T, NP, ET | IHC |
| 11 | M | 9 | B | Bronchointerstitial pneumonia | NA | L, SI, LI, T, NP, ET | IHC |
| 12 | F | 9 | C | Bronchointerstitial pneumonia; Rhinitis | LC, N, Mo | L, SI, LI, T, NC, ET | IHC |
| 13 | F | 9 | C | Bronchointerstitial pneumonia; Nasopharyngitis | NSF | L, SI, LI, T, NP | IHC, qPCR |
| 14 | M | 9 | B | Bronchointerstitial pneumonia; Nasopharyngitis | LC, N, Mo | L, SI, LI, T, NP | IHC, qPCR |
| 15 | M | 9 | B | Bronchiolitis; Nasopharyngitis; Tracheitis | LC, N | L, SI, LI, T, NP | IHC |
| 16 | M | 9 | B | Bronchointerstitial pneumonia; Nasopharyngitis | M | L, SI, LI, T, NP, ME | IHC |
| 17 | M | 9 | B | Bronchointerstitial pneumonia; Nasopharyngitis | LC, N, Mo | L, SI, LI, T, NP | IHC, qPCR |
| 18 | M | 9 | C | Bronchiolitis; Nasopharyngitis; Tracheitis | LC, N, Mo | L, SI, LI, T, NP, ME | IHC |
| 19 | M | 8 | C | Bronchointerstitial pneumonia; Nasopharyngitis | LC, N, Mo | L, SI, LI, T, NP, ME | IHC, qPCR, IFA, TEM |
Abbreviations: NSG, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ; IHC, immunohistochemistry; ISH, in situ hybridization; qPCR, quantitative polymerase chain reaction; F, female; C, cohousing; LC, leukocytosis; N, neutrophilia; L, lung; SI, small intestine; LI, large intestine; T, trachea; NP, nasopharynx; NC, nasal cavity; ET, Eustachian tube; Mo, monocytosis; IFA, indirect immunofluorescence assay; ME, middle ear; V, vagina; U, uterus; O, oviduct; M, male; B, soiled bedding; NA, not applicable; NSF, no significant findings; TEM, transmission electron microscopy.
At euthanasia (months).
Figure 1.
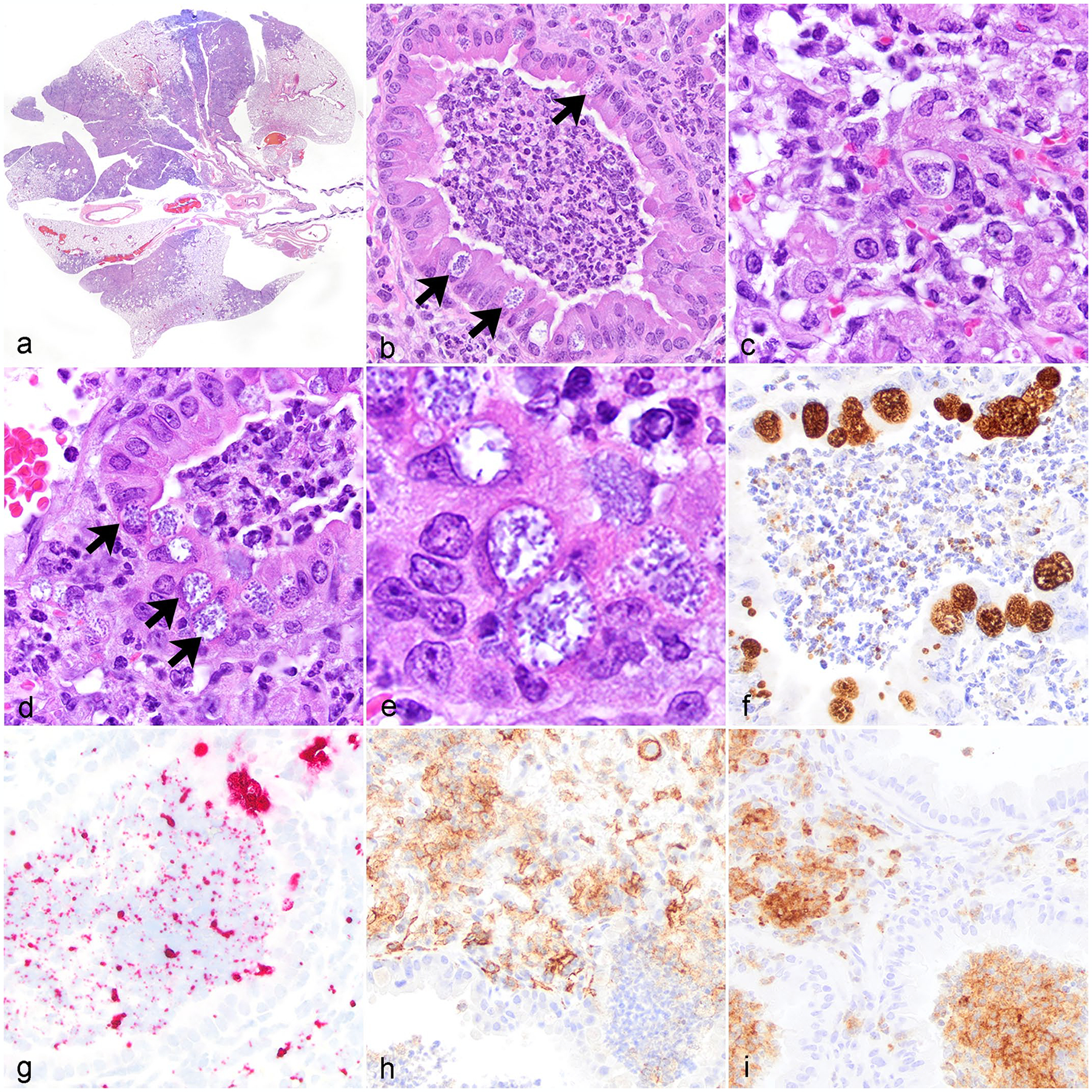
Chlamydia muridarum associated bronchointerstitial pneumonia, lung, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice. (a) Multifocal to regional bronchointerstitial pneumonia. Hematoxylin and eosin stain (HE). (b) Histiocytic and neutrophilic bronchointerstitial pneumonia with intracytoplasmic chlamydial inclusions (CIs) in bronchiolar epithelium (arrows). HE. (c) Interstitial and alveolar spaces are expanded by histiocytic and neutrophilic inflammation admixed with fibrin, pyknotic and karyorrhectic debris, and intralesional CI. HE. (d) Histiocytic and neutrophilic bronchointerstitial pneumonia with necrotic debris, degenerate neutrophils, bronchiolar epithelial degeneration, and intracytoplasmic CI in bronchiolar epithelium (arrows). HE. (e) High magnification CI within bronchiolar epithelial cells. HE. (f) Chlamydia antigen in inclusions within bronchiolar epithelial cells and the bronchiolar lumen. Immunohistochemistry (IHC) for Chlamydia major outer membrane protein antigen (MOMP). (g) Chlamydia nucleic acid in inclusions in bronchiolar epithelial cells and extracellularly the bronchiolar lumen. In situ hybridization (ISH) for Chlamydia RNA. (h) Immunolabeling of macrophages within areas of inflammation in the interstitium and peribronchiolar regions. IHC for F4/80. (i) Immunolabeling of neutrophils in bronchioles and throughout the alveolar interstitium. IHC for Ly6G.
Large numbers of distended, occasionally vacuolated, macrophages were detected by membranous and cytoplasmic expression of F4/80 IHC (Fig. 1h). Admixed with these macrophages were many viable and degenerate neutrophils, detected by Ly6G IHC, that filled both bronchiolar lumens and alveolar spaces and infiltrated the alveolar septa (Fig. 1i). Inflammatory cells were admixed with strands of fibrin, eosinophilic proteinaceous fluid interpreted as edema, and necrotic pyknotic karyorrhectic debris. Throughout the inflammatory foci, type I pneumocytes showed varying degrees of damage including necrosis with occasional type II pneumocyte hyperplasia (data not shown). Clear acicular clefts were observed in the alveolar spaces in a few cases. In a subset of severe cases, alveolar septa were markedly obliterated by foamy macrophages mixed with spindle-shaped mononuclear cells, which were negative with a Masson’s trichrome stain.
To better assess the cellular tropism of the CI within the lungs, IF for CI with F4/80 and pancytokeratin were performed in 3 mice with bronchointerstitial pneumonia (Fig. 2a–d). Chlamydial organisms were frequently localized within bronchiolar epithelium and extracellularly (Fig. 2a, b). Occasionally, CIs were within pneumocytes lining alveolar septa (Fig. 2b). With IF, CIs were not detected in macrophages, but multifocally, macrophages within alveolar spaces exhibited trace amounts of Chlamydia suggesting internalization of nonviable antigen inside macrophages (Fig. 2c, d).
Figure 2.
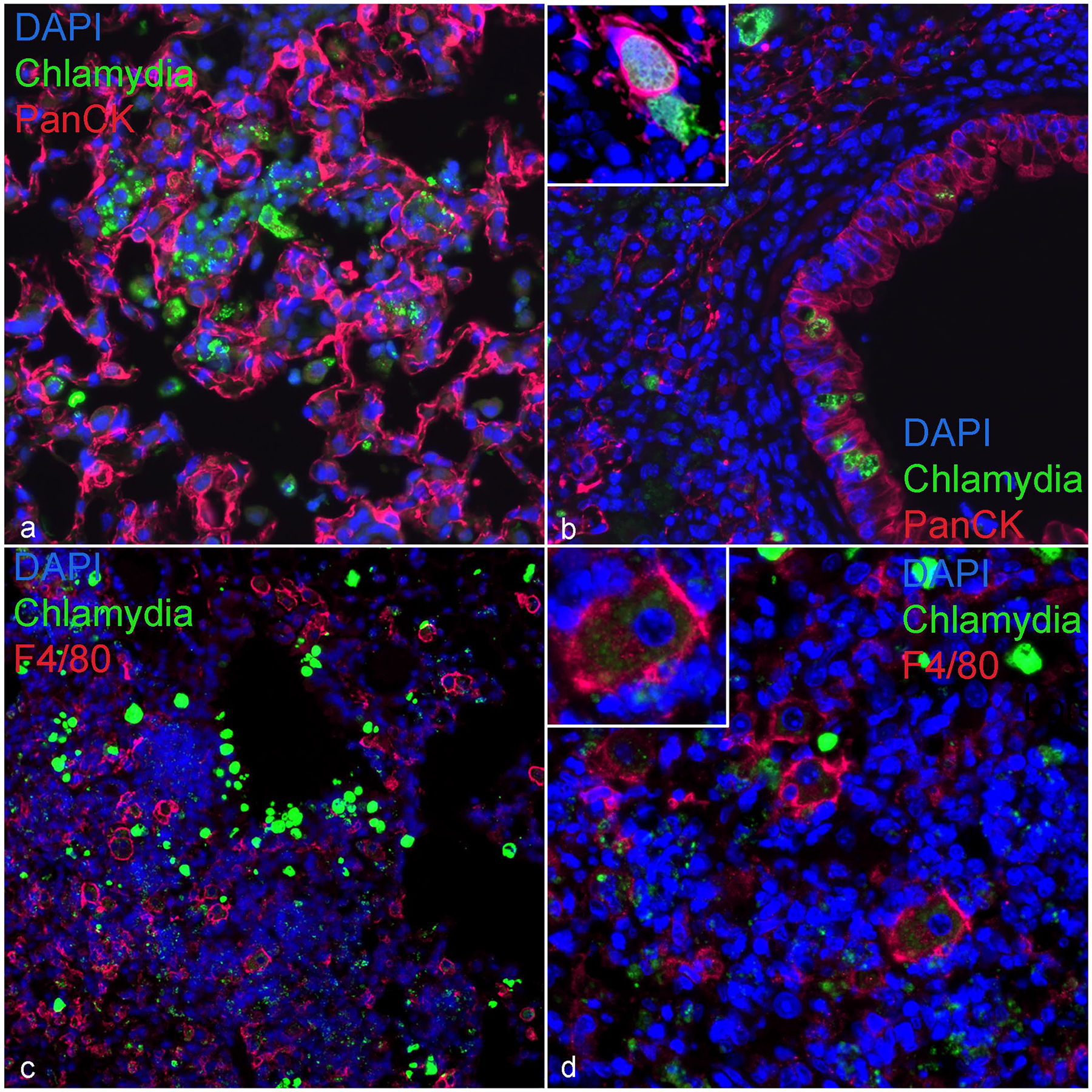
Immunofluorescence. Chlamydia muridarum inclusions in bronchiolar and alveolar epithelial cells, lung, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mouse. DAPI (blue) highlights nuclei in all figures. (a) Alveolar epithelial cells (pancytokeratin; red) associated with extracellular and intraepithelial Chlamydia (MOMP; green). (b) A bronchiole with chlamydial inclusions (CIs) (MOMP; green) in bronchiolar (pancytokeratin; red) and alveolar epithelial cells (high magnification inset), and extracellular organisms. (c) Representative section of lung highlighting CI (MOMP; green) and macrophages (F4/80; red) within areas of peribronchiolar inflammation. (d) An area of peribronchiolar inflammation composed of macrophages (F4/80; red) with intracytoplasmic specks of Chlamydia antigen (MOMP; green; high magnification inset). DAPI, 4’,6-diamidino-2-phenylindole; MOMP, major outer membrane protein antigen.
To characterize the inclusions and demonstrate the presence of reticulate and elementary bodies, TEM was performed on the lungs of 2 mice with bronchopneumonia. Ultrastructural analysis of a representative alveoli demonstrated both elementary and reticulate bodies within the cytoplasm of 2 epithelial cells and in the alveolar space (Fig. 3a–c). Within the cells, each organism was located within an inclusion membrane. Elementary bodies ranged from 200 to 300 nm in diameter and contain electron dense compacted nucleoid structures (Fig. 3b, c). Reticulate bodies were larger, measuring 500 to 700 nanometers in diameter (Fig. 3b, c). Occasionally, intermediate bodies were observed, which contained a condensed nucleoid that had not yet compacted to the dimensions of the elementary bodies.
Figure 3.
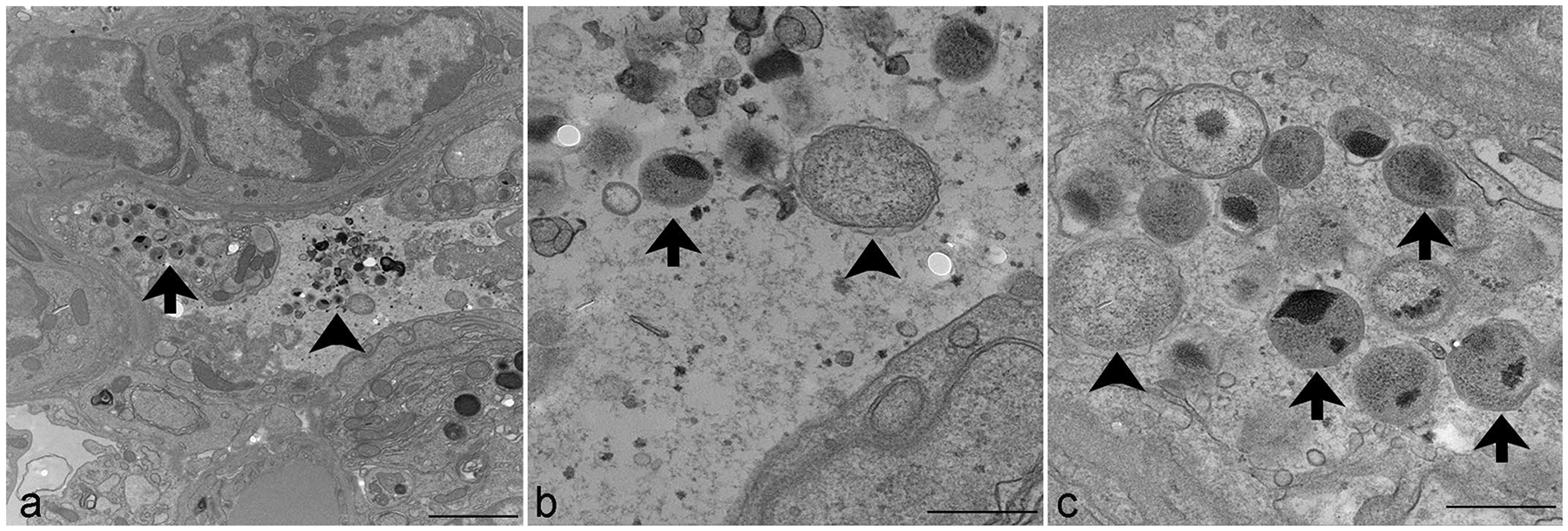
Transmission electron microscopy. Chlamydia muridarum, lung, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mouse. (a) Numerous extracellular (arrowhead) and intraepithelial (arrow) Chlamydia elementary and reticulate bodies. Scale bar = 2 μm. (b) Higher magnification of extracellular elementary bodies (arrow), measuring 200 to 300 nm in diameter, and reticulate bodies (arrowhead), measuring 500 to 700 nm in diameter. Scale bar = 500 nm. (c) Higher magnification of elementary bodies (arrows), measuring 200 to 300 nm in diameter, and reticulate bodies (arrowhead), measuring 500 to 700 nm in diameter, expanding the cytoplasm of an epithelial cell. Scale bar = 500 nm.
Cm Infection in the Upper Respiratory Tract, Ears, Reproductive Tract, and Gastrointestinal Tract
Due to prominent pulmonary lesions, the nasal cavity, nasopharynx, and trachea were thoroughly examined. Three of 19 mice had neutrophilic rhinitis with luminal exudate and intraepithelial CI (Fig. 4a, b, Table 1). Eight mice had CI noted in the nasal mucosal epithelium with IHC, with or without inflammation (Fig. 4b, Table 1). Sixteen mice had CI observed in the nasopharyngeal epithelium with IHC and 9 of these mice had associated inflammation (Fig. 4c, d, Table 1). Although CIs were observed in the trachea of all mice with IHC (19/19), only 4 had a neutrophilic tracheitis accompanied by inclusions (Fig. 4e, f, Table 1). Seven mice had otitis media consisting of histiocytic to suppurative inflammation with no epithelial inclusions noted. IHC for Cm revealed intraepithelial CI in the middle ear of 6 mice, although these were not associated with inflammation. The same IHC revealed CI within the eustachian tube epithelium in 4 mice. One of the 9 female NSG mice examined had lesions in the lower and upper reproductive tract associated with chlamydial infection including a neutrophilic vaginitis, cervicitis, endometritis, and salpingitis (Fig. 4g–j, Table 1). Each of these sites contained multifocal intraepithelial CI and positive ISH signal within epithelial cells and extracellularly (Fig. 4g–j, Table 1). The inflammation in the uterus was associated with endometrial gland ectasia, degeneration, and necrosis. In all 10 male NSG mice, histopathologic changes consistent with chlamydial infection and evidence of Chlamydia antigen in urogenital tissues were not observed.
Figure 4.
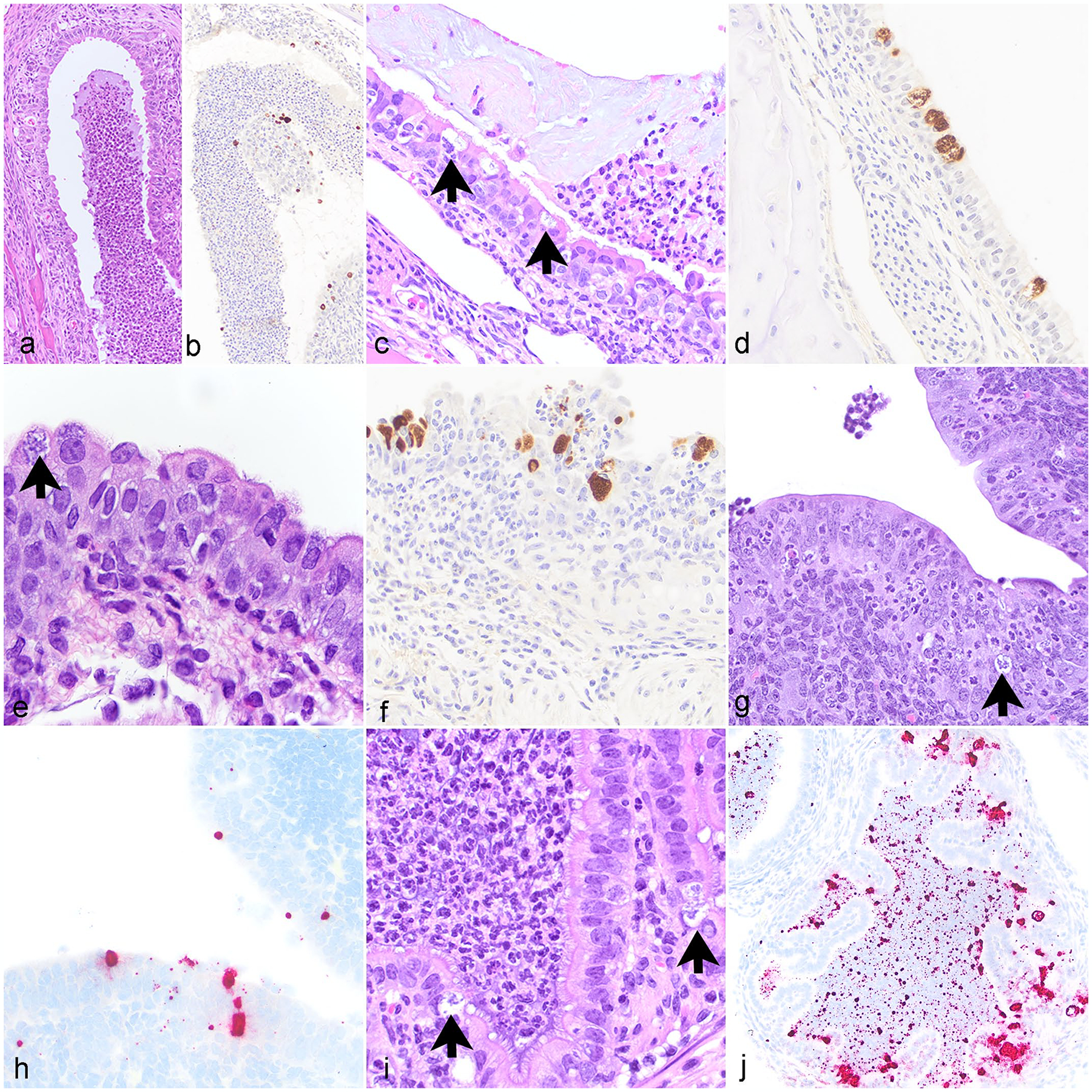
Chlamydia muridarum associated lesions in various tissues, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice. (a) Nasal cavity. Neutrophilic rhinitis with luminal neutrophilic exudate. Hematoxylin and eosin stain (HE). (b) Nasal cavity. Chlamydia antigen within epithelial cells and in luminal exudate. Immunohistochemistry (IHC) for Chlamydia major outer membrane protein antigen (MOMP). (c) Nasopharynx. Neutrophilic nasopharyngitis with intraepithelial Chlamydial inclusions (CIs) (arrows). HE. (d) Nasopharynx. Chlamydia antigen within epithelial cells. IHC for MOMP. (e) Trachea. Neutrophilic, histiocytic, and lymphocytic tracheitis with intraepithelial CI (arrow) and epithelial hyperplasia. HE. (f) Trachea. Chlamydia antigen within epithelial cells. IHC for MOMP. (g) Uterus. Neutrophilic endometritis with epithelial cell apoptosis and intralesional CI (arrow). HE. (h) Uterus. CI in epithelial cells lining the uterus. ISH for Chlamydia RNA. (i) Oviduct. Neutrophilic salpingitis with CI (arrows). HE. (j) Oviduct. CI within epithelial cells and free nucleic acid in the oviduct lumen. ISH for Chlamydia RNA.
Numerous multifocal CIs were widely distributed throughout the apical cytoplasm of cecal and colonic enterocytes in all 19 cases (Fig. 5a–d, Table 1). Similar inclusions were also observed within mucosal epithelial cells throughout the small intestinal villi (Fig. 5e, f, Table 1). These inclusions were not associated with intestinal pathology. Three mice had focal to multifocal neutrophilic and histiocytic typhlocolitis with sub-mucosal edema and crypt abscesses. These changes were not colocalized with CI. Aerobic and anaerobic culture of the cecum was undertaken in 7 of the 19 mice. Anaerobic culture was negative, while aerobic culture revealed mixed commensal enteric flora.
Figure 5.
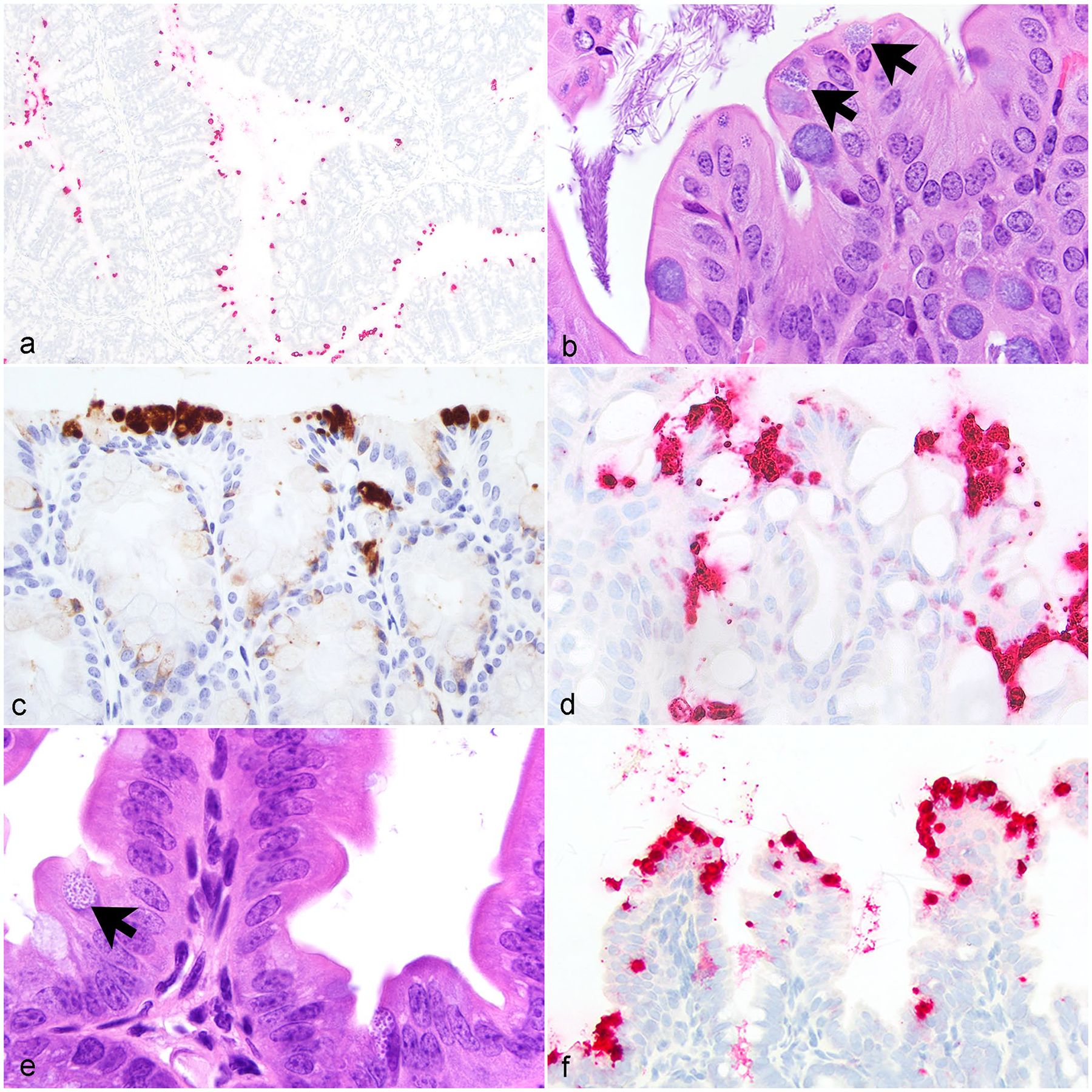
Chlamydia muridarum infection in the gastrointestinal tract, small and large intestine, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice. (a) Representative section of colon with Chlamydia nucleic acid widely distributed throughout. In situ hybridization (ISH) for Chlamydia RNA. (b) Chlamydial inclusions (CIs) within apical cytoplasm of colonic epithelium (arrows). HE. (c) Chlamydia antigen within apical cytoplasm of colonic epithelium. Immunohistochemistry (IHC) for Chlamydia major outer membrane protein antigen (MOMP). (d) Chlamydia nucleic acid within apical cytoplasm of colonic epithelium. ISH for Chlamydia RNA. (e) Normal jejunum with CI (arrow). HE. (f) CI within the apical cytoplasm of jejunum enterocytes and small amounts of free nucleic acid within the jejunum lumen. ISH for Chlamydia RNA. HE, hematoxylin and eosin stain.
Discussion
What is currently known about the pathology of Cm infection in laboratory mice is primarily extrapolated from experimental models of chlamydial infections, which often use high infectious doses and artificial routes of inoculation. Limited data exist in the literature regarding lesions associated with Cm infection when laboratory mice are exposed to Cm-shedding mice. In this study, we characterized the lesions of Cm in a severely immunodeficient mouse strain, NSG, which is commonly used in biomedical research. NSG mice were highly susceptible to Cm infection, demonstrated by the presence of CI associated with pulmonary lesions ranging from bronchiolitis to bronchointerstitial pneumonia in all exposed mice, and neutrophilia in most cases. Cm also colonized the mucosal epithelium of the small and large intestine in all NSG mice, although no lesions were observed in association with the inclusions. In addition, in a subset of cases, Chlamydia colonized the surface epithelium of the trachea, nasopharynx, nasal cavity, and middle ear with variable inflammation in these tissues.
The bronchointerstitial pneumonia and/or bronchiolitis in these NSG mice is consistent with the pulmonary lesions seen in the acute phase of Cm infection in previous studies using BALB/c and Tlr2 knockout mice following intranasal challenge.1,12,23 We have also observed similar pulmonary lesions in a subset of IL12rb2 knockout mice naturally infected with Cm in a research mouse colony at our facility (data not shown). The severity of the pulmonary lesions seen in NSG mice was likely exacerbated by the absence of CD4+ T and NK cells in this mouse strain. These immune cells are important producers of interferon-gamma (IFN-γ), which is an essential effector cytokine involved in the resolution of Cm infections in experimental models of murine chlamydiosis.25 Furthermore, NK cell depletion in mice alters pulmonary macrophage polarization from an M1 to M2 phenotype, which in turn results in higher bacterial loads and more severe pulmonary inflammation than controls during Cm infection.28 Interestingly, the first 11 mice in our experiment developed clinical disease including dyspnea, lethargy, hunched posture, and weight loss, but the subsequent 8 NSGs infected at a later timepoint had minimal to no clinical signs, even though the majority possessed pulmonary lesions. A cause for this is uncertain although it could be related to differences in the infectious dose shed by the donor mice or present in dirty bedding material used in the 2 separate cohousing experiments (November/December vs April/May) or differences in the virulence of the field Cm strains infecting the lungs of NSG mice for each experiment. Although naïve NSG mice were not included as controls for this study, and this may be a limitation, we did not observe acidophilic macrophage pneumonia, pulmonary adenomas, or Rodentibacter pneumotropicus associated pneumonia, which are diseases previously observed in our NSG colonies.20 We have also observed spontaneous alveolar histiocytosis in adult NSG mice, but these changes were often mild and not associated with tissue damage.20 In this study, the mice exhibited a mixed neutrophilic and histiocytic bronchointerstitial pneumonia associated with CI and/or extracellular elementary bodies. These microscopic findings are consistent with our ongoing experiments studying Cm biology in NSG mice.
Cm exhibited widespread colonization of the small and large intestine in all NSG mice, characterized by inclusions within the apical cytoplasm of mucosal epithelial cells, without eliciting histopathological changes in the lamina propria. The nonpathogenic colonization in the intestine of NSG mice suggests that Cm likely achieved complete fitness in the intestine, as demonstrated in other experimental murine models of Cm infection in the gastrointestinal tract.29 Despite the lack of association of Cm infection with intestinal lesions, we cannot rule out the possibility that Cm infection alters mucosal homeostasis in specific tissue compartments and/or immune cell subsets in the intestine of NSG mice. Overall, our results are consistent with phenotypes seen in immunocompetent mouse strains, such as BALB/c, DBA/2, and C57BL/6 mice infected orally.27 In these mice, Cm colonized both the small and large intestines in the absence of histopathology.27 In this gastrointestinal model, Cm is eventually cleared from the small intestine but establishes long-lasting colonization in the large intestine of mice.25 The clearance of Cm from the small intestine in mouse strains derived from a C57BL/6J background was mediated by IFN-γ produced by antigen-specific CD4+ T-cells, while Cm colonization in the large intestine was modulated by IFN-γ produced by type 3 innate lymphoid cells.12,25 In another gastrointestinal model using Rag1 knockout and Cd4 knockout mice, He et al8 showed that Chlamydia spread was mediated by CD4+ T-cells in the small and large intestines, as these cells were important in maintaining the intestinal barrier function. Recent studies using different Cm mutant strains revealed Chlamydia has plasmid-encoded proteins or chromosomal proteins that are important for its colonization in the upper gastrointestinal tract or the large intestine, respectively.29 Given that NSG mice do not have functional CD4+ T-cells, we suspect Cm may establish long-lasting colonization of the intestinal mucosa and persistent shedding in feces. Although establishing the kinetics of Cm infections in the gastrointestinal tract of NSG mice was not within the scope of this study, additional investigations may elucidate the persistence of Cm infections in tissues of NSG mice. There is a precedent as athymic nude mice inoculated intravaginally with Cm remained infected for over 260 days.19
Our results demonstrate intraepithelial cytoplasmic CI in the upper respiratory tract, lung, nasopharynx, intestines, and middle ear with multiple techniques: immunohistochemistry, ISH, immunofluorescence (IF), and electron microscopy. Previous studies have shown that Chlamydia MOMP is expressed in both elementary and reticulate bodies.2 Our MOMP IHC results also highlighted Chlamydia antigen in extracellular spaces, demonstrating infectious elementary bodies in tissues, easily noticeable in the lungs and intestines. Cm mRNA was also detected by ISH in cells and extracellular spaces and correlated well with the tissue compartments and extracellular spaces that tested positive by MOMP IHC. The presence of CI in the nasal cavity, nasopharynx, trachea, and bronchi/bronchioles suggests that the naïve NSG mice inhaled elementary bodies from mice shedding Cm in their feces. The size and high frequency of the CI in bronchiolar epithelium compared to pulmonary macrophages and the bronchiolar pattern of the pulmonary lesions suggest aerogenous spread of Cm to the lungs of NSG mice. Our ultrastructural findings are similar to findings from a previous Chlamydia pneumoniae study, in which CIs were frequently seen in bronchiolar epithelial cells and all developmental forms of chlamydial organisms were detected in the bronchiolar lumen and alveolar space, but it should be noted that this study was performed with immunocompetent Swiss Webster mice and the route of inoculation was different.26 Although Cm was detected in the nasal epithelium in NSG mice, it did not spread into the olfactory nerves as reported in experimental models of Cm infection of the central nervous system.15 In addition, the widespread distribution of CI in the intestines suggests that naïve NSG mice were likely infected by ingestion of Cm-positive fecal material from shedding mice. Recent experimental studies demonstrate that mice shedding Cm in their feces failed to auto-inoculate Cm into the genital tract and induce ascending urogenital infections.24,30 Our results showed the urogenital tract of only 1 female NSG mouse out of 9 was infected with Cm. A possible explanation for this result is related to the design of our cohousing experiment as naïve female NSG mice were not cohoused with Cm-infected male NSG mice and/or the low frequency of ascending urogenital infections in naturally infected, Cm-shedding mice. We cannot rule out the possibility that Cm infection in the genital tract of this female NSG mouse was caused by the systemic spread of Cm through the blood as demonstrated in experimental murine models of Cm.18,30
This study characterized the pulmonary pathology and extent of extrapulmonary lesions associated with Cm infection in male and female NSG mice after being cohoused with Cm-shedding mice and/or exposed to dirty bedding. Our findings illustrate the significant impact Cm infections could have in immunocompromised mouse strains if the agent is present in research mouse colonies in academic institutions. Therefore, Cm should be added as an excluded agent in standard mouse health surveillance testing panels in immunocompromised mouse colonies.
Acknowledgment
We thank the staff of the Laboratory of Comparative Pathology, in particular Sockie Jiao and Irina Dobtsis, as well as Panagiota Momtsios at Charles River Laboratories and the Weill Cornell Medicine Microscopy and Imaging Core Facility, including Lee Cohen-Gould and Juan Pablo Jimenez, for their technical assistance.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Memorial Sloan Kettering (MSK’s) Core Facilities are supported by MSK’s National Cancer Institute (NCI) Cancer Center Support Grant P30 CA008748.
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Beckett EL, Phipps S, Starkey MR, et al. TLR2, but not TLR4, is required for effective host defence against Chlamydia respiratory tract infection in early life. PLoS ONE. 2012;7(6):e39460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cevenini R, Donati M, Sambri V, et al. Reactivity of elementary and reticulate bodies of Chlamydia trachomatis LGV2 with monoclonal antibodies specific for the major outer membrane protein. FEMS Microbiol Lett. 1987;42(1):47–51. [Google Scholar]
- 3.De Clercq E, Kalmar I, Vanrompay D. Animal models for studying female genital tract infection with Chlamydia trachomatis. Infect Immun. 2013;81(9):3060–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elwell C, Mirrashidi Engel J. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol. 2016;14(6):385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foreman O, Kavirayani AM, Griffey SM, et al. Opportunistic bacterial infections in breeding colonies of the NSG mouse strain. Vet Pathol. 2011;48(2):495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gönnert R. Bronchopneumonia, a new disease of mice. Zentralbl Bakteriol B. 1941;147(3):161–174. [Google Scholar]
- 7.Gordon FB, Freeman G, Clampit JM. A pneumonia-producing filtrable agent from stock mice. Proc Soc Exp Biol Med. 1938;39(3):450–453. [Google Scholar]
- 8.He C, Xu Y, Huo Z, et al. Regulation of Chlamydia spreading from the small intestine to the large intestine via an immunological barrier. Immunol Cell Biol. 2021;99(6):611–621. [DOI] [PubMed] [Google Scholar]
- 9.He X, Nair A, Mekasha S, et al. Enhanced virulence of Chlamydia muridarum respiratory infections in the absence of TLR2 activation. PLoS ONE. 2011;6(6):e20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilleman MR. Immunological studies on the psittacosis-lymphogranuloma group of viral agents. J Infect Dis. 1945;76(2):96–114. [Google Scholar]
- 11.Institute for Laboratory Animal Research. Guide for the Care and Use of Laboratory Animals. Washington, DC: National Academies Press; 2011. [Google Scholar]
- 12.Lin H, He C, Koprivsek JJ, et al. Antigen-specific CD4+ T cell-derived gamma interferon is both necessary and sufficient for clearing Chlamydia from the small intestine but not the large intestine. Infect Immun. 2019;87(6):e00055–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marée AF, Komba M, Finegood DT, et al. A quantitative comparison of rates of phagocytosis and digestion of apoptotic cells by macrophages from normal (BALB/c) and diabetes-prone (NOD) mice. J Appl Physiol. 2008;104:157–169. [DOI] [PubMed] [Google Scholar]
- 14.Mishkin N, Ricart Arbona RJ, Carrasco SE, et al. Reemergence of the murine bacterial pathogen Chlamydia muridarum in research mouse colonies. Comp Med. 2022;72(4):230–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nazareth L, Walkden H, Chacko A, et al. Chlamydia muridarum can invade the central nervous system via the olfactory and trigeminal nerves and infect peripheral nerve glial cells. Front Cell Infect Microbiol. 2021;10:607779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien BA, Huang Y, Geng X, et al. Phagocytosis of apoptotic cells by macrophages from NOD mice is reduced. Diabetes. 2002;51:2481–2488. [DOI] [PubMed] [Google Scholar]
- 17.Perry LL, Hughes S. Chlamydial colonization of multiple mucosae following infection by any mucosal route. Infect Immun. 1999;67(7):3686–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poston TB, O’Connell CM, Girardi J, et al. T cell-independent gamma interferon and B cells cooperate to prevent mortality associated with disseminated Chlamydia muridarum genital tract infection. Infect Immun. 2018;86(7):e00143–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rank RG, Soderberg LS, Barron AL. Chronic chlamydial genital infection in congenitally athymic nude mice. Infect Immun. 1985;48(3):847–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santagostino SF, Arbona RJ, Nashat MA, et al. Pathology of aging in NOD scid gamma female mice. Vet Pathol. 2017;54(5):855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shultz LD, Lyons BL, Burzenski LM, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174(10):6477–6489. [DOI] [PubMed] [Google Scholar]
- 22.Stair MI, Carrasco SE, Annamalai D, et al. The epidemiology of invasive, multiple antibiotic resistant Klebsiella pneumoniae infection in a breeding colony of immunocompromised NSG mice. Comp Med. 2022;72(4):220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Virok DP, Raffai T, Kókai D, et al. Indoleamine 2,3-dioxygenase activity in Chlamydia muridarum and Chlamydia pneumoniae infected mouse lung tissues. Front Cell Infect Microbiol. 2019;9:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Zhang Q, Zhang T, et al. The Chlamydia muridarum organisms fail to auto-inoculate the mouse genital tract after colonization in the gastrointestinal tract for 70 days. PLoS ONE. 2016;11(5):e0155880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winner H, Friesenhahn A, Wang Y, et al. Regulation of chlamydial colonization by IFNγ delivered via distinct cells. Trends Microbiol. 2023;31:270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang ZP, Cummings PK, Patton DL, Kuo CC. Ultrastructural lung pathology of experimental Chlamydia pneumoniae pneumonitis in mice. J Infect Dis. 1994;1:464–467. [DOI] [PubMed] [Google Scholar]
- 27.Yeruva L, Spencer N, Bowlin AK, et al. Chlamydial infection of the gastrointestinal tract: a reservoir for persistent infection. Pathog Dis. 2013;68(3):88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L, Li J, Zhou X, et al. Natural killer cells regulate pulmonary macrophages polarization in host defense against chlamydial respiratory infection. Front Cell Infect Microbiol. 2022;11:775663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong G Chlamydia overcomes multiple gastrointestinal barriers to achieve long-lasting colonization. Trends Microbiol. 2021;29(11):1004–10012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong G Chlamydia spreading from the genital tract to the gastrointestinal tract – a two-hit hypothesis. Trends Microbiol. 2018;26(7):611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]


