Abstract
Background
Rare disorders comprise of ~ 7500 different conditions affecting multiple systems. Diagnosis of rare diseases is complex due to dearth of specialized medical professionals, testing labs and limited therapeutic options. There is scarcity of data on the prevalence of rare diseases in different populations. India being home to a large population comprising of 4600 population groups, of which several thousand are endogamous, is likely to have a high burden of rare diseases. The present study provides a retrospective overview of a cohort of patients with rare genetic diseases identified at a tertiary genetic test centre in India.
Results
Overall, 3294 patients with 305 rare diseases were identified in the present study cohort. These were categorized into 14 disease groups based on the major organ/ organ system affected. Highest number of rare diseases (D = 149/305, 48.9%) were identified in the neuromuscular and neurodevelopmental (NMND) group followed by inborn errors of metabolism (IEM) (D = 47/305; 15.4%). Majority patients in the present cohort (N = 1992, 61%) were diagnosed under IEM group, of which Gaucher disease constituted maximum cases (N = 224, 11.2%). Under the NMND group, Duchenne muscular dystrophy (N = 291/885, 32.9%), trinucleotide repeat expansion disorders (N = 242/885; 27.3%) and spinal muscular atrophy (N = 141/885, 15.9%) were the most common. Majority cases of β-thalassemia (N = 120/149, 80.5%) and cystic fibrosis (N = 74/75, 98.7%) under the haematological and pulmonary groups were observed, respectively. Founder variants were identified for Tay-Sachs disease and mucopolysaccharidosis IVA diseases. Recurrent variants for Gaucher disease (GBA:c.1448T > C), β-thalassemia (HBB:c.92.+5G > C), non-syndromic hearing loss (GJB2:c.71G > A), albinism (TYR:c.832 C > T), congenital adrenal hyperplasia (CYP21A2:c.29–13 C > G) and progressive pseudo rheumatoid dysplasia (CCN6:c.298T > A) were observed in the present study.
Conclusion
The present retrospective study of rare disease patients diagnosed at a tertiary genetic test centre provides first insight into the distribution of rare genetic diseases across the country. This information will likely aid in drafting future health policies, including newborn screening programs, development of target specific panel for affordable diagnosis of rare diseases and eventually build a platform for devising novel treatment strategies for rare diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13023-024-03300-z.
Keywords: Rare diseases, Diagnosis, IEM, Neuromuscular, Neurodevelopmental disorders, Lysosomal storage diseases, Prevalence, Common variant, Founder variant, India
Background
Rare diseases affect multiple organ systems and present with a range of phenotypes, majority (~ 50–75%) of them affecting children. Presently, there are ~ 7500 rare diseases for which molecular basis is known that collectively are estimated to affect ~ 300 million people worldwide [1]. Nearly 80% of all rare diseases are estimated to have an underlying genetic aetiology [2].
The diagnosis and treatment of rare genetic diseases is complex due to poor awareness among healthcare providers [3, 4], dearth of testing laboratories and unavailability of therapy for majority of them. Advancements in genomic technologies, especially, emergence of high throughput technologies like DNA microarray analysis and next generation sequencing (NGS) have identified several novel genes which are associated with rare genetic diseases that has led to improvement in diagnostic yields. In addition, international initiatives such as NIH Undiagnosed Diseases Program and Network and The International Rare Diseases Research Consortium have been undertaken [5, 6] in order to delineate aetiology of rare genetic diseases, and accelerate research for discovery of novel genetic conditions and possible therapeutics.
Statistics on the prevalence of rare genetic diseases in different populations is scarce. Walker et al. 2016 in Western Australia did one of the first pioneering attempts to estimate the collective prevalence and burden of rare diseases in a large population. The study observed that ~ 2% of the population of Western Australia is affected by a rare disease [7]. Similarly, a rare disease prevalence of 1.5% was reported in Hong Kong [8]. Interestingly, a study by Hsu et al. 2018 in Taiwan showed that the estimated prevalence of rare diseases increased at an average rate of 19.46% per year [9].
The available epidemiology data for rare diseases is mostly from data by national registries or small cohorts on single diseases or disease groups [1]. A recent study on neuromuscular patient cohort from Lebanon identified 62 rare genetic neuromuscular disorders (NMD). The group reported a high incidence of approximately 1 in 7500 births for spinal muscular atrophy (SMA) in Lebanon [10]. Likewise, a retrospective study carried out in Australia found a combined incidence of lysosomal storage disorders (LSDs) to be approximately 1 per 4,800 live births, with Fabry disease observed as the most common LSD [11].
India has the largest population in the world comprising of 4600 population groups which is stratified into several tribes and castes based on the socio-cultural background and geographical location [12, 13]. The strict inbreeding practices among certain communities has resulted in high prevalence of several genetic rare diseases and associated founder variants [14]. Despite this, limited information is presently available on the prevalence of rare genetic diseases in India due to the lack of a centralized patient registry, diagnostic facility, affordable therapeutic interventions and awareness. The Foundation for Research on Rare Diseases and Disorders has estimated that about 70 million people are affected with rare genetic diseases in India [15]. The Indian Council of Medical Research (ICMR) has recently set up a National Registry for Rare Diseases, which compiles epidemiological data on rare diseases and estimates that there are 4,001 identified rare diseases [16]. This data and the observations by several groups working on rare diseases in India have shown the most prevalent among rare diseases to be haematological disorders, lysosomal storage disorders (LSDs), neuromuscular and neurodegenerative disorders (NMND), primary immunodeficiency diseases (PID) and mitochondrial diseases [17]. For example, Yadav et al. 2022 has shown a high prevalence of β-thalassemia (β-thal) trait in central India, ranging between 1.4 and 3.4% [18]. Likewise, the prevalence of haemophilia A in India is estimated to be 0.9 per 100,000 people [19]. Furthermore, recent studies have estimated the incidence of SMA to be 1 in 10,000 newborns [20] and that of cystic fibrosis (CF) to be between 1 in 40,000 and 1 in 100,000 [21].
Considering the large and heterogeneous population in India, studying the epidemiology of rare genetic disorders is important in order to provide appropriate, timely, cost-effective diagnosis and targeted therapeutic interventions to affected patients. Till date, no single study has assessed the collective prevalence or burden of different rare diseases at a national/ state or a tertiary centre level. The present study provides a 22-year retrospective overview of a cohort of patients with rare genetic diseases identified at a tertiary genetic centre in India. To the best of our knowledge this is a first of its kind systematic study within the Indian population.
Methods
Patient cohort detail
This is a retrospective study carried out at FRIGE Institute of Human Genetics, a tertiary referral centre, from 2000 to 2022. Patients with a strong clinical presentation, positive primary investigations and a confirmatory molecular or biochemical test were included in the present study. The institutional ethics committee of FRIGE’s Institute of Human Genetics approved this study. All procedures performed in studies were in accordance with the ethical morals of the 1975 Helsinki declaration.
For all patients, 4–5 millilitres of peripheral blood sample was collected in an EDTA vacutainer after obtaining an informed consent. This was used for isolating genomic DNA using the salting-out method [22]. Genomic DNA was quantified using QIAexpert (Qiagen, Germany) and stored at -20 ֯C until further molecular studies. For patients suspected with inborn errors of metabolism, in addition to the peripheral blood sample, urine and/or plasma samples were also collected. For the purpose of biochemical investigations in patients suspected with LSDs, leukocytes were isolated from whole blood.
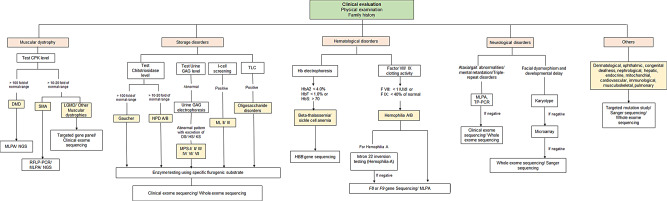
Briefly, 1 ml of blood sample was added to acid-citrate-dextrose (ACD) solution and incubated for 1 h. The supernatant containing the leukocyte was collected in a separate vial and subjected to centrifugation to settle the pellet. Two rounds of normal saline washes were given to the pellet and stored at -20 ֯C until further testing. The line of investigation strategy utilized for the diagnosis was dictated by the preliminary clinical suspicion or the type of rare genetic disorder suspected by the referring clinician (Fig. 1).
Fig. 1.
Overview of the diagnostic test pathway used for testing rare genetic diseases. CPK = creatine phosphokinase; DMD = Duchenne muscular dystrophy; SMA; spinal muscular atrophy; LGMD = Limb girdle muscular dystrophy; MLPA = Multiplex ligation probe dependent amplification; NGS = Next generation sequencing; RFLP-PCR = Restriction fragment length polymorphism- polymerase chain reaction; GAG = Glycosaminoglycan; NPD = Niemman-Pick disease; MPS = Mucopolysaccharidosis; ML = Mucolipidosis; TP-PCR = Triplet prime repeat polymerase chain reaction
Muscular dystrophy
Patients with a primary suspicion of muscular dystrophy were first tested for their serum creatine phosphokinase (CPK) levels [23]. Patient samples with elevated CPK levels were then processed for molecular testing of DMD/ Becker muscular dystrophy (BMD). For this, DNA samples were subjected to multiplex PCR to detect hemizygous deletion in the DMD gene [24]. Reflex testing by multiplex ligation-dependent probe amplification (MLPA) was performed in cases whereby hemizygous deletion in the DMD gene was not observed [25] in accordance with the manufacturer’s protocol of SALSA MLPA probemix P034-A3/P035-A3 DMD/Becker (MRC Holland, Netherlands). Patient samples that remained undiagnosed after carrying out the above tests and those with a clinical suspicion of muscular dystrophies were subjected to NGS based test (Details provided in the following section titled clinical exome sequencing/ whole exome sequencing).
Motor neuron disease
For patients with a primary suspicion of SMA, their DNA was subjected to Restriction fragment length polymorphism (RFLP)-PCR as per the protocol described previously [26] or MLPA using SALSA MLPA Probe mixes; P021-A2 or P021-B1 (MRC-Holland, Netherlands). DNA of patients suspected with spinal and bulbar muscular atrophy were subjected to Sanger sequencing to assess the trinucleotide CAG repeats in the AR gene as described by Spada et al. 1991 [27].
IEM disorders
Preliminary screening tests were performed for different LSDs based on the clinical differential diagnosis as described in Fig. 1. Plasma chitotriosidase testing was performed using 4-methyl umbelliferyl (4-MU) fluorometric assay [28, 29]. Glycosaminoglycan levels in urine samples were tested (quantitative and qualitative) in MPS suspected patients [30]. I-cell screening and thin layer chromatography was performed from plasma and urine sample, respectively, using previously described methodology [31].
Enzyme assay from leukocytes and/ or plasma of screening positive patients was performed by standard protocol for a given enzyme using 4-MU fluorometric assay or para-nitrocatechol sulfate (PNCS) spectrophotometric synthetic substrate as outlined previously [32, 33].
Molecular genetic study in patients with a positive enzymatic assay result was carried out by Sanger sequencing. The region of interest was amplified using specific primers designed with Primer3 tool (https://bioinfo.ut.ee/primer3-0.4.0/) [34]. Following this, bi-directional Sanger sequencing using ABI SeqStudio platform (Thermo Fisher Scientific, USA) was performed. List of genes for which Sanger sequencing was performed is mentioned in Additional file 1.
For the detection of other IEM disorders like amino acid disorders, organic acidemias and fatty acid oxidation disorders, clinical exome sequencing (CES) study was performed as described in the following section. Patients with IEM disorders identified by tandem mass spectrometry (TMS) or gas chromatography mass spectrometry (GCMS) were excluded from the present study.
Haematological disorders
A preliminary Hb electrophoresis test was performed in patients with a suspicion and/or family history of β-thalassemia and/or sickle cell anaemia [35]. DNA samples of patients with positive Hb electrophoresis test result were processed for the detection of variants in the HBB gene by Sanger sequencing using exon specific primers (Additional file 1).
Factor VIII/ IX clotting activity was assessed in plasma samples of patients suspected with haemophilia. Molecular genetic study was performed for the detection of the common variant in haemophilia A patients: intron 22 inversion of the F8 gene using the inverse PCR protocol described previously [38]. Sanger sequencing using exon-specific primers was performed for the detection of other variants in the F8 and F9 gene (Additional file 1).
Trinucleotide repeat expansion disorders
For the molecular genetic diagnosis of myotonic dystrophy type I and type II and Fragile-X, triplet repeat primed PCR followed by fragment length analysis was performed as per the protocol described previously [39, 40]. End-point PCR was performed for the diagnosis of Huntington’s disease, Friedreich’s ataxia and spinocerebellar ataxias (1, 2, 3, 6, 7, 12 and 17) using region specific primers [41–44].
Cystic fibrosis
For the detection of the common variant c.1521_1523del in the CFTR gene, ARMS-PCR protocol as described previously was performed in all patients that were referred with a clinical suspicion of cystic fibrosis [45]. If the patient was not detected with the CFTR:c.1521_1523del variant, Sanger sequencing using exon-specific primers was performed for the detection of causative variants in the CFTR gene (Additional file 1).
Clinical exome sequencing/ whole exome sequencing
For patients with unusual clinical presentation and/ or a strong clinical suspicion for a rare genetic disease which are mentioned above, clinical exome sequencing (CES) was performed. Whole exome sequencing was performed in cases with overlapping, complex and uncertain phenotypes. CES included exon and splice site regions of > 6670 genes associated with known inherited diseases, captured using a custom capture kit (Additional file 2). Paired-end 150 bp sequencing was carried out on Illumina sequencing platforms (Illumina, USA) in accordance with the manufacturer’s protocols, with an average coverage of the target regions with ~ 100x. For whole exome sequencing (WES), genomic DNA of the proband was subjected to selective capture and sequencing of the protein coding regions that included exons and exon-intron boundaries of genes using either Agilent SureSelect v6 enrichment kit (Agilent, USA) or Twist Human Core Exome kit (Twist Biosciences, USA). The prepared library was subjected to paired-end sequencing with a mean coverage of ~ 100x on the Illumina HiSeq 2500 or NovaSeq 6000 platform (Illumina, USA).
Sequences from CES or WES obtained as FASTQ files were aligned using BWA MEM v0.7.12 [46] to the human reference genome (GRCh37/hg19). SNVs and indels were called using GATK Haplotype caller [47]. Additionally, copy number variants (CNVs) were detected from the data using the ExomeDepth v1.1.10 [48]. Variant annotation, filtration and prioritization was performed based on a given set of Human Phenotype Ontology coded phenotype terms using Exomiser using hiPHIVE prioritisation methodology [49]. Common variants were filtered based on minor allele frequency > 1% in the 1000Genome Phase 3, TopMed and/or gnomAD databases. Only non-synonymous variants in the coding region and canonical splice site variants with a depth of > 20x were used for analysis and clinical correlation. Various in-silico prediction tools such as PolyPhen-2 [50], SIFT [51], MutationTaster2 [52] and CADD [53] were used to predict pathogenicity of non-synonymous and indel variants. Post-gross filtering, variants were prioritised based on the following: (a) known disease causing variant previously reported in databases like ClinVar, and; (b) novel variants in known genes based on the Z-score for missense and pLOF or LOEUF score for loss of function variants available in the gnomAD database. In the case of candidate CNVs, variants were primarily screened for population frequency and known disease associations using publicly available databases like gnomAD, DGV, DECIPHER and OMIM. All the variants identified were classified in accordance with the American College of Medical Genetics-American College of Pathologists (ACMG-AMP) and ClinGen classification system [54, 55].
Results
Patient cohort details
A total of 7481 patients were referred for genetic test between the year 2000 and 2022. Of this, 3294 patients, including 2141 males (65%) and 1153 females (35%) were diagnosed with a rare genetic disorder. The age of the patients at the time of diagnosis ranged from 1 day to 74 years. Table 1 provides age-group wise distribution of all patients along with the male to female ratio under each group. State-wise distribution of positive cases based on their referral clinic is presented in Additional file 3. The highest number of diagnosed cases were referred from Gujarat (N = 1601; 48.6%), followed by Maharashtra (N = 825; 25%), Karnataka (N = 226; 6.9%), Kerala (N = 164; 4.9%) and Punjab (N = 163; 4.9%). A poor representation of only 33 diagnosed cases was from the eastern states of the country- namely West Bengal, Odisha, and Chhattisgarh.
Table 1.
Stratification of disease groups by age and sex
| Disease group | < 1 year | > 1 to 12 years | > 12 to 18 years | > 18 years and above | Male/ Female |
Total |
|---|---|---|---|---|---|---|
| Inborn errors of metabolism | 504 | 1404 | 42 | 42 | 1270/722 | 1992 |
| Neuromuscular and neurodegenerative disorder | 87 | 395 | 47 | 356 | 627/258 | 885 |
| Haematological | 7 | 121 | 2 | 19 | 96/53 | 149 |
| Pulmonary | 44 | 28 | 1 | 2 | 43/32 | 75 |
| Musculoskeletal | 18 | 23 | 0 | 2 | 25/18 | 43 |
| Dermatological | 6 | 12 | 2 | 10 | 13/17 | 30 |
| Congenital hearing loss | 0 | 12 | 0 | 17 | 10/19 | 29 |
| Ophthalmic | 0 | 4 | 0 | 16 | 9/11 | 20 |
| Endocrine | 4 | 14 | 0 | 2 | 10/10 | 20 |
| Hepatic | 6 | 6 | 0 | 3 | 12/3 | 15 |
| Nephrological | 1 | 0 | 3 | 11 | 11/4 | 15 |
| Mitochondrial | 0 | 5 | 2 | 5 | 9/3 | 12 |
| Cardiological | 2 | 0 | 0 | 3 | 3/2 | 5 |
| Immunological | 0 | 4 | 0 | 0 | 3/1 | 4 |
| Total | 679 | 2028 | 99 | 488 | 2141/1153 | 3294 |
Technological impact on diagnostic yield
Overall, 305 genetic diseases in 3294 patients were identified in the present study cohort from years 2000 to 2022. The disorders were classified into 14 groups based on the major organ system or organ affected (Table 2). These included neuromuscular and neurodevelopmental disorder (NMND), inborn errors of metabolism (IEM), haematological, pulmonary, musculoskeletal, dermatological, congenital hearing loss, ophthalmic, endocrine, hepatic, nephrological, mitochondrial, cardiological and immunological. Majority of the genetic disorders identified in the present study were under the NMND group (D = 149), followed by the IEM group with 47 disorders. Majority of the disorders identified in the study were autosomal recessive (D = 180, 59.2%) followed by autosomal dominant (D = 107, 35.1%,). X-linked disorders constituted 5.7% of the total cases (D = 16) and two disorders showed mitochondrial mode of inheritance (Additional file 4 and 5).
Table 2.
Stratification of diagnosed patients by disease groups across 22 year period
| 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | 2020 | 2021 | 2022 | Total cases | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inborn errors of metabolism | 3 | 10 | 10 | 19 | 9 | 15 | 13 | 39 | 39 | 38 | 60 | 71 | 81 | 98 | 124 | 184 | 186 | 176 | 188 | 176 | 90 | 176 | 187 | 1992 |
| NMND | 19 | 24 | 41 | 26 | 51 | 69 | 61 | 63 | 61 | 71 | 38 | 82 | 63 | 86 | 130 | 885 | ||||||||
| Haematological | 8 | 16 | 20 | 22 | 22 | 16 | 9 | 8 | 7 | 3 | 4 | 4 | 7 | 3 | 149 | |||||||||
| Pulmonary | 5 | 2 | 9 | 3 | 4 | 4 | 12 | 6 | 7 | 1 | 1 | 4 | 8 | 9 | 75 | |||||||||
| Musculoskeletal | 1 | 2 | 1 | 2 | 4 | 2 | 8 | 2 | 2 | 8 | 11 | 43 | ||||||||||||
| Dermatological | 1 | 1 | 2 | 2 | 1 | 2 | 1 | 5 | 4 | 1 | 10 | 30 | ||||||||||||
| Congenital deafness | 1 | 1 | 1 | 3 | 3 | 10 | 10 | 29 | ||||||||||||||||
| Ophthalmic | 1 | 1 | 5 | 2 | 6 | 5 | 20 | |||||||||||||||||
| Endocrine | 1 | 1 | 2 | 2 | 2 | 2 | 4 | 4 | 2 | 20 | ||||||||||||||
| Hepatic | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 2 | 3 | 15 | ||||||||||||||
| Nephrological | 2 | 2 | 3 | 3 | 3 | 2 | 15 | |||||||||||||||||
| Mitochondrial | 1 | 3 | 1 | 1 | 1 | 1 | 2 | 2 | 12 | |||||||||||||||
| Cardiological | 1 | 1 | 3 | 5 | ||||||||||||||||||||
| Immunological | 1 | 2 | 1 | 4 | ||||||||||||||||||||
| Total | 3 | 10 | 10 | 19 | 9 | 15 | 13 | 39 | 63 | 72 | 126 | 121 | 161 | 197 | 217 | 272 | 274 | 263 | 247 | 285 | 182 | 313 | 374 | 3294 |
| Total cases referred | 10 | 27 | 24 | 37 | 28 | 41 | 45 | 145 | 192 | 180 | 303 | 341 | 367 | 403 | 546 | 592 | 583 | 635 | 618 | 662 | 415 | 517 | 770 | 7481 |
| Diagnostic yield (%) | 30.0 | 37.0 | 41.7 | 51.4 | 32.1 | 36.6 | 28.9 | 26.9 | 32.8 | 40.0 | 41.6 | 35.5 | 43.9 | 48.9 | 39.7 | 45.9 | 47.0 | 41.4 | 40.0 | 43.1 | 43.9 | 60.5 | 48.6 |
It is noteworthy that before year 2015, the proportion of patients were mainly identified under the IEM, NMND and haematological group (Table 2; Additional file 6 and 7). It is only after the introduction of CES and WES based tests in 2015 that a significant number of patients with rare genetic diseases were identified under the NMND, congenital hearing loss, dermatological, hepatic and nephrological disorder groups. Additionally, introduction of the NGS technology impacted the diagnostic yield as we observed an increase from 30% in 2000 to 49.1% in 2022 (Table 2; Additional file 6 and 7) with an average diagnostic rate of 40.8% was achieved in the present study cohort throughout the 22-year period. A total of 186 cases and 134 cases were diagnosed using CES and WES, respectively, in the present study. Critically, whilst NMND group constituted on an average 60.5% of all cases diagnosed across 14 disease groups each year using CES/WES between 2015 and 2022, the proportion of all NMND cases diagnosed with these technologies increased from 15.9% in 2015 to 54.6% in 2022. This suggests a significant rise in detection of rare neurological and neurodevelopmental disorders that were missed previously by conventional approaches (Additional file 5–8).
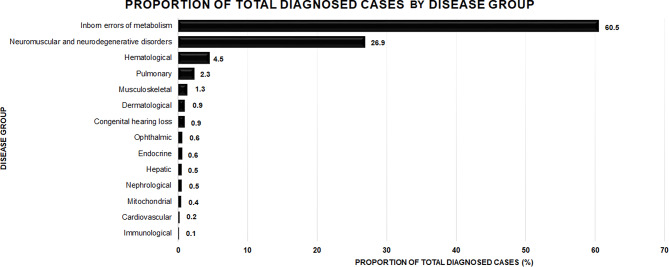
Genetic disorder prevalence by disease groups
In the present cohort, we observed highest number of cases diagnosed under the IEM (N = 1992; 60.5%) and NMND (N = 885; 26.9%) groups. Figure 2 gives the percentage distribution of patients under the 14 disease groups. Interestingly, majority of the patients diagnosed under the NMND group were referred from Gujarat whereas, patients diagnosed under the IEM group were referred from across India (Additional file 3) as our centre represents one of the major referral centres for lysosomal storage disorders across the country. Excluding the NMND and IEM group, 13% of the total diagnosed cases were distributed across the remaining 12 disease groups. Major centres that referred these patients were from Gujarat followed by Maharashtra and Rajasthan (Additional file 3).
Fig. 2.
Percentage distribution of patients with rare genetic diseases diagnosed under the 14 disease groups
The number of patients diagnosed with DMD (OMIM#310200; N = 291/885; 32.9%) and SMA (N = 141/885; 15.9%) were the highest under the NMND group. Notably, 242 patients (27%) were diagnosed with one of the trinucleotide repeat expansion disorders, the highest of which was spinocerebellar ataxia (SCA) (N = 85/885; 9.6%) (Table 3). SCA2 cases (OMIM#183090) were the most common amongst SCA types. Less common muscular dystrophies identified in this cohort included limb-girdle muscular dystrophy (LGMD) (N = 23/885, 2.6%) with causative variants identified in 6 different genes (Additional file 4). Spastic paraplegia 11 (OMIM#604360) was the most common (N = 6/12; 50%) among hereditary spastic paraplegia in the present study. In total, 82 patients were diagnosed with neurodevelopmental disorders (Additional file 4). Among IEM disorders, majority of the patients were diagnosed with Gaucher disease (N = 224/1992, 12%) followed by other LSDs (Table 3) and small molecule IEM disorders (Additional file 5).
Table 3.
Common genetic variants identified across rare genetic disorders
| Disorder group | Disease name | OMIM | Number of patients | Gene | Common mutation identified | Percentage of cases |
|---|---|---|---|---|---|---|
| Neuromuscular and neurodevelopmental disorders | Duchenne muscular dystrophy | #310200 | 291 | DMD | Exon 45 to exon 52 deletion | 58.2% (N = 169/291) |
| Spinal muscular atrophy | #253300 | 141 | SMN1 | Exon 7 and exon 8 deletion | 100% (N = 141/141) | |
| Myoyonic dystrophy type I | #160900 | 82 | DMPK | NA | NA | |
| Spinocerebellar ataxia 2 | #183090 | 38 | ATXN2 | NA | NA | |
| Spinocerebellar ataxia 3 | #109150 | 29 | ATXN3 | NA | NA | |
| Spinocerebellar ataxia 1 | #164400 | 10 | ATXN1 | NA | NA | |
| Huntington’s disease | #143100 | 37 | HTT | NA | NA | |
| Friedrich ataxia | #229300 | 19 | FXN | NA | NA | |
| Fragile-X | #300624 | 19 | FMR1 | NA | NA | |
| Limb girdle muscular dystrophy type 1 | #253600 | 8 | CAPN3 | No common variant identified | NA | |
| GNE myopathy | #605820 | 9 | GNE | c.2179G > A (p.Val727Met) | 100% (N = 9/9) *Heterozygous | |
| Spastic paraplegia 11 | #604360 | 6 | SPG11 | c.267G > A (p.Trp89Ter) | 50% (N = 3/6) | |
| Inborn errors of metabolism | Gaucher disease | #230800 | 224 | GBA | c.1448T > C (p.Leu483Pro) | 60% (N = 134/223) |
| MPS IVA | #253000 | 209 | GALNS | c.230C > G (p.Pro77Arg) | 56% (N = 14/25) | |
| Niemann-Pick disease A/B | #257200/ #607616 | 167 | SMPD1 | No common variant identified | NA | |
| GM1 gangliosidosis | #230500 | 141 | GM1 | No common variant identified | NA | |
| MPS II | #309900 | 133 | IDS | No common variant identified | NA | |
| Metachromatic leukodystrophy | #250100 | 136 | ARSA | No common variant identified | NA | |
| Tay-Sachs disease | #272800 | 126 | HEXA | c.1385A > T (p.Glu462Val ) | 22% (N = 11/48) | |
| #272800 | HEXA | c.1278insTATC | 16% (N = 8/48) | |||
| #272800 | HEXA | c.964G > T (p.Asp322Tyr) | 6% (N = 3/48) | |||
| Mucolipidosis II/III | #252500/ #252600 | 79 | GNPTAB |
c.3503_3504delTC (p.Leu1168GlnfsTer5) |
34% (N = 12/35) | |
| Hematologic disorders | Beta-thalassemia | #613985 | 120 | HBB |
c.92 + 5G > C, c.316 − 149_*342delinsAAGTAGA, c.126_129del, c.27dupG, c.92 + 1G > T |
85% (N = 102/120) |
| Hemophilia A | #306700 | 16 | F8 | Intron 22 inversion | 42% (N = 5/12) | |
| Pulmonary disorders | Cystic fibrosis | #219700 | 74 | CFTR | c.1521_1523delCTT (deltaF508) | 66% (N = 49/74) |
| Musculoskeletal disorders | Achondroplasia | #100800 | 21 | FGFR3 | c.1138G > A | 71% (N = 15/21) |
| FGFR3 | c.1138G > C | 29% (N = 6/21) | ||||
| Osteogenesis imperfecta type I/II/III/IV |
#166200, #166210, #259420, #166220 |
1 | COL1A1 | No common variant identified | NA | |
| Osteogenesis imperfecta type I/II/III/IV |
#166200, #166210, #259420, #166220 |
2 | COL1A2 | No common variant identified | NA | |
| Osteogenesis imperfecta type V | #610967 | 2 | IFITM5 | No common variant identified | NA | |
| Progressive pseudorheumatoid dysplasia | #208230 | 3 | CCN6 | c.298T > A (p.Cys100Ser) | 100% (N = 3/3) | |
| Dermatological disorders | Oculocutaneous albinism type IA/ IB | #203100/ #606952 | 20 | TYR | c.832C > T (p.Arg278Ter) | 30% (N = 6/20) |
| Congenital ichthyosis-1 | #242300 | 4 | TGM1 | No common variant identified | ||
| Congenital hearing loss | Deafness, autosomal recessive | #220290 | 8 | GJB2 | c.71G > A (p.Trp24Ter) | 37% (N = 3/8) |
| Usher syndrome type IIA | #276901 | 4 | USH2A | NA | NA | |
| Ophthalmic disorders | Retinitis pigmentosa-54 | #613428 | 3 | PCARE | No common variant identified | NA |
| Endocrinological disorders | Congenital adrenal hyperplasia | #201910 | 16 | CYP21A2 | c.29–13C > G | 43% (N = 7/16) |
| Hepatic disorders | Gilbert syndrome | #143500 | 13 | UGT1A1 | A [TA]7 TAA | 46.2% (N = 6/13) |
| Crigler-Najjar syndrome, type I | #218800 | |||||
| Nephrological disorders | Alport syndrome | #301050 | 5 | COL4A5 | c.2918-11G > A | 40% (N = 2/5) |
| Mitochondrial disorders | Leber’s Hereditary Optic Neuropathy | #535000 | 3 | MTND4 | G11778A | 100% (N = 3/3) |
| Leigh syndrome | #500017 | 3 | MTATP6 | No common variant identified | NA |
We observed majority of the patients diagnosed with β-thalassemia (OMIM#613985; N = 120/149; 80.5%), cystic fibrosis (OMIM#219700; N = 74/75; 98.7%) and achondroplasia (OMIM#100800; N = 21/43; 48.8%) under the haematological, pulmonary, and musculoskeletal disease groups, respectively (Table 3). Likewise, oculocutaneous albinism type IA (OMIM#203100; N = 6/30; 20%), congenital hearing loss due to biallelic GJB2 gene variant (OMIM#220290; N = 8/29; 27.6%), retinitis pigmentosa (N = 5/20; 25%) and congenital adrenal hyperplasia (CAH) (OMIM#201910; N = 16/20; 80%) represented the highest cases under the dermatological, congenital deafness, ophthalmic and endocrine groups, respectively. Gilbert syndrome (OMIM#143500; N = 6/15; 40%) and Alport syndrome (N = 5/15; 33.3%) were common disorders identified under the hepatic and nephrological groups, respectively (Table 3).
Variant distribution amongst genetic disorders
Assessment of the genotype and variant details of the patients diagnosed with rare genetic disorders revealed commonly occurring and founder variants for particular disorders. In 58.2% (N = 169) of the patients diagnosed with DMD disease, hemizygous deletion encompassing exons 45 to exon 52 of the DMD gene was observed. Another common variant c.1448T > C (p.L483P) in the GBA gene was detected in approximately 60% (N = 134) of all patients diagnosed with Gaucher disease. Table 3 summarises information on the common variants detected in corresponding genes in patients diagnosed with rare disorders under the 14 disease groups. In addition to this, 82 distinct variants in 63 genes were identified in patients with neurodevelopmental disorders (Additional file 4). Variant and genotype details of patients diagnosed under haematological, pulmonary, musculoskeletal, dermatological, congenital hearing loss, ophthalmic, endocrine, hepatic, nephrological, mitochondrial, cardiological and immunological groups have been described in Additional file 5.
Discussion
In recent years, increased use of DNA sequencing in clinical settings has resulted in identifying the genetic basis of several rare genetic diseases [2, 56]. Improvement in technology has contributed towards development of high throughput sequencing technologies, at an affordable cost [57]. Coupled with this, several therapeutic options are now available for some of these rare diseases (Table 4). Currently, there are 35 major clinical research institutions in India, including our institute, which are working on different rare disease groups [17]. Commonly addressed among these are dermatological disorders, mitochondrial disorders, neurological, neuromuscular, LSDs, IEMs, haematological disorders, musculoskeletal and ophthalmic disorders. The aim of the present study was to understand the burden and genetic epidemiology of these rare genetic diseases diagnosed at a tertiary genetic centre, in India over a period of 22 years.
Table 4.
Therapeutic modalities currently available for rare genetic diseases
| Treatment modality | Principle | Conditions for which there is clinical approval | Reference |
|---|---|---|---|
| Substrate reduction therapy |
Inhibiting the biosynthesis of Storage metabolites in the lysosomes by using small molecules. |
Gaucher disease, Fabry disease, Niemann-Pick type C, MPS-IIIB | [124] |
| Enzyme replacement therapy |
Recombinant enzymes that are modified to provide a longer half-life, more potent activity, resistance to degradation or targeting to a specific organ, tissue or cell type |
Gaucher disease, Fabry disease, MPS I, MPS II, Pompe disease, MPS VI, Wolman disease, Batten disease, MPS IVA, MPS VII and α-mannosidosis | [125] |
| Oligonucleotide therapies | Targeting RNA to reduce the production of a specific disease-associated protein by promoting degradation of its mRNA. |
SMA and DMD (Nusinersen, Eteplirsen) |
[126] |
| Gene therapy |
A vector is used to express a transgene (with the endogenous sequence or codon optimized) that encodes the desired protein, under the control of an appropriate promoter |
SMA, hemophilia A, hemophilia B, adrenoleukodystrophy, β-thalassemia, sickle cell disease | [127] |
| Hematopoietic stem cell therapy | The ability of the transplanted cells and/or their progeny to contribute to fixed-tissue macrophage populations in the affected tissues and to become local permanent sources of functional lysosomal enzymes. | MPS I, MPS II, metachromatic leukodystrophy, Krabbe, β-thalassemia, sickle cell disease, Gaucher disease, MPS IVA, epidermolysis bullosa | [128] |
| Iron chelation therapy | To maintain safe levels of body iron at all times, by balancing iron intake from blood transfusion with iron excretion by chelation | β-thalassemia | [129] |
Neuromuscular and neurological disease group
A high burden of DMD and SMA cases that were documented in the present cohort was also observed previously in a neuromuscular cohort study in Lebanon, demonstrating SMA in 40.3%, followed by DMD in 17% of its patients [10]. Of note, a recent study by Nilay et al. 2020 showed carrier frequency of 1 in 38 for SMA in the North Indian population [20], higher than that previously reported in the US, Australia, Europe and UK [58, 59]. This is probably one of the reasons for the high SMA cases in the country and in the present cohort. Interestingly, the mutation spectrum seen for DMD in the present cohort is similar to that described previously in the DMD global database, with exon 45 deletion identified in 68% cases, being the most common [60]. Likewise, exon 7–8 deletion of the SMN1 gene was reported in 90.7% of the total SMA cases in the NMD cohort in Lebanon [10], which is in concordance with our present observations and earlier study [61].
Of note, we observed CAPN3 and DYSF as recurrently mutated genes among the LGMD patients in our cohort. This is in congruence with the observations of Nallamilli et al. whereby 17% and 16% of the 4656 LGMD patients presented with variant(s) in the CAPN3 and DYSF genes, respectively [62]. Importantly, the founder variant c.2338G > C in the CAPN3 gene, previously reported in the Indian Agarwal community [63], was seen in only two patients in a compound heterozygous state in the present cohort, suggesting that the mutation spectrum for CAPN3 gene is likely to be distinct among different sub-populations in the country. Contrary to this, variant c.2179G > A (p.Val727Met) in the GNE gene was observed in all patients with GNE myopathy (OMIM#605820) in the present study in compound heterozygous state. This variant has previously been reported to be a common variant amongst Indian patients of GNE myopathy [64]. This adds further evidence of probability of a founder effect for GNE myopathy in India, although haplotype analysis is beyond the scope of the current study.
Trinucleotide repeat expansion disorders
Of the NMND disease group, trinucleotide repeats expansion disorders were identified in 27% cases (N = 242). A significantly high proportion of SCA cases, particularly SCA2 (N = 37) in the present cohort is likely to be due to the founder effect [65]. Similarly, whilst the SCA12 founder mutation has been reported in the Indian Agarwal community [66], a significantly low numbers of SCA12 cases were observed in the present study. This could be due to limited number of referrals from the eastern geographical region of the country, where the Agarwal community predominantly resides. Following the study of Bhowmik et al. 2016, we present here the largest series of patients (N = 82) with myotonic dystrophy type I from India [67]. Likewise, we observed a significant number of Huntington disease cases in the present cohort. This observation is supported by another study where Chheda et al. 2018 showed a high prevalence of 49% in 503 pan-India patients suspected with Huntington disease [68]. The average range of CAG repeats on the expanded allele of the HTT gene observed in the Indian patients with Huntington’s disease is 41–59 [69], which was also observed in present cohort.
Neurodevelopmental disorders
Strikingly high proportion of neurodevelopmental disorder (NDD) cases were reported in a recent population based study across five regions in India [70]. The high proportion of NDD cases observed in the present cohort further supports prior observation. Furthermore, we observe an increase in detection of NDD due to the use of NGS based CES/ WES approaches, with causative variants in genes associated with autosomal dominant or X-linked phenotypes [71, 72]. Primary mode of inheritance for these rare NDDs is de novo with the variant reported to be occurring on the paternal allele in majority of the cases [73] However, it is beyond the scope of the present study to record the paternal age at proband’s conception. Interestingly, the genes in which causative variants were identified in present study encode transcriptional and chromatin regulators, translation initiator factors, ion channels, synaptic proteins and neuronal migration machinery, which is in congruence with the reported literature [71, 74].
Inborn errors of metabolism group
The distribution of different LSDs in the present study is similar to that previously reported by our and other groups in the country [75, 76], with Gaucher disease being the most common LSD followed by the MPS disease group. Also, common variants identified in present study cohort for LSDs like Gaucher disease, MPS I, MPS II and Mucolipidosis II/III is in concordance with data reported by other groups in India [77–79]. Critically, several variants for Niemann-Pick A/B, GM1 gangliosidosis, Krabbe, Tay-Sachs and MPS IVA were enriched in certain sub-populations, with two variants being shown to be founder variants from 2 communities in Gujarat [80, 81].
Pilot studies for newborn screening across India detected high prevalence of small molecule IEMs such as glucose-6-phosphate dehydrogenase (G6PD) deficiency, amino acid disorders and organic acidemias [82–84]. However, low proportion of patients with this disease were detected on the current study which is likely to be due to exclusion of cases diagnosed with tandem mass spectrometry and/or gas chromatography mass spectrophotometry only.
Haematological group
India contributes significantly to the global burden for β-thalassemia with estimates suggesting that approximately 32,400 children each year are born with hemoglobinopathies. Interestingly however, we observed a higher proportion of β-thalassemia carriers (75%) compared to affected cases (13%) in the present cohort. Indeed, the proportion of overall cases stratified in the haematological group increased from 0% in 2000–2004, peaked at 11.7% in 2010–2014 and reduced to 1.6% in 2020–2022 (Additional file 6). This inverted U shaped curve follows the national and state government initiatives on increasing disease awareness of thalassemia which led to the paradigm shift from proband diagnosis to parental screening and prenatal diagnosis. Additionally, the multicentre Jai Vigyan programme established by the Indian Council of Medical Research helped to strengthen screening testing centres in different states for community control of thalassemia [85]. Moreover, the number of centres offering testing services for β-thalassemia has increased over the past decade, which is also a probable reason for the poor representation of these cases at our centre. Nonetheless, the mutation spectrum observed in these patients was similar to that reported by other groups in the country, with c.92 + 5G > C being the most common HBB gene variant [86].
Pulmonary group
Population prevalence estimates of 1 in 43,321 to 1 in 100,323 have been carried out previously for cystic fibrosis (CF) [87]. The present cohort had 98% (N = 74) of patients diagnosed with CF under the pulmonary group. Of all the variants reported in the CFTR gene, 66% of the patients with CF were detected with the CFTR:c.1521_1523del variant, which has an estimated frequency of 19–34% in the Indian CF patients [88, 89]. This observation suggests that patients suspected with CF could be tested for CFTR:c.1521_1523del variant only in the beginning, due to the high probability of detection, with a subsequent reflex testing for the entire gene if the variant is not detected in the patient.
Musculoskeletal group
A high proportion of patients in the musculoskeletal group with a diagnosis of achondroplasia or osteogenesis imperfecta in the present cohort is in congruence with prior observations in Indian patients [90]. Of note, three unrelated patients but belonging to the same ethnic community- Gujarati Patni- were diagnosed with progressive pseudo rheumatoid dysplasia (PPD; OMIM#208230) due to the presence of the same variant c.298T > A in the CCN6 gene [91]. However, the aforementioned variant was not detected in a large series of 79 patients with PPD from southern India, which primarily consisted of two common variants c.233G > A and c.1010G > A in the CCN6 gene [92]. This suggests possible founder variants in ethnic communities residing in two distinct geographical locations, however, the assessment of the same is beyond the scope of the current study.
Dermatological group
Observation of oculocutaneous albinism type 1 A (OMIM#203100) being commonly observed dermatological group disease in the current study is in congruence with the observation by another study from India [93]. Pathogenic variants in the TYR gene were detected in 84.7% of the total cases in their cohort, which is similar to observation in present study. A founder variant c.832C > T in the TYR gene, reported in the Tilli population in eastern India was also observed at a high frequency (27%) in the present study cohort. Surprisingly, this variant has been reported in several other populations [94, 95] indicating that this is a recurrently occurring variant across multiple ethnic populations. Although, it is possible that this variant has also accumulated as a regional founder mutation because of its high allele frequency in the gnomAD database for the South Asian population as compared to African and European populations.
Congenital hearing loss group
More than 125 genes are associated with non-syndromic hearing loss, of which 70 genes have been associated with autosomal recessive non-syndromic hearing loss (AR-NSHL). GJB2-related AR-NSHL is the most common genetic aetiology in Asian and European population [96]. This is also evident by the highest percentage of GJB2 gene variants identified in the present study cohort. Furthermore, a probable founder variant c.71G > A in the GJB2 gene [97] was detected in 26% of the patients in our cohort. This variant is also a common variant in the Pakistani population as well as in the Roma population of Slovakia probably because of close correlation to the Indian origin of these populations [98].
Ophthalmic group
Retinitis pigmentosa (RP) has been reported as the most common ophthalmic group disorder in Indian population previously [99]. A comparative study to assess the prevalence of inherited retinal dystrophies (IRDs) in patients from India and USA [100] showed RP, Leber congenital amaurosis (LCA) and Stargardt’s disease to be the most common IRDs. Whilst patients within ophthalmic group represented a small proportion of the overall number of patients across the 14 disease groups, majority of the patients within ophthalmic group were diagnosed with RP and LCA. Critically, a significant proportion of patients with RP had variants in the PCARE and RPE65 genes, which is in congruence with prior literature observation [100].
Endocrine disorder group
Congenital adrenal hyperplasia due to 21-hydroxylase deficiency (CAH; OMIM#201910) represented the largest proportion of patients within endocrine disorder group, similar to the prior report from India [82]. Of note, regional variation in the CAH prevalence has been noted in India and prior genetic study in these patients presented c.293–13 C > G variant in the CYP21A2 gene as a common variant; similar observation was made in our cohort too [101, 102]. This presents an opportunity to assess the common variant on the outset followed by CYP21A2 gene sequencing as reflex test, if required.
Hepatic disease group
Gilbert syndrome (OMIM#143500) has been reported to be the most common hereditary unconjugated hyperbilirubinemia due to a common promoter variant (TA)7/7 of the UGT1A1 gene [103]. Within the hepatic disease group of the current study, 46% (N = 6/13) of the patients presented with the aforementioned variant during genetic test. Critically, stronger association between the presence of homozygous (TA)7/7 allele in the UGT1A1 gene and risk of Gilbert syndrome has been observed in Indian population compared to other ethnic groups [104, 105]. This in part could explain the high incidence of Gilbert syndrome within the current cohort.
Nephrological group
Inherited kidney diseases account for ~ 10–15% of adult patients undergoing kidney replacement therapy [106, 107]. In present cohort, among patients with nephrological disorders, 33% of cases were diagnosed with Alport syndrome followed by polycystic kidney disease (PKD). Previously, Alport syndrome and PKD have been shown to be a common cause of hereditary nephropathy in an Indian cohort of patients with chronic renal failure as a key phenotypic feature [96]. Additionally, Alport syndrome was also one of the common genetic diagnoses (14%) in a cohort of 76 Indian children suspected with kidney disorders [108]. This suggests an overall high prevalence of Alport syndrome in patients with chronic kidney diseases in India.
Mitochondrial disease group
An interesting observation within this group was that majority of the adult onset disease patients presented with variants in the mtDNA genes compared to nuclear DNA genes, which is in concordance with prior literature observation [109]. Critically, several recent studies have shown a high diagnostic yield in patients suspected with mitochondrial disease using WES [110]. However, not all patients suspected of mitochondrial disease were subjected to WES, which in turn could likely explain the low diagnostic yield within the mitochondrial disease group.
Cardiological group
Inherited cardiac disorders, broadly classified into cardiomyopathies and channelopathies have a prevalence of 3% worldwide [111]. The American Heart Association guidelines for the diagnosis and treatment of hypertrophic cardiomyopathy (HCM) in 2011 recommended genetic testing for the management of HCM as ~ 40% of the cardiomyopathies are likely to be genetic in origin [112]. Literature evidence suggests that a causative variant is identified in genes encoding a sarcomeric protein in up to 60% of individuals with HCM, with MYBPC3 or MYH7 being the most common genes [113]. Likewise, available data from India showed that variants in the MYH7, TNNT2 and MYBPC3 genes accounted for one third of the total reported variants [114]. Currently however, clinical application of genetic testing in cardiology practice especially in cases with sudden cardiac death is at a nascent stage in comparison to that in paediatric clinics for metabolic and other neurological conditions. This likely to be one of the reasons for the poor presentation of this group in present cohort. Despite our small cohort of 5 patients, we could also identify one patient with a pathogenic variant in the MYBPC3 gene. Thus, increase in uptake of genetic testing is necessary to delineate the genetic architecture for Indian patients with cardiomyopathy.
Immunological disease group
The present cohort had a very poor representation of inherited immunological disorders. One contributing factor can be lack of awareness of inherited immunological disorders, especially among general practitioners, paediatricians and adult specialists. The second critical reason could be high mortality rate in these patients [115] before a diagnosis is achieved. Hence, there is a dearth of referrals for genetic testing in patients presenting with recurrent and severe infections, as most of them are likely misdiagnosed and treated as cases of infectious diseases [116]. Recent review from India suggests X-linked Agammaglobulinemia, severe combined immunodeficiency and Wiskott-Aldrich syndrome to be commonly observed immunodeficiency disorders [116]. With increase in awareness and subsequent update in genetic testing there will likely be improved epidemiological estimates of these diseases in India.
Future perspectives
Several countries have used different approaches such as administrative hospitalization data or cohort-based study statistics to estimate the prevalence of rare diseases [117]. The present retrospective data here provides information on the distribution of different genetic conditions diagnosed at a tertiary genetic clinic over 22 years’ duration. The data shows a high prevalence of monogenic conditions namely DMD, SMA, β-thalassemia and sickle cell anaemia, as well as, LSDs such as Gaucher disease and MPS disorders. Recently, a national rare disease registry established by the ICMR, Government of India has included all of the aforementioned disorders to assess the national burden. The awareness of different rare genetic disorders, its prevalence, the available diagnostic tests and treatment options is important in order to help draft future health policies, including newborn screening programs as well as choice of diseases for screening in premarital/ preconception stages.
Secondly, information on founder and common mutations identified for several diseases within certain ethnic communities in the present study cohort can be used for the development of affordable, targeted and scalable genetic testing pathways, similar to that developed for the Ashkenazi Jewish population [118]. In contrast, application of “hypothesis free” based assays such as DNA microarray and NGS is critical for genetic diagnosis of disorders with a broad genetic aetiology, such as NDD, intellectual disability and autism spectrum disorder [119]. Even targeted gene panels could be critical in improvement of diagnostic yields in diseases with overlapping phenotypes but different genetic aetiologies such as LSDs [120].
In view of upcoming treatment modalities available for several rare diseases, it is imperative to have molecular epidemiology data of these patients. Lastly, prevention by prenatal genetic screening in families with a history of rare genetic diseases has gained importance and molecular epidemiological data could help in providing targeted interventions in certain communities and/or geographical regions with a high burden of genetic disorders.
Limitations
Despite the unbiased retrospective methodology used in the present study, several limitations are to be noted. First, a high proportion of patients were diagnosed with LSDs, which is likely to be due to referral bias since the institute is one of the national referral centres for LSDs. Despite this, several impactful work on the distribution of LSD burden, diagnostic assay development and therapeutic interventions in India have been carried out by the institute [121]. Second, due to referral bias, referrals for patients suspected with other disease groups were significantly fewer compared to LSDs. Due to this, accurate and reliable estimates of disease burden pertaining to these disease groups would be untenable. Nonetheless, majority of the observations on disease prevalence and molecular epidemiology are in congruence with the prior literature data as described above. Third, patients with chromosomal abnormalities, sporadic cancer and newborns with inborn errors of metabolism diagnosed only with TMS/ GCMS were excluded. Since their aetiology is primarily sporadic or de novo in origin, accurate estimation of disease incidence and burden within the population is beyond the scope of the current study. In contrast, a high proportion of patients with an autosomal recessive disorder within the current cohort could be attributed to the practice of endogamy amongst several ethnic groups in India [12, 122, 123]. Fourth, genetic diagnosis of a given disorder is based upon the sensitivity, specificity and availability of certain genetic diagnostic assays that are available at a given time period. The availability application of NGS based assay at the institute post 2015 suggests underestimation of NDD within the current cohort. Nonetheless, the genetic architecture of the NDD disease patients within the current cohort is in congruence with the data from other populations, suggesting a shared genetic aetiology of these diseases.
Conclusion
The present 22 years retrospective study of patients diagnosed with rare genetic diseases at a tertiary genetic centre in India, showed the distribution of genetic diseases under different disorder groups. We observed high burden of IEM based disorders such as LSDs followed NMND group consisting of DMD and SMA. The data provides valuable insights into plausible avenues of implementation of affordable diagnostic pathways, implementation of NGS based assays in improving the diagnostic yield of NDD, development of treatment modalities for commonly observed variants and deployment of awareness, genetic counselling and disease prevention programs in communities with a high burden of a certain genetic disorder. With the high number of diseases being observed with a common genetic aetiology, efforts could be directed for research in avenues of novel drug development for these disorders. Lastly, collaboration among the clinical, research and diagnostic communities is needed to address the challenges of diagnosis, management, treatment and prevention of rare diseases in the country.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to all the patient families for their kind co-operation and permission.
Abbreviations
- IEM
Inborn errors of metabolism
- LSDs
Lysosomal storage disorders
- NMND
Neuromuscular and neurodevelopmental disorders
- CES
Clinical exome sequencing
- WES
Whole exome sequencing
- TMS
Tandem mass spectrometry
- GCMS
Gas chromatography mass spectrometry
Authors’ contribution
Conceived and designed experiments: Jayesh Sheth, Harsh Sheth, Frenny Sheth Patient recruitment and clinical analysis: Jayesh Sheth, Frenny Sheth, Koumudi Godbole, Chaitanya Datar, Sheela Nampoothiri, Inusha Panigrahi, Anupriya Kaur, Seema Kapoor, Neerja Gupta, Sunita Bijarnya-Mahay, Ajit Gandhi, Ashish Bavdekar, Sandeep Kadam, Premal Naik, Maulin Shah, Dhaval Solanki, Soham Desai, Anand Iyer, Ketan Patel, Harsh Patel, Raju C Shah, Shalmi Mehta, Ruchi Shah, Bhagyadhan Patel, Sudhir Shah, Heli Shah, Shalin Shah, Siddharth Shah, Nilam Thaker, Umesh Kalane, Mahesh Kamte, Vykunta Raju KN, Mamta Muranjan, Shruti Bajaj, Naresh Tayade, Sujatha Jagadeesan, Deepika Jain, Mitesh Chandanara, Jitendra Singh, Sanjiv Mehta, Nishith Vora, Beena Suresh. Molecular genetic testing: Jayesh Sheth, Harsh Sheth, Frenny Sheth, Tejasvi Dhondekar, Manali Ajagekar, Aadhira Nair, Riddhi Bhavsar, Jhanvi Shah, Mili Pandya Biochemical testing: Jayesh Sheth, Aadhira Nair, Riddhi Bhavsar Writing the first draft and data collation: Jayesh Sheth, Tejasvi Dhondekar, Manali Ajagekar, Aadhira Nair, Riddhi Bhavsar, Jhanvi Shah, Mili Pandya, Harsh Sheth Data analysis: Aadhira Nair, Manali Ajagekar, Tejasvi Dhondekar, Harsh Sheth, Jayesh Sheth.
Funding
We sincerely acknowledge the research grant by the Indian Council of Medical Research (BMS: 54/2010), ICMR-DHR-NTF (GIA/ 31 (ii)/ 2014-DHR), Department of Biotechnology, Government of India (BT/PR4112/MED/12/654/2014) and (BT/PR39587/MED/12/851/2020) and Gujarat State Biotechnology Mission (GSBTM/JDR and D/608/2020/459–461) for funding this work. We also acknowledge the generous contribution towards free genetic diagnosis offered to several patients by the FRIGE-Bakeri Rare Disease Fund which is supported jointly by the Foundation for Research in Genetics and Endocrinology and the Bakeri Group. The funders had no role in the study design, sample collection and data analysis, decision to publish or preparation of the manuscript.
Data availability
All data supporting the findings of this study are available within the paper and it’s Supplementary Information.
Declarations
Ethics approval and consent to participate
All procedures followed were in accordance with the ethical standards of the institutional ethics committee of FRIGE’s Institute of Human Genetics (Reg No- E/13237) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients before inclusion in the study.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jayesh Sheth, Email: jayesh.sheth@frige.co.in.
Harsh Sheth, Email: harsh.sheth@frige.co.in.
References
- 1.Nguengang Wakap S, Lambert DM, Olry A, Rodwell C, Gueydan C, Lanneau V, et al. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur J Hum Genet EJHG. 2020;28(2):165–73. 10.1038/s41431-019-0508-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee CE, Singleton KS, Wallin M, Faundez V. Rare genetic diseases: Nature’s experiments on Human Development. iScience. 2020;23(5):101123. 10.1016/j.isci.2020.101123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramalle-Gómara E, Domínguez-Garrido E, Gómez-Eguílaz M, Marzo-Sola ME, Ramón-Trapero JL. Gil-de-Gómez J. Education and information needs for physicians about rare diseases in Spain. Orphanet J Rare Dis. 2020;15(1):18. 10.1186/s13023-019-1285-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandeborne L, van Overbeeke E, Dooms M, De Beleyr B, Huys I. Information needs of physicians regarding the diagnosis of rare diseases: a questionnaire-based study in Belgium. Orphanet J Rare Dis. 2019;14(1):99. 10.1186/s13023-019-1075-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gahl WA, Mulvihill JJ, Toro C, Markello TC, Wise AL, Ramoni RB, et al. The NIH Undiagnosed Diseases Program and Network: applications to modern medicine. Mol Genet Metab. 2016;117(4):393–400. 10.1016/j.ymgme.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuehn BM. NIH’s Undiagnosed Diseases Program reports on successes, challenges. JAMA. 2011;306(24):2657–8. 10.1001/jama.2011.1836 [DOI] [PubMed] [Google Scholar]
- 7.Walker CE, Mahede T, Davis G, Miller LJ, Girschik J, Brameld K, et al. The collective impact of rare diseases in Western Australia: an estimate using a population-based cohort. Genet Med off J Am Coll Med Genet. 2017;19(5):546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu ATG, Chung CCY, Wong WHS, Lee SL, Chung BHY. Healthcare burden of rare diseases in Hong Kong - adopting ORPHAcodes in ICD-10 based healthcare administrative datasets. Orphanet J Rare Dis. 2018;13(1):147. 10.1186/s13023-018-0892-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu JC, Wu HC, Feng WC, Chou CH, Lai ECC, Lu CY. Disease and economic burden for rare diseases in Taiwan: a longitudinal study using Taiwan’s National Health Insurance Research Database. PLoS ONE. 2018;13(9):e0204206. 10.1371/journal.pone.0204206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Megarbane A, Bizzari S, Deepthi A, Sabbagh S, Mansour H, Chouery E, et al. A 20-year clinical and genetic neuromuscular cohort analysis in Lebanon: An International Effort. J Neuromuscul Dis. 2022;9(1):193–210. 10.3233/JND-210652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin SJ, Fuller M. Prevalence of lysosomal storage disorders in Australia from 2009 to 2020. Lancet Reg Health West Pac. 2022;19:100344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moorjani P, Thangaraj K, Patterson N, Lipson M, Loh PR, Govindaraj P, et al. Genetic evidence for recent Population mixture in India. Am J Hum Genet. 2013;93(3):422–38. 10.1016/j.ajhg.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majumder PP, Basu A. A genomic view of the peopling and population structure of India. Cold Spring Harb Perspect Biol. 2014;7(4):a008540. 10.1101/cshperspect.a008540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakatsuka N, Moorjani P, Rai N, Sarkar B, Tandon A, Patterson N, et al. The promise of discovering population-specific disease-associated genes in South Asia. Nat Genet. 2017;49(9):1403–7. 10.1038/ng.3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rare Diseases India [Internet]. [cited 2024 Jul 20]. https://www.rarediseasesindia.org/
- 16.ICMR Rare Disease Registry [Internet]. [cited 2024 Jul 19]. https://rdrdb.icmr.org.in/registry/
- 17.GUaRDIAN Consortium, Sivasubbu S, Scaria V. Genomics of rare genetic diseases-experiences from India. Hum Genomics. 2019;14(1):52. 10.1186/s40246-019-0215-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yadav SS, Panchal P, Menon KC. Prevalence and management of β-Thalassemia in India. Hemoglobin. 2022;46(1):27–32. 10.1080/03630269.2021.2001346 [DOI] [PubMed] [Google Scholar]
- 19.Kar A, Phadnis S, Dharmarajan S, Nakade J. Epidemiology & social costs of haemophilia in India. Indian J Med Res. 2014;140(1):19–31. [PMC free article] [PubMed] [Google Scholar]
- 20.Nilay M, Moirangthem A, Saxena D, Mandal K, Phadke SR. Carrier frequency of SMN1-related spinal muscular atrophy in north Indian population: the need for population based screening program. Am J Med Genet A. 2021;185(1):274–7. 10.1002/ajmg.a.61918 [DOI] [PubMed] [Google Scholar]
- 21.da Silva Filho LVRF, Zampoli M, Cohen-Cymberknoh M, Kabra SK. Cystic fibrosis in low and middle-income countries (LMIC): a view from four different regions of the world. Paediatr Respir Rev. 2021;38:37–44. [DOI] [PubMed] [Google Scholar]
- 22.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. 10.1093/nar/16.3.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okinaka S, Kumagai H, Ebashi S, Sugita H, Momoi H, Toyokura Y, et al. Serum creatine phosphokinase. Activity in progressive muscular dystrophy and neuromuscular diseases. Arch Neurol. 1961;4:520–5. 10.1001/archneur.1961.00450110050006 [DOI] [PubMed] [Google Scholar]
- 24.Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16(23):11141–56. 10.1093/nar/16.23.11141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lalic T, Vossen RHAM, Coffa J, Schouten JP, Guc-Scekic M, Radivojevic D, et al. Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet. 2005;13(11):1231–4. 10.1038/sj.ejhg.5201465 [DOI] [PubMed] [Google Scholar]
- 26.Dastur RS, Gaitonde PS, Khadilkar SV, Udani VP, Nadkarni JJ. Correlation between deletion patterns of SMN and NAIP genes and the clinical features of spinal muscular atrophy in Indian patients. Neurol India. 2006;54(3):255–9. 10.4103/0028-3886.27147 [DOI] [PubMed] [Google Scholar]
- 27.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352(6330):77–9. 10.1038/352077a0 [DOI] [PubMed] [Google Scholar]
- 28.Aerts JM, van Breemen MJ, Bussink AP, Ghauharali K, Sprenger R, Boot RG, et al. Biomarkers for lysosomal storage disorders: identification and application as exemplified by chitotriosidase in Gaucher disease. Acta Paediatr Oslo nor 1992. 2008;97(457):7–14. [DOI] [PubMed] [Google Scholar]
- 29.Sheth J, Sheth F, Oza N, Gambhir P, Dave U, Shah R. Plasma chitotriosidase activity in children with lysosomal storage disorders. Indian J Pediatr. 2009;77:203–5. 10.1007/s12098-009-0249-0 [DOI] [PubMed] [Google Scholar]
- 30.Hopwood JJ, Harrison JR. High-resolution electrophoresis of urinary glycosaminoglycans: an improved screening test for the mucopolysaccharidoses. Anal Biochem. 1982;119(1):120–7. 10.1016/0003-2697(82)90674-1 [DOI] [PubMed] [Google Scholar]
- 31.Sheth J, Mistri M, Kamate M, Vaja S, Sheth FJ. Diagnostic strategy for mucolipidosis II/III. Indian Pediatr. 2012;49(12):975–7. 10.1007/s13312-012-0247-6 [DOI] [PubMed] [Google Scholar]
- 32.Shapiro LJ, Aleck KA, Kaback MM, Itabashi H, Desnick RJ, Brand N, et al. Metachromatic leukodystrophy without arylsulfatase A deficiency. Pediatr Res. 1979;13(10):1179–81. 10.1203/00006450-197910000-00021 [DOI] [PubMed] [Google Scholar]
- 33.Sheth J, Patel P, Sheth F, Shah R. Lysosomal storage disorders. Indian Pediatr. 2004;41(3):260–5. [PubMed] [Google Scholar]
- 34.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wajcman H, Moradkhani K. Abnormal haemoglobins: detection & characterization. Indian J Med Res. 2011;134(4):538–46. [PMC free article] [PubMed] [Google Scholar]
- 36.Soloviov OO, Pampukha VM, Livshits LA. Development of ARMS PCR tests for detection of common CFTR gene mutations. Biopolym Cell. 2010;26(5):378–83. 10.7124/bc.00016C [DOI] [Google Scholar]
- 37.Mishra KK, Patel P, Bhukhanvala DS, Shah A, Ghosh K. A multiplex ARMS PCR approach to detection of common β-globin gene mutations. Anal Biochem. 2017;537:93–8. 10.1016/j.ab.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 38.Rossetti LC, Radic CP, Larripa IB, De Brasi CD. Genotyping the hemophilia inversion hotspot by use of inverse PCR. Clin Chem. 2005;51(7):1154–8. 10.1373/clinchem.2004.046490 [DOI] [PubMed] [Google Scholar]
- 39.Singh S, Zhang A, Dlouhy S, Bai S. Detection of large expansions in myotonic dystrophy type 1 using triplet primed PCR. Front Genet. 2014;5:94. 10.3389/fgene.2014.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajan-Babu IS, Chong SS. Triplet-repeat primed PCR and Capillary Electrophoresis for characterizing the Fragile X Mental Retardation 1 CGG repeat hyperexpansions. Methods Mol Biol Clifton NJ. 2019;1972:199–210. 10.1007/978-1-4939-9213-3_14 [DOI] [PubMed] [Google Scholar]
- 41.Akbas F, Erginel-Unaltuna N. DNA testing for Huntington disease in the Turkish population. Eur Neurol. 2003;50(1):20–4. 10.1159/000070854 [DOI] [PubMed] [Google Scholar]
- 42.Ciotti P, Di Maria E, Bellone E, Ajmar F, Mandich P. Triplet repeat primed PCR (TP PCR) in molecular diagnostic testing for Friedreich ataxia. J Mol Diagn JMD. 2004;6(4):285–9. 10.1016/S1525-1578(10)60523-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basu P, Chattopadhyay B, Gangopadhaya PK, Mukherjee SC, Sinha KK, Das SK, et al. Analysis of CAG repeats in SCA1, SCA2, SCA3, SCA6, SCA7 and DRPLA loci in spinocerebellar ataxia patients and distribution of CAG repeats at the SCA1, SCA2 and SCA6 loci in nine ethnic populations of eastern India. Hum Genet. 2000;106(6):597–604. 10.1007/s004390000320 [DOI] [PubMed] [Google Scholar]
- 44.Origone P, Gotta F, Lamp M, Trevisan L, Geroldi A, Massucco D, et al. Spinocerebellar ataxia 17: full phenotype in a 41 CAG/CAA repeats carrier. Cerebellum Ataxias. 2018;5:7. 10.1186/s40673-018-0086-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245(4922):1066–73. 10.1126/science.2475911 [DOI] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinforma Oxf Engl. 2012;28(21):2747–54. 10.1093/bioinformatics/bts526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smedley D, Jacobsen JOB, Jäger M, Köhler S, Holtgrewe M, Schubach M, et al. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat Protoc. 2015;10(12):2004–15. 10.1038/nprot.2015.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chap. 7:Unit7.20. [DOI] [PMC free article] [PubMed]
- 51.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452–457. [DOI] [PMC free article] [PubMed]
- 52.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–2. 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- 53.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–94. 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biesecker LG, Harrison SM, ClinGen Sequence Variant Interpretation Working Group. The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet Med off J Am Coll Med Genet. 2018;20(12):1687–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med off J Am Coll Med Genet. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balasar Ö, Başdemirci M. Assessment of whole-exome sequencing results in neurogenetic diseases. J Hum Genet. 2023;68(12):797–804. 10.1038/s10038-023-01185-7 [DOI] [PubMed] [Google Scholar]
- 57.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–51. 10.1038/nrg.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Al Jumah M, Al Rajeh S, Eyaid W, Al-Jedai A, Al Mudaiheem H, Al Shehri A, et al. Spinal muscular atrophy carrier frequency in Saudi Arabia. Mol Genet Genomic Med. 2022;10(11):e2049. 10.1002/mgg3.2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shawky RM, El-Sayed NS. Clinico-epidemiologic characteristics of spinal muscular atrophy among egyptians. Egypt J Med Hum Genet. 2011;12(1):25–30. 10.1016/j.ejmhg.2011.02.015 [DOI] [Google Scholar]
- 60.Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K, et al. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat. 2015;36(4):395–402. 10.1002/humu.22758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheth J, Patel H, Mehta S, Tewari S, Sheth F. Clinical and molecular characterization of patients with gross hypotonia and impaired lower motor neuron function. Indian Pediatr. 2013;50(6):591–3. 10.1007/s13312-013-0168-z [DOI] [PubMed] [Google Scholar]
- 62.Nallamilli BRR, Chakravorty S, Kesari A, Tanner A, Ankala A, Schneider T, et al. Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients. Ann Clin Transl Neurol. 2018;5(12):1574–87. 10.1002/acn3.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ankala A, Kohn JN, Dastur R, Gaitonde P, Khadilkar SV, Hegde MR. Ancestral founder mutations in calpain-3 in the Indian Agarwal community: historical, clinical, and molecular perspective. Muscle Nerve. 2013;47(6):931–7. 10.1002/mus.23763 [DOI] [PubMed] [Google Scholar]
- 64.Bhattacharya S, Khadilkar SV, Nalini A, Ganapathy A, Mannan AU, Majumder PP, et al. Mutation spectrum of GNE Myopathy in the Indian Sub-continent. J Neuromuscul Dis. 2018;5(1):85–92. 10.3233/JND-170270 [DOI] [PubMed] [Google Scholar]
- 65.Sonakar AK, Shamim U, Srivastava MP, Faruq M, Srivastava AK. SCA2 in the Indian population: unified haplotype and variable phenotypic patterns in a large case series. Parkinsonism Relat Disord. 2021;89:139–45. 10.1016/j.parkreldis.2021.07.011 [DOI] [PubMed] [Google Scholar]
- 66.Bahl S, Virdi K, Mittal U, Sachdeva MP, Kalla AK, Holmes SE, et al. Evidence of a common founder for SCA12 in the Indian population. Ann Hum Genet. 2005;69(Pt 5):528–34. 10.1046/j.1529-8817.2005.00173.x [DOI] [PubMed] [Google Scholar]
- 67.Das Bhowmik A, Rangaswamaiah S, Srinivas G, Dalal AB. Molecular genetic analysis of trinucleotide repeat disorders (TRDs) in Indian population and application of repeat primed PCR. Eur J Med Genet. 2015;58(3):160–7. 10.1016/j.ejmg.2014.12.010 [DOI] [PubMed] [Google Scholar]
- 68.Chheda P, Chanekar M, Salunkhe Y, Dama T, Pais A, Pande S, et al. A study of Triplet-primed PCR for identification of CAG repeat expansion in the HTT gene in a cohort of 503 Indian cases with Huntington’s disease symptoms. Mol Diagn Ther. 2018;22(3):353–9. 10.1007/s40291-018-0327-y [DOI] [PubMed] [Google Scholar]
- 69.Kaur J, Parveen S, Shamim U, Sharma P, Suroliya V, Sonkar AK, et al. Investigations of Huntington’s Disease and Huntington’s Disease-Like syndromes in Indian Choreatic patients. J Huntingt Dis. 2020;9(3):283–9. 10.3233/JHD-200398 [DOI] [PubMed] [Google Scholar]
- 70.Arora NK, Nair MKC, Gulati S, Deshmukh V, Mohapatra A, Mishra D, et al. Neurodevelopmental disorders in children aged 2–9 years: Population-based burden estimates across five regions in India. PLoS Med. 2018;15(7):e1002615. 10.1371/journal.pmed.1002615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pande S, Majethia P, Nair K, Rao LP, Mascarenhas S, Kaur N et al. De novo variants underlying monogenic syndromes with intellectual disability in a neurodevelopmental cohort from India. Eur J Hum Genet EJHG. 2023. [DOI] [PMC free article] [PubMed]
- 72.Brunet T, Jech R, Brugger M, Kovacs R, Alhaddad B, Leszinski G, et al. De novo variants in neurodevelopmental disorders-experiences from a tertiary care center. Clin Genet. 2021;100(1):14–28. 10.1111/cge.13946 [DOI] [PubMed] [Google Scholar]
- 73.Goldmann JM, Wong WSW, Pinelli M, Farrah T, Bodian D, Stittrich AB, et al. Parent-of-origin-specific signatures of de novo mutations. Nat Genet. 2016;48(8):935–9. 10.1038/ng.3597 [DOI] [PubMed] [Google Scholar]
- 74.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–15. 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agarwal S, Lahiri K, Muranjan M, Solanki N. The face of lysosomal storage disorders in India: a need for early diagnosis. Indian J Pediatr. 2015;82(6):525–9. 10.1007/s12098-014-1628-8 [DOI] [PubMed] [Google Scholar]
- 76.Verma PK, Ranganath P, Dalal AB, Phadke SR. Spectrum of Lysosomal storage disorders at a medical genetics center in northern India. Indian Pediatr. 2012;49(10):799–804. 10.1007/s13312-012-0192-4 [DOI] [PubMed] [Google Scholar]
- 77.Agrawal N, Verma G, Saxena D, Kabra M, Gupta N, Mandal K, et al. Genotype-phenotype spectrum of 130 unrelated Indian families with mucopolysaccharidosis type II. Eur J Med Genet. 2022;65(3):104447. 10.1016/j.ejmg.2022.104447 [DOI] [PubMed] [Google Scholar]
- 78.Pasumarthi D, Gupta N, Sheth J, Jain SJMN, Rungsung I, Kabra M, et al. Identification and characterization of 30 novel pathogenic variations in 69 unrelated Indian patients with mucolipidosis type II and type III. J Hum Genet. 2020;65(11):971–84. 10.1038/s10038-020-0797-8 [DOI] [PubMed] [Google Scholar]
- 79.Bisariya V, Mistry PK, Liu J, Chaudhari MR, Gupta N, Kabra M. The mutation spectrum in Indian patients with gaucher disease. Genome Biol. 2011;12(Suppl 1):P26. 10.1186/gb-2011-12-s1-p26 [DOI] [Google Scholar]
- 80.Mistri M, Tamhankar PM, Sheth F, Sanghavi D, Kondurkar P, Patil S, et al. Identification of novel mutations in HEXA gene in children affected with Tay Sachs disease from India. PLoS ONE. 2012;7(6):e39122. 10.1371/journal.pone.0039122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheth H, Naik P, Shah M, Bhavsar R, Nair A, Sheth F, et al. The GALNS p.P77R variant is a probable gujarati-indian founder mutation causing Mucopolysaccharidosis IVA syndrome. BMC Genomics. 2022;23(1):458. 10.1186/s12864-022-08693-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rama Devi AR, Naushad SM. Newborn screening in India. Indian J Pediatr. 2004;71(2):157–60. 10.1007/BF02723099 [DOI] [PubMed] [Google Scholar]
- 83.Kapoor S, Thelma BK. Status of newborn screening and inborn errors of metabolism in India. Indian J Pediatr. 2018;85(12):1110–7. 10.1007/s12098-018-2681-5 [DOI] [PubMed] [Google Scholar]
- 84.Nagaraja D, Mamatha SN, De T, Christopher R. Screening for inborn errors of metabolism using automated electrospray tandem mass spectrometry: study in high-risk Indian population. Clin Biochem. 2010;43(6):581–8. 10.1016/j.clinbiochem.2009.12.009 [DOI] [PubMed] [Google Scholar]
- 85.Colah R, Italia K, Gorakshakar A. Burden of Thalassemia in India: the road map for control. Pediatr Hematol Oncol J. 2017;2(4):79–84. 10.1016/j.phoj.2017.10.002 [DOI] [Google Scholar]
- 86.Kulkarni GD, Kulkarni SS, Kadakol GS, Kulkarni BB, Kyamangoudar PH, Lakkakula BVKS, et al. Molecular basis of β-thalassemia in Karnataka, India. Genet Test Mol Biomark. 2012;16(2):138–41. 10.1089/gtmb.2011.0035 [DOI] [PubMed] [Google Scholar]
- 87.Kabra SK, Kabra M, Lodha R, Shastri S. Cystic fibrosis in India. Pediatr Pulmonol. 2007;42(12):1087–94. 10.1002/ppul.20677 [DOI] [PubMed] [Google Scholar]
- 88.Schwarz MJ, Super M, Wallis C, Beighton P, Newton C, Heptinstall LE, et al. Delta F508 testing of the DNA Bank of the Royal Manchester Children’s Hospital. Hum Genet. 1990;85(4):428–30. 10.1007/BF02428298 [DOI] [PubMed] [Google Scholar]
- 89.Kabra SK, Kabra M, Lodha R, Shastri S, Ghosh M, Pandey RM, et al. Clinical profile and frequency of delta f508 mutation in Indian children with cystic fibrosis. Indian Pediatr. 2003;40(7):612–9. [PubMed] [Google Scholar]
- 90.Nampoothiri S, Yesodharan D, Sainulabdin G, Narayanan D, Padmanabhan L, Girisha KM, et al. Eight years experience from a skeletal dysplasia referral center in a tertiary hospital in Southern India: a model for the diagnosis and treatment of rare diseases in a developing country. Am J Med Genet A. 2014;164A(9):2317–23. 10.1002/ajmg.a.36668 [DOI] [PubMed] [Google Scholar]
- 91.Sheth H, Shah J, Nair A, Naik P, Sheth J. Case Report: recurrent variant c.298 TA in CCN6 gene found in Progressive Pseudorheumatoid Dysplasia patients from Patni Community of Gujarat: a report of three cases. Front Genet. 2021;12:724824. 10.3389/fgene.2021.724824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bhavani GS, Shah H, Dalal AB, Shukla A, Danda S, Aggarwal S, et al. Novel and recurrent mutations in WISP3 and an atypical phenotype. Am J Med Genet A. 2015;167A(10):2481–4. 10.1002/ajmg.a.37164 [DOI] [PubMed] [Google Scholar]
- 93.Kohli S, Saxena R, Puri RD, Bijarnia Mahay S, Pal S, Dubey S et al. The molecular landscape of oculocutaneous albinism in India and its therapeutic implications. Eur J Hum Genet EJHG. 2023. [DOI] [PMC free article] [PubMed]
- 94.Gv MS. H, S A, S L, I D, S H, Tyrosinase (TYR) gene sequencing and literature review reveals recurrent mutations and multiple population founder gene mutations as causative of oculocutaneous albinism (OCA) in Pakistani families. Eye Lond Engl [Internet]. 2019 Aug [cited 2024 Jun 30];33(8). https://pubmed.ncbi.nlm.nih.gov/30996339/ [DOI] [PMC free article] [PubMed]
- 95.Z C, Y Y et al. S H, Q Z, B Z, X F,. Mutation Analysis of 63 Northwest Chinese Probands with Oculocutaneous Albinism. Curr Eye Res [Internet]. 2021 Jan [cited 2024 Jun 30];46(1). https://pubmed.ncbi.nlm.nih.gov/32552135/ [DOI] [PubMed]
- 96.Smith RJ, Azaiez H, Booth K. GJB2-Related Autosomal Recessive Nonsyndromic Hearing Loss. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJ, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993 [cited 2024 Jul 3]. http://www.ncbi.nlm.nih.gov/books/NBK1272/ [PubMed]
- 97.RamShankar M, Girirajan S, Dagan O, Ravi Shankar HM, Jalvi R, Rangasayee R, et al. Contribution of connexin26 (GJB2) mutations and founder effect to non-syndromic hearing loss in India. J Med Genet. 2003;40(5):e68. 10.1136/jmg.40.5.e68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Neagu A, Mocanu AI, Bonciu A, Coadă G, Mocanu H. Prevalence of GJB2 gene mutations correlated to presence of clinical and environmental risk factors in the etiology of congenital sensorineural hearing loss of the Romanian population. Exp Ther Med. 2021;21(6):612. 10.3892/etm.2021.10044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sen P, Bhargava A, George R, Ve Ramesh S, Hemamalini A, Prema R, et al. Prevalence of retinitis pigmentosa in South Indian population aged above 40 years. Ophthalmic Epidemiol. 2008;15(4):279–81. 10.1080/09286580802105814 [DOI] [PubMed] [Google Scholar]
- 100.Yohe S, Sivasankar M, Ghosh A, Ghosh A, Holle J, Murugan S, et al. Prevalence of mutations in inherited retinal diseases: a comparison between the United States and India. Mol Genet Genomic Med. 2019;8(2):e1081. 10.1002/mgg3.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gangodkar P, Khadilkar V, Raghupathy P, Kumar R, Dayal AA, Dayal D, et al. Clinical application of a novel next generation sequencing assay for CYP21A2 gene in 310 cases of 21- hydroxylase congenital adrenal hyperplasia from India. Endocrine. 2021;71(1):189–98. 10.1007/s12020-020-02494-z [DOI] [PubMed] [Google Scholar]
- 102.Newborn Screening for Congenital Hypothyroidism and Congenital Adrenal Hyperplasia. Indian J Pediatr. 2018;85(11):935–40. 10.1007/s12098-018-2645-9 [DOI] [PubMed] [Google Scholar]
- 103.Weber SN, Lammert F. Genetics in liver diseases: from diagnostics to precise therapy. Clin Liver Dis. 2017;9(1):1–4. 10.1002/cld.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kamisako T. What is Gilbert’s syndrome? Lesson from genetic polymorphisms of UGT1A1 in Gilbert’s syndrome from Asia. J Gastroenterol Hepatol. 2004;19(9):955–7. 10.1111/j.1440-1746.2004.03524.x [DOI] [PubMed] [Google Scholar]
- 105.Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci U S A. 1998;95(14):8170–4. 10.1073/pnas.95.14.8170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Devuyst O, Knoers NVAM, Remuzzi G, Schaefer F, Board of the Working Group for Inherited Kidney Diseases of the European Renal Association and European Dialysis and Transplant Association. Rare inherited kidney diseases: challenges, opportunities, and perspectives. Lancet Lond Engl. 2014;383(9931):1844–59. 10.1016/S0140-6736(14)60659-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vivante A, Hildebrandt F. Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol. 2016;12(3):133–46. 10.1038/nrneph.2015.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saha A, Kapadia SF, Vala KB, Patel HV. Clinical utility of genetic testing in Indian children with kidney diseases. BMC Nephrol. 2023;24(1):212. 10.1186/s12882-023-03240-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gorman GS, Schaefer AM, Ng Y, Gomez N, Blakely EL, Alston CL, et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol. 2015;77(5):753–9. 10.1002/ana.24362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pronicka E, Piekutowska-Abramczuk D, Ciara E, Trubicka J, Rokicki D, Karkucińska-Więckowska A, et al. New perspective in diagnostics of mitochondrial disorders: two years’ experience with whole-exome sequencing at a national paediatric centre. J Transl Med. 2016;14(1):174. 10.1186/s12967-016-0930-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cecchi F, Tomberli B, Olivotto I. Clinical and molecular classification of cardiomyopathies. Glob Cardiol Sci Pract. 2012;2012(1):4. 10.5339/gcsp.2012.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Charron P, Elliott PM, Gimeno JR, Caforio ALP, Kaski JP, Tavazzi L, et al. The Cardiomyopathy Registry of the EURObservational Research Programme of the European Society of Cardiology: baseline data and contemporary management of adult patients with cardiomyopathies. Eur Heart J. 2018;39(20):1784–93. 10.1093/eurheartj/ehx819 [DOI] [PubMed] [Google Scholar]
- 113.Kaski JP, Syrris P, Esteban MTT, Jenkins S, Pantazis A, Deanfield JE, et al. Prevalence of sarcomere protein gene mutations in preadolescent children with hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2009;2(5):436–41. 10.1161/CIRCGENETICS.108.821314 [DOI] [PubMed] [Google Scholar]
- 114.Vaidya V, Dhiman RS, Mittal A, Khullar M, Sharma M, Bahl A. Genotyping Indian patients with primary cardiomyopathies-analysis of database. Indian Heart J. 2023;75(1):43–6. 10.1016/j.ihj.2022.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Belaid B, Lamara Mahammed L, Drali O, Oussaid AM, Touri NS, Melzi S, et al. Inborn errors of immunity in Algerian children and adults: a single-center experience over a period of 13 years (2008–2021). Front Immunol. 2022;13:900091. 10.3389/fimmu.2022.900091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jindal AK, Pilania RK, Rawat A, Singh S. Primary Immunodeficiency disorders in India-A situational review. Front Immunol. 2017;8:714. 10.3389/fimmu.2017.00714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ward MM. Estimating rare disease prevalence from administrative hospitalization databases. Epidemiol Camb Mass. 2005;16(2):270–1. 10.1097/01.ede.0000153643.88019.92 [DOI] [PubMed] [Google Scholar]
- 118.Leib JR, Gollust SE, Hull SC, Wilfond BS. Carrier screening panels for Ashkenazi jews: is more better? Genet Med off J Am Coll Med Genet. 2005;7(3):185–90. [DOI] [PubMed] [Google Scholar]
- 119.Sheth F, Shah J, Jain D, Shah S, Patel H, Patel K, et al. Comparative yield of molecular diagnostic algorithms for autism spectrum disorder diagnosis in India: evidence supporting whole exome sequencing as first tier test. BMC Neurol. 2023;23(1):292. 10.1186/s12883-023-03341-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sheth H, Nair A, Bhavsar R, Kamate M, Gowda VK, Bavdekar A, et al. Development, validation and application of single molecule molecular inversion probe based novel integrated genetic screening method for 29 common lysosomal storage disorders in India. Hum Genomics. 2024;18(1):46. 10.1186/s40246-024-00613-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sheth J, Nair A, Jee B. Lysosomal storage disorders: from biology to the clinic with reference to India. Lancet Reg Health - Southeast Asia [Internet]. 2023 Feb 1 [cited 2024 Feb 6];9. 10.1016/j.lansea.2022.100108 [DOI] [PMC free article] [PubMed]
- 122.Basu A, Sarkar-Roy N, Majumder PP. Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc Natl Acad Sci U S A. 2016;113(6):1594–9. 10.1073/pnas.1513197113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vadivelu MK. Emergence of sociocultural norms restricting intermarriage in large social strata (endogamy) coincides with foreign invasions of India. Proc Natl Acad Sci U S A. 2016;113(16):E2215–2217. 10.1073/pnas.1602697113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sheth J, Nair A. Treatment for Lysosomal Storage Disorders. Curr Pharm Des. 2020;26(40):5110–8. 10.2174/1381612826666201015154932 [DOI] [PubMed] [Google Scholar]
- 125.Li M. Enzyme replacement therapy: a review and its role in treating lysosomal Storage diseases. Pediatr Ann. 2018;47(5):e191–7. 10.3928/19382359-20180424-01 [DOI] [PubMed] [Google Scholar]
- 126.Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov. 2020;19(10):673–94. 10.1038/s41573-020-0075-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Papaioannou I, Owen JS, Yáñez-Muñoz RJ. Clinical applications of gene therapy for rare diseases: a review. Int J Exp Pathol. 2023;104(4):154–76. 10.1111/iep.12478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tucci F, Galimberti S, Naldini L, Valsecchi MG, Aiuti A. A systematic review and meta-analysis of gene therapy with hematopoietic stem and progenitor cells for monogenic disorders. Nat Commun. 2022;13(1):1315. 10.1038/s41467-022-28762-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Borgna-Pignatti C, Marsella M. Iron Chelation in Thalassemia Major. Clin Ther. 2015;37(12):2866–77. 10.1016/j.clinthera.2015.10.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and it’s Supplementary Information.