ABSTRACT
Mucormycoses are emerging fungal infections caused by a variety of heterogeneous species within the Mucorales order. Among the Mucor species complex, Mucor circinelloides is the most frequently isolated pathogen in mucormycosis patients and despite its clinical significance, there is an absence of established genome manipulation techniques to conduct molecular pathogenesis studies. In this study, we generated a spontaneous uracil auxotrophic strain and developed a genetic transformation procedure to analyze molecular mechanisms conferring antifungal drug resistance. With this new model, phenotypic analyses of gene deletion mutants were conducted to define Erg3 and Erg6a as key biosynthetic enzymes in the M. circinelloides ergosterol pathway. Erg3 is a C-5 sterol desaturase involved in growth, sporulation, virulence, and azole susceptibility. In other fungal pathogens, erg3 mutations confer azole resistance because Erg3 catalyzes the production of a toxic diol upon azole exposure. Surprisingly, M. circinelloides produces only trace amounts of this toxic diol and yet, it is still susceptible to posaconazole and isavuconazole due to alterations in membrane sterol composition. These alterations are severely aggravated by erg3Δ mutations, resulting in ergosterol depletion and, consequently, hypersusceptibility to azoles. We also identified Erg6a as the main C-24 sterol methyltransferase, whose activity may be partially rescued by the paralogs Erg6b and Erg6c. Loss of Erg6a function diverts ergosterol synthesis to the production of cholesta-type sterols, resulting in resistance to amphotericin B. Our findings suggest that mutations or epimutations causing loss of Erg6 function may arise during human infections, resulting in antifungal drug resistance to first-line treatments against mucormycosis.
IMPORTANCE
The Mucor species complex comprises a variety of opportunistic pathogens known to cause mucormycosis, a potentially lethal fungal infection with limited therapeutic options. The only effective first-line treatments against mucormycosis consist of liposomal formulations of amphotericin B and the triazoles posaconazole and isavuconazole, all of which target components within the ergosterol biosynthetic pathway. This study uncovered M. circinelloides Erg3 and Erg6a as key enzymes to produce ergosterol, a vital constituent of fungal membranes. Absence of any of those enzymes leads to decreased ergosterol and consequently, resistance to ergosterol-binding polyenes such as amphotericin B. Particularly, losing Erg6a function poses a higher threat as the ergosterol pathway is channeled into alternative sterols similar to cholesterol, which maintain membrane permeability. As a result, erg6a mutants survive within the host and disseminate the infection, indicating that Erg6a deficiency may arise during human infections and confer resistance to the most effective treatment against mucormycoses.
KEYWORDS: genetic tranformation, ergosterol, antifungal resistance, Erg3, Erg6, Mucorales, fungi
INTRODUCTION
Mucorales comprise an order of human pathogenic filamentous fungi that includes Mucor, Rhizopus, and Lichtheimia species. These species cause infections known as mucormycoses, which typically occur when individuals inhale the spores of these molds or are infected through skin injuries, burns, or other traumatic wounds (1). Immunocompromised patients, such as those with cancer and/or undergoing immunosuppressive treatment, are particularly susceptible to invasive mucormycoses. Individuals suffering from poorly controlled diabetes mellitus are also at higher risk. This disease is associated with poor prognoses and dramatically high morbidity, resulting in mortality rates ranging from 23% to 80% (2). Because Mucorales are understudied pathogens, it is challenging to gather information on global incidence rates. Nevertheless, the trend over the past decade has shown an increase in the number of cases, especially those associated with COVID-19 infections (3, 4), which has led the World Health Organization to include Mucoralean pathogens on their fungal priority pathogens list (5).
Recent global practice guidelines discourage the use of fluconazole, voriconazole (6), or the echinocandins (7) as therapeutic options for mucormycoses due to inherent resistance to those antifungal drugs. Instead, first-line treatment with liposomal formulations of the polyene amphotericin B is recommended at high doses, followed by salvage treatment with the triazoles posaconazole or isavuconazole (2). However, the applicability of these treatments can be limited by toxicity and resistance. Due to the limited availability of effective antifungal drugs, the emergence of resistant strains in the environment or in the host poses a global threat to public health. Antifungal drug resistance mechanisms in Mucoralean clinical isolates are poorly understood due to the lack of proper diagnostic tools and the heterogeneity of infections (5). Recent studies have revealed gain-of-function mutations in erg11 confer resistance to azoles (6). Additionally, a novel mechanism for antifungal drug resistance based on RNA interference, called epimutation, has been discovered in Mucor species (8). This mechanism could represent a source of transient antifungal drug resistance during host infection (9). The epimutation mechanism operates through RNAi-mediated gene silencing, repressing the expression of drug targets. Therefore, identifying loss-of-function mutations conferring antifungal drug resistance might elucidate new targets of this epigenetic process.
Understanding the mechanisms of action of the recommended antifungal therapies against mucormycoses may help to identify drug targets involved in drug resistance. Polyenes bind directly to ergosterol and both increase membrane permeability and sequester ergosterol (10, 11), while azoles block the main ergosterol biosynthetic pathway by inhibiting the Erg11 enzyme (12). By antagonistically binding Erg11, azoles can divert ergosterol biosynthesis into a different pathway that results in toxic sterol intermediates (13, 14). Thus, the mechanisms of action of both polyenes and azoles converge on ergosterol production and conceivably, altering the ergosterol biosynthetic pathway may lead to changes in drug susceptibility. Indeed, loss-of-function mutations in two key ergosterol biosynthetic genes, erg3 and erg6, have been reported to result in antifungal drug resistance in other fungal pathogens. Erg3 is a C-5 sterol desaturase involved in the main ergosterol biosynthetic pathway, but also in the toxic sterol pathway (13). Therefore, loss of Erg3 function prevents the formation of toxic sterol intermediates produced during azole treatment, enabling evasion of the fungistatic effect of the drug (14, 15). On the other hand, Erg6 is a C-24 sterol methyltransferase that either converts zymosterol into fecosterol or lanosterol into eburicol (16), two alternative ergosterol precursors. Mutations affecting Erg6 function favor the production of cholesta-type sterols, conferring polyene resistance (17–23).
Research on molecular mechanisms leading to drug resistance in Mucorales is of paramount importance, albeit challenging given current genetic and molecular models and tools. Mucor is a genus of related fungal species within the order Mucorales. M. circinelloides is the Mucor species most frequently isolated from clinical sources and is associated with mucormycotic infections (24–26). Previously, the Mucor circinelloides species complex was delineated into formae but recent taxonomic and phylogenetic analyses have defined a minimum of 16 distinct phylogenetic species, 7 of which are known to cause human disease (27). As a consequence, research has advanced genetics, genomics, and host-microbe interaction models for this understudied group of pathogens, leading to the development of Mucor as a model for mucormycoses over the past decade (8, 28–31). Most of these studies relied on M. lusitanicus because it was the only species amenable to genetic transformation and it has a well-assembled genome (32, 33). Yet among Mucor species, M. lusitanicus is not a frequent cause of mucormycosis, probably due to its maximum growth temperature being 35–37°C, compared to M. circinelloides, which grows at 37–39°C (27). In this study, we introduce a new model to study mucormycosis pathogenesis and antifungal drug susceptibility in M. circinelloides. We developed a novel transformation method to characterize the erg3 and erg6 genes in a M. circinelloides pathogenic isolate, leading to the discovery of new mechanisms for amphotericin B resistance and azole susceptibility in Mucorales.
RESULTS
A genetic transformation procedure for Mucor circinelloides pathogenic species
M. circinelloides is the most frequent Mucor species isolated from mucormycosis patients, and these isolates are usually thermotolerant and highly virulent in host-pathogen infection models, yet reliable genetic transformation procedures are not available for this species. To develop a genetic model for virulence and antifungal susceptibility studies, we aimed at generating a M. circinelloides strain that is competent for genetic transformation. Because ensuring full virulence and thermotolerance in our model was a priority, M. circinelloides 1006PhL human isolate (hereinafter M. circinelloides) was selected to conduct studies as it has both attributes (34). Also, its genome has been sequenced (35) and is publicly available (https:// www.ncbi.nlm.nih.gov/datasets/genome/GCA_000401635.1/), providing an advantage to genetic manipulation. Traditionally in mucoralean genetics, the application of dominant resistance markers is not possible due to high intrinsic resistance to these drugs. M. circinelloides is not an exception, as it exhibits residual growth upon exposure to drugs frequently utilized for transformant selection, complicating the discrimination between resistant transformants and residually growing colonies (Fig. S1).
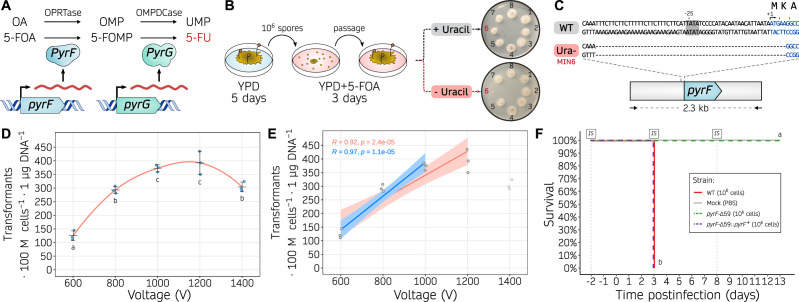
As a consequence, we designed a workflow to obtain an auxotrophic mutant that could serve as a transformation recipient strain based on successful previous studies (36). 5-Fluoroorotic acid (5-FOA) is sequentially converted into a cytotoxic 5-fluorouracil (5-FU), by the action of the orotate phosphoribosyltransferase PyrF and orotidine-5′-monophosphate decarboxylase PyrG. As Mucorales are haploid fungi, and these two enzymes catalyze essential steps in the uracil biosynthetic pathway, a single loss-of-function mutation in either of their encoding genes results in resistance to 5-FOA and uracil auxotrophy that can be rescued by uridine and/or uracil supplementation. Following this rationale (Fig. 1A), we selected spontaneous uracil auxotrophic mutants by utilizing 5-FOA but avoiding additional mutagenic agents to prevent undesired secondary mutations. A total of 237 5-FOA-resistant colonies were isolated and 8 were tested for uracil auxotrophy by comparing their ability to grow on media with or without uridine and uracil. Among these, one isolate named MIN6 was unable to grow without uridine and uracil indicative of uracil auxotrophy (Fig. 1B, isolate 6). Subsequent Sanger-sequencing of the pyrF and pyrG genes revealed a 59-basepair (bp) microdeletion within the pyrF locus (pyrF-Δ59). This microdeletion comprises the last 54 bp of the promoter region, including a putative TATA-box element whose deletion may impair transcription; and the first 5 bp of the coding sequence, leading to a start loss and a consequent frameshift mutation assuming translation would start at the next available AUG triplet (Fig. 1C).
Fig 1.
Uracil auxotrophy enables M. circinelloides transformation based on a pyrF selectable marker. (A) Role of PyrF (orotate phosphoribosyltransferase, OPRTase) and PyrG (orotidine monophosphate decarboxylase, OMPDCase) in the uracil biosynthetic pathway, converting orotic acid (OA) to orotidine monophosphate (OMP), and OMP into uridine monophosphate (UMP), respectively. Alternatively, the same enzymes catalyze the conversion of 5-fluoroorotic acid (5-FOA) into fluoro-OMP (FOMP), and FOMP into 5-fluorouracil (5-FU), respectively. (B) Experimental design followed to isolate uracil auxotrophic strains. Briefly, wild-type spores were collected from YPD medium, counted, and one million spores were plated onto YPD containing 5-FOA. After 3 days, 5-FOA resistant colonies were isolated and transferred to new 5-FOA containing medium to confirm the resistance and then inoculated onto minimal medium either with or without uracil to confirm the auxotrophy. (C) pyrF sequences from the wild-type (WT) and uracil auxotrophic (Ura-, MIN6) strains, focusing on the 5′-region of the gene. The TATA box motif (gray background around position −25), transcription start site (arrow, position +1), pyrF coding sequence (blue font), and encoded one-letter amino acids are shown. (D, E) Transformation efficiency is measured as transformants per 100 million protoplasts per microgram of DNA using different voltages in the electroporation procedure. Individual values (dots) from biological triplicates were used to determine the mean and SD values as black lines. In (D), a smoothed curve is drawn as a red line. Significant differences among voltages were determined by a one-way ANOVA and Tukey HSD test, and voltages showing different letters indicate significant differences (P-value ≤ 0.05). In (E), two linear regression models are displayed showing a positive correlation for voltages from 600 to 1,000 V (blue line) or to 1,200 V (red line). Pearson’s correlation coefficients (R) are shown for each regression. (F) Kaplan-Meier survival curves of immunosuppressed mice after infection with M. circinelloides wildtype and transformable strains (color-coded). Intraperitoneal immunosuppressive (IS) treatments are shown. Strains showing significant differences in virulence are indicated as different letters (P-value ≤ 0.0001), assessed by a Log-rank test.
Next, we complemented MIN6 uracil auxotrophy by integrating the pyrF marker, a 2.3-kilobase (kb) DNA fragment comprising the wild-type pyrF promoter, coding sequence, and terminator regions aiming at favoring homologous recombination and subsequent replacement of the pyrF-Δ59 allele with its wildtype sequence. To integrate the pyrF marker, we improved a reliable protoplast transformation procedure involving electroporation. For optimal protoplast preparation, the spore germination rate in M. circinelloides was comparatively slower than in M. lusitanicus; this delay may be attributed to the pre-swollen and larger size of M. lusitanicus spores (37). DNA was introduced into the protoplasts by exponential decay waveform electroporation with constant capacitance and resistance, and transformation efficiency was assessed at different voltages (Fig. 1D). The most efficient transformation rate was obtained with 1,000 and 1,200 V. A linear regression modeling indicates a positive correlation between transformation efficiency and voltage, particularly significant at voltages ranging from 600 to 1,000 V (Fig. 1E). Therefore, subsequent electroporations were carried out at 1,000 V to ensure maximum efficiency while avoiding unnecessary protoplast lethality. The successful complementation of the pyrF-Δ59 allele was verified by PCR-amplification and restriction fragment length polymorphism (RFLP, Fig. S2).
Next, we determined the effect of the pyrF-Δ59 allele on virulence and tested whether pyrF complementation could rescue a fully virulent phenotype (Fig. 1F). To do so, the wild-type 1006PhL strain, the pyrF-Δ59 uracil auxotrophic strain, and the pyrF-Δ59::pyrF+ complemented strain were analyzed in a survival assay using immunosuppressed mice. The uracil auxotrophic strain showed a complete and significant lack of virulence, whereas the wildtype 1006PhL and pyrF+ complemented strain displayed full virulence without significant differences. Similarly, fungal virulence correlated with animal weight loss (Fig. S3). These results demonstrate that uracil auxotrophy impairs the pathogenic potential of M. circinelloides, which can be restored by pyrF+ complementation. Overall, these findings highlight this procedure as a highly efficient, reliable method to obtain uracil auxotrophic strains, transform by electroporation, and replace genes by homologous recombination in M. circinelloides. But more importantly, they support the application of this genetic model to study virulence processes and antimicrobial drug resistance within the Mucor species complex, as opposed to other thermosensitive, partially virulent, and clinically less relevant species such as M. lusitanicus.
Erg3 and Erg6 distribution across the mucoromycota
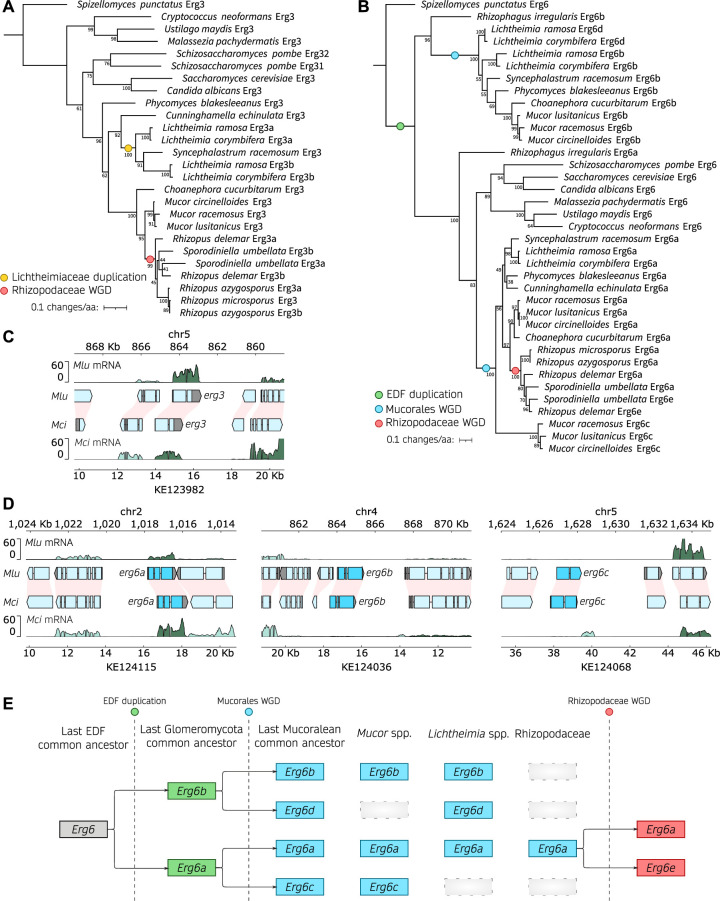
We aimed to delete the erg3 and erg6 genes in M. circinelloides and M. lusitanicus to illustrate the potential of our newly transformable M. circinelloides strain for standardized antifungal susceptibility testing and virulence assays. A protein-protein similarity search identified one Erg3 and three Erg6 homologs whose evolutionary history was examined in several pathogenic species from the Mucoromycota, as well as in model organisms with single-copy erg genes for reference (Fig. 2; Table S1 for Erg3 and Table S2 for Erg6 sequences). Erg3 protein sequences from Mucoromycota fungi form a robust clade within fungal Erg3 proteins, highly consistent with their species phylogeny (Fig. 2A). This phylogeny suggests two independent duplication events in the Lichtheimiaceae and Rhizopodaceae families as most of their members harbor two Erg3 paralogs, which could be explained by recent segmental duplication events in these families (38–40). Besides these specific duplication events, most Mucoromycota species harbor a single Erg3 ortholog and particularly, the Mucor clade. The Erg3 ortholog exhibits high synteny among Mucor species (Fig. 2C; and Fig. S4A), confirming our phylogenetic inference. In addition, transcriptomic analysis revealed that erg3 is actively transcribed in both Mucor species. Taken together, these results indicate that erg3 deletion will suffice to confer loss of Erg3 function.
Fig 2.
Erg3 and Erg6 evolutionary history across the Mucoromycota. (A, B) Phylogenetic tree depicting the Erg3 (A) and Erg6 (B) amino acid sequence alignments in clinically relevant fungi, with a specific focus on Mucorales. The Erg protein sequences from the chytrid Spizellomyces punctatus were employed as outgroups to root the trees. Additionally, the Erg6 sequence from Rhizophagus irregularis, a member of the Glomeromycota and the closest non-Mucoromycota sequence, was included to provide clarity on duplication events in (B). Noteworthy branching events, including whole-genome duplications (WGD) or gene duplications, are denoted by color-coded circles. Node support is represented as a percentage based on 1,000 bootstrapping iterations. The branch length is scaled to 0.1 substitutions per amino acid, as indicated in the legend. (C, D) Plots illustrating gene transcription across M. lusitanicus (Mlu) and M. circinelloides (Mci) genomic regions containing erg3 (C) and erg6 orthologs (D). The genomic coordinates for both species are displayed above (Mlu) and below (Mci) each plot. Gene annotation is depicted as arrowed blocks, indicating the direction of transcription and the boundaries of exons and introns. The coloring scheme assigns cyan blue to erg coding sequences, light blue to the remaining coding sequences, and gray to untranslated regions. Interspecies synteny among genes is depicted as pink shading. Gene transcription is shown as rRNA-depleted RNA read coverage, with the forward strand represented in dark green and the reverse strand in light green, enabling assessment of sense and antisense transcription. In some instances, the x-axes were inverted, and in those cases, the forward and reverse strand orientations were also inverted to enhance visualization and clarification. (E) Model illustrating the inferred evolutionary history of Erg6 across the early-diverging fungi (EDF).
On the other hand, the evolutionary history of Erg6 is more complex (Fig. 2B). We identified a robustly supported, monophyletic clade encompassing the orthologs from well-known model organisms as well as Glomeromycota and Mucoromycota Erg6a paralogs. This clade exhibits two different Erg6 paralogs from Mucor species: one closer to the clade root and the reference Erg6 sequences (Fig. 2B, Erg6a) and another considerably divergent (Fig. 2B, Erg6c, bottom clade). In addition, most species from the Mucoromycota harbor another Erg6 paralog, that is more divergent compared to the reference Erg6 sequences (Fig. 2B, Erg6b). We developed a model (Fig. 2E) that offers the most plausible interpretation of the phylogenetic and synteny patterns observed across all of the Mucoromycota species analyzed, explaining the origin of these paralogs (Fig. S4D through H). An ancient duplication event predating the Glomeromycota and Mucoromycota common ancestor resulted in two highly divergent Erg6 paralogs in species like Rhizozophagus irregularis and the Mucoromycota (Fig. 2B, EDF duplication, Erg6a and Erg6b). Subsequent genome duplications in the mucoralean common ancestor (32), along with gene loss, led to most Mucoromycota species harboring two to three Erg6 paralogs. Interestingly, gene loss did not occur uniformly in all clades: e.g., the third paralog found in Mucor species (Erg6c) is different from that of Lichtheimia species (Erg6d), and several species harbor only two paralogs (Erg6a and Erg6b). Moreover, Rhizopodiaceae species exhibit an additional, more recent duplication (Erg6e), which is consistent with another round of whole-genome duplication and subsequent gene loss in this lineage (38, 39). All three Erg6 paralogs found in Mucor species, which also show conserved synteny (Fig. 2D; Fig. S4D through H), are explained by this model. Interestingly, only erg6a is actively expressed (Fig. 2D), which together with the phylogenetic data suggests that erg6b and erg6c may not be as biologically relevant during standard growth conditions. Therefore, we named the Mucor paralogs Erg6a, Erg6b, and Erg6c in agreement with our evolutionary model (Fig. 2E), phylogenetic, and transcriptomic data, and we reasoned that erg6a deletion may suffice to confer loss of Erg6 function.
To explore the role of the erg3 and erg6a genes in mucoralean ergosterol biosynthesis, antifungal susceptibility, and pathogenesis, we generated deletion mutant strains in M. circinelloides and M. lusitanicus (Fig. S5). The genetic loci were successfully replaced through homologous recombination with uracil prototrophy markers: pyrF for M. circinelloides (Fig. S5A and B), utilizing the newly designed MIN6 strain and genetic transformation via electroporation procedure; and pyrG for M. lusitanicus (Fig. S5C and D), using a previously described transformation method (41). Because Mucorales have non-septate hyphae and multinucleated spores, transformed protoplasts frequently harbor wild-type and mutant nuclei simultaneously, a phenomenon known as heterokaryosis. To lose the wild-type nuclei and obtain exclusively homokaryotic deletion mutants, transformants must be cultured for several asexual generations in a medium that positively selects the mutant nuclei. After this process of mutant nuclei enrichment, erg3Δ and erg6aΔ homokaryotic mutations were verified by PCR-amplified fragment length polymorphism (AFLP, Fig. S5), and two independently generated homokaryotic mutants were selected for each deletion to ensure phenotypic specificity in subsequent studies.
erg3 and erg6a deletions affect growth and virulence in M. circinelloides
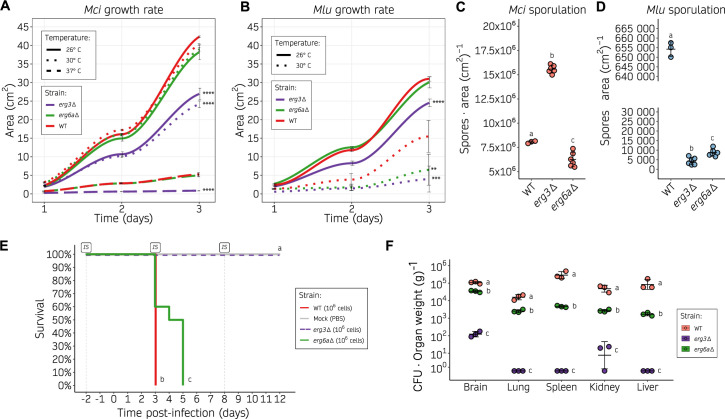
Alterations in ergosterol composition may affect the stability of biological membranes and usually lead to fitness impairments. Consequently, we examined the effects of loss-of-function mutations in the erg3 and erg6a genes of M. circinelloides and M. lusitanicus, specifically focusing on evaluating defects in various aspects of fitness, including growth rate, sporulation, and virulence. For these and subsequent analyses, data from each pair of independently generated mutants harboring the same gene deletion were grouped for simplicity when necessary for primary figures, after ensuring that there were not significant differences between them as can be observed in the supplementary information (Fig. S6 to S8; Data set S1). The growth rate was evaluated at different temperatures, including optimal growth and human physiological temperatures (Fig. 3A and B ). The M. circinelloides erg3 mutant exhibited a significant growth delay under all conditions tested (26°C, 30°C, and 37°C) compared to the wild type, whereas the M. circinelloides erg6aΔ mutation did not affect growth significantly (Fig. 3A). The same growth trends were observed in minimal media (Fig. S6A). Similarly, the M. lusitanicus erg3Δ mutant displayed growth defects under every condition (26°C and 30°C), while the erg6aΔ mutant showed a growth delay only at 30°C (Fig. 3B). As M. lusitanicus is not able to withstand human physiological temperatures (37°C), and exhibits a considerably lower optimal growth temperature (26°C), this could explain the interspecies differences observed. Notably, increasing the temperature range beyond the optimal growth temperature for each species (37°C for M. circinelloides and 30°C for M. lusitanicus) severely affected the growth rate in every strain and wild-type controls but, more importantly, rendered the phenotype confirmed by erg gene deletions more severe. All mutant strains exhibited full uracil prototrophy after pyrF or pyrG insertion, and the leucine auxotrophy in M. lusitanicus did not affect their growth rates, as no differences were observed upon uridine, uracil, and leucine supplementation (Fig. S6C and E).
Fig 3.
Alterations in the ergosterol biosynthetic pathway result in decreased growth rate, sporulation, and virulence. (A, B) Growth rate of M. circinelloides (A) and M. lusitanicus (B) erg3Δ and erg6aΔ (shown as color-coded lines) on YPD medium at different temperatures (shown as distinct line types), measured as colony area across 24 hour intervals. Individual values were determined either in six biological replicates for the wild type or in three biological replicates of two independently generated mutants (six total values, grouped together for simplicity as no significant differences were detected), and used to plot a smoothed curve and SD values as black lines. Significant area differences in depicted strains compared to the wild-type growth rate across the whole time-course were determined by a one-way ANOVA and Tukey HSD test for each temperature group and shown as asterisks (**P-value ≤ 0.01, ***P-value ≤ 0.001, and ****P-value ≤ 0.0001). (C, D) Sporulation rate of M. circinelloides (C) and M. lusitanicus (D) erg3Δ and erg6aΔ after a 72 hour incubation at room temperature on YPG medium, determined as the number of spores per cm2. Values from individual measurements were obtained from three biological replicates of either the wild-type strain or two independently generated mutants (grouped together as no significant differences were found). These values are depicted in a dot plot, with standard deviation (SD) values represented as black lines. Strains are grouped by letters showing significant sporulation differences determined by a one-way ANOVA and Tukey HSD test (P-value ≤ 0.01). (E) Kaplan-Meier survival curves of immunosuppressed mice after infection with M. circinelloides wild-type and ergosterol mutants (color-coded). For erg3Δ and erg6aΔ, two independently generated mutants were injected into groups of five mice. For simplicity, after observing no significant differences in the results, survival curves were grouped together per gene deletion in groups of 10. Intraperitoneal immunosuppressive (IS) treatments are shown. Strains showing significant differences in virulence are indicated as different letters (P-value ≤ 0.01), assessed by a Log-rank test. (F) Fungal burden at 3 days post-infection in immunosuppressed mice, measured as colony-forming units (CFU) per gram of five different organs. SD and mean (black lines) were assessed from three biological triplicates. For each organ category, strains are grouped by letters showing significant fungal burden differences (one-way ANOVA and Tukey HSD test, P-value ≤ 0.01).
Asexual sporulation was also affected by erg mutations. In M. circinelloides, the erg3Δ mutant displayed a significant increase in sporulation, as opposed to the decrease observed in the erg6a mutant compared to the wild-type strain (Fig. 3C). On the other hand, M. lusitanicus erg3Δ and erg6aΔ mutants showed a severe and significant reduction in sporulation (Fig. 3D).
The observed differences in both growth and sporulation imply that deletion of the erg3 and erg6a genes could impact fitness under various stress conditions, particularly temperature. To investigate whether these defects are relevant during the infection process, a survival assay was conducted using M. circinelloides strains, as they withstand murine physiological temperatures and exhibit a more reliable pathogenic potential. Immunosuppressed mice were infected with two independently generated mutants for each gene, and with the wild-type control strain (Fig. 3E; Fig. S7). Although erg6aΔ mutants exhibited attenuated virulence compared to the wild-type control, they were able to establish a fully disseminated infection in every organ analyzed for fungal burden resulting in mortality among all infected mice. Conversely, animals infected with erg3 mutants survived, possibly due to the significant and drastic reduction in fungal burden within vital organs (Fig. 3E through F). These findings suggest that erg3 deletion abolishes virulence in M. circinelloides, whereas erg6a deletion may not suffice to prevent dissemination and mortality.
Taken together, these results confirm that deletion of erg3 and erg6a leads to a reduction in fitness in specific settings and particularly at higher than optimal temperatures, observing a higher fitness cost for M. lusitanicus, and confirming interspecies differences between M. circinelloides and M. lusitanicus. As expected, this erg deletion-related loss of fitness impacts the virulence potential of M. circinelloides, more profoundly as a consequence of erg3 deletion, and slightly due to erg6a deletion. Although erg6a deletion leads to delayed infection, it is noteworthy that this strain is still able to cause full mortality.
erg6a loss-of-function mutations result in antifungal drug resistance
The antifungal drug susceptibility profiles of erg3 and erg6a mutants were conducted following the CLSI and EUCAST standard methodology for filamentous fungi, determining the minimal inhibitory concentration values required to inhibit growth (MIC) by broth microdilution (Table 1). Susceptibility testing focused on the only three clinical antifungal drugs recommended for mucormycosis treatment: the liposomal formulation of amphotericin B (Ambisome), posaconazole (Noxafil), and isavuconazonium sulfate (Cresemba). Currently, clinical breakpoints have not been established for any of these treatments for Mucorales, and therefore, it is not possible to strictly assess resistance or susceptibility. However, for clarity, the term “resistance” will henceforth refer to decreased susceptibility in our isolates. Both M. circinelloides and M. lusitanicus exhibited similar trends in MIC values, indicating generalizable alterations in antifungal drug susceptibilities. First, erg3 deletion caused a minor increase in MIC for amphotericin B, with a two- and fourfold increase observed in M. circinelloides and M. lusitanicus, respectively. Conversely, erg3Δ mutants also exhibited increased susceptibility to azoles, as evidenced by a fourfold decrease in MIC for posaconazole, and a twofold decrease for isavuconazole in both M. circinelloides and M. lusitanicus. Second, the erg6aΔ mutants showed substantial resistance to amphotericin B, specifically a 4- and 16-fold MIC increase in M. circinelloides and M. lusitanicus, respectively. These erg6aΔ mutants also exhibited a slightly increased susceptibility to azoles, manifested by a twofold MIC decrease for posaconazole and isavuconazole in M. lusitanicus, and a twofold decrease for posaconazole in M. circinelloides.
TABLE 1.
Amphotericin B (AMB), posaconazole (POS), and isavuconazole (ISA) minimal inhibitory concentration (MIC) for M. circinelloides and M. lusitanicus erg3Δ and erg6aΔ mutants
| Strain | Amphotericin B (mg/L) | Posaconazole (mg/L) | Isavuconazole (mg/L) |
|---|---|---|---|
| M. circinelloides | |||
| Wild-type 1006PhL | 0.125 | 2 | 8 |
| erg3Δ−1 | 0.25 | 0.5 | 4 |
| erg3Δ−2 | 0.25 | 0.5 | 4 |
| erg6aΔ−1 | 0.5 | 1 | 8 |
| erg6aΔ−2 | 0.5 | 1 | 8 |
| M. lusitanicus | |||
| Wild-type MU636 | 0.03125 | 0.5 | 4 |
| erg3Δ−1 | 0.125 | 0.125 | 2 |
| erg3Δ−2 | 0.125 | 0.125 | 2 |
| erg6aΔ−1 | 0.5 | 0.25 | 2 |
| erg6aΔ−2 | 0.5 | 0.25 | 2 |
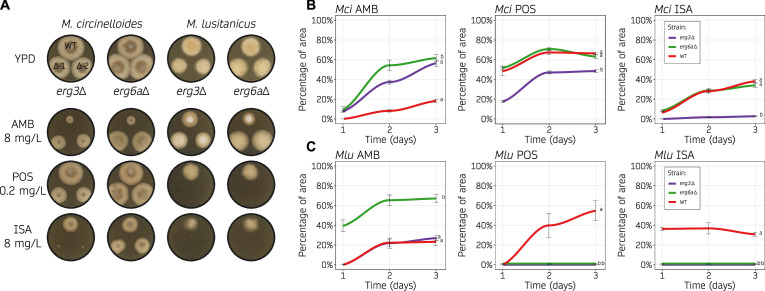
Antifungal drug susceptibility testing in molds by broth microdilution is challenging and subject to qualitative interpretation. More importantly, it is difficult to account for the inherent growth defects that are present in erg3 and erg6a deletion strains. An alternative method was employed to address this issue, measuring antifungal drug susceptibility as the area of inhibited growth on solid media, normalizing to growth without drug. To obtain a dynamic quantification of drug susceptibility, area measurements were taken at different time points (Fig. 4; Fig. S8). The differences in drug susceptibility became more evident on day 2 (Fig. 4A), as indicated by a plateau in the growth inhibition curves between days 2 and 3 (Fig. 4B and C). Growth inhibition was stronger on day 1 in every condition tested and stabilized after day 2, suggesting that amphotericin B and azoles also restrict germination in addition to growth. The erg3Δ mutants of M. circinelloides and M. lusitanicus showed an increased susceptibility to both azoles—posaconazole and isavuconazole—and this hypersusceptibility was more severe under isavuconazole stress. Similarly, amphotericin B resistance was confirmed in erg6aΔ mutants for both Mucor species, and in M. circinelloides erg3Δ mutants. M. lusitanicus erg6aΔ mutants also showed an increased susceptibility to azole drugs, overall confirming that these differences in susceptibility observed in the MIC tests (Table 1) were significant and independent of their inherent growth defects. Taken together, our results indicate that modifying the ergosterol biosynthetic pathway, and consequently, the content of ergosterol and intermediate sterols, may contribute to the observed drug susceptibility changes that we aim to address further.
Fig 4.
Loss of Erg3 or Erg6a function leads to changes in azole and polyene susceptibility. (A, B, C) Amphotericin B (AMB, 8 mg/L), posaconazole (POS, 0.2 mg/mL), and isavuconazole (ISA, 8 mg/mL) susceptibility testing of wild type and ergosterol mutants on solid YPD media. Each species was assayed at its optimal temperature, 30°C and 26°C for M. circinelloides (Mci) and M. lusitanicus (Mlu), respectively. In (A), representative images of growth after 48 hours in all of the drug containing media and the control medium without drug (YPD). Two independently generated mutants were assayed for each gene deletion (Δ−1 and Δ−2). In (B, C), Mci (B) and Mlu (C) growth rates from each mutant across time (24 hour intervals) were determined as the percentage of growth area from the same strain cultured on YPD medium with and without drug. Individual values were determined either in six biological replicates for the wild type or in three biological replicates from two independently generated mutants (six total values, grouped together for simplicity as no significant differences were detected) and used to plot a smoothed curve and SD values as black lines. Strains were grouped by letters showing significant growth rate differences across the whole time-course (one-way ANOVA and Tukey HSD test, P-value ≤ 0.01).
Ergosterol profile alteration as the underlying mechanism of antifungal drug resistance
The C-5 desaturase Erg3 and C-24 methyltransferase Erg6 are indispensable components of the ergosterol biosynthetic pathway, responsible for converting lanosterol into ergosterol through a series of catalytic reactions performed by these and other Erg enzymes. Loss-of-function of either enzyme has been shown to alter the ergosterol and related sterol composition in other organisms (42, 43), and we hypothesized that deletion of the erg3 or erg6a genes in Mucor species might similarly modify their sterol profiles, explaining the observed changes in antifungal drug susceptibility. To test this hypothesis, we analyzed the sterol profiles of M. circinelloides wild-type control, erg3Δ, and erg6aΔ strains by gas chromatography and mass spectrometry (Fig. 5A and Data set S1). Sterols were classified into different categories: C-5(6)-desaturated ergosta-type sterols, C-5(6)-saturated ergosta-type sterols, C-14-methylated sterols, and cholesta-type sterols. This classification aims to simplify sterol characterization and emphasize the accumulation of sterols that may result from Erg3 or Erg6 loss-of-function, as well as exposure to polyene or azole treatments. It is important to note that several sterols classified as cholesta-type may also exhibit C-5(6) desaturation or C-14 methylation.
Fig 5.
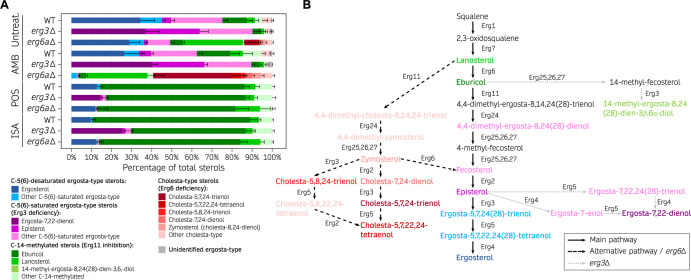
Ergosterol composition upon exposure to first-line mucormycosis treatments is altered by Erg3 and Erg6a deficiencies. (A) Percentage of total sterols in M. circinelloides wildtype (WT), erg3Δ, and erg6aΔ strains when exposed to different antifungal treatments (amphotericin B, posaconazole, and isavuconazole) and untreated RPMI cultures, represented in a stacked bar plot. Mean and SD (only upper bound is shown) values were determined from six replicates in the WT strain; for each mutation and condition, two independently generated mutants were tested in triplicate (total of 6 replicates). Sterols are classified into four main categories that reflect the expected end products from the main ergosterol pathway [C-5(6)-desaturated sterols], Erg3 deficiency [C-5(6)-saturated sterols], Erg6 deficiency (cholesta-type sterols), and Erg11 inhibition due to azole exposure (14-methylated sterols). Within each category, the most abundant sterols are color-coded to facilitate visualization. (B) Model of the ergosterol biosynthetic pathway in Mucorales. The main pathway is depicted by straight, black arrows. Alternative pathways that are relevant in erg3 (dashed, black arrows) or erg6a deletions (dotted, gray arrows) are also shown. For better illustration, sterol compounds are classified and color-coded as in (A).
First, we focused on the sterol profile of M. circinelloides cultured in untreated conditions, beginning with the wild-type strain and then the erg3Δ and erg6aΔ mutants. The relative ergosterol content was lower in M. circinelloides wild type (34.3% ± 7.2%) than in other fungi (18, 22, 44, 45), yet it remains the most abundant sterol (Fig. 5A; Data set S1). Intriguingly, we found relatively high levels of C-5(6)-saturated sterols [fecosterol, episterol, ergosta-7,22,24 (28)-trienol, and others], as well as C-14-methylated sterols (lanosterol, eburicol, and others). These results indicate that other ergosta-type sterols may act as essential components of M. circinelloides membranes in addition to ergosterol. M. circinelloides erg3 deletion resulted in a >90% increase in C-5(6)-saturated sterols; notably, these include elevated levels of ergosta-7,22-dienol (37.0% ± 6.7%), which was not detected in the wild-type strain. More importantly, erg3Δ mutation led to complete depletion of C-5(6)-desaturated ergosta-type sterols, including ergosterol, confirming a complete loss of Erg3 function. On the other hand, erg6a deletion caused a decrease in ergosterol and an increase in C-24-non-methylated sterols (>40% lanosterol and cholesta-type sterols), confirming Erg6a activity as a C-24 methyltransferase and suggesting a partial loss of Erg6 function. Surprisingly, M. circinelloides erg6aΔ mutants still synthesize C-24-methylated sterols, such as ergosterol, which suggests a compensatory upregulation of additional erg6 genes. Following this hypothesis, RT-qPCR transcription analysis revealed that upon erg6a deletion, erg6b is upregulated in regular growth conditions, while erg6c is induced upon exposure to amphotericin B (Fig. S10B). Overall, these findings are consistent with Erg6a serving as the primary Erg6 enzyme, as Erg6a loss-of-function significantly redirects the ergosterol biosynthetic pathway toward the production of cholesta-type sterols, which is indicative of a severe Erg6 deficiency (Fig. 5B; Fig. S9, S10A and C). Erg6 deficiency caused by erg6a deletion may be partially compensated by erg6b and/or erg6c upregulation (Fig. S10B), though not completely. The relative abundance of eburicol and absence of zymosterol in the wild-type strain, together with higher relative amounts of lanosterol compared to zymosterol in erg6aΔ mutants, evidence that Erg6 precedes Erg11 activity on lanosterol, delimiting the preferred ergosterol biosynthetic pathway in M. circinelloides (Fig. 5B; Fig. S9, center lane).
Next, we analyzed the sterol profile of the wild-type and mutant strains after amphotericin B treatment. Polyenes, such as amphotericin B, binding to ergosterol leads to fungal cell death and is usually correlated with a drastic decrease in ergosterol (10, 11). Indeed, the normalized amount of ergosterol was considerably lower in every strain treated with amphotericin B (Fig. S11). The relative sterol contents of amphotericin B-treated M. circinelloides wild-type and erg3Δ mutant strains were similar to their untreated cultures, except for a marked decrease in ergosterol in the wild type. On the other hand, the relative sterol profile of the erg6aΔ mutant was substantially altered by amphotericin B treatment, resulting in a total absence of ergosterol production, abolishment of most C-5(6)-saturated sterols except for fecosterol, which exhibited a drastic decrease, and more importantly, a near 60% relative abundance of cholesta-5,7,24-trienol and other cholesta-type sterols (Fig. 5A and Data set S1). M. circinelloides erg6aΔ mutants exhibit an increased resistance to amphotericin B, suggesting that cholesta-type sterol-based membranes are sufficiently stable and that producing these sterols as an alternative to ergosterol may protect the fungus against amphotericin B treatment.
Finally, the sterol content in these strains was also assessed after exposure to two different azoles: posaconazole and isavuconazole. These drugs are the only azoles recommended for salvage therapy against mucormycosis. Azoles target the Erg11 enzyme, inhibiting the crucial C-14 sterol demethylase activity responsible for converting lanosterol or eburicol into the subsequent C-14-demethylated sterol intermediate, ultimately leading to ergosterol (Fig. 5B; Fig. S9 and S10C). In many fungal species, this blockade in the ergosterol biosynthetic pathways redirects the sterol flow into a different pathway that leads to the accumulation of the toxic sterol 14-methyl-ergosta-8,24 (28)-dien-3,6-diol, hereinafter toxic 3,6-diol, which is the cause of the azole antifungal effect. Indeed, the sterol profile revealed that exposure to either azole drug led to the overaccumulation of C-14-methylated sterols, mainly eburicol (>70%); and a substantial decrease in ergosterol and other ergosta-type sterols (Fig. 5A; Data set S1). But surprisingly, azole treatment did not result in an overaccumulation of the toxic 3,6-diol or its precursor 14-methyl-fecosterol, both frequently found in large proportions in other fungal species (12). Only trace amounts of the toxic 3,6-diol (0.2% ± 0.1%) were found in the wild-type strain during exposure to both azoles, while erg3Δ mutants did not produce the toxic 3,6-diol at all (Fig. 5A and Data set S1). These findings reveal that although M. circinelloides Erg3 is capable of catalyzing this step, there is a limited utilization of the pathway responsible for converting eburicol into toxic sterols during azole exposure (Fig. S10C and D) and are particularly intriguing given the susceptibility of M. circinelloides to posaconazole and isavuconazole. Notably, the hypersusceptibility of the erg3Δ mutants to these azoles implies that the antifungal effect results from the absence of ergosterol and other stable sterols in the cellular membranes (Fig. 5A; Data set S1).
Taken together, our findings define the preferred ergosterol biosynthetic pathway in M. circinelloides (Fig. 5B; Fig. S9, center lane) and reveal that Erg3 and Erg6a play critical roles in antifungal drug susceptibility. The erg6a deletion neither impairs growth nor abolishes virulence completely but results in decreased Erg6 activity and,therefore, an abundance of stable cholesta-type sterols over ergosterol during exposure to amphotericin B that accounts for the antifungal resistance observed in erg6aΔ mutants. In addition, despite erg3Δ mutation leading to increased susceptibility to azoles, we could not detect an abundance of toxic 3,6-diol during azole exposure, suggesting that lacking critical sterols for proper membrane function is driving posaconazole and isavuconazole antifungal effect in Mucorales.
DISCUSSION
Mucormycoses are fungal infections that present with various clinical manifestations, respond differently to therapy, and result in varying outcomes. These infections are as diverse as the distinct species responsible for causing the disease. Indeed, mucoralean fungi display interspecies differences in antifungal drug susceptibility (2, 46) and interactions with host immunity (47), which can explain disease heterogeneity, and these considerations emphasize the importance of specific pathogenicity models in mucormycosis research. However, these species remain understudied with limited genetic models available to study pathogenesis and therapeutics. Recent taxonomic advances revealed that many M. circinelloides strains actually belong to different phylogenetic species within the M. circinelloides species complex (27), with M. circinelloides being the most prevalent in mucormycosis patients. However, research on pathogenicity mechanisms has been conducted with M. lusitanicus, a sister species previously classified as M. circinelloides f. lusitanicus, for which no clinical isolates have been found to date (27). This work addresses these limitations by developing a new molecular biology model based on the pathogenic human isolate M. circinelloides strain 1006PhL. As a result of this study, this strain that exhibits full virulence in our murine model is now amenable to genetic transformation and mortality rates and fungal burden are consistently high and reproducible, partially due to this pathogenic isolate thermotolerance to the range of mammalian physiological temperatures. This thermotolerance also allows for standardized antifungal drug susceptibility testing at recommended temperatures. Our research has demonstrated that loss-of-function mutations of key target genes, such as pyrF, results in a completely avirulent phenotype, which can be reversed by genetic complementation via genomic integration. The necessity of the pyrimidine biosynthetic pathway for virulence is in agreement with research in other fungal model organisms (36, 48–50), demonstrating that this model is suitable for virulence factor characterization and establishing a foundation for novel molecular pathogenesis research to follow.
The only available antifungal agents effective against mucormycosis, namely azoles and liposomal amphotericin B, modify the ergosterol content, emphasizing the crucial role of ergosterol biosynthesis in antifungal drug susceptibility. For instance, gain-of-function mutations affecting Erg11, the target of azole drugs, typically confer azole drug resistance in many fungal pathogens (51). Similarly, Mucorales exhibit intrinsic resistance to voriconazole and fluconazole due to an Erg11 duplication with an amino acid substitution affecting the azole-binding site (6). Mucorales can develop antifungal drug resistance not only through classical genetic point mutations (6) and insertional mutations (52) but also via epigenetic modifications that lead to transient loss of drug target function (8, 52, 53). Our findings elucidate the preferred ergosterol biosynthetic pathway in M. circinelloides, indicating that C-24 methylation catalyzed by Erg6a occurs prior to C-14 demethylation by Erg11. This result is consistent with our understanding of ergosterol biosynthesis in other filamentous fungi (44), as well as in Candida species (18, 42, 54) and Cryptococcus neoformans (55), which deviates from the ergosterol pathway proposed in Saccharomyces cerevisiae (56). However, the relative content of ergosterol in M. circinelloides is reduced compared to these other fungal species, while other sterols such as episterol, lanosterol, and eburicol are more abundant. There is a common misconception that ergosterol is the sole sterol found in all fungi and is essential for fungal membrane stability. While it may be the dominant sterol in many of the Dikarya fungi, there is substantial evidence of alternative sterols serving the same role, especially early-diverging fungi (EDF) (57). Most EDF lack Erg5 and even Erg11 and instead harbor a different set of sterol biosynthetic enzymes, such as 7-dehydrocholesterol reductase (DHCR7), resulting in a wider diversity of sterols and a complete absence of ergosterol (57, 58). While this is not the case in most Mucoromycota species, our research reveals that they possess at least two Erg6 paralogs, and Mucor species harbor three: Erg6a, Erg6b, and Erg6c. These additional Erg6 paralogs, which are putative C-24 methyltransferases, may be responsible for M. circinelloides unique sterol composition, particularly the abundance of C-24-methylated sterols. As a sister clade to the Dikarya, Mucoromycota represents their closest EDF relatives and, therefore, displays a hybrid sterol composition profile between more phylogenetically distant EDF and well-studied Dikarya. Taken together, our findings provide new insights into the evolution of membrane sterol composition in fungi, showing evidence of alternative sterol pathways.
Dissecting the role of the C-5 sterol desaturase Erg3 in the ergosterol pathway of M. circinelloides uncovered unexpected peculiarities. Despite a total absence of ergosterol in the erg3Δ mutants, our findings indicate that Mucor species are able to survive on alternative sterol intermediates, as has been previously reported in C. glabrata erg11 mutants (59), and similar to EDF species naturally lacking ergosterol (57, 58). The altered sterol composition may explain the growth delay observed in erg3Δ mutants at optimal temperatures, which becomes more pronounced at higher temperatures, as well as the mutants’ complete loss of virulence. Reasonably, the changes in sterol content, possibly accompanied by compensating alterations in other membrane lipids, can cause an unstable equilibrium that enables growth in non-stressful conditions but collapses upon harsh environmental changes, particularly during thermal stress and host interactions. These findings align with the observed erg3 loss-of-function mutations in Candida species, which exhibit increased susceptibility to physiological stresses (60, 61) and reduced hyphal growth under certain conditions (62). Consequently, many of these erg3 null mutants of Candida species have been associated with reduced pathogenicity in animal models, albeit these azole-resistant, erg3 mutant isolates originated from human patients (60, 62, 63), and some compensatory mutations can restore their pathogenic potential (64). In addition to these defects, erg3 loss-of-function mutations confer azole resistance in nearly every Candida species (13, 54, 60, 61, 63–67), which might be the reason they are so frequently identified from clinical isolates. Azoles block ergosterol synthesis by inhibiting the C-14 demethylase Erg11, causing an accumulation of Erg11 substrates: lanosterol and eburicol. Typically, these substrates are redirected to a different pathway characterized by the production of 14-methyl-fecosterol (through the subsequent action of Erg25, Erg26, and Erg27), which is then turned into the toxic 3,6-diol through Erg3 C-6 hydroxylase activity. The absence of Erg3 activity prevents the formation of the toxic 3,6-diol and allows the accumulation of 14-methyl-fecosterol, accounting for the observed resistance in the Candida erg3 null mutants (42, 68). Because the loss of Erg3 function prevents ergosterol synthesis in Candida species, polyenes are not able to bind the cell membrane nor exert their antifungal effects. As a result, these mutants often exhibit resistance not only to azoles but also to polyenes such as amphotericin B (15, 54, 69, 70). M. circinelloides erg3Δ mutants are also more resistant to amphotericin B and, as in Candida species, this is likely due to a complete absence of ergosterol that prevents polyene binding. However, these mutants are hypersusceptible to azole drugs instead of more resistant, and neither the wild-type nor the mutant strains produce the toxic 3,6-diol. This finding suggests that Erg3 may exhibit a limited capability for C-6 hydroxylation of C-14-methylated sterols compared to other fungi. Nevertheless, the overaccumulation of eburicol in M. circinelloides erg3Δ mutants during azole exposure, together with the absence of 14-methyl-fecosterol and 4,14-dimethyl-cholesta-8,24-dienol, imply that the Erg25, Erg26, and Erg27 enzymes may prefer different substrates other than eburicol or are simply unable to initiate C-4 demethylation on C-14-methylated sterols as more likely scenarios. This sterol composition with abundant eburicol and absence of 14-methyl-fecosterol or ergosterol could hinder growth and cell membrane function, explaining the hypersusceptibility to azoles in M. circinelloides erg3Δ mutants and the differences compared to Candida species.
This study further investigates Erg6, a C-24 methyltransferase that catalyzes an essential step in the biosynthesis of ergosterol. Erg6 function has been studied in several fungal pathogens and revealed as important for growth and development under various conditions, adaptation to cell wall stresses, virulence (18–23), and in some instances, essential for cell viability (44). Despite these defects, the absence of Erg6 activity can provide a selective advantage by conferring amphotericin B drug resistance, as seen in multiple Candida species and Cryptococcus neoformans (17–23). Indeed, a recently conducted large-scale study, involving a collection of over four hundred C. auris strains evolved from clinical isolates, reported that mutations in erg6 were the most frequent cause of amphotericin B resistance (71). Exploring the function of Erg6 in Mucor species presented additional challenges due to the presence of three different Erg6 paralogs. Our phylogenetic and transcriptomic analyses identified Erg6a as the primary Erg6 enzyme involved in Mucor species ergosterol biosynthesis. This hypothesis was confirmed by a substantial reduction in C-24-methylated sterols in the erg6aΔ mutants although some residual Erg6 activity persisted. Erg6b and Erg6c are upregulated upon loss of Erg6a function and, therefore, might serve as functionally redundant C-24 methyltransferases, accounting for the residual Erg6 activity. As in other fungal species, Mucor species erg6aΔ mutants exhibit defects in asexual sporulation, a significant delay in virulence, and decreased host dissemination. More importantly, erg6aΔ mutants are highly resistant to amphotericin B. The sterol profile of erg6aΔ mutants under amphotericin B exposure provided insights into the mechanism of drug resistance. It revealed an abundance of cholesta-type sterols, which bear a closer resemblance to cholesterol found in animal cell membranes. A similar sterol profile was observed in amphotericin B-resistant C. auris (18) and C. glabrata (20) clinical isolates harboring erg6 mutations, indicating that cholesta-type sterols are a viable alternative to ergosterol.
Our findings raise questions with respect to a recent study of M. lusitanicus erg6 mutants suggesting the fungus utilizes cholesta-type sterols to produce desmosterol or cholesterol (72), a process requiring DHCR7 activity. Neither their analysis nor ours identified any C7-saturated sterols and although many EDF harbor DHCR7, mucoralean genomes do not encode this enzyme (58). Notably, the authors observed no changes in susceptibility to amphotericin B in their reported erg6a mutants, whereas we identified clear resistance to amphotericin B in multiple independently generated erg6aΔ mutants of two different Mucor species. We attribute these seemingly conflicting results to potential methodological issues in gene deletion verification, as Bauer et al. (72) presented no DNA-based evidence confirming the complete and homokaryotic deletion of erg6 gene sequences. Our findings, supported by rigorous genetic methodology, define the most plausible pathway for cholesta-type sterol production and their role in amphotericin B resistance in Mucor erg6aΔ mutants, corroborated by research in other fungal pathogens.
Given the growing susceptible patient population and dearth of available antifungal drugs, mucormycoses pose substantial challenges to global public health. We propose that mutations and epimutations leading to the loss of Erg6a function may provide a selective advantage to Mucorales by conferring amphotericin B resistance. Although M. circinelloides erg6aΔ mutants exhibit minor defects in virulence, they can sustain mammalian physiological temperatures and effectively disseminate the infection. It is likely that the residual Erg6 activity observed in the erg6aΔ mutants can rescue the most deleterious consequences of loss of Erg6 function, while still conferring amphotericin B resistance due to cholesta-type sterols production. A comparable scenario was recently reported in C. auris evolved isolates, where erg6 mutations conferring amphotericin B resistance emerged in vivo, occasionally accompanied by other mechanisms that compensate for fitness trade-offs (71). Similarly, the loss of Erg6a function in mucoralean pathogens may arise in vivo and lead to mucormycosis infections that do not respond to first-line antifungal therapies and may challenge recent and exciting developments in polyene antifungal drug discovery (73). Currently, it is complicated to identify resistant clinical isolates because there are no clinical breakpoints defined for antifungal susceptibility testing in Mucorales (74). Therefore, improving current guidelines to identify clinically amphotericin B-resistant isolates from mucormycosis patients should be a priority. Antifungal susceptibility testing may pose a challenge in identifying epimutant clinical isolates due to the transient and unstable nature of epigenetic changes, which can occur but remain undetected. This adds an additional layer of complexity to the identification of resistant clinical isolates but opens new and exciting avenues for future research.
MATERIALS AND METHODS
Fungal strains and culture conditions
M. lusitanicus mutant strains generated in this work derive from the double auxotrophic (Ura-, Leu-) MU402 strain (75), using the selectable marker pyrG to complement uracil auxotrophy through genetic transformation. The MU636 strain (28) was used as the wildtype, complemented control (pyrG−::pyrG+ leuA−) for phenotypic screening of M. lusitanicus mutants. Similarly, M. circinelloides mutant strains derive from M. circinelloides 1006PhL (34). A uracil auxotroph amenable to genetic transformation, MIN6 (Ura-) was generated in this work. Uracil auxotrophy was complemented by the selectable marker pyrF. For phenotypic analysis, the CPA6 strain (pyrF−::pyrF+) was used as a control. All the strains generated in this study are listed in Table S3.
Spores to conduct experiments were harvested and counted from YPD solid cultures after 4–5 days. The growth rates were quantified by spot inoculating 500 spores onto rich yeast-peptone-dextrose (YPD) medium and minimal yeast-nitrogen-base (YNB) medium at 26°C, 30°C, and 37°C, and analyzed with Fiji (76).
Isolation of uracil auxotrophic strain MIN6
106 spores of the wild-type 1006PhL strain were spread-plated on YPD-5-FOA (3 g/L) supplemented with uracil (100 mg/L) and uridine (200 mg/L) and screened for resistant isolates after 3 days. Resistant isolates were rechallenged and those exhibiting stable 5-FOA resistance were inoculated onto YNB media with or without uracil and uridine to determine uracil auxotrophy. Genomic DNA from isolates exhibiting uracil auxotrophy was purified using the MasterPure Complete DNA and RNA Purification Kit (Lucigen). The promoter, ORF, and terminator containing sequences from pyrG and pyrF were PCR-amplified and Sanger-sequenced using primers listed in Table S4.
Protoplast generation and transformation by electroporation
M. lusitanicus transformation was performed following previously established protocols (41). For M. circinelloides transformation, MIN6 spores were incubated in yeast-peptone-glucose (YPG) medium at 26°C until germlings appeared. Cell walls were digested with lysing enzymes from Trichoderma harzianum (L-1412, Sigma-Aldrich) and chitosanase (C-0794, Sigma-Aldrich) at 30°C for 1.5 hours (41). Protoplasts were washed, centrifuged, and resuspended in isotonic buffer. After that, protoplasts were mixed with linear double-stranded DNA (3–6 μg) and subjected to exponential decay waveform electroporation (1,000 V, 25 µF, and 400 Ω). Protoplasts were recovered in YPG at 26°C for 1 hour, and spread-plated on minimal medium with casamino acids for Mucor (MMC) pH 3.2 to select prototroph transformants after 2–5 days incubation in dark conditions.
For gene deletions, we designed linear DNA constructs containing the selectable marker flanked by 1 kb upstream and downstream regions of the target gene. Either pyrG or pyrF were used as selectable markers for M. lusitanicus and M. circinelloides, respectively. In addition, a M. circinelloides complemented control strain named CPA6 (pyrF−Δ59::pyrF+) was generated by integrating the wildtype pyrF marker into MIN6, replacing the pyrF mutated allele (pyrF-Δ59). Gene deletions as well as homokaryosis were verified by AFLP; first, after two vegetative passages in selective medium MMC, and subsequently, after every additional passage until homokaryosis was achieved.
Ortholog search and phylogenetic tree inference
Erg3 and Erg6 protein sequences from Saccharomyces cerevisiae were used as queries in a PSI-BLAST v2.12.0 search (77) against a proteomic database of selected species (see Table S5 for a comprehensive list) (32–34, 38–40, 78–90). Every match was subjected to a reciprocal BLASTp search against the S. cerevisiae proteome. These putative orthologs (Tables S1 and S2) were aligned using MAFFT v7.475 (91), trimmed with TrimAl v1.4.rev15 (92), and phylogenetic trees with 1,000 ultrafast bootstraps and SH-aLRT replicates were inferred by IQ-TREE v2.2.0.3 (93).
RNA isolation, sequencing, and data analyses
Duplicate YPD liquid cultures were grown for 16 hours at 26°C and 250 rpm. Total RNA was purified with a QIAGEN miRNeasy Mini Kit. rRNA-depleted RNA libraries were prepared using Illumina Stranded Total RNA Prep with Ribo-Zero Gold rRNA Removal Kit, and sequenced to obtain 150 bp paired-end reads. In addition to these, M. lusitanicus similar and publicly available data were used for gene expression analyses (52). Quality was assessed by FASTQC v0.11.9 and adapters and low-quality reads removed by TrimGalore! V0.6.7. Processed reads were aligned to either M. circinelloides 1006PhL (https://fungidb.org/fungidb/app/record/dataset/ DS_8b08c1c31d) or M. lusitanicus MU402 genome (https://mycocosm.jgi.doe.gov/Muccir1_3/ Muccir1_3.info.html) employing STAR v.2.7.10a (94). Coverage files were generated using bamCompare from Deeptools2 v3.5.1 (95) to merge duplicates into a single bigWig file.
Synteny analysis
Pairwise synteny among closely related mucoralean species was assessed using the JCVI toolkit MCScan pipeline (96). Genomic plots were generated by Deeptools2 pyGenomeTracks v3.7 (97).
Virulence assays and fungal burden
Four-week-old BALB/c mice (Charles River) weighing 20–25 g were immunosuppressed with cyclophosphamide via intraperitoneal injection (200 mg/kg of body weight), 2 days prior to infection and every 5 days thereafter. Groups of 10 mice were intravenously challenged via retro-orbital injection with 1 × 106 spores from each of the mutant and control strains.
Fungal burden was quantified in five organs (brain, lung, spleen, kidney, and liver) of three infected mice meeting end-point criteria per group. Organ homogenates were plated onto YPD supplemented with 1 µg/mL of FK506 to induce yeast growth. Colony-forming units (CFU) were quantified after 4 days and normalized per volume plated and organ weight.
Antifungal drug susceptibility testing
Antifungal drug susceptibility was determined by broth microdilution using the CLSI and EUCAST standard methodology for molds. Minimal inhibitory concentrations (MIC) were evaluated for amphotericin B, posaconazole, and isavuconazole. 105 spores/mL were incubated in Roswell Park Memorial Institute (RPMI) 1640 medium at different drug concentrations and 35°C for 24 and 48 hours. Wells were evaluated for visible growth or lack therein. Additionally, 500 spores were spot-inoculated onto solid YPD solid at specific drug concentrations: 8 mg/L of liposomal amphotericin B (Ambisome, Gilead Sciences), 0.2 mg/L of posaconazole (Noxafil, Merck), and 8 mg/L of isavuconazole (Cresemba, Astellas Pharma). The area of inhibited growth was determined as the ratio of growth area in treated compared to untreated plates using Fiji.
Ergosterol profile quantification
104 spores were cultured at 35°C for 48 hours and 60 rpm in RPMI media (untreated) and RPMI supplemented with half the MIC for each corresponding strain for amphotericin B, posaconazole, and isavuconazole drugs. Non-saponifiable lipids were extracted from lyophilized mycelia as previously described (12), with cholesterol added as an internal standard. Sterols were derivatized using 0.1 mL N,O-Bis(trimethylsilyl)trifluoroacetamide and trimethylsilyl chloride [BSTFA and TMCS, (99:1)] and 0.3 mL anhydrous pyridine and heating at 80°C for 2 hours (98). TMS-derivatized sterols were analyzed using gas chromatography–mass spectrometry (GS/MS) (Thermo 1300 GC coupled to a Thermo ISQ mass spectrometer, Thermo Scientific) and identified with reference to relative retention times, mass ions, and fragmentation spectra. GC/MS data files were analyzed using Xcalibur software (Thermo Scientific). Sterol composition was calculated from peak areas, as a mean of three replicates per independently generated mutant (six replicates per gene deletion) or six replicates for the wildtype strain 1006PhL. The relative quantity of sterols present was determined from the peak areas of the sterol and the internal standard and divided by the dry weight of the sample.
RT-qPCR analysis
Cultures were obtained using the same conditions as for ergosterol profiling. Total RNA was isolated as previously described, and cDNA synthesized using Maxima H Minus cDNA Synthesis Kit (Thermo Scientific). qPCRs were prepared with SYBR green PCR master mix (Applied Biosystems) using primers that specifically amplified erg6b and erg6c paralogs, and the vma1 gene served as the endogenous control (Table S4), in triplicate, and performed in a QuantStudio three real-time PCR system.
ACKNOWLEDGMENTS
We thank Anna Floyd Averette for her constant technical support. We also extend our gratitude to Marcus Hull for providing technical support during the ergosterol profiling process. We commend Dr. Devjanee Swain Lenz and Duke’s Sequencing and Genomic Technologies Core Facility for their assistance, as well as Thomas Milledge and the Duke Computer Cluster team for their computing resources.
This study was supported by Astellas Pharma Investigator Sponsored Research ISR005855/CRES-21B01, and NIH/National Institute of Allergy and Infectious Diseases R01 Grant AI170543 both awarded to J.H. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication. J.H. is Codirector and Fellow of the CIFAR program Fungal Kingdom: Threats & Opportunities.
Footnotes
This article is a direct contribution from Joseph Heitman, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Anna Selmecki, University of Minnesota Twin Cities, and Theodore White, University of Missouri Kansas City.
Contributor Information
María Isabel Navarro-Mendoza, Email: maribel.navarro@duke.edu.
Carlos Pérez-Arques, Email: carlos.parq@duke.edu.
Joseph Heitman, Email: heitm001@duke.edu.
Anuradha Chowdhary, Vallabhbhai Patel Chest Institute, Delhi, India.
DATA AVAILABILITY
Raw rRNA-depleted RNA-sequencing data sets obtained from the M. circinelloides 1006PhL strain are accessible under PRJNA1046487 NCBI’s Sequence Read Archive (SRA) project accession number. RNA-seq data sets derived from the M. lusitanicus MU402 strain were similarly generated for a prior study (PRJNA903107) and are publicly available (52).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01661-24.
Figures S1-S6.
Figures S7-S11.
Detailed overview of materials and methods.
Tables S1-S5.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Petrikkos G, Skiada A, Lortholary O, Roilides E, Walsh TJ, Kontoyiannis DP. 2012. Epidemiology and clinical manifestations of mucormycosis. Clin Infect Dis 54 Suppl 1:S23–34. doi: 10.1093/cid/cir866 [DOI] [PubMed] [Google Scholar]
- 2. Cornely OA, Alastruey-Izquierdo A, Arenz D, Chen SCA, Dannaoui E, Hochhegger B, Hoenigl M, Jensen HE, Lagrou K, Lewis RE, et al. 2019. Global guideline for the diagnosis and management of mucormycosis: an initiative of the European confederation of medical mycology in cooperation with the mycoses study group education and research consortium. Lancet Infect Dis 19:e405–e421. doi: 10.1016/S1473-3099(19)30312-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoenigl M, Seidel D, Carvalho A, Rudramurthy SM, Arastehfar A, Gangneux J-P, Nasir N, Bonifaz A, Araiza J, Klimko N, Serris A, Lagrou K, Meis JF, Cornely OA, Perfect JR, White PL, Chakrabarti A, ECMM and ISHAM collaborators . 2022. The emergence of COVID-19 associated mucormycosis: a review of cases from 18 countries. Lancet Microbe 3:e543–e552. doi: 10.1016/S2666-5247(21)00237-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gold JAW, Adjei S, Gundlapalli AV, Huang Y-LA, Chiller T, Benedict K, Toda M. 2023. Increased hospitalizations involving fungal infections during COVID-19 pandemic, United States, January 2020–December 2021. Emerg Infect Dis 29:1433–1437. doi: 10.3201/eid2907.221771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization . 2022. WHO fungal priority pathogens list to guide research, development and public health action. Geneva. [Google Scholar]
- 6. Caramalho R, Tyndall JDA, Monk BC, Larentis T, Lass-Flörl C, Lackner M. 2017. Intrinsic short-tailed azole resistance in mucormycetes is due to an evolutionary conserved aminoacid substitution of the lanosterol 14α-demethylase. Sci Rep 7:15898. doi: 10.1038/s41598-017-16123-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Almyroudis NG, Sutton DA, Fothergill AW, Rinaldi MG, Kusne S. 2007. In vitro susceptibilities of 217 clinical isolates of zygomycetes to conventional and new antifungal agents. Antimicrob Agents Chemother 51:2587–2590. doi: 10.1128/AAC.00452-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calo S, Shertz-Wall C, Lee SC, Bastidas RJ, Nicolás FE, Granek JA, Mieczkowski P, Torres-Martínez S, Ruiz-Vázquez RM, Cardenas ME, Heitman J. 2014. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature 513:555–558. doi: 10.1038/nature13575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang Z, Heitman J. 2019. Drug-resistant epimutants exhibit organ-specific stability and induction during murine infections caused by the human fungal pathogen Mucor circinelloides. mBio 10:e02579–19. doi: 10.1128/mBio.02579-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gray KC, Palacios DS, Dailey I, Endo MM, Uno BE, Wilcock BC, Burke MD. 2012. Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci U S A 109:2234–2239. doi: 10.1073/pnas.1117280109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palacios DS, Dailey I, Siebert DM, Wilcock BC, Burke MD. 2011. Synthesis-enabled functional group deletions reveal key underpinnings of amphotericin B ion channel and antifungal activities. Proc Natl Acad Sci U S A 108:6733–6738. doi: 10.1073/pnas.1015023108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Kelly DE. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14 alpha-methylergosta-8,24(28)-dien-3 beta,6 alpha-diol. Biochem Biophys Res Commun 207:910–915. doi: 10.1006/bbrc.1995.1272 [DOI] [PubMed] [Google Scholar]
- 13. Watson PF, Rose ME, Ellis SW, England H, Kelly SL. 1989. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem Biophys Res Commun 164:1170–1175. doi: 10.1016/0006-291x(89)91792-0 [DOI] [PubMed] [Google Scholar]
- 14. Parker JE, Warrilow AGS, Price CL, Mullins JGL, Kelly DE, Kelly SL. 2014. Resistance to antifungals that target CYP51. J Chem Biol 7:143–161. doi: 10.1007/s12154-014-0121-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AGS, Rolley N, Kelly DE, Kelly SL. 2010. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob Agents Chemother 54:4527–4533. doi: 10.1128/AAC.00348-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCammon MT, Hartmann MA, Bottema CD, Parks LW. 1984. Sterol methylation in Saccharomyces cerevisiae. J Bacteriol 157:475–483. doi: 10.1128/jb.157.2.475-483.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jensen-Pergakes KL, Kennedy MA, Lees ND, Barbuch R, Koegel C, Bard M. 1998. Sequencing, disruption, and characterization of the Candida albicans sterol methyltransferase (ERG6) gene: drug susceptibility studies in ERG6 mutants. Antimicrob Agents Chemother 42:1160–1167. doi: 10.1128/AAC.42.5.1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rybak JM, Barker KS, Muñoz JF, Parker JE, Ahmad S, Mokaddas E, Abdullah A, Elhagracy RS, Kelly SL, Cuomo CA, Rogers PD. 2022. In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin Microbiol Infect 28:838–843. doi: 10.1016/j.cmi.2021.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Young LY, Hull CM, Heitman J. 2003. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob Agents Chemother 47:2717–2724. doi: 10.1128/AAC.47.9.2717-2724.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahmad S, Joseph L, Parker JE, Asadzadeh M, Kelly SL, Meis JF, Khan Z. 2019. ERG6 and ERG2 are major targets conferring reduced susceptibility to amphotericin B in clinical Candida glabrata isolates in Kuwait. Antimicrob Agents Chemother 63:e01900–18. doi: 10.1128/AAC.01900-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vandeputte P, Tronchin G, Larcher G, Ernoult E, Bergès T, Chabasse D, Bouchara J-P. 2008. A nonsense mutation in the ERG6 gene leads to reduced susceptibility to polyenes in a clinical isolate of Candida glabrata. Antimicrob Agents Chemother 52:3701–3709. doi: 10.1128/AAC.00423-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Joseph-Horne T, Hollomon D, Loeffler RS, Kelly SL. 1995. Cross-resistance to polyene and azole drugs in Cryptococcus neoformans. Antimicrob Agents Chemother 39:1526–1529. doi: 10.1128/AAC.39.7.1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oliveira FFM, Paes HC, Peconick LDF, Fonseca FL, Marina CLF, Bocca AL, Homem-de-Mello M, Rodrigues ML, Albuquerque P, Nicola AM, Alspaugh JA, Felipe MSS, Fernandes L. 2020. Erg6 affects membrane composition and virulence of the human fungal pathogen Cryptococcus neoformans. Fungal Genet Biol 140:103368. doi: 10.1016/j.fgb.2020.103368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walther G, Pawłowska J, Alastruey-Izquierdo A, Wrzosek M, Rodriguez-Tudela JL, Dolatabadi S, Chakrabarti A, de Hoog GS. 2013. DNA barcoding in mucorales: an inventory of biodiversity. Persoonia 30:11–47. doi: 10.3767/003158513X665070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richardson M. 2009. The ecology of the Zygomycetes and its impact on environmental exposure. Clin Microbiol Infect 15 Suppl 5:2–9. doi: 10.1111/j.1469-0691.2009.02972.x [DOI] [PubMed] [Google Scholar]
- 26. Mousavi B, Costa JM, Arné P, Guillot J, Chermette R, Botterel F, Dannaoui E. 2018. Occurrence and species distribution of pathogenic Mucorales in unselected soil samples from France. Med Mycol 56:315–321. doi: 10.1093/mmy/myx051 [DOI] [PubMed] [Google Scholar]
- 27. Wagner L, Stielow JB, de Hoog GS, Bensch K, Schwartze VU, Voigt K, Alastruey-Izquierdo A, Kurzai O, Walther G. 2020. A new species concept for the clinically relevant Mucor circinelloides complex. Persoonia 44:67–97. doi: 10.3767/persoonia.2020.44.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pérez-Arques C, Navarro-Mendoza MI, Murcia L, Lax C, Martínez-García P, Heitman J, Nicolás FE, Garre V. 2019. Mucor circinelloides thrives inside the phagosome through an ATF-mediated germination pathway. mBio 10:1–15. doi: 10.1128/mBio.02765-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Navarro-Mendoza MI, Pérez-Arques C, Murcia L, Martínez-García P, Lax C, Sanchis M, Capilla J, Nicolás FE, Garre V. 2018. Components of a new gene family of ferroxidases involved in virulence are functionally specialized in fungal dimorphism. Sci Rep 8:7660. doi: 10.1038/s41598-018-26051-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pérez-Arques C, Navarro-Mendoza MI, Murcia L, Navarro E, Garre V, Nicolás FE. 2020. A non-canonical RNAi pathway controls virulence and genome stability in Mucorales. PLoS Genet 16:e1008611. doi: 10.1371/journal.pgen.1008611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee SC, Li A, Calo S, Heitman J. 2013. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog 9:e1003625. doi: 10.1371/journal.ppat.1003625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Corrochano LM, Kuo A, Marcet-Houben M, Polaino S, Salamov A, Villalobos-Escobedo JM, Grimwood J, Álvarez MI, Avalos J, Bauer D, et al. 2016. Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr Biol 26:1577–1584. doi: 10.1016/j.cub.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Navarro-Mendoza MI, Pérez-Arques C, Panchal S, Nicolás FE, Mondo SJ, Ganguly P, Pangilinan J, Grigoriev IV, Heitman J, Sanyal K, Garre V. 2019. Early diverging fungus Mucor circinelloides lacks centromeric histone CENP-A and displays a mosaic of point and regional centromeres. Curr Biol 29:3791–3802. doi: 10.1016/j.cub.2019.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Kong HH, Segre JA, NIH Intramural Sequencing Center Comparative Sequencing Program . 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. doi: 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee SC, Billmyre RB, Li A, Carson S, Sykes SM, Huh EY, Mieczkowski P, Ko DC, Cuomo CA, Heitman J. 2014. Analysis of a food-borne fungal pathogen outbreak: virulence and genome of a Mucor circinelloides isolate from yogurt. mBio 5:e01390–14. doi: 10.1128/mBio.01390-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lax C, Navarro-Mendoza MI, Pérez-Arques C, Navarro E, Nicolás FE, Garre V. 2021. Stable and reproducible homologous recombination enables CRISPR-based engineering in the fungus Rhizopus microsporus. Cell Rep Methods 1:100124. doi: 10.1016/j.crmeth.2021.100124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li CH, Cervantes M, Springer DJ, Boekhout T, Ruiz-Vazquez RM, Torres-Martinez SR, Heitman J, Lee SC. 2011. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog 7:e1002086. doi: 10.1371/journal.ppat.1002086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma L-J, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M, Elias M, Idnurm A, Lang BF, Sone T, et al. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet 5:e1000549. doi: 10.1371/journal.pgen.1000549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gryganskyi AP, Golan J, Dolatabadi S, Mondo S, Robb S, Idnurm A, Muszewska A, Steczkiewicz K, Masonjones S, Liao H-L, Gajdeczka MT, Anike F, Vuek A, Anishchenko IM, Voigt K, de Hoog GS, Smith ME, Heitman J, Vilgalys R, Stajich JE. 2018. Phylogenetic and phylogenomic definition of Rhizopus species. G3 (Bethesda) 8:2007–2018. doi: 10.1534/g3.118.200235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwartze VU, Winter S, Shelest E, Marcet-Houben M, Horn F, Wehner S, Linde J, Valiante V, Sammeth M, Riege K, Nowrousian M, Kaerger K, Jacobsen ID, Marz M, Brakhage AA, Gabaldón T, Böcker S, Voigt K. 2014. Gene expansion shapes genome architecture in the human pathogen Lichtheimia corymbifera: an evolutionary genomics analysis in the ancient terrestrial Mucorales (Mucoromycotina). PLoS Genet 10:e1004496. doi: 10.1371/journal.pgen.1004496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nicolás FE, Navarro-Mendoza MI, Pérez-Arques C, López-García S, Navarro E, Torres-Martínez S, Garre V. 2018. Molecular tools for carotenogenesis analysis in the mucoral Mucor circinelloides. Methods Mol Biol 1852:221–237. doi: 10.1007/978-1-4939-8742-9_13 [DOI] [PubMed] [Google Scholar]
- 42. Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S. 2002. Molecular basis of resistance to azole antifungals. Trends Mol Med 8:76–81. doi: 10.1016/s1471-4914(02)02280-3 [DOI] [PubMed] [Google Scholar]
- 43. Robbins N, Caplan T, Cowen LE. 2017. Molecular evolution of antifungal drug resistance. Annu Rev Microbiol 71:753–775. doi: 10.1146/annurev-micro-030117-020345 [DOI] [PubMed] [Google Scholar]
- 44. Xie J, Rybak JM, Martin-Vicente A, Guruceaga X, Thorn HI, Nywening AV, Ge W, Parker JE, Kelly SL, Rogers PD, Fortwendel JR. 2024. The sterol C-24 methyltransferase encoding gene, erg6, is essential for viability of Aspergillus species. Nat Commun 15:4261. doi: 10.1038/s41467-024-48767-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luna-Tapia A, Parker JE, Kelly SL, Palmer GE. 2021. Species-specific differences in C-5 sterol desaturase function influence the outcome of azole antifungal exposure. Antimicrob Agents Chemother 65:e0104421. doi: 10.1128/AAC.01044-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wagner L, de Hoog S, Alastruey-Izquierdo A, Voigt K, Kurzai O, Walther G. 2019. A revised species concept for opportunistic Mucor species reveals species-specific antifungal susceptibility profiles. Antimicrob Agents Chemother 63:1–8. doi: 10.1128/AAC.00653-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nicolás FE, Murcia L, Navarro E, Navarro-Mendoza MI, Pérez-Arques C, Garre V. 2020. Mucorales species and macrophages. JoF 6:94. doi: 10.3390/jof6020094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Varma A, Edman JC, Kwon-Chung KJ. 1992. Molecular and genetic analysis of URA5 transformants of Cryptococcus neoformans. Infect Immun 60:1101–1108. doi: 10.1128/iai.60.3.1101-1108.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Retallack DM, Heinecke EL, Gibbons R, Deepe GS, Woods JP. 1999. The URA5 gene is necessary for Histoplasma capsulatum growth during infection of mouse and human cells. Infect Immun 67:624–629. doi: 10.1128/IAI.67.2.624-629.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. D’Enfert C, Diaquin M, Delit A, Wuscher N, Debeaupuis JP, Huerre M, Latge JP. 1996. Attenuated virulence of uridine-uracil auxotrophs of Aspergillus fumigatus. Infect Immun 64:4401–4405. doi: 10.1128/iai.64.10.4401-4405.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fisher MC, Alastruey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, Bromley M, Brüggemann R, Garber G, Cornely OA, Gurr SJ, Harrison TS, Kuijper E, Rhodes J, Sheppard DC, Warris A, White PL, Xu J, Zwaan B, Verweij PE. 2022. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol 20:557–571. doi: 10.1038/s41579-022-00720-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Navarro-Mendoza MI, Pérez-Arques C, Heitman J. 2023. Heterochromatin and RNAi act independently to ensure genome stability in Mucorales human fungal pathogens. Proc Natl Acad Sci U S A 120:e2220475120. doi: 10.1073/pnas.2220475120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Calo S, Nicolás FE, Lee SC, Vila A, Cervantes M, Torres-Martinez S, Ruiz-Vazquez RM, Cardenas ME, Heitman J. 2017. A non-canonical RNA degradation pathway suppresses RNAi-dependent epimutations in the human fungal pathogen Mucor circinelloides. PLoS Genet 13:e1006686. doi: 10.1371/journal.pgen.1006686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kelly SL, Lamb DC, Kelly DE, Manning NJ, Loeffler J, Hebart H, Schumacher U, Einsele H. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta5,6-desaturation. FEBS Lett 400:80–82. doi: 10.1016/s0014-5793(96)01360-9 [DOI] [PubMed] [Google Scholar]
- 55. Nes WD, Zhou W, Ganapathy K, Liu J, Vatsyayan R, Chamala S, Hernandez K, Miranda M. 2009. Sterol 24-C-methyltransferase: an enzymatic target for the disruption of ergosterol biosynthesis and homeostasis in Cryptococcus neoformans. Arch Biochem Biophys 481:210–218. doi: 10.1016/j.abb.2008.11.003 [DOI] [PubMed] [Google Scholar]
- 56. Bhattacharya S, Esquivel BD, White TC. 2018. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio 9:e01291–18. doi: 10.1128/mBio.01291-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weete JD, Abril M, Blackwell M. 2010. Phylogenetic distribution of fungal sterols. PLoS One 5:e10899. doi: 10.1371/journal.pone.0010899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Amses KR, Simmons DR, Longcore JE, Mondo SJ, Seto K, Jerônimo GH, Bonds AE, Quandt CA, Davis WJ, Chang Y, et al. 2022. Diploid-dominant life cycles characterize the early evolution of fungi. Proc Natl Acad Sci U S A 119:e2116841119. doi: 10.1073/pnas.2116841119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hull CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AGS, Kelly DE, Kelly SL. 2012. Facultative sterol uptake in an ergosterol-deficient clinical isolate of Candida glabrata harboring a missense mutation in ERG11 and exhibiting cross-resistance to azoles and amphotericin B. Antimicrob Agents Chemother 56:4223–4232. doi: 10.1128/AAC.06253-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Papp C, Bohner F, Kocsis K, Varga M, Szekeres A, Bodai L, Willis JR, Gabaldón T, Tóth R, Nosanchuk JD, Vágvölgyi C, Gácser A. 2020. Triazole evolution of Candida parapsilosis results in cross-resistance to other antifungal drugs, influences stress responses, and alters virulence in an antifungal drug-dependent manner. mSphere 5:e00821-20. doi: 10.1128/mSphere.00821-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sanglard D, Ischer F, Parkinson T, Falconer D, Bille J. 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob Agents Chemother 47:2404–2412. doi: 10.1128/AAC.47.8.2404-2412.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chau AS, Gurnani M, Hawkinson R, Laverdiere M, Cacciapuoti A, McNicholas PM. 2005. Inactivation of sterol Δ 5,6 -desaturase attenuates virulence in Candida albicans. Antimicrob Agents Chemother 49:3646–3651. doi: 10.1128/AAC.49.9.3646-3651.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ksiezopolska E, Schikora-Tamarit MÀ, Beyer R, Nunez-Rodriguez JC, Schüller C, Gabaldón T. 2021. Narrow mutational signatures drive acquisition of multidrug resistance in the fungal pathogen Candida glabrata. Curr Biol 31:5314–5326. doi: 10.1016/j.cub.2021.09.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vale-Silva LA, Coste AT, Ischer F, Parker JE, Kelly SL, Pinto E, Sanglard D. 2012. Azole resistance by loss of function of the sterol Δ5,6-desaturase gene (ERG3) in Candida albicans does not necessarily decrease virulence. Antimicrob Agents Chemother 56:1960–1968. doi: 10.1128/AAC.05720-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rybak JM, Dickens CM, Parker JE, Caudle KE, Manigaba K, Whaley SG, Nishimoto AT, Luna-Tapia A, Roy S, Zhang Q, Barker KS, Palmer GE, Sutter TR, Homayouni R, Wiederhold NP, Kelly SL, Rogers PD. 2017. Loss of C-5 sterol desaturase activity results in increased resistance to azole and echinocandin antifungals in a clinical isolate of Candida parapsilosis. Antimicrob Agents Chemother 61:e00651-17. doi: 10.1128/AAC.00651-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scott NE, Edwin Erayil S, Kline SE, Selmecki A. 2023. Rapid evolution of multidrug resistance in a Candida lusitaniae infection during micafungin monotherapy. Antimicrob Agents Chemother 67:e0054323. doi: 10.1128/aac.00543-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ben Abid F, Salah H, Sundararaju S, Dalil L, Abdelwahab AH, Salameh S, Ibrahim EB, Almaslmani MA, Tang P, Perez-Lopez A, Tsui CKM. 2023. Molecular characterization of Candida auris outbreak isolates in Qatar from patients with COVID-19 reveals the emergence of isolates resistant to three classes of antifungal drugs. Clin Microbiol Infect 29:1083. doi: 10.1016/j.cmi.2023.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shapiro RS, Robbins N, Cowen LE. 2011. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 75:213–267. doi: 10.1128/MMBR.00045-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nolte FS, Parkinson T, Falconer DJ, Dix S, Williams J, Gilmore C, Geller R, Wingard JR. 1997. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob Agents Chemother 41:196–199. doi: 10.1128/AAC.41.1.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Robbins N, Cowen LE. 2021. Antifungal drug resistance: deciphering the mechanisms governing multidrug resistance in the fungal pathogen Candida glabrata. Curr Biol 31:R1520–R1523. doi: 10.1016/j.cub.2021.09.071 [DOI] [PubMed] [Google Scholar]
- 71. Carolus H, Sofras D, Boccarella G, Sephton-Clark P, Romero CL, Vergauwen R, Yazdani S, Pierson S, Jacobs S, Vandecruys P, Wijnants S, Meis J, Berg P van den, Rybak J, Cuomo C, Dijck PV. 2023. Acquired amphotericin B resistance and fitness trade-off compensation in Candida auris. Res Sq. doi: 10.21203/rs.3.rs-3621420/v1 [DOI]
- 72. Bauer K, Rafael B, Vágó B, Kiss-Vetráb S, Molnár A, Szebenyi C, Varga M, Szekeres A, Vágvölgyi C, Papp T, Nagy G. 2023. Characterization of the sterol 24-C-methyltransferase genes reveals a network of alternative sterol biosynthetic pathways in Mucor lusitanicus. Microbiol Spectr 11:e0031523. doi: 10.1128/spectrum.00315-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Maji A, Soutar CP, Zhang J, Lewandowska A, Uno BE, Yan S, Shelke Y, Murhade G, Nimerovsky E, Borcik CG, et al. 2023. Tuning sterol extraction kinetics yields a renal-sparing polyene antifungal. Nature 623:1079–1085. doi: 10.1038/s41586-023-06710-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Espinel-Ingroff A, Chakrabarti A, Chowdhary A, Cordoba S, Dannaoui E, Dufresne P, Fothergill A, Ghannoum M, Gonzalez GM, Guarro J, Kidd S, Lass-Flörl C, Meis JF, Pelaez T, Tortorano AM, Turnidge J. 2015. Multicenter evaluation of MIC distributions for epidemiologic cutoff value definition to detect amphotericin B, posaconazole, and Itraconazole resistance among the most clinically relevant species of Mucorales. Antimicrob Agents Chemother 59:1745–1750. doi: 10.1128/AAC.04435-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nicolás FE, de Haro JP, Torres-Martínez S, Ruiz-Vázquez RM. 2007. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet Biol 44:504–516. doi: 10.1016/j.fgb.2006.09.003 [DOI] [PubMed] [Google Scholar]
- 76. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Triana S, González A, Ohm RA, Wösten HAB, de Cock H, Restrepo S, Celis A. 2015. Draft genome sequence of the animal and human pathogen Malassezia pachydermatis strain CBS 1879. Genome Announc 3:e01197-15. doi: 10.1128/genomeA.01197-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lebreton A, Corre E, Jany J-L, Brillet-Guéguen L, Pèrez-Arques C, Garre V, Monsoor M, Debuchy R, Le Meur C, Coton E, Barbier G, Meslet-Cladière L. 2020. Comparative genomics applied to Mucor species with different lifestyles. BMC Genomics 21:135. doi: 10.1186/s12864-019-6256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chen ECH, Morin E, Beaudet D, Noel J, Yildirir G, Ndikumana S, Charron P, St‐Onge C, Giorgi J, Krüger M, Marton T, Ropars J, Grigoriev IV, Hainaut M, Henrissat B, Roux C, Martin F, Corradi N. 2018. High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytologist 220:1161–1171. doi: 10.1111/nph.14989 [DOI] [PubMed] [Google Scholar]
- 81. Mondo SJ, Lastovetsky OA, Gaspar ML, Schwardt NH, Barber CC, Riley R, Sun H, Grigoriev IV, Pawlowska TE. 2017. Bacterial endosymbionts influence host sexuality and reveal reproductive genes of early divergent fungi. Nat Commun 8:1843. doi: 10.1038/s41467-017-02052-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. 1996. Life with 6000 genes. Science 274:546, doi: 10.1126/science.274.5287.546 [DOI] [PubMed] [Google Scholar]
- 83. Wood V, Gwilliam R, Rajandream M-A, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880. doi: 10.1038/nature724 [DOI] [PubMed] [Google Scholar]
- 84. Russ C, Lang BF, Chen Z, Gujja S, Shea T, Zeng Q, Young S, Cuomo CA, Nusbaum C. 2016. Genome sequence of Spizellomyces punctatus. Genome Announc 4:e00849-16. doi: 10.1128/genomeA.00849-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mondo SJ, Dannebaum RO, Kuo RC, Louie KB, Bewick AJ, LaButti K, Haridas S, Kuo A, Salamov A, Ahrendt SR, et al. 2017. Widespread adenine N6-methylation of active genes in fungi. Nat Genet 49:964–968. doi: 10.1038/ng.3859 [DOI] [PubMed] [Google Scholar]
- 86. Kämper J, Kahmann R, Bölker M, Ma L-J, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Müller O, et al. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101. doi: 10.1038/nature05248 [DOI] [PubMed] [Google Scholar]
- 87. Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S. 2004. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A 101:7329–7334. doi: 10.1073/pnas.0401648101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chang Y, Wang Y, Mondo S, Ahrendt S, Andreopoulos W, Barry K, Beard J, Benny GL, Blankenship S, Bonito G, et al. 2022. Evolution of zygomycete secretomes and the origins of terrestrial fungal ecologies. iScience 25:104840. doi: 10.1016/j.isci.2022.104840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Janbon G, Ormerod KL, Paulet D, Byrnes EJ, Yadav V, Chatterjee G, Mullapudi N, Hon C-C, Billmyre RB, Brunel F, et al. 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10:e1004261. doi: 10.1371/journal.pgen.1004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Linde J, Schwartze V, Binder U, Lass-Flörl C, Voigt K, Horn F. 2014. De novo whole-genome sequence and genome annotation of Lichtheimia ramosa. Genome Announc 2:e00888-14. doi: 10.1128/genomeA.00888-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Dobin A, Gingeras TR. 2015. Mapping RNA‐seq reads with STAR. Curr Protoc Bioinformatics 51:11. doi: 10.1002/0471250953.bi1114s51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44:W160–W165. doi: 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. 2008. Synteny and collinearity in plant genomes. Science 320:486–488. doi: 10.1126/science.1153917 [DOI] [PubMed] [Google Scholar]
- 97. Lopez-Delisle L, Rabbani L, Wolff J, Bhardwaj V, Backofen R, Grüning B, Ramírez F, Manke T. 2021. pyGenomeTracks: reproducible plots for multivariate genomic datasets. Bioinformatics 37:422–423. doi: 10.1093/bioinformatics/btaa692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Parker JE, Warrilow AGS, Cools HJ, Fraaije BA, Lucas JA, Rigdova K, Griffiths WJ, Kelly DE, Kelly SL. 2013. Prothioconazole and prothioconazole-desthio activities against Candida albicans sterol 14-α-demethylase. Appl Environ Microbiol 79:1639–1645. doi: 10.1128/AEM.03246-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S6.
Figures S7-S11.
Detailed overview of materials and methods.
Tables S1-S5.
Data Availability Statement
Raw rRNA-depleted RNA-sequencing data sets obtained from the M. circinelloides 1006PhL strain are accessible under PRJNA1046487 NCBI’s Sequence Read Archive (SRA) project accession number. RNA-seq data sets derived from the M. lusitanicus MU402 strain were similarly generated for a prior study (PRJNA903107) and are publicly available (52).