Abstract
We report the discovery of a novel gene in the varicella-zoster virus (VZV) genome, designated open reading frame (ORF) S/L. This gene, located at the left end of the prototype VZV genome isomer, expresses a polyadenylated mRNA containing a splice within the 3′ untranslated region in virus-infected cells. Sequence analysis reveals significant differences between the ORF S/Ls of wild-type and attenuated strains of VZV. Antisera raised to a bacterially expressed portion of ORF S/L reacted specifically with a 21-kDa protein synthesized in cells infected with a VZV clinical isolate and with the original vaccine strain of VZV (Oka-ATCC). Cells infected with other VZV strains, including a wild-type strain that has been extensively passaged in tissue culture and commercially produced vaccine strains of Oka, synthesize a family of proteins ranging in size from 21 to 30 kDa that react with the anti-ORF S/L antiserum. MeWO cells infected with recombinant VZV harboring mutations in the C-terminal region of the ORF S/L gene lost adherence to the stratum and adjacent cells, resulting in an altered plaque morphology. Immunohistochemical analysis of VZV-infected cells demonstrated that ORF S/L protein localizes to the cytoplasm. ORF S/L protein was present in skin lesions of individuals with primary or reactivated infection and in the neurons of a dorsal root ganglion during virus reactivation.
Varicella-zoster virus (VZV) caused approximately 4 million cases of chickenpox each year in the United States prior to the introduction of the attenuated vaccine (4). Following primary infection, the virus remains latent in the ganglia of the infected individual and can reactivate later in life, causing shingles, a painful and potentially debilitating disease (reviewed in references 17 and 18). The natural history of this infection is typical of infections caused by members of the Alphaherpesvirinae. This subfamily, which also includes herpes simplex virus (HSV) and pseudorabies virus, is characterized by viruses that are neurotropic, reactivate to cause a secondary disease, and have with the exception of VZV a wide host range in culture (44). Recent comparison of the nucleic acid and protein sequences of these viruses has confirmed their phylogenetic relationship, a relationship originally based on their similar biological characteristics (20).
Analyses of VZV DNA have shown that the overall structure of the genomic DNA is similar to that of HSV. The linear genomic DNAs of HSV and VZV contain a single unpaired nucleotide at the 3′ end of each strand (10, 37). Two regions of unique sequence, the long (UL) and short (US) components, comprise the majority of the 125-kbp VZV genome. Two sets of inverted repeats, designated TRL/IRL and IRS/TRS, bracket UL and US, respectively (Fig. 1A) (10, 14, 46, 47). Recombination between these inverted repeat segments yields a population of four genomic isomers, which differ in the relative orientation of UL and US. Unlike HSV, which generates an equimolar population of the four isomers, over 90% of the viral progeny of a VZV-infected cell have one particular orientation of UL, the prototype isomer (10, 22).
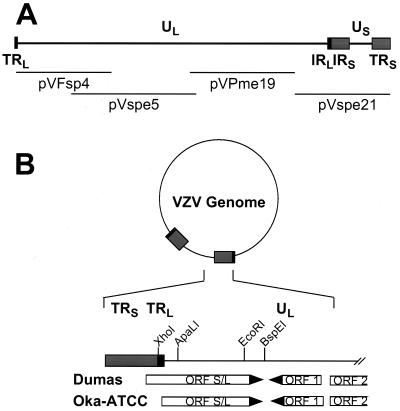
FIG. 1.
Location of ORF S/L in the VZV genome. (A) Schematic representation of the linear VZV genome. The repeat sequences TRL/IRL (■) and IRS/TRS (░⃞) are depicted. The four overlapping cosmids are indicated relative to their map positions beneath the genomic diagram. (B) The VZV genome is circularized by adjoining TRL and TRS and accounting for the one unpaired nucleotide. The region of the genome near the TRL/TRS junction is expanded to show the presence of ORFs and selected restriction sites in both Dumas and Oka. The ORF 1 and ORF 2 designations are consistent with those of Davison and Scott (12). ORF S/L Dumas and ORF S/L Oka initiate at different positions.
In addition to the similar genomic structures of VZV and HSV, hybridization and DNA sequence analyses have demonstrated the extensive colinearity of both nucleotide and predicted amino acid sequences (12, 13, 33, 34). Over 90% of the 71 open reading frames (ORFs) described for VZV have a significantly similar counterpart in the HSV genome. Several of the homologous gene products, including VZV ORF 61p and HSV ICP0, ORF 62p and HSV ICP4, and ORF 51p and the HSV origin binding protein, can complement one another in functional assays (15, 38, 50).
One significant difference between the VZV and HSV genomes is the size of the repeats bracketing UL. VZV TRL and IRL are only 89 bp in length, whereas the inverted repeats bordering HSV UL, designated b and b′, are approximately 9,000 bp each (10, 44). The b and b′ repeats harbor many genes that can influence the replication cycle and neurovirulence of HSV (2, 7, 27, 51). For example, ICP0 regulates transcription of several classes of viral genes (3, 5). The product of VZV gene 61, located at the right side of UL, is the homologue of HSV ICP0. Although the primary sequences of these two genes have diverged significantly, their polypeptide products can functionally complement each other (38).
The observation that the right edge of VZV UL encodes the homologue of HSV ICP0 led us to hypothesize that homologues of other HSV genes resident in the b/b′ repeats may be located near the edges of VZV UL. Here we document the existence of ORF S/L, a new gene in the VZV genome. A spliced mRNA from this gene was expressed in cells infected with Oka-ATCC, and a protein migrating between 21 and 30 kDa was detected in cells infected with a variety of VZV isolates. The protein localized to the cytoplasm of cells infected in tissue culture and was detected in skin lesions of individuals with primary or recurrent infection as well as in neurons of a dorsal root ganglion (DRG) during reactivation. Analyses of other strains of VZV revealed heterogeneity in the sequence encoding ORF S/L, resulting in proteins that migrated with different mobilities in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Recombinant viruses encoding altered ORF S/L genes demonstrated that the protein influences adherence of the infected cells to neighboring cells and the stratum in tissue culture.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (CCL-81) and the vaccine strain of VZV (Oka-ATCC) (VR-795) were obtained from the American Type Culture Collection (Manassas, Va.). Strains Oka-SK and Oka-Merck were obtained from Smith-Kline/Beecham (Philadelphia, Pa.) and Merck Research Laboratories (West Point, Pa.), respectively. The Merck strain is from a commercial production run. VZV (Ellen) was originally a wild-type isolate that has been passaged over 100 times in tissue culture. VZV (Jones), a recent isolate from a patient, has undergone fewer than 10 passages in tissue culture. The human melanoma cell line MeWO was a gift from Charles Grose (University of Iowa, Iowa City); human foreskin fibroblast (HF) cells were a gift from Ed Mocarski (Stanford University, Stanford, Calif.). Human embryonal lung fibroblasts (HELF) were obtained from Bio-Whittaker (Walkersville, Md.).
Preparation and cloning of viral DNA.
Viral DNA was prepared as described previously (21). In brief, lysates of infected cells were digested with DNase and RNase prior to the extraction of viral DNA.
Cosmid clones of VZV (Oka-ATCC) were produced by digestion of purified viral DNA with FspI, PmeI, or SpeI (New England Biolabs, Beverly, Mass.), and the ends of the fragments were filled by incubation with T4 DNA polymerase in the presence of the four deoxynucleoside triphosphates. The Asc-Bam adapter (5′-PO4-TGG CGC GCC G-3′ and 5′-HO-GAT CCG GCG CGC CA-3′) was ligated to the ends of these restriction fragments. After ligation, the high-molecular-weight products were purified on a 5-ml column of Sepharose CL-4B equilibrated with 10 mM Tris (pH 7.5)–1 mM EDTA–150 mM NaCl. The DNA was ligated to SuperCos-1 (Stratagene, La Jolla, Calif.) that had been digested with XbaI, dephosphorylated with shrimp alkaline phosphatase (AP; United States Biochemicals, Cleveland, Oh), and digested with BamHI. The ligated mixture was packaged into lambda heads and introduced into Escherichia coli XLI-MR cells using the conditions suggested by the supplier (Stratagene). The appropriate cosmid clones were identified by restriction enzyme digestion. Four overlapping cosmids representing the entire VZV genome, designated pVFsp4, pVSpe5, pVPme19, and pVSpe21, were isolated (Fig. 1A).
Plasmid clones representing the TRL, TRS, and IRL/IRS regions were derived from insertion of BamHI-digested cosmid DNA into the BamHI site of pGEM3zf+. The appropriate clones were identified by restriction enzyme digestion. These double-stranded plasmid DNAs were sequenced using synthetic oligonucleotides generated at Aviron in order to define the sequence of VZV (Oka-ATCC) in the region of interest. A plasmid representing the TRS/TRL junction of VZV (Oka-ATCC) was constructed by amplifying viral DNA with primers flanking this region (5′-GCCGCCATGGGATGAAAAAAGTGTCTGTCTGTCTGTGCG-3′ and 5′-GCCGCCATGGTCATGTAGTTGAGTTGGGAGGTTCC-3′). The PCR product was digested with NcoI and inserted into the NcoI site of pGEM5zf+. The resulting plasmid was designated pORFS/LC3.
Plasmid pVXLeft4 was created by digestion of pVFsp4 with XhoI and PmeI; the resulting VZV fragment was inserted into pGEM3zf+ that was digested with SacI, made blunt, and digested with XhoI. A deletion of ORF S/L was made by partial digestion of pVXLeft4 with ApaLI, followed by complete digestion with BspEI. The appropriate size DNA fragment was gel purified, made blunt, and ligated. This plasmid lacking ORF S/L, pVXLeftΔ4, was identified by restriction enzyme digestion. A cytomegalovirus (CMV) gB epitope was inserted in frame at the C terminus of ORF S/L, using a double-stranded oligonucleotide (5′-CTAGGGTACCTTAGTGGCGATATCCGTTCTTGCGGTGGCGGAGGCGGTCGAGGAGGTTGGGCTTCTGCCCCTT-3′ and 5′-AATTAAGGGGCAGAAGCCCAACCTCCTCGACCGCCTCCGCCACCGCAAGAACGGATATCGCCACTAAGGTACC-3′) (29). This oligonucleotide was inserted into pGS/LC3 digested with EcoRI and XbaI, and the resulting plasmid pGORFS/LgB was identified by restriction enzyme digestion and DNA sequencing. ORF S/L with the glycoprotein B (gB) epitope insertion was placed into pVXLeft4. pGORFS/LgB was digested with KpnI, made blunt with T4 polymerase, and digested with XhoI. This fragment was purified and ligated to pVXLeft4 that had been digested with BspEI, made blunt, and subsequently digested with XhoI. The resulting plasmid pVXLeft4-gB contained the left 1.2 kbp of VZV (Oka-ATCC) with the nucleotides between the EcoRI and BspEI sites replaced by the CMV gB oligonucleotide.
Both ORF S/L mutations were introduced into cosmid pVFsp4 by digestion of pVXLeftΔ4 or pVXLeft4-gB with XhoI and SgrAI (Boehringer Mannheim, Indianapolis, Ind.) and insertion of the fragments into XhoI/SgrAI-digested pVFsp4. This step yielded the intermediate plasmids pVdSgrΔ4 and pVdSgrgB that include the ORF S/L sequences with the deletion or gB tag to nucleotide (nt) 777 of VZV. The 28-kbp SgrAI fragment corresponding to nt 777 to 28734 of VZV was isolated from pVFsp4 and inserted into SgrAI-digested pVdSgrΔ4 and pVdSgrgB to yield cosmids pVFspΔ4 and pVFspgB.
Construction of VZV recombinants.
Recombinant VZV genomes were constructed in a manner similar to the method described by Cohen and Seidel (8). Aliquots of 1 to 2 μg of AscI-restricted pVFsp4 (or its derivatives), pVSpe5, and pVPme19 were mixed with 0.5 to 1 μg of AscI-restricted pVSpe21 and transfected into MeWO cells by CaPO4 precipitation as described elsewhere (23). Plasmid pCMV-62, containing VZV gene 62 under transcriptional control of the CMV major immediate-early promoter/enhancer, a generous gift from John Hay (State University of New York, Buffalo), was included but found not to be essential. Three to five days after transfection of a 25-cm2 monolayer of MeWO cells, the cells were trypsinized and replated on a 75-cm2 vessel. Plaques were evident approximately 7 to 10 days posttransfection. Infected MeWO cells were harvested and replated on Vero cells to generate large quantities of infected cells.
RNA preparation and analysis.
Freshly plated HF cells were infected with one-fourth as many VZV-infected HF cells; 3 days after infection, whole-cell RNA was isolated (6). Poly(A)-enriched RNA was obtained by fractionation on oligo(dT)-cellulose (Boehringer Mannheim) as described elsewhere (24). Northern analysis was performed by separating 10 μg of either whole-cell or 1 μg of oligo(dT)-fractionated RNA on 1% agarose–2.2 M formaldehyde gels electrophoresed in 20 mM MOPS (morpholinepropanesulfonic acid)–10 mM sodium acetate–1 mM EDTA (pH 7.0). The RNA was transferred to Hybond N+ (Amersham, Arlington Heights, Ill.) and hybridized to the indicated 32P-labeled in vitro-transcribed RNA probe in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1× Denhardt's solution–30% formamide at 58°C for 16 to 20 h. After hybridization, the blots were washed in 1× SSC–0.1% SDS and 0.1× SSC–0.1% SDS for 15 min each at 58°C. Following rinsing with 2× SSC, the blots were incubated with RNase A (1 μg/ml) for 15 min at room temperature to remove any probe that was not annealed. The blots were rinsed again in 0.1× SSC–0.1% SDS for 45 min at 50°C and subjected to autoradiography.
Rapid amplification of cDNA ends (RACE) was performed to establish the 3′ end of the transcript. In brief, 1 μg of whole-cell RNA from either infected or uninfected HF cells was reverse transcribed using an oligo(dT) amplification primer, and the cDNA was detected by PCR using the universal amplification primer (Gibco BRL, Gaithersburg, Md.) and an oligonucleotide (5′-CGTCCACCCCTCGTTTACTG-3′) corresponding to nt 319 to 338 (Fig. 2).
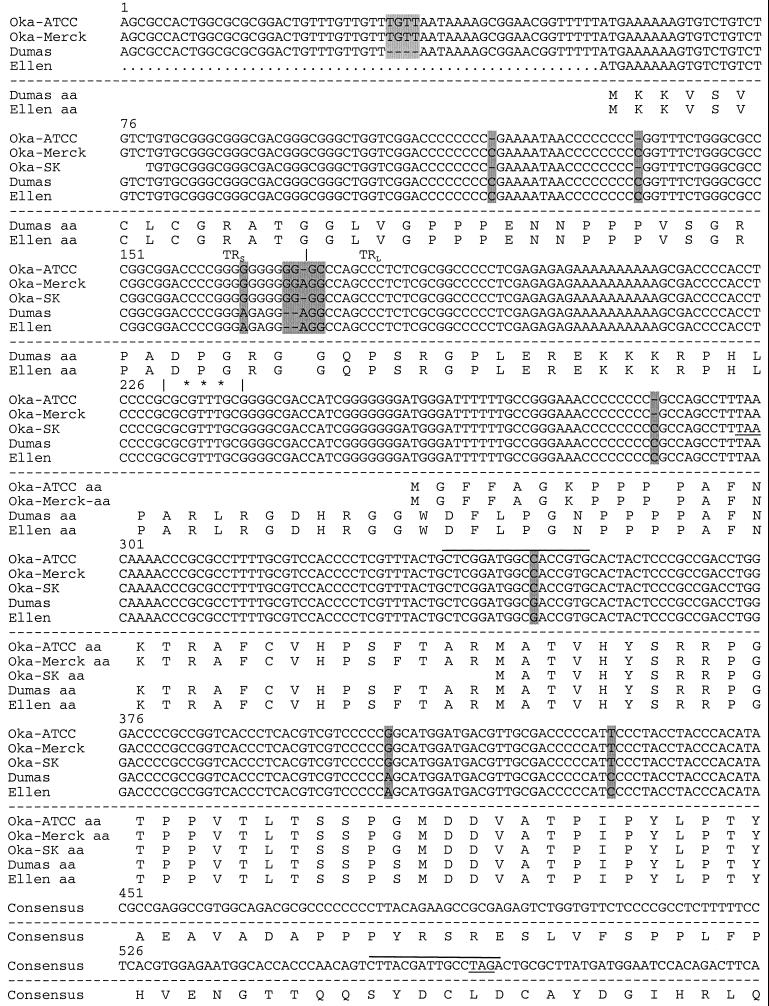
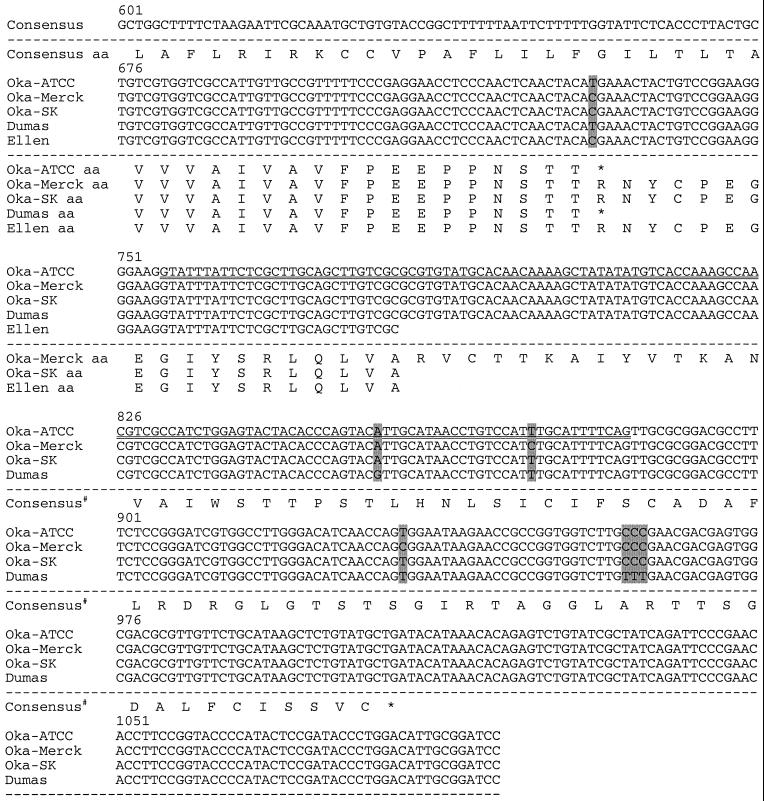
FIG. 2.
Sequence comparison of VZV genomes near ORF S/L. The nucleotide sequences of Oka-ATCC, Oka-Merck, Oka-SK, Dumas, and Ellen as displayed all begin within TRS; the TRS/TRL boundary is identified with a vertical line, and nucleotide deletions (−) are indicated. Sequence differences between the strains are shaded. The predicted amino acid sequences are displayed beneath the nucleotide sequences. Consensus# represents the Oka-Merck and Oka-SK consensus amino acids. Positions of the transcription initiation site (∗∗∗), the 130-nt intron (⩵⩵), potential termination sites (underline), and primer sites used to generate anti-ORF S/L serum (overline) are indicated. The terminator at position 567 would truncate the S/L protein encoded by Oka-Merck if it were to initiate at the same ATG as Dumas.
RNase mapping to identify the 5′ end of the transcript was performed by incubating 30 μg of the RNA with the 32P-labeled in vitro-transcribed RNA probe in 80% formamide–40 mM MOPS–400 mM sodium acetate–1 mM EDTA (pH 6.5). The samples were heated to 90°C for 5 min and hybridized overnight at 50°C. Unhybridized RNA was removed by the addition of 30 U of RNase ONE (Promega, Madison, Wis.) in 10 mM Tris (pH 7.5)–200 mM sodium acetate–5 mM EDTA for 60 min at 35°C. Nuclease digestion was terminated by the addition of 0.2% SDS, and nucleic acids were collected by ethanol precipitation. The products were suspended in 80% formamide–0.1% SDS, denatured at 95°C for 5 min, and separated on a 6% polyacrylamide–8 M urea gel in 1× Tris-borate-EDTA. The gel was visualized by autoradiography.
Generation of anti-ORF S/L antibody.
The DNA region between nt 339 and 570 of ORF S/L from VZV (Ellen) (Fig. 2) was amplified by PCR using Vent polymerase (New England Biolabs). The amplimer was cloned in the bacterial expression vector pALEX (41), which places glutathione S-transferase (GST) at the amino terminus and a six-histidine moiety at the carboxyl terminus of the VZV peptide. The fusion protein was expressed in E. coli strain BL21(DE3) and purified to apparent homogeneity by affinity chromatography on glutathione-Sepharose (41). The purity of the protein was determined by SDS-PAGE, and the protein was used to immunize rabbits. Antibodies that cross-reacted with E. coli proteins, GST, and mammalian cell (Vero and HELF) proteins were removed by adsorption on columns containing these proteins (31). Western blotting of whole-cell lysates was performed as described elsewhere (48).
Tissue specimens.
DRG from two seropositive patients without clinical evidence of zoster, from one fetus without maternal history of varicella, and from one patient with zoster were obtained at autopsy and examined by immunohistochemistry for the presence of ORF S/L. The DRG from the seropositive patients without zoster were shown to have latent VZV by in situ hybridization and immunohistochemistry, while the DRG from the fetus were negative (30, 31). In situ hybridization and immunohistochemistry demonstrated that the DRG from the patient with zoster contained reactivated VZV (30, 31).
Skin biopsies from patients with clinical evidence of chickenpox or zoster that were positive for VZV by immunohistochemistry using an antibody to gC (data not shown) were examined for the presence of ORF S/L protein. A skin biopsy specimen that was negative for VZV gC by immunohistochemistry from a patient with Grover's disease, a dermatologic disorder that may be confused with chickenpox clinically but with different histopathologic features, was included as a negative control (1).
Immunohistochemistry.
Tissue sections were deparaffinized with xylenes, rinsed twice with ethyl alcohol, and treated with Serotec target unmasking fluid (Harlan Bioproducts for Science, Indianapolis, Ind.) according to the manufacturer's recommendations. DRG sections were then blocked with 1% goat serum in phosphate-buffered saline (PBS) for 20 min and incubated with a 1:100 dilution of purified polyclonal rabbit anti-ORF S/L antibodies. After washing, the DRG specimens were incubated for 30 min with an AP-labeled goat anti-rabbit antibody (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) diluted 1:150 in PBS containing 1% goat serum. The slides were washed in AP buffer (100 mM Tris [pH 9.5], 100 mM NaCl, 10 mM MgCl2), and the signal was visualized by light microscopy after staining for 20 min with 9 μl of nitroblue tetrazolium chloride and 3.5 μl of 5-bromo-4-chloro-3-indolylphosphate (Boehringer Mannheim) in 2 ml of AP buffer. Immunohistochemistry of cultured HELF and skin biopsy specimens was performed in the same manner except that Tris-buffered saline was substituted for PBS. AP detection was done in the presence of levamisole to block endogenous AP activity using an AP substrate kit (Vector Laboratories, Burlingame, Calif.) as recommended by the manufacturer. Immunohistochemistry of cultured HELF for gC was performed in the same manner using rabbit polyclonal antibodies generated to VZV gC (31).
DNA sequence analysis.
VZV genomic DNAs from strains Oka-ATCC, Oka-SK, and Ellen were prepared from infected cells or the appropriate cosmid clones, and the regions of interest were amplified by PCR using Vent polymerase and the appropriate primers. DNAs were sequenced in the Columbia University Cancer Center DNA Sequencing Facility or at Aviron, using oligonucleotide primers specific for ORF S/L and the surrounding regions from both TRS and TRL. The sequence for Oka-Merck was generously provided by Daniel DiStefano and Nikolai Kraiouchkine, Virus and Cell Biology Division of Merck Research Laboratories; the Dumas sequence was obtained from GenBank (accession no. X04370). Sequences were aligned and compared using DNAsis 3.5 (Hitachi Biosystems, Santa Clara, Calif.).
Nucleotide sequence accession numbers.
The Oka-ATCC sequence reported in this paper can be obtained from GenBank as accession no. U68702; the Oka-SK sequence has been assigned accession no. AF272392, the Ellen sequence has been assigned accession no. AF272391.
RESULTS
Identification of ORF S/L.
A search for previously undescribed ORFs near the edges of VZV UL was undertaken based on the hypothesis that this region may harbor genes similar to those located in the HSV b/b′ repeats. The genomic sequence of VZV (Dumas) was circularly permuted after accounting for the unpaired nucleotides present at the 3′ end of each strand of genomic DNA. We searched both edges of UL for potential ORFs of significant size. One ORF, designated ORF S/L, initiated within TRS, proceeded through TRL, and terminated 562 nts into the left side of UL (Fig. 1B). This 224-amino-acid ORF shares no similarity with any other ORF in the current databases.
Sequence analysis of VZV termini.
To demonstrate the presence of this ORF in other strains of VZV, we analyzed the termini of VZV strains Oka, Ellen, and Dumas. The Oka strains were chosen because of their wide use as a vaccine for chickenpox. Four overlapping cosmid clones from VZV (Oka-ATCC), designated pVFsp4, pVSpe5, pVPme19, and pVSpe21 (Fig. 1A), were isolated and mapped relative to the genome using restriction enzyme and Southern blot analyses (data not shown). Plasmids representing TRL, TRS, and the IRL/IRS junction were subcloned from the appropriate cosmid (Fig. 1A). Comparison of the sequence of the TRL and TRS plasmids with the sequence of the IRL/IRS junction demonstrated the lack of one base at the end of the terminal clones. The cosmids were prepared in a manner that should either preserve an unpaired 5′ nucleotide or delete an unpaired 3′ nucleotide present on the viral DNA. The lack of a single base in the terminal clones, compared to the IRL/IRS junction, indicated that an unpaired C was present at the 3′ end of the top viral strand and an unpaired G was present at the 3′ end of the bottom viral strand. The presence of the unpaired nucleotide at the 3′ end of the viral DNA was similar to that established for Dumas as well as other herpesvirus genomes (10, 36, 37). The sequences of the ORF S/L regions of Ellen and Oka-SK were determined. The sequences of the ORF S/L region of Dumas, Ellen, Oka-ATCC, Oka-Merck, and Oka-SK were aligned and compared. Position 1 was arbitrarily assigned within TRS for the sake of this alignment.
The termini of the strains analyzed were similar though they contained several nucleotide differences. A deletion of two nucleotides in TRS of Oka-ATCC and Oka-SK (nt 119 and 136 [Fig. 2]), relative to Dumas, altered ORF S/L (Fig. 1B and 2). Unlike Dumas, whose ORF S/L has an initiating ATG codon within TRS, ORF S/L from Oka-ATCC is likely to initiate at the ATG codon at nt 259 (Fig. 2). This ATG, just past the TRL/UL junction, contains a consensus initiation sequence (26). The Oka-Merck sequence does not contain the deletion at positions 119 and 136. However, an insertion of two nucleotides at positions 170 and 171 relative to Dumas places a premature stop codon in the reading frame at position 567. For this reason, we believe that ORF S/L of Oka-Merck probably initiates at nt 259 as does Oka-ATCC. A single nucleotide deletion at position 288 of Oka-ATCC and Oka-Merck (Fig. 2) brings their ORF S/Ls and that of Dumas into register with one another. ORF S/L of Dumas and Oka-ATCC then continue into UL, terminating 570 nt from the left edge of the prototype isomer of the viral genome. The 157-amino-acid ORF S/L Oka-ATCC is 96% identical to the C-terminal 157 amino acids of ORF S/L Dumas (Fig. 2). The Oka-Merck sequence has a single base pair substitution at position 731 that changes the stop codons found in Oka-ATCC and Dumas to a codon encoding Arg. ORF S/L of Oka-Merck has a stop codon at position 1007 (Fig. 2). ORF S/L of Oka-SK has deletions at nt 119 and 136. However, the nucleotide deletion at position 288 in Oka-ATCC was not present in Oka-SK, and the sequence of Oka-SK contains a stop codon in ORF S/L at position 298. Oka-SK ORF S/L most likely initiates at position 344. As in Oka-Merck, a single base pair substitution at nucleotide position 731 of Oka-SK changed the stop codon found in Dumas and Oka-ATCC to a codon encoding an Arg, and ORF S/L of Oka-SK terminates at position 1007 (Fig. 2). ORF S/L of Ellen and ORF S/L of Dumas are identical until nucleotide position 731 (Fig. 2). The Ellen sequence has the substitution at position 731 that was found in Oka-SK and Oka-Merck. No stop codon was identified in the ORF S/L region from Ellen that was sequenced as of this report.
Structure of ORF S/L RNA.
RNA from Oka-ATCC-infected HF cells was analyzed to determine if the DNA encoding ORF S/L was transcribed. Northern analysis was performed using in vitro-transcribed 32P-labeled RNA probes derived from plasmid pORFS/LC3, containing the Oka-ATCC TRS/TRL junction (Fig. 3A). Probes representing both strands of the viral DNA were hybridized to immobilized whole-cell RNA harvested from Oka-ATCC-infected or uninfected fibroblasts. Only the probe derived from the T7 promoter of XhoI-digested pORFS/LC3 hybridized specifically to VZV-infected cell RNA; an Sp6-transcribed, EcoRI-terminated probe representing the complementary strand did not hybridize (Fig. 3A and B). The RNA species detected was approximately 900 nt in length and corresponded to the strand predicted to encode ORF S/L. Because the T7-transcribed, XhoI-terminated probe contained approximately 60 nt present in TRL and could potentially hybridize to RNA from either side of UL, we generated a T7-transcribed, ApaLI-terminated probe consisting entirely of UL sequence from the ORF S/L region (Fig. 3A). This smaller probe also specifically detected the 900-nt transcript (data not shown). Oligo(dT) chromatography of the whole-cell RNA prior to hybridization demonstrated that the 900-nt transcript was polyadenylated (Fig. 3B).
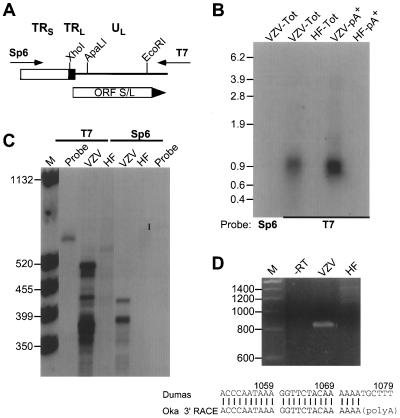
FIG. 3.
Mapping the ORF S/L mRNA. (A) Depiction of pGS/LC3, a plasmid clone of a PCR product spanning the TRL/TRS junction of Oka-ATCC. Directions of the T7 and Sp6 transcripts generated from this insert are indicated; the Sp6-transcribed RNA would be the same sense as ORF S/L mRNA. (B) Northern analysis of RNA from ORF S/L. Ten micrograms of whole-cell (Tot) or 1 μg of poly(A)+ (pA+) RNA isolated from uninfected (HF) or VZV-infected (VZV) fibroblasts was subjected to Northern analysis with either the T7/XhoI (T7) or Sp6/EcoRI (Sp6) probe. The autoradiographic image of this analysis is shown, with sizes of the RNA markers indicated in kilobases. (C) RNase protection analysis of ORF S/L RNA. The T7/XhoI (T7) in vitro-transcribed probe was hybridized to whole-cell RNA isolated from VZV-infected (VZV) or uninfected (HF) fibroblasts and digested with RNase ONE for 60 min. The products were separated by electrophoresis in a denaturing polyacrylamide gel, and the gel was exposed to X-ray film. Positions and sizes (in nucleotides) of the molecular weight standards (lane M) are indicated at the left. The closed circle adjacent to the lane marked probe identifies the position of unhybridized probe. (D) 3′ RACE analysis of ORF S/L RNA. Whole-cell RNAs from uninfected (HF) and infected (VZV) cells were reverse transcribed and subjected to RT-PCR. The products of these reactions and a control reaction that was done without reverse transcriptase (−RT) were electrophoresed on a 4% 3:1 agarose gel and subsequently stained with ethidium bromide. The RT-PCR product shown in the VZV lane was directly sequenced. The junction of the Oka ORF S/L mRNA and the poly(A) region are depicted and aligned with the Dumas sequence (accession no. X04370). The numbers refer to Dumas genomic DNA. Sizes of the DNA markers (lane M) are shown in nucleotides at the left. Autoradiograms and negatives of stained gels were scanned with a GS-250 imaging densitometer (Bio-Rad, Hercules, Calif.). Photographic quality images were generated using Molecular Analyst (Bio-Rad) and Adobe Photoshop (Adobe Systems, Mountain View, Calif.) software.
The boundaries of the RNA transcript were mapped by both RNase protection and RACE analyses. The 5′ end of the transcript was mapped by hybridizing a 580-nt T7-transcribed 32P-labeled RNA from XhoI-digested pORFS/LC3 (Fig. 3A). The probe was hybridized to total RNAs extracted from Oka-ATCC-infected or uninfected HF cells and subsequently digested with RNase to remove any nonhybridized regions of the probe. Autoradiography of the gel after electrophoresis demonstrated a protected species of approximately 500 nt in the sample hybridized to RNA from infected cells (Fig. 3C). The protected species was not detected when hybridized to cell RNA or when a sense-strand probe was used (Fig. 3C). The protected species mapped the 5′ end of the viral mRNA within TRL and approximately 25 nt upstream of the initiator ATG codon (Fig. 2).
The 3′ end was established by synthesizing oligo(dT)-primed cDNA and amplifying this material with one primer that hybridized to the 5′ end of the synthetic dT primer and one primer specific for ORF S/L (nt 319 to 338 [Fig. 2]). A reverse transcription-PCR (RT-PCR) product of approximately 800 bp was produced (Fig. 3D). The specificity of this product was confirmed by demonstrating its absence from reactions that contained only RNA from uninfected cells or from reactions incubated without reverse transcriptase or without RNA. Sequence analysis of the amplified product located the 3′ end of the transcript upstream of the ORF 2 initiation codon (Fig. 1B). Poly(A) addition began 27 nts downstream of the consensus AATAAA sequence within a G/T-rich region, both hallmarks of a polyadenylation signal (35, 42). In addition, a 130-nt intron, spanning nt 756 to 885, was identified (Fig. 2). Splice donor and splice acceptor sites that were consistent with the consensus sequences (39) flanked this intron.
Construction of mutant VZV.
Recombinant viruses were constructed with altered forms of ORF S/L to evaluate the expression and function of this gene. Overlapping cosmids spanning the VZV genome can reconstitute infectious VZV following cotransfection into MeWO cells (8). A cosmid representing the left edge of Oka-ATCC, pVFsp4, was used to introduce mutations into the ORF S/L region. The DNA between the ApaLI (nt 333) and BspEI (nt 722) sites in ORF S/L was deleted, forming pVFspΔ4 (Fig. 4B). In this construct, the sequence between the codon for amino acid 32 through the termination codon for ORF S/L was deleted. A second mutation in ORF S/L was constructed by inserting an oligonucleotide encoding a CMV gB epitope between the EcoRI and BspEI sites of ORF S/L (Fig. 4B). This construct replaced the 40 C-terminal amino acids of ORF S/L from Oka-ATCC with 27 amino acids of a defined CMV gB epitope.
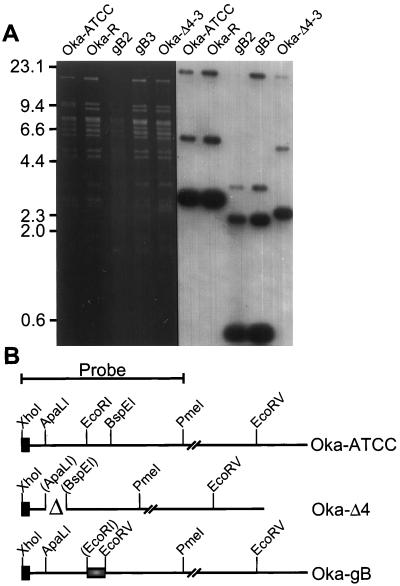
FIG. 4.
Southern analysis of recombinant VZVs. (A) Ethidium
bromide-stained gel and Southern analysis of EcoRV-digested
DNA prepared from Vero cells infected with either Oka-ATCC, Oka-R, or
the three recombinant viruses Oka-gB2, Oka-gB3, and Oka-Δ4-3.
Positions of molecular weight markers are shown in kilobases at the
left. (B) Diagram depicting the ORF S/L region from the Oka-ATCC,
Oka-Δ4-3, Oka-gB2, and Oka-gB3 viruses. Oka-ATCC is depicted on the
top line, and the 1.2-kbp XhoI-to-PmeI probe is
indicated. OkaΔ4 has a deletion of 400 nt between the
ApaLI and BspEI sites (▵); and the gB mutants
have a 60-nt CMV gB sequence
( ) containing a novel
EcoRV site replacing the 120 bp between the EcoRI
and BspEI sites of Oka. Images of the autoradiograms and
stained gels were generated as for Fig. 3.
) containing a novel
EcoRV site replacing the 120 bp between the EcoRI
and BspEI sites of Oka. Images of the autoradiograms and
stained gels were generated as for Fig. 3.
Four cosmids (pVSpe5, pVPme19, pVSpe21, and either pVFsp4, pVFspΔ4-3, pVFsp4-gB2, or pVFsp4-gB3) were transfected into MeWO cells to generate the desired mutant viruses as described in Materials and Methods. Viral DNA was isolated, digested with EcoRV, and hybridized to a XhoI-PmeI probe encompassing the left 1.2 kbp (Fig. 4). DNA prepared from Oka-ATCC or regenerated from overlapping cosmids, designated Oka-R, hybridized to the expected 2.9-kbp band representing TRL of the prototype isomer (Fig. 4A). In addition, we detected three slower-migrating bands of approximately 5.9, 18, and 23 kbp, which represented either the IRL/IRS junction of the prototype isomer (23 kbp) or the TRL (18 kbp) and IRL/IRS junction (5.9 kbp) formed by inversion of the UL component. The reduced intensity of the large 18- and 23-kbp bands, which comigrated in this gel, was due to hybridization to only 89 bp of the probe. The reduced intensity of the 5.9-kbp band reflected the proportion of the viral progeny that existed as either a circle or inverted UL isomer (22). Phosphorimager analysis of this blot revealed that only 10% of the molecules were in this configuration.
The recombinant viruses contained the expected fragments when hybridized to the XhoI-PmeI probe. The gB2 and gB3 recombinants represented two viruses derived from independent transfections using cosmid pVFsp4-gB. Southern analysis of DNA from cells infected with the gB recombinants showed two predominant bands of 2.4 and 0.5 kbp, the latter representing TRL (Fig. 4A). These two bands were derived from the 2.9-kbp TRL band of Oka by virtue of the new EcoRV site introduced along with the CMV gB epitope. Southern analysis of the recombinant lacking sequences between the ApaLI and BspEI sites of ORF S/L, designated Oka-Δ4-3, revealed a 2.5-kbp TRL fragment as expected (Fig. 4A). This band was derived by deleting approximately 400 bp between the ApaLI and BspEI sites within the 2.9-kbp TRL band of Oka-ATCC (Fig. 4B). All of the viruses contained the expected restriction fragments, including those fragments representative of the inverted UL isomer.
Plaque phenotype of VZV mutants.
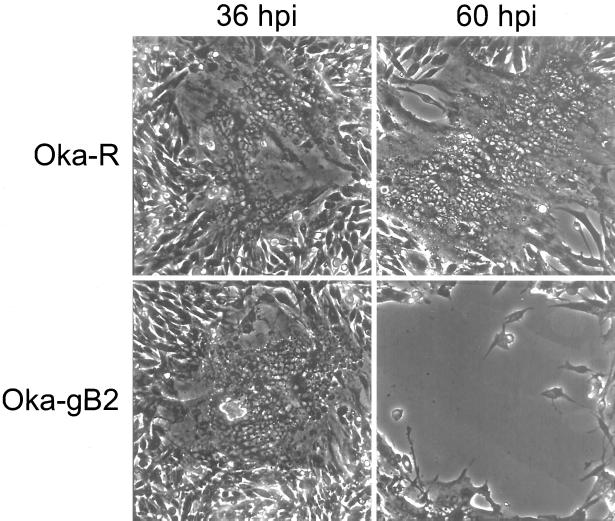
While there was no detectable difference in yield for Oka-ATCC or its derivatives, a difference was detected in plaque morphology between the wild-type and ORF S/L mutant viruses. Oka-ATCC formed typical, syncytial plaques on MeWO cells as did Oka-R. The ORF S/L mutant viruses displayed an altered plaque phenotype (Fig. 5). Plaques from Oka-gB2, Oka-gB3, or Oka-Δ4-3 first appeared on MeWO cells as syncytia indistinguishable from plaques formed by Oka-ATCC 36 h postinfection (hpi) (Fig. 5). However, at 48 hpi the plaques formed by cells infected by mutant virus differed significantly from those formed by Oka-ATCC. In contrast to the typical increase in syncytium size caused by Oka-ATCC, the cells infected with mutant viruses lost the ability to adhere to the surrounding stratum and lifted off the dish leaving large clear zones at the location of each plaque (Fig. 5, 60 hpi). This alteration in plaque morphology required only disruption of ORF S/L near amino acid 120 (Fig. 1B), as insertion of a 27-amino-acid gB epitope in frame at this point had the same effect as deletion of approximately 400 bp between the ApaLI and BspEI sites.
FIG. 5.
Plaque morphology of Oka-R and Oka-gB2 on MeWO cells. Syncytial plaques formed by Oka-R are shown at 36 and 60 hpi on MeWO cells. Plaques formed by Oka-gB2 were photographed at the same points in time as the Oka-R-infected cells. Note the cleared area at 60 hpi on the Oka-gB2-infected MeWO cells.
Identification of the S/L protein.
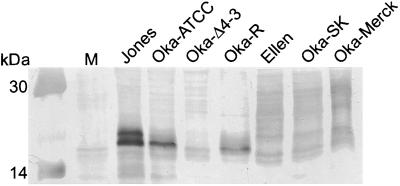
Rabbit polyclonal antibody was generated to a portion of ORF S/L from the region of consensus in the wild-type and Oka strains as described in Materials and Methods. Western blot analysis using purified anti-ORF S/L antibody detected a protein of approximately 21 kDa, in lysates of HELF infected with Jones, Oka-ATCC, or Oka-R; this band was not present on Western blots of Oka-Δ4-3 or uninfected HELF lysates (Fig. 6). The Jones ORFS/L protein migrated as two distinct bands, while the Oka-ATCC ORF S/L migrated as a single band. ORF S/L of Ellen, Oka-SK, and Oka-Merck appeared as a family of proteins migrating from 21 to 30 kDa. The mechanism responsible for the heterogeneity of size is under investigation.
FIG. 6.
Identification of S/L proteins in extracts of infected cells. Whole-cell extracts of uninfected HELF (M) or HELF infected with the VZV strains noted above the lanes were subjected to Western blot analysis using an anti-S/L rabbit serum diluted 1:500, followed by goat anti-rabbit antibody conjugated to horseradish peroxidase. The molecular masses of prestained size markers (Amersham Pharmacia Biotech, Piscataway, N.J.) are indicated at the left. The 20.1-kDa marker was too faint to see in this reproduction.
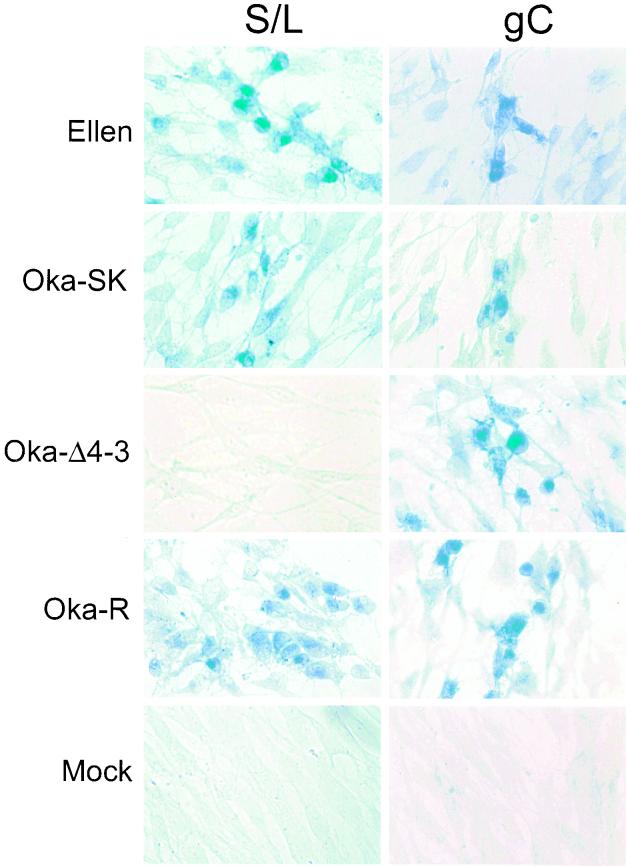
Having established that ORF S/L encodes a virus-specified protein, we next examined the distribution of this protein in infected cells. The anti-ORF S/L antibody was used in immunohistochemical analyses of HELF cells infected with Ellen, Oka-SK, Oka-Δ4-3, or Oka-R. The S/L protein was detected predominantly in the cytoplasm of infected cells that synthesized it, and its distribution was similar to that of VZV gC as detected in HELF cells infected with all of the VZV strains (Fig. 7).
FIG. 7.
Immunohistochemical detection of VZV proteins in HELF. Cells were infected with VZV strains Ellen and Oka-SK and the recombinant viruses Oka-Δ4-3 and Oka-R. Immunohistochemical analysis of mock-infected cells is also shown. The products of ORF S/L and gC were detected using anti-ORF S/L or anti-gC antibodies. The signal was visualized after addition of AP-conjugated goat anti-rabbit immunoglobulin secondary antibody and developed with AP substrate in the presence of levamisole. The viruses infecting the monolayers are noted at the left; VZV proteins that were analyzed are identified at the top.
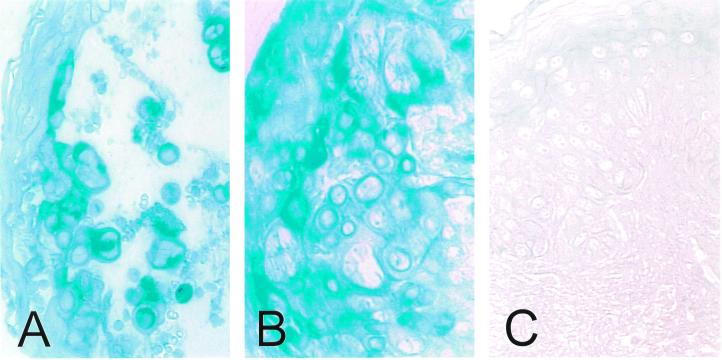
Immunohistochemical analysis of skin biopsy specimens obtained from patients with chickenpox and zoster demonstrated that S/L protein was present in the cytoplasm of epithelial cells during both primary infection and reactivation (Fig. 8A and B). A skin biopsy obtained from a patient with Grover's disease, a dermatologic disorder characterized by a vesicular eruption that is not caused by VZV infection, was negative for S/L protein (Fig. 8C).
FIG. 8.
Immunohistochemical analysis of ORF S/L protein in skin biopsy specimens. Sections of formalin-fixed, paraffin-embedded skin biopsy specimens from patients with chickenpox (A), zoster (B), and Grover's disease (C) were deparaffinized with xylene and rehydrated with successive alcohol washes. ORF S/L protein was detected using anti-ORF S/L antibody. The signal was visualized as described in the legend to Fig. 7.
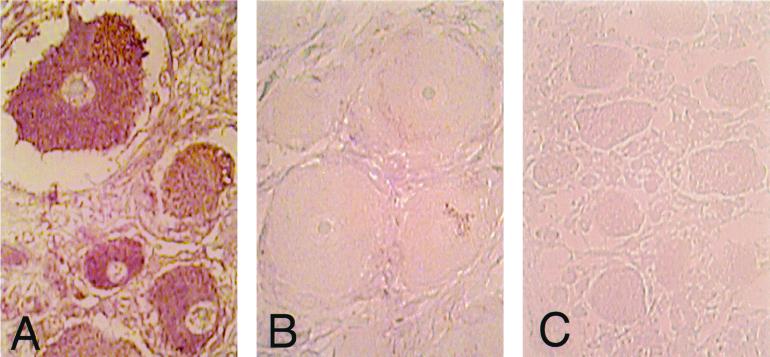
Human DRG harvested at autopsy from three adults and one fetus were also analyzed for S/L protein (Fig. 9). One individual had zoster and the other two had no clinical evidence of VZV reactivation at the time of death. The DRG analyzed from the patient with zoster innervated one of the affected dermatomes; ORF S/L protein was detected in the cytoplasm of neurons in this DRG (Fig. 9A). The DRG obtained from the patients who did not have zoster at the time of death were found to contain latent virus, as indicated by in situ hybridization (30); S/L protein was not detected in these DRG (Fig. 9B). The fetal DRG was also negative by immunohistochemical analysis for S/L (Fig. 9C). Therefore, S/L protein is not detected in latently infected ganglia and appears in neurons only during active replication.
FIG. 9.
Immunohistochemical analysis of ORF S/L in DRG. Sections of formalin-fixed, paraffin-embedded DRG harboring reactivated (A) or latent (B) VZV and a control uninfected fetal DRG (C) were processed as described in the legend to Fig. 8. ORF S/L was detected by immunohistochemistry essentially as described in the legend to Fig. 7 except that levamisole was omitted from the AP detection buffer and nitroblue tetrazolium chloride was the substrate.
DISCUSSION
We have demonstrated the existence of a new gene, ORF S/L, in the VZV genome. This ORF was probably not detected in earlier analyses because of its location. Genes spanning the termini of a herpesvirus are not unique to VZV; Epstein-Barr virus and bovine herpesvirus 1 (BHV) have genes that span their termini (16, 28). BHV has a genome structure similar to that of VZV; short repeats bracket UL, and only two isomeric forms predominate among the progeny (44). BHV circ is located entirely within UL and is produced from a spliced transcript that spans the termini. Although the circ gene has no homology to ORF S/L (16), its presence indicates that other herpesviruses may also have genes spanning the termini.
We identified a 900-nt, spliced, poly(A)-containing RNA in cells infected with Oka-ATCC that maps to the S/L junction. Previous transcription maps of VZV did not document a mRNA near ORF S/L. However, a 0.8-kb RNA directed leftward and located near the IRL/IRS junction could encode S/L, as it hybridizes to probes that cross the TRL/IRL repeats (32, 40, 43). In addition, Maguire and Hyman reported that a 0.8-kb poly(A)+, cytoplasmic RNA hybridized to the large EcoRI C fragment of strain 80-2. The C-terminal portion of ORF S/L is encoded in this fragment. However, the directionality and map position of this transcript were not reported (32). In general, spliced messages are rare in alphaherpesviruses, and this is the first example of a spliced RNA in VZV. Only five HSV genes are spliced, and only one VZV gene (ORF 42) is likely to be generated from a spliced RNA (11, 45). The location of the splice in the 3′ untranslated region of Oka-ATCC is unusual for mRNA. 3′ untranslated introns are found in the cellular prostaglandin E receptor gene and some forms of cellular H-ras (19, 25). The 3′ untranslated intron in c-H-ras can affect expression of the gene by modulating nearby sequences. The sequences of Oka-Merck and Oka-SK are conserved through the intronic region; however, the absence of a termination codon in these genomes allows for the expression of larger S/L proteins. Further sequence and RNA analyses will be required to determine the termination sites for these proteins and whether the RNAs encoded by these S/L genes are spliced.
VZV ORF 1 is expressed in productively infected cells (9). A 470-nt transcript was mapped to this portion of the genome. This transcript is antisense to the ORF S/L transcript, and we predict that the transcripts overlap one another. This scenario is similar to that recently observed for the HSV transcripts encoding ORF P and ICP34.5 as well as UL43 and UL43.5 (27, 49). In the case of ORF P, increased transcription of its cognate promoter leads to a decrease in accumulation of ICP34.5.
ORF S/L exhibits an unusually high degree of polymorphism for VZV. The five VZV strains analyzed in this investigation were heterogeneous with respect to their S/L reading frames. The DNA sequence analysis in this study included strains that were passaged many times in tissue culture. Western blot analysis suggests that the predominant S/L protein from VZV (Jones), a clinical isolate, had the same electrophoretic mobility as that encoded by Oka-ATCC and other clinical isolates (data not shown). The mutations within the Oka strains that have attenuated these viruses for use as vaccines have not been identified. Therefore, any differences between Oka strains and wild-type strains such as Dumas or Ellen must be considered possible attenuating mutations. The vast majority of restriction sites in Oka-ATCC, Oka-Merck, and Dumas are conserved; of the 1,095 nts reported here, we identified only 24 differences between the Oka strains and Dumas. It remains unclear whether these differences between VZV strains affect replication or pathogenicity. The lack of a model system in which the attenuation of Oka can be measured makes the analysis of these mutations difficult.
A 21-kDa protein in lysates of cells infected with VZV strains Jones and Oka-ATCC reacts specifically with anti-ORF S/L by Western blotting. Lysates of cells infected with Ellen, Oka-SK, or Oka-Merck contain protein moieties that react with anti-ORF S/L and migrate more slowly in SDS-PAGE. This difference in mobilities is consistent with the missing stop codon in these three strains, although it may also result from differences in protein modification. The lack of a discrete band when lysates from cells infected with Ellen, Oka-SK, and Oka-Merck are examined by Western blot analysis suggests that the larger S/L protein either is degraded or undergoes posttranslational modification. During primary infection and reactivation, S/L protein accumulates in the cytoplasm of infected epithelial cells. It is also present in the cytoplasm of neurons during reactivation. In contrast, S/L was not detected in the two DRGs harboring latent VZV that were analyzed. It appears that this protein is expressed only during lytic infection.
The altered plaque morphology on MeWO cells was brought about by a change in adherence of the infected cells. Although the plaques derived from infection with the mutant initially appeared similar to plaques produced by the parental virus, the adherence of the cells infected with mutant virus changed with time. Cells infected with the mutant were much more likely to detach from the surrounding cells. Those cells at the edges of the plaque, probably the most recently infected, remained adherent to the surrounding stratum. The distinct phenotype of the ORF S/L deletion mutant suggests that this protein may play a role in altering cell adhesion molecules in infected cells, thus changing their ability to adhere. Whether this function is relevant to the pathogenesis of infection in human hosts remains to be investigated.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
This work was supported by NIH grants AI-01409 (to P.A.) and AI-124021 (to S.J.S. and A. A. Gershon) and by Aviron. P.A. is the recipient of an Irving Scholar Award.
REFERENCES
- 1.Annunziato P W, Lungu O, Panagiotidis C, Zhang J H, Silvers D N, Gershon A A, Silverstein S J. Varicella-zoster virus proteins in skin lesions: implications for a novel role of ORF29p in chickenpox. J Virol. 2000;74:2005–2010. doi: 10.1128/jvi.74.4.2005-2010.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohenzky R A, Papavassiliou A G, Gelman I H, Silverstein S. Identification of a promoter mapping within the reiterated sequences that flank the herpes simplex virus type 1 ULregion. J Virol. 1993;67:632–642. doi: 10.1128/jvi.67.2.632-642.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol. 1992;66:2904–2915. doi: 10.1128/jvi.66.5.2904-2915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Varicella-related deaths among adults—United States 1997. Morbid Mortal Wkly Rep. 1997;46:409–412. [PubMed] [Google Scholar]
- 5.Chen J, Silverstein S. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J Virol. 1992;66:2916–2927. doi: 10.1128/jvi.66.5.2916-2927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Chou J, Kern E R, Whitley R J, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to γ1 34.5, a gene nonessential for growth in culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 8.Cohen J I, Seidel K E. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc Natl Acad Sci USA. 1993;90:7376–7380. doi: 10.1073/pnas.90.15.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen J I, Seidel K E. Varicella-zoster virus open reading frame 1 encodes a membrane protein that is dispensable for growth of VZV in vitro. Virology. 1995;206:835–842. doi: 10.1006/viro.1995.1006. [DOI] [PubMed] [Google Scholar]
- 10.Davison A J. Structure of the genome termini of varicella-zoster virus. J Gen Virol. 1984;65:1969–1977. doi: 10.1099/0022-1317-65-11-1969. [DOI] [PubMed] [Google Scholar]
- 11.Davison A J. Varicella-zoster virus. J Gen Virol. 1991;65:475–486. doi: 10.1099/0022-1317-72-3-475. [DOI] [PubMed] [Google Scholar]
- 12.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 13.Davison A J, Wilkie N M. Location and orientation of homologous-sequences in the genomes of five herpesviruses. J Gen Virol. 1983;64:1927–1942. doi: 10.1099/0022-1317-64-9-1927. [DOI] [PubMed] [Google Scholar]
- 14.Ecker J R, Hyman R W. Varicella zoster virus DNA exists as two isomers. Proc Natl Acad Sci USA. 1982;79:156–160. doi: 10.1073/pnas.79.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felser J M, Straus S E, Ostrove J M. Varicella-zoster virus complements herpes simplex virus type 1 temperature-sensitive mutants. J Virol. 1987;61:225–228. doi: 10.1128/jvi.61.1.225-228.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraefel C, Wirth U V, Vogt B, Schwyzer M. Immediate-early transcription over covalently joined genome ends of bovine herpesvirus 1: the circgene. J Virol. 1993;67:1328–1333. doi: 10.1128/jvi.67.3.1328-1333.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelb L. Varicella-zoster virus. In: Fields B N, Knipe D M, editors. Virology. 2nd ed. New York, N.Y: Raven Press; 1990. pp. 2011–2054. [Google Scholar]
- 18.Gilden D H, Kleinschmidt-DeMasters B K, LaGuardia J J, Mahalingam R, Cohrs R J. Neurologic complications of the reactivation of varicella-zoster virus. N Engl J Med. 2000;342:635–645. doi: 10.1056/NEJM200003023420906. [DOI] [PubMed] [Google Scholar]
- 19.Hwang D Y, Cohen J B. A splicing enhancer in the 3′-terminal c-H-rasexon influences mRNA abundance and transforming activity. J Virol. 1997;71:6416–6426. doi: 10.1128/jvi.71.9.6416-6426.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlin S, Mocarski E S, Schachtel G A. Molecular evolution of herpesviruses: genomic and protein sequence comparisons. J Virol. 1994;68:1886–1902. doi: 10.1128/jvi.68.3.1886-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemble G, Duke G, Winter R, Spaete R. Defined large scale alteration of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J Virol. 1996;70:2044–2048. doi: 10.1128/jvi.70.3.2044-2048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinchington P R, Reinhold W C, Casey T A, Straus S E, Hay J, Ruyechan W T. Inversion and circularization of the varicella-zoster virus genome. J Virol. 1985;56:194–200. doi: 10.1128/jvi.56.1.194-200.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingston R. Calcium phosphate transfection. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. Brooklyn, N.Y: Greene Publishing Associates, Inc., and John Wiley & Sons, Inc.; 1993. pp. 9.1.4–9.1.11. [Google Scholar]
- 24.Kingston R E. Preparation of poly (A)+ RNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. Brooklyn, N.Y: Greene Publishing Associates, Inc., and John Wiley & Sons, Inc.; 1993. pp. 4.5.1–4.5.3. [Google Scholar]
- 25.Kotani M, Tanaka I, Ogawa Y, Usui T, Mori K, Ichikawa A, Narumiya S, Yoshimi T, Nakao K. Molecular cloning and expression of multiple isoforms of human prostaglandin E receptor EP3 subtype generated by alternative messenger RNA splicing: multiple second messenger systems and tissue-specific distributions. Mol Pharmacol. 1995;48:869–879. [PubMed] [Google Scholar]
- 26.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 27.Lagunoff M, Roizman B. Expression of a herpes simplex virus 1 open reading frame antisense to the γ134.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J Virol. 1994;68:6021–6028. doi: 10.1128/jvi.68.9.6021-6028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laux G, Perricaudet M, Farrell P J. A spliced Epstein-Barr virus gene expressed in immortalized lymphocytes is created by circularization of the linear viral genome. EMBO J. 1988;7:769–774. doi: 10.1002/j.1460-2075.1988.tb02874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F, Roizman B. The promoter, transcriptional unit, and coding sequence of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J Virol. 1991;65:206–212. doi: 10.1128/jvi.65.1.206-212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lungu O, Annunziato P, Gershon A, Stegatis S, Josefson D, LaRussa P, Silverstein S. Reactivated and latent varicella-zoster virus in human dorsal root ganglia. Proc Natl Acad Sci USA. 1995;92:10980–10984. doi: 10.1073/pnas.92.24.10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lungu O, Panagiotidis C A, Annunziato P W, Gershon A A, Silverstein S J. Aberrant intracellular localization of varicella-zoster virus regulatory proteins during latency. Proc Natl Acad Sci USA. 1998;95:7080–7085. doi: 10.1073/pnas.95.12.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maguire H F, Hyman R W. Polyadenylated, cytoplasmic transcripts of varicella-zoster virus. Intervirology. 1986;26:181–191. doi: 10.1159/000149700. [DOI] [PubMed] [Google Scholar]
- 33.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 34.McGeoch D J, Dolan A, Donald S, Rixon F J. Sequence determination and genetic content of the short unique region of herpes simplex virus type 1. J Mol Biol. 1985;181:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 35.McLauchlan J, Gaffney D, Whitton J L, Clements J B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res. 1985;13:1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mocarski E S, Liu A C, Spaete R R. Structure and variability of the a sequence in the genome of human cytomegalovirus (Towne strain) J Gen Virol. 1987;68:2223–2230. doi: 10.1099/0022-1317-68-8-2223. [DOI] [PubMed] [Google Scholar]
- 37.Mocarski E S, Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization, and generation of virion DNA. Cell. 1982;31:89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- 38.Moriuchi H, Moriuchi M, Smith H A, Straus S E, Cohen J I. Varicella-zoster virus open reading frame 61 protein is functionally homologous to herpes simplex virus type 1 ICP0. J Virol. 1992;66:7303–7308. doi: 10.1128/jvi.66.12.7303-7308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mount S M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostrove J M, Reinhold W, Fan C-H, Zom S, Hay J, Straus S E. Transcription mapping of the varicella-zoster virus genome. J Virol. 1985;56:600–606. doi: 10.1128/jvi.56.2.600-606.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panagiotidis C, Silverstein S. pAlex, a dual-tag prokaryotic expression vector for the purification of full-length proteins. Gene. 1995;164:45–47. doi: 10.1016/0378-1119(95)00417-5. [DOI] [PubMed] [Google Scholar]
- 42.Proudfoot N J, Brownlee G G. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 43.Reinhold W C, Straus S E, Ostrove J M. Directionality and further mapping of varicella zoster virus transcripts. Virus Res. 1988;9:249–261. doi: 10.1016/0168-1702(88)90034-2. [DOI] [PubMed] [Google Scholar]
- 44.Roizman B. The family Herpesviridae. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press, Ltd.; 1993. pp. 1–9. [Google Scholar]
- 45.Roizman B, Sears A. Herpes simplex viruses and their replication. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press, Ltd.; 1993. pp. 11–68. [Google Scholar]
- 46.Straus S E, Aulakh H S, Ruyechan W T, Hay J, Casey T A, vandeWoude G F, Owens J, Smith H A. Structure of varicella-zoster virus DNA. J Virol. 1981;40:516–525. doi: 10.1128/jvi.40.2.516-525.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Straus S E, Owens J, Ruyechan W T, Takiff H E, Casey T A, vande Woude G F, Hay J. Molecular cloning and physical mapping of varicella-zoster virus DNA. Proc Natl Acad Sci USA. 1982;79:993–997. doi: 10.1073/pnas.79.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward P L, Barker D E, Roizman B. A novel herpes simplex virus 1 gene, UL43.5, maps antisense to the UL43 gene and encodes a protein which colocalizes in nuclear structures with capsid proteins. J Virol. 1996;70:2684–2690. doi: 10.1128/jvi.70.5.2684-2690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webster C B, Chen D, Horgan M, Olivo P D. The varicella-zoster virus origin-binding protein can substitute for the herpes simplex virus origin-binding protein in a transient origin-dependent DNA replication assay in insect cells. Virology. 1995;206:655–660. doi: 10.1016/s0042-6822(95)80084-0. [DOI] [PubMed] [Google Scholar]
- 51.Yeh L, Schaffer P A. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. J Virol. 1993;67:7373–7382. doi: 10.1128/jvi.67.12.7373-7382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]