Abstract
The herpes simplex virus type 1 (HSV-1) immediate-early gene IE63 (ICP27), the only HSV-1 regulatory gene with a homologue in every mammalian and avian herpesvirus sequenced so far, is a multifunctional protein which regulates transcriptional and posttranscriptional processes. One of its posttranscriptional effects is the inhibition of splicing of viral and cellular transcripts. We previously identified heterogeneous nuclear ribonucleoprotein (hnRNP) K and casein kinase 2 (CK2) as two protein partners of IE63 (H. Bryant et al., J. Biol. Chem. 274:28991–28998, 1999). Here, using a yeast two-hybrid assay, we identify another partner of IE63, the cellular protein p32. Confirmation of this interaction was provided by coimmunoprecipitation from virus-infected cells and recombinant p32 binding assays. A p32-hnRNP K-CK2 complex, which required IE63 to form, was isolated from HSV-1-infected cells, and coimmunoprecipitating p32 was phosphorylated by CK2. Expression of IE63 altered the cytoplasmic distribution of p32, with some now colocalizing with IE63 in the nuclei of infected and transfected cells. As p32 copurifies with splicing factors and can inhibit splicing, we propose that IE63 together with p32, possibly with other IE63 partner proteins, acts to disrupt or regulate pre-mRNA splicing. As well as contributing to host cell shutoff, this effect could facilitate splicing-independent nuclear export of viral transcripts.
A key regulatory protein of herpes simplex type 1 (HSV-1) lytic infection is the 63-kDa nuclear phosphoprotein IE63 (also known as ICP27). IE63 is essential for viral replication (23, 36–39) and is required for the switch from early to late virus gene expression (21). It has been shown to perform multiple functions at both transcriptional and posttranscriptional levels (reviewed in reference 33). Acting posttranscriptionally, IE63 binds RNA in vivo with a reported specificity for intronless viral transcripts (40), enhances pre-mRNA 3′ processing (22), and contributes to the shutoff of host protein synthesis by inhibiting splicing of viral and cellular transcripts (11, 12). IE63 colocalizes with nuclear antigens such as snRNPs (31) and causes the nuclear retention of intron-containing viral transcripts (34). More recently, IE63 has been shown to be capable of shuttling from the nucleus to the cytoplasm (24, 32, 46) and may facilitate the nuclear export of intronless RNAs, which form the majority of viral transcripts (40). IE63 mediates the export of some viral RNAs via a Crm-1-dependent pathway, whereas other viral RNAs are exported via a Crm-1-independent pathway (47).
In HSV-1-infected cells, IE63 interacts with heterogeneous nuclear ribonucleoprotein (hnRNP) K and with casein kinase 2 (CK2), the latter activity being able to phosphorylate both IE63 and hnRNP K, possibly to alter their activities (4). Here, we show that, consistent with its multiple functions, IE63 interacts with another cellular protein, p32. First isolated as a protein tightly associated with ASF/SF2 purified from HeLa cells (16), p32 regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation (30). p32 is reported to have a mitochondrial distribution (19, 28) but can also be found in the nucleus as granules and tubules (19). The distribution of p32 is altered during adenovirus infection, where, with viral core protein V, it redistributes to the nucleus (19). Numerous interactions between cellular and viral proteins and p32 have been reported, including with lamin B receptor (44), transcription factor TFIIB (52), HSV-1 open reading frame (ORF) P protein (3), Epstein-Barr virus (EBV) EBNA I protein (5, 51), adenovirus polypeptide V (19), and the human immunodeficiency virus (HIV) proteins Rev and Tat (17, 49, 52). Both cell location and interactions have suggested a role for p32 not only in splicing (17, 30, 49, 53) but also in nucleocytoplasmic transport (18, 19, 29) to and from the mitochondria (13, 19) and in maintaining oxidative phosphorylation (28).
Using the yeast two-hybrid system, immunoprecipitation from HSV-1-infected cells, and in vitro binding assays, we show that IE63 interacts with p32. The IE63 partner proteins hnRNP K and CK2 also were found in the complex, which required IE63 for its formation. We demonstrate that p32 coimmunoprecipitated with IE63 is phosphorylated in vitro by coimmunoprecipitating CK2 activity. The intracellular distribution of p32 is altered by IE63 during HSV-1 infection to show some nuclear staining which colocalizes with IE63. The interaction between IE63 and p32 suggests that in HSV-1-infected cells, p32 is involved in splicing inhibition. As well as contributing to host cell shutoff, this inhibition could facilitate nucleocytoplasmic transport of viral transcripts by uncoupling splicing from RNA nuclear export.
MATERIALS AND METHODS
Plasmids and antisera.
For transient expression of IE63, plasmid pCMV63 containing an EcoRI/BamHI fragment with the entire ORF of IE63 and an additional 400 bp at the C terminus was ligated into the expression vector pCMV-10 (48). IE63 constructs used in the yeast two-hybrid screen have been described previously (4). Anti-IE63 antiserum H1113 was a mouse monoclonal antibody (MAb) (1) supplied by the Goodwin Institute for Cancer Research. Anti-hnRNP K antiserum was a rabbit antibody (50) generously supplied by K. Bomsztyk (University of Washington). For Western blots, a mouse p32 MAb was raised against purified recombinant p32. For indirect immunofluorescence, rabbit antiserum raised against recombinant p32 (19) was used.
Cells and viruses.
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 2.5% fetal calf serum, 2.5% newborn calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). Stocks of wild-type (wt) HSV-1 strain 17+, the IE63 insertion mutant HSV-1 27-lacZ (45), and the HSV-1 gE− insertion mutant (14) were grown as described previously (22).
Infection of cells and preparation of extracts.
Ninety percent confluent HeLa cell monolayers (4 × 107 cells) were infected with HSV-1 wt or 27-lacZ at a multiplicity of 10 PFU/cell or left uninfected (mock infected). After 1 h of absorption at 37°C, medium was added and cells were left for 16 h. For preparation of cell extracts, monolayers were washed with phosphate-buffered saline (PBS) and cells were lysed by suspension in 1 ml of cell extract buffer (50 mM HEPES, 50 mM NaCl, 0.1% NP-40 [pH 7.5]) containing a protease inhibitor cocktail (Boehringer Mannheim). Extracts were sonicated on ice, cell debris was pelleted, and the protein concentration was determined by the Bradford assay (Bio-Rad). Extracts for recombinant p32 and recombinant glucose oxidase (rMp32 and rMpGO) column pull-down assays were prepared as for coimmunoprecipitation except that monolayers were resuspended in 500 μl of extract buffer. For immunofluorescence, 13-mm-diameter coverslips were seeded at 0.5 × 105 HeLa cells per well in 1 ml of normal HeLa medium and incubated overnight at 37°C prior to infection at a multiplicity of 10 PFU/cell. Alternatively, 5 × 106 HeLa cells were transfected with the IE63 expression plasmid pCMV63 by electroporation with 20 μg of DNA and left for 24 h before extracts were prepared as described above or were fixed for immunofluorescence.
Preparation of rMp32 and rMpGO columns; pull-down assays.
Recombinant p32 was expressed in Escherichia coli, purified by fast protein liquid chromatography, and coupled to activated Sepharose to generate rMp32 columns as previously described (19). Control columns of glucose oxidase (rMpGO) using protein from Sigma were prepared using the same protocol. Pull-down assays were performed with 40 μl of rMp32 or rMpGO column material and 100 μl of cell extract as described previously (19). After washing, bound proteins were assayed for CK2 activity or were removed (along with p32 and glucose oxidase) by boiling prior to analysis by Western blotting.
Immunoprecipitation and Western blotting.
A 100-μg aliquot of cell extract was mixed with 5 μl of MAb H1113 monoclonal antibody (and 1 μl of sheep anti-mouse immunoglobulin G) in 50 μl of binding buffer (100 mM Tris HCl, 5 mM EDTA, 1% Triton X-100 [pH 7.4]) for 3 h at 4°C; 75 μl of protein A-Sepharose was then added, and mixing was for 1 h as described previously (4). After pelleting of the beads and multiple washes, the bound proteins were eluted, and proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and detected by Western blotting. Western blotting was performed as described previously (4), using primary antibody dilutions for IE63 of 1:2,000, for hnRNP K of 1:10,000, and for p32 of 1:200.
CK2 activity assay.
Following washing, proteins bound to rMp32 or rMpGO columns were resuspended in 30 μl of CK2 reaction buffer (50 mM Tris–20 mM MgCl2 [pH 8.2] containing 10 μCi of [γ-32P]ATP per reaction), either with or without 0.1 mM CK2 peptide substrate (Arg-Arg-Arg-Glu-Glu-Glu-Thr-Glu-Glu-Glu), and CK2 activity was detected as described elsewhere (4).
Phosphorylation assay.
After washing, coimmunoprecipitates obtained with anti-IE63 antiserum were washed with 50 mM Tris (pH 7.4) and then resuspended in 20 μl of 50 mM Tris (pH 7.4); 5 μl of radioactive solution (50 mM Tris–20 mM MgCl2 [pH 7.4] containing 10 μM ATP and 2.5 μCi of [γ-32P]ATP per reaction) was added, and phosphorylation was allowed to take place for 15 min at 25°C in either the presence or absence of 100 μM DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole), a specific inhibitor of CK2 activity, as described elsewhere (4). Immunoprecipitated proteins were separated by electrophoresis on a 10% polyacrylamide gel containing 0.1% SDS, and phosphorylated proteins were visualized using a phosphorimaging system (Molecular Dynamics) and by Western blotting for p32.
Yeast two-hybrid screen and mapping the regions of IE63 involved in the p32 interaction.
The yeast two-hybrid screen was performed using IE63 amino acids (aa) 10 to 512 as bait and target plasmids containing a HeLa cell cDNA library (Clontech) as described previously (4). Mapping the IE63 regions involved in the interaction utilized the truncated IE63 constructs described in reference 4; these were mated into yeast cells transfected with a full-length p32 clone identified from the library screen, using a β-galactosidase (β-Gal) filter assay. Results from each cotransformation represented an analysis of some 200 individual colonies. The time taken for the positive cells to turn blue varied between experiments, depending on factors such as colony size. However, a positive interaction was identified by the presence of blue colonies seen after 3 h on β-Gal filter assays which, when compared to interacting standards assayed by liquid β-Gal assays, had activities of between 10 to 50 U, while standards showing lack of interaction had activities of <1 U. Filter assays and liquid β-Gal assays were performed according to the manufacturer's instructions (Clontech protocol PT3024-1).
Indirect immunofluorescence.
Infected, transfected, or mock-infected cell monolayers were fixed for 10 min at 20°C with 2% sucrose–5% formaldehyde in PBS. After three washes in PBS, cells were permeabilized for 10 min at 20°C with 0.5% NP-40–10% sucrose in PBS. After further washes, primary antibody diluted to the appropriate concentration (IE63, 1:100; p32, 1:50) in PBS with 1% calf serum was added for 60 min at 20°C. Cells were once again washed before incubation with secondary antibody coupled to fluorescein isothiocyanate or Cy5 (Sigma) diluted 1:100 in PBS was carried out for 30 min at 20°C. After a final wash, cells were examined with a Zeiss LSM 510 confocal microscope system with two lasers giving excitation at 488 nm (fluorescein isothiocyanate) and 633 nm (Cy5) and a Zeiss Axioplan microscope using a 63× oil immersion objective lens (nuclear aperture, 1.4). Data were processed with LSM 510 software and then exported for preparation using Photoshop.
RESULTS
p32 interacts with IE63 using the yeast two-hybrid screen.
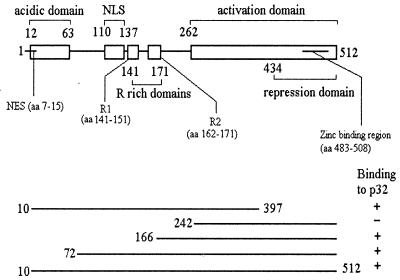
To identify cellular proteins capable of interacting with IE63 in the yeast two-hybrid assay, we screened a HeLa cell library fused to the GAL4 activation domain and expressed in pGADGH using IE63 aa 10 to 512 as bait. From a total of 2.3 × 106 transformants screened, 82 clones fulfilled the criteria for interaction of gene products. These were sequenced and checked against the GenEMBL database. For 12 clones, the portions of inserts sequenced all had high sequence homology (maximum, 99.3% homology in 446 bp) with the splicing factor p32, with inserts around 1.1 kb consistent with full-length cDNA of the 1.16-kb p32 gene. To map the regions of IE63 required for interaction with p32, a series of IE63 truncations expressed as hybrids with the GAL4 DNA-binding domain (4) was mated into cells transformed with a full-length p32 clone identified from the library screen. The results (Fig. 1) showed that sequences within IE63 aa 166 to 242 were involved in the interaction with p32.
FIG. 1.
Schematic representation (not to scale) of IE63 protein showing the different functional regions as described elsewhere (4). NLS, nuclear localization signal; NES, leucine-rich nuclear export signal; R1 and R2, arginine-rich regions. Shown below are the IE63 truncations used in the yeast two-hybrid assay to map the region involved in interaction with p32.
Coimmunoprecipitation of IE63 and p32.
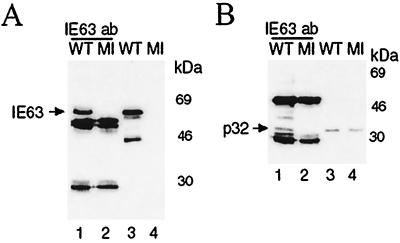
Extracts of HeLa cells infected with HSV-1 wt or mock infected were subjected to immunoprecipitation with anti-IE63 monoclonal antibody. The immunoprecipitated proteins following separation by SDS-PAGE were transferred to nitrocellulose membranes and analyzed by Western blotting using antiserum directed against IE63 or p32. IE63 MAb precipitated IE63 from the wt-infected extract (Fig. 2A, lane 1). p32 was coimmunoprecipitated with IE63 (Fig. 2B, lane 1) and was present as a single band in the wt- and mock-infected extracts used for immunoprecipitations (Fig. 2B, lanes 3 and 4). By contrast, p32 was not precipitated by IE63 MAb from mock-infected extracts (Fig. 2B, lane 2). Prominent bands located below IE63 (Fig. 2A, lanes 1 and 2) and straddling p32 (Fig. 2B, lanes 1 and 2) are the antibody heavy and light chains. A more rapidly migrating degradation product of IE63 was sometimes present in whole cell extracts used for immunoprecipitation (Fig. 2A, lane 3), and the band above p32 present in immunoprecipitates (Fig. 2B, lane 1) is a cross-reacting protein of unknown origin.
FIG. 2.
In vivo coimmunoprecipitation of IE63 and p32, using antibodies directed against IE63. HSV-1 wt-infected (WT) or mock-infected (MI) HeLa cell extracts were immunoprecipitated with IE63 MAb. Aliquots of the precipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose, and analyzed by Western blotting using p32 MAb (B, lanes 1 and 2) or IE63 MAb (A, lanes 1 and 2). A 100-μg aliquot of total protein was added to each immunoprecipitation, and half was loaded in lanes 1 and 2; 20 μg of total protein from extracts was loaded in lanes 3 and 4.
IE63 interacts with rMp32-Sepharose.
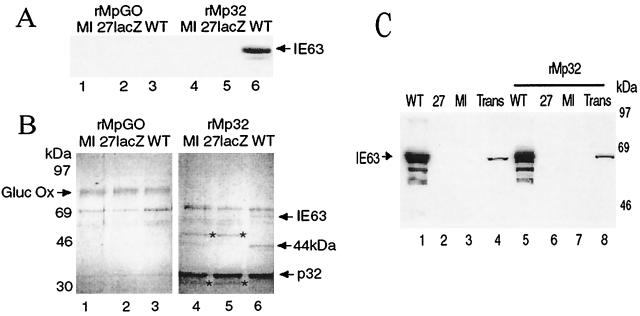
rMp32 and rMpGO were coupled to Sepharose beads and used in pull-down assays with extracts from HeLa cells infected with HSV-1 wt or the IE63 mutant 27-lacZ or mock infected. After washing, bound proteins and p32 or glucose oxidase GO were eluted off the Sepharose beads. Following separation by SDS-PAGE, Western blotting of the proteins revealed that IE63 from the wt-infected extract alone was bound to rMp32 (Fig. 3A, lane 6). By contrast, IE63 did not bind to the rMpGO control column containing glucose oxidase, a protein with a pI similar to that of p32 (Fig. 3A, lane 3). Enough IE63 was bound to the rMp32 column to be visualized by Coomassie blue staining (Fig. 3B, lane 6). Other bands visualized by Coomassie blue staining corresponded to glucose oxidase (Fig. 3B, lanes 1 to 3) and p32 (Fig. 3B, lanes 4 to 6). A 44-kDa band that interacted with p32 was detected, but only when infected cell extracts were used (Fig. 3B, lane 6); the identity of this band is under investigation. Bands of around 50 and 30 kDa (Fig. 3B, lanes 4 and 5) were seen to interact with p32 and not with glucose oxidase, using 27-lacZ- or mock-infected extracts, but were not seen with wt-infected extract; these may reflect interactions of p32 in uninfected cells which are disrupted by the presence of IE63. Use of extracts made from cells transiently transfected with the IE63 expression construct pCMV63 demonstrated expression of IE63 (Fig. 3C, lane 4), and use of the rMp32 column (Fig. 3C, lane 8) showed that p32 and IE63 were capable of interacting in the absence of any other viral proteins (Fig. 3C, lane 8).
FIG. 3.
Interaction of IE63 with p32 Sepharose column. HSV-1 wt- or 27-lacZ-infected, pCMV63-transfected, or mock-infected HeLa cell extracts were mixed with rMpGO-Sepharose or rMp32-Sepharose. After washing, proteins were boiled off the Sepharose, separated by SDS-PAGE, transferred to nitrocellulose, and Coomassie blue stained or analyzed by Western blotting using IE63 MAb. (A) Western blot using IE63 MAb of proteins from wt-infected (WT), 27-lacZ-infected, or mock-infected (MI) cell extracts, bound and then removed from an rMpGO (lanes 1 to 3) or rMp32 (lanes 4 to 6) column. (B) Coomassie blue staining of the bound proteins shown in panel A. (C) Western blot using IE63 MAb of proteins from wt-infected (WT), mock-infected (MI), or pCMV63-transfected (Trans) cell extracts, bound and then removed from an rMp32 column (lanes 5 to 8); cell extracts used (lanes 1 to 4).
The complex with IE63 and p32 also includes hnRNP K and CK2.
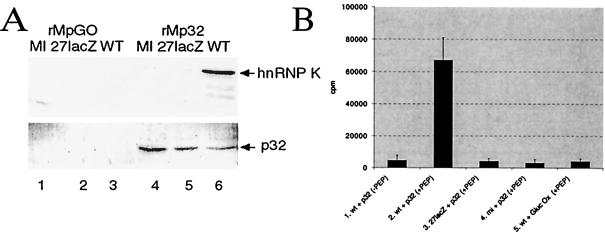
We have shown that IE63 forms a complex which includes hnRNP K and CK2 (4). Thus, following separation by SDS-PAGE, aliquots of the proteins from wt-infected, 27-lacZ-infected, or mock-infected cells which interacted with rMp32- or rMpGO-Sepharose beads were analyzed by Western blotting for hnRNP K (Fig. 4A, top). Similar amounts of column were used, as shown by direct Western blotting for p32 (Fig. 4A, bottom, lanes 4 to 6). hnRNP K was seen to bind strongly to rMp32 in wt-infected extracts, with binding significantly lowered in the absence of IE63 but the presence of other viral IE proteins; there was no binding with uninfected extracts (Fig. 4A, top, compare lane 6 with lanes 4 and 5). hnRNP K did not bind to glucose oxidase (Fig. 4A, top, lanes 1 to 3).
FIG. 4.
CK2 and hnRNP K from HSV-1-infected extracts interact with p32 attached to a Sepharose column. Aliquots of bound and subsequently eluted proteins, as shown in Fig. 3, were separated by SDS-PAGE, transferred to nitrocellulose, and analyzed by Western blotting using anti-hnRNP K serum or p32 MAb and also analyzed for CK2 activity using a peptide assay. (A) Western blot analysis using anti-hnRNP K serum (top) or p32 MAb (bottom) of proteins from wt-infected (WT), 27-lacZ-infected, or mock-infected (MI) cell extracts. Proteins were bound and then removed from an rMpGO (lanes 1 to 3) or rMp32 (lanes 4 to 6) column. (B) CK2 activity, as measured by phosphorylation of a specific peptide substrate, by proteins which bound to rMpGO from wt-infected extracts (lane 5) and to rMp32 from wt-infected (lanes 1 and 2), 27-lacZ-infected (lane 3), and mock-infected (lane 4) extracts. CK2 assays were performed in the presence (lanes 2 to 5) or absence (lane 1) of peptide substrate.
The presence of CK2 activity in cell extracts following interaction with rMp32 or rMpGO was examined using a specific CK2 peptide substrate assay. CK2 activity was found associated only with the p32-IE63-hnRNP K complex from wt-infected cells (Fig. 4B, lane 2). Strikingly, CK2 activity was not associated with proteins bound to rMp32 from 27-lacZ-infected or mock-infected extracts (Fig. 4B, lanes 3 and 4), nor was it bound to rMpGO beads when wt-infected cell extract was used (Fig. 4B, lane 5). No CK2 activity was observed when the peptide substrate was absent (Fig. 4B, lane 1). These results indicate that a complex of proteins which includes p32, hnRNP K, and CK2 is formed during HSV-1 infection and that IE63 is required for this process.
Coimmunoprecipitating CK2 activity can phosphorylate p32.
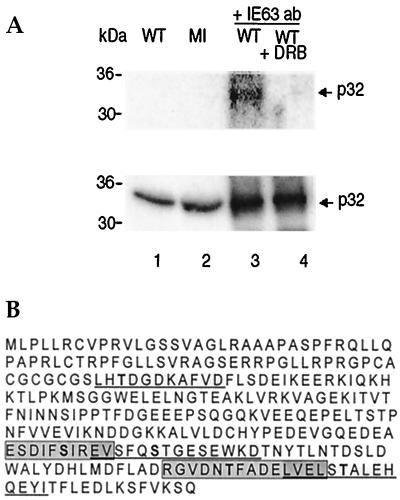
CK2 activity found in immunoprecipitates with anti-IE63 serum has been shown to phosphorylate IE63 and hnRNP K present in the complex (4). Thus, immunoprecipitates generated with anti-IE63 antiserum were incubated with [γ-32P]ATP, and phosphorylation of proteins in the complex by kinase activity present was examined in the presence or absence of the specific CK2 inhibitor DRB. A single band in the size range of 30 to 36 kDa was phosphorylated as visualized by phosphorimaging, and its phosphorylation was inhibited by DRB, indicating CK2 activity (Fig. 5A, top, compare lanes 3 and 4). Western blotting of this protein gel with anti-p32 serum revealed a single band of 32 kDa that coimmunoprecipitated with anti-IE63 serum (Fig. 5A, bottom, lanes 3 and 4) present in wt-infected and mock-infected extracts (Fig. 5A, bottom, lanes 1 and 2). In the p32 amino acid sequence shown in Fig. 5B, the five CK2 consensus phosphorylation sites (8) are indicated.
FIG. 5.
The CK2 inhibitor DRB inhibits phosphorylation of p32. (A) Using IE63 MAb, coimmunoprecipitation was performed as for Fig. 2. The coimmunoprecipitate from wt-infected cell extract (WT) was incubated with [γ-32P]ATP, with (lane 4) or without (lane 3) the CK2 inhibitor DRB. Proteins were separated by SDS-PAGE and transferred to nitrocellulose; the same gel was analyzed by phosphorimaging (top) and then Western blotted for p32 (bottom). Cell extracts were used in lanes 1 and 2. (B) CK2 phosphorylation consensus sites in p32. The sequence of a 282-aa protein was obtained from the GenBank database. CK2 consensus sites (8) are alternately underlined and boxed to distinguish between overlapping sites; serine and threonine (S/T) residues potentially phosphorylated by CK2 are in boldface.
IE63 causes redistribution of p32.
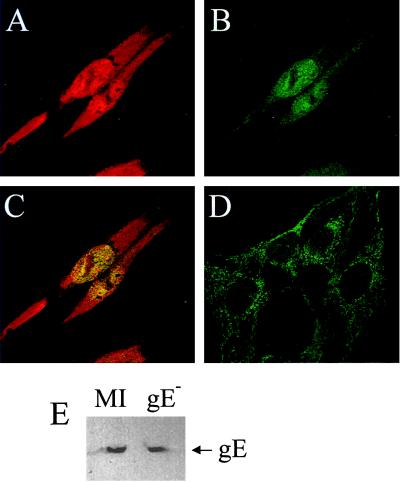
Infection with HSV-1 gE− was necessary due to a cross-reaction between the rabbit polyclonal p32 antibody and HSV-1 glycoprotein E, which can act as a low-affinity Fc receptor (2, 9, 14). Immunofluorescence studies have indicated that p32 is found predominantly in the cell cytoplasm (19, 28). Consistent with this, in cells that were not infected with HSV-1, p32 staining was cytoplasmic and was almost entirely excluded from the nucleus (Fig. 6D). However, in cells infected with HSV-1 gE−, and also transfected with pCMV-63 (data not shown), the pattern of p32 distribution was altered (Fig. 6B). Although still present in the cytoplasm, p32 in both infected and transfected cells expressing IE63 was now also present in the nucleus, where it colocalized with IE63 in a speckled pattern. Western blot analysis using equivalent amounts of protein from mock-infected and HSV-1 gE−-infected cells showed that p32 levels in both extracts were similar (Fig. 6E) at 6 h postinfection, indicating that there was no up- or down-regulation of p32 at this time.
FIG. 6.
IE63 causes p32 to redistribute in cells. Immunofluorescence was performed on HeLa cells infected with HSV-1 gE− and on mock-infected cells using anti-IE63 MAb and anti-p32 serum. A, B, and C show the same field of vision. (A) HSV-1 gE−-infected HeLa cells 6 h postinfection, stained for IE63; (B) HSV-1 gE−-infected HeLa cells 6 h postinfection, stained for p32; (C) HSV-1 gE−-infected HeLa cells 6 h postinfection, stained for IE63 and p32; (D) mock-infected cells stained for p32; (E) extracts of HSV-1 gE−-infected and mock-infected cells Western blotted for p32.
DISCUSSION
Using the yeast two-hybrid assay, in vitro binding assays, and coimmunoprecipitation from virus-infected cells, we have shown that HSV-1 IE63 interacts with the cellular protein p32. Two previously identified IE63 partner proteins, hnRNP K and CK2 (4), were shown by in vitro binding and CK2 activity assays to be present in the complex involving IE63 and p32. However, the partner proteins have not been shown to be interact simultaneously with IE63 in vivo; differential interactions of IE63 with regard to timing, subcellular location, and function will be the subject of further study. As recombinant p32 interacts with IE63 and hnRNP K and CK2 are found in the complex, these data show that all three partner proteins are capable of interaction with IE63 at the same time, at least in vitro. Previous studies have shown that p32 interacts with several proteins of viral and cellular origin. The use of glucose oxidase, a similarly highly charged protein, as a control in the pull-down assay, the alteration of p32 cellular distribution on virus infection and in the presence of IE63, and the demonstration that p32 can regulate splicing, a function which IE63 disrupts (11, 12, 42, 43), all point to this interaction being specific and functionally important.
The different partner proteins were detected with the recombinant p32 column, and IE63 was required for the complex to form. This implies that IE63 is capable of contacting all partner proteins either simultaneously on one molecule or via IE63 oligomers which require the C-terminal zinc finger region to form (4, 54). From the yeast two-hybrid assay, sequences within IE63 aa 166 to 242 were involved in the interaction with p32. This region also contributed to the interaction of IE63 with hnRNP K and with CK2 (4); however, the 80-aa stretch is large enough to allow different protein interactions at discrete points within it. Alternatively, removing this region may alter protein structure and disrupt interactions that occur throughout the protein. The ability of IE63 protein to oligomerize could also facilitate interactions with multiple partner proteins which recognize similar regions of it.
Identifying the biological function of p32 has proven to be challenging. Initially identified as a component of the ASF/SF2 splicing factor (16), subsequent work demonstrated that the p33 subunit alone contained all the functional properties of a splicing factor and that p32 was not required (20). However, p32 has been shown to inhibit ASF/SF2 from acting as a splicing enhancer/repressor and to interact with other SR proteins (30), suggesting that the p32 interaction could be the mechanism by which IE63 inhibits pre-mRNA splicing.
The cellular location of p32 has added to the debate on its function. Using immunofluorescence with a standard fixation method, we saw p32 predominantly in the cytoplasm of uninfected cells. During HSV-1 infection and with expression of IE63 alone, this distribution was altered, with p32 now present in the nucleus and in the cytoplasm. A similar change in p32 distribution was observed in adenovirus-infected cells, where p32 has been suggested to play a role in viral splicing regulation (19). In different cell types, alterations in the relative concentration of key splicing factors acts to regulate alternative RNA splicing (10, 15), and p32 has been proposed to modulate splicing by interacting with one of these factors (30). The p32 protein prevents ASF/SF2 from binding RNA and can inhibit the initiation of prespliceosome formation as well as blocking ASF/SF2 phosphorylation (30). Perhaps, under most circumstances only a small amount of p32 is present in the nucleus, allowing ASF/SF2 to function in splicing. However, under certain conditions, such as HSV-1 infection, p32 may redistribute from the cytoplasm to the nucleus, disrupting splicing by interacting with ASF/SF2.
As well as its role in splicing, p32 has been identified as a transcriptional activator (53) and is implicated in transcriptional regulation of viral gene expression in both HIV-1 (52, 53) and EBV (51) infection. Thus, a potential role in HSV and host gene transcriptional regulation should not be ignored.
A protein of the size of p32 size that reacted with p32 antiserum was coimmunoprecipitated with IE63 and was phosphorylated by coimmunoprecipitating CK2 activity. Although p32 contains CK2 consensus phosphorylation sites, we showed that CK2 activity did not copurify with p32 in uninfected cell extracts, and previous studies have found no evidence of CK2 phosphorylation of p32 (8). While the in vitro kinase assay does not directly demonstrate that CK2 and p32 are present in the same complex, their copurification only in the presence of IE63 clearly shows they can be found in association with each other. Our in vitro data therefore suggest a model in which IE63 introduces CK2 to p32 and via phosphorylation may alter p32 activity. Phosphorylation is key to the functionality of splicing factors, with cycles of phosphorylation and dephosphorylation regulating SR protein function and in turn splicing (25–27). IE63 acts to increase the phosphorylation of the U1 snRNP 70-kDa component found at the 5′ splice site of pre-mRNA (41); thus, changes in phosphorylation appear important for splicing inhibition in HSV-1-infected cells.
During HIV infection, Rev binds the Rev response element (RRE) and up-regulates the cytoplasmic appearance of partially spliced and unspliced viral RNAs (7). p32 has been shown to interact directly with the basic domain of Rev (49) to form a ternary complex of p32, Rev, and RRE-containing RNA in which p32 is proposed to function as a link between Rev and the cellular splicing apparatus and to inhibit splicing. In HIV-1 infection, it is suggested that the stalled spliceosome, containing the p32-Rev complex, enables Rev to export unspliced and partially spliced viral transcripts (49). Acting similarly, the p32-IE63 complex could facilitate splicing-independent export of viral RNAs. In this regard, Cheung et al. (6) have shown that IE63 facilitates cytoplasmic accumulation of spliced and unspliced α-globin transcripts, and they propose that IE63 induces a splicing-independent pathway for α-globin RNA accumulation and nuclear export; equally, this pathway could facilitate export of HSV-1 RNAs which are likely to contain cryptic splicing signals.
Another HSV-1 product, ORF P, interacts with p32 (3). ORF P expression caused the decreased accumulation of proteins encoded by spliced ICP0 and ICP22 mRNAs relative to the protein products of intronless viral mRNAs (3, 35). Consistent with an interaction involving p32 affecting splicing, ORF P protein colocalized with splicing factor SC35, although the concurrent presence of p32 was not examined (3).
IE63 acts to inhibit splicing (11, 12), and we show that it interacts with and alters the cellular distribution of p32. As p32 copurifies with splicing factors (16) and can inhibit splicing (30). Irrespective of p32 function in uninfected cells, these data are consistent with a scheme in which IE63 interacts with p32, and possibly other factors such as CK2, to disrupt splicing. As well as causing host cell shutoff, disruption of splicing could facilitate the nuclear export of viral RNAs.
ACKNOWLEDGMENTS
We thank John McLauchlan for comments on the manuscript.
This work was supported by an award from the Medical Research Council (G9826324); sequencing provision was provided by an award from the Wellcome Trust (046745/2/96).
REFERENCES
- 1.Ackerman M, Braun D K, Pereira L, Roizman B. Characterization of herpes simplex 1 α proteins 0, 4, and 27 with monoclonal antibodies. J Virol. 1984;52:108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell S, Cranage M, Borysiewicz L, Minson T. Induction of immunoglobulin G Fc receptors by recombinant vaccinia viruses expressing glycoproteins E and I of herpes simplex type 1. J Virol. 1990;64:2181–2186. doi: 10.1128/jvi.64.5.2181-2186.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruni R, Roizman B. Open reading frame P—a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc Natl Acad Sci USA. 1996;93:10423–10427. doi: 10.1073/pnas.93.19.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant H, Wadd S, Filhol O, Scott J E, Hsieh T, Everett R, Clements J B. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J Biol Chem. 1999;274:28991–28998. doi: 10.1074/jbc.274.41.28991. [DOI] [PubMed] [Google Scholar]
- 5.Chen M R, Yang J F, Wu C W, Middeldorp J M, Chen J Y. Physical association between the EBV protein EBNA 1 and P32/TAP/hyaluronectin. J Biomed Sci. 1998;5:173–179. doi: 10.1007/BF02253466. [DOI] [PubMed] [Google Scholar]
- 6.Cheung P, Ellison K S, Verity R, Smiley J R. Herpes simplex virus ICP27 induces cytoplasmic accumulation of unspliced polyadenylated α-globin pre-mRNA in infected HeLa cells. J Virol. 2000;74:2913–2919. doi: 10.1128/jvi.74.6.2913-2919.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen B R. Posttranscriptional regulation by the HIV-1 Rev protein. Semin Virol. 1998;8:327–334. [Google Scholar]
- 8.Deb T B, Datta K. Molecular cloning of human fibroblast hyaluronic acid-binding protein confirms its identity with p32, a protein co-purified with splicing factor SF2. J Biol Chem. 1996;271:2206–2212. doi: 10.1074/jbc.271.4.2206. [DOI] [PubMed] [Google Scholar]
- 9.Dubin G, Frank I, Friedman H M. Herpes simplex virus type 1 encodes two Fc receptors which have different binding characteristics for monomeric immunoglobulin G (IgG) and IgG complexes. J Virol. 1990;64:2725–2731. doi: 10.1128/jvi.64.6.2725-2731.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanamura A, Caceres J F, Mayeda A, Franza B R, Krainer A R. Regulated tissue-specific expression of antagonist pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 11.Hardwicke M A, Sandri-Goldin R M. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang J Z, Zhang Y, Krainer A R, Xu R M. Crystal structure of human p32 a doughnut-shaped acidic mitochondrial matrix protein. Proc Natl Acad Sci USA. 1999;96:3572–3577. doi: 10.1073/pnas.96.7.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson D C, Frame M C, Ligas M W, Cross A M, Stow N D. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol. 1988;62:1347–1354. doi: 10.1128/jvi.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krainer A R, Conway G C, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 16.Krainer A R, Conway G C, Kozak D. Purification and characterisation of SF2, a human pre-mRNA splicing factor. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 17.Luo Y, Yu H, Peterlin B M. Cellular protein modulates effects of human immunodeficiency virus type 1 Rev. J Virol. 1994;68:3850–3856. doi: 10.1128/jvi.68.6.3850-3856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma J, Ptashne M. Converting a eukaryotic transcriptional inhibitor into an activator. Cell. 1988;55:443–446. doi: 10.1016/0092-8674(88)90030-x. [DOI] [PubMed] [Google Scholar]
- 19.Matthews D A, Russell W C. Adenovirus core protein V interacts with p32—a protein which is associated with both the mitochondria and the nucleus. J Gen Virol. 1998;79:1677–1685. doi: 10.1099/0022-1317-79-7-1677. [DOI] [PubMed] [Google Scholar]
- 20.Mayeda A, Zahler A M, Krainer A R, Roth M B. Two members of a conserved family of phosphoproteins are involved in pre-mRNA splicing. Proc Natl Acad Sci USA. 1992;89:1301–1304. doi: 10.1073/pnas.89.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGregor F, Phelan A, Dunlop J, Clements J B. Regulation of herpes simplex virus type 1 poly(A) site usage and the action of the immediate-early protein IE63 in the early-late switch. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMahan L, Schaffer P A. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J Virol. 1990;64:3471–3485. doi: 10.1128/jvi.64.7.3471-3485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mears W E, Rice S A. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology. 1998;242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- 25.Mermound J E, Cohen P T W, Lamond A I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mermound J E, Cohen P T W, Lamond A I. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 1992;20:5263–5269. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misteli T, Spector D L. Protein phosphorylation and the nuclear organisation of pre-mRNA splicing. Trends Cell Biol. 1997;7:135–138. doi: 10.1016/S0962-8924(96)20043-1. [DOI] [PubMed] [Google Scholar]
- 28.Muta T, Kang D, Kitajima S, Fujiwara T, Hamaski N. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J Biol Chem. 1997;272:24363–24370. doi: 10.1074/jbc.272.39.24363. [DOI] [PubMed] [Google Scholar]
- 29.Peterson K L, Zhang W, Lu P D, Keilbaugh S A, Peerschke E I, Ghebrehiwet B. The C1q-binding cell membrane proteins cC1q-R and gC1q-R are released from activated cells: subcellular distribution and immunochemical characterisation. Clin Immunol Immunopathol. 1997;84:17–26. doi: 10.1006/clin.1997.4374. [DOI] [PubMed] [Google Scholar]
- 30.Peterson-Mahrt S K, Estmer C, Ohrmalm C, Mathews D A, Russell W C, Akusjarvi G. The splicing factor associated protein p32 regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 1999;18:1014–1024. doi: 10.1093/emboj/18.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phelan A, Carmo-Fonseca M, McLauchlan J, Lamond A I, Clements J B. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci USA. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phelan A, Clements J B. Herpes simplex virus type 1 immediate-early protein IE63 shuttles between nuclear compartments and the cytoplasm. J Gen Virol. 1997;78:3327–3331. doi: 10.1099/0022-1317-78-12-3327. [DOI] [PubMed] [Google Scholar]
- 33.Phelan A, Clements J B. Posttranscriptional regulation in herpes simplex virus. Semin Virol. 1998;8:309–318. [Google Scholar]
- 34.Phelan A, Dunlop J, Clements J B. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J Virol. 1996;70:5255–5265. doi: 10.1128/jvi.70.8.5255-5265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Randall G, Roizman B. Transcription of the depressed open reading frame P of herpes simplex virus 1 precludes the expression of the antisense γ134.5 gene and may account for the attenuation of the mutant virus. J Virol. 1997;71:7750–7757. doi: 10.1128/jvi.71.10.7750-7757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice S A, Knipe D M. Gene-specific transactivation by herpes simplex virus type 1 α protein ICP27. J Virol. 1988;62:3814–3823. doi: 10.1128/jvi.62.10.3814-3823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice S A, Knipe D M. Genetic evidence for two distinct transactivation functions of the herpes simplex virus α protein ICP27. J Virol. 1990;64:1704–1715. doi: 10.1128/jvi.64.4.1704-1715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice S A, Su L, Knipe D M. Herpes simplex virus α protein ICP27 possesses separable positive and negative regulatory activities. J Virol. 1989;63:3399–3407. doi: 10.1128/jvi.63.8.3399-3407.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sacks W R, Greene C C, Aschman D P, Schaffer P A. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandri-Goldin R M, Hibbard M K. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J Virol. 1996;70:108–118. doi: 10.1128/jvi.70.1.108-118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandri-Goldin R M, Mendoza G E. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 44.Simos G, Georgatos S D. The lamin B receptor-associated protein p34 shares sequence homology and antigenic determinants with the splicing factor 2-associated protein p32. FEBS Lett. 1994;346:225–228. doi: 10.1016/0014-5793(94)00479-x. [DOI] [PubMed] [Google Scholar]
- 45.Smith I L, Hardwicke M A, Sandri-Goldin R M. Evidence that the herpes simplex virus immediate-early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 46.Soliman T M, Sandri-Goldin R M, Silverstein S J. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and the cytoplasm mediates the expression of late proteins. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soliman T M, Silverstein S J. Herpesvirus mRNAs are sorted for export via Crm-1 dependent and -independent pathways. J Virol. 2000;74:2814–2825. doi: 10.1128/jvi.74.6.2814-2825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stow N D, Hammarsten O, Arbukle M I, Elias P. Inhibition of HSV-1 DNA replication by mutant forms of the origin binding protein. Virology. 1993;196:413–419. doi: 10.1006/viro.1993.1496. [DOI] [PubMed] [Google Scholar]
- 49.Tange T O, Jensen T H, Kjems J. In vitro interaction between human immunodeficiency virus type 1 Rev protein and splicing factor ASF/SF2-associated protein p32. J Biol Chem. 1996;271:10066–10072. doi: 10.1074/jbc.271.17.10066. [DOI] [PubMed] [Google Scholar]
- 50.Van Seuningen I, Ostrowski J, Bomsztyk K. Description of an Il-1-responsive kinase that phosphorylates the K-protein—enhancement of phosphorylation by selective DNA and RNA motifs. Biochemistry. 1995;34:5644–5650. doi: 10.1021/bi00016a040. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y L, Finan J E, Middeldorp J M, Hayward S D. p32/Tap, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology. 1997;236:18–29. doi: 10.1006/viro.1997.8739. [DOI] [PubMed] [Google Scholar]
- 52.Yu L, Loewenstein P M, Zhang Z, Green M. In vitro interaction of the human immunodeficiency virus type 1 Tat transactivator and the general transcription factor TFIIB with the cellular protein TAP. J Virol. 1995;69:3017–3023. doi: 10.1128/jvi.69.5.3017-3023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu L, Zhang Z, Loewenstein P M, Desai K, Tang Q, Mao D, Symington J S, Green M. Molecular cloning and characterization of a cellular protein that interacts with the human immunodeficiency virus type 1 Tat transactivator and encodes a strong transcriptional activation domain. J Virol. 1995;69:3007–3016. doi: 10.1128/jvi.69.5.3007-3016.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhi Y, Sciabica K F, Sandri-Goldin R M. Self-interaction of the herpes simplex virus type 1 regulatory protein ICP 27. Virology. 1999;257:341–351. doi: 10.1006/viro.1999.9698. [DOI] [PubMed] [Google Scholar]