ABSTRACT
A substantial percentage of the population remains at risk for cervical cancer due to pre-existing human papillomavirus (HPV) infections, despite prophylactic vaccines. Early diagnosis and treatment are crucial for better disease outcomes. The development of new treatments heavily relies on suitable preclinical model systems. Recently, we established a mouse papillomavirus (MmuPV1) model that is relevant to HPV genital pathogenesis. In the current study, we validated the use of Papanicolaou (Pap) smears, a valuable early diagnostic tool for detecting HPV cervical cancer, to monitor disease progression in the MmuPV1 mouse model. Biweekly cervicovaginal swabs were collected from the MmuPV1-infected mice for viral DNA quantitation and cytology assessment. The Pap smear slides were evaluated for signs of epithelial cell abnormalities using the 2014 Bethesda system criteria. Tissues from the infected mice were harvested at various times post-viral infection for additional histological and virological assays. Over time, increased viral replication was consistent with higher levels of viral DNA, and it coincided with an uptick in epithelial cell abnormalities with higher severity scores noted as early as 10 weeks after viral infection. The cytological results also correlated with the histological evaluation of tissues harvested simultaneously. Both immunocompromised and immunocompetent mice with squamous cell carcinoma (SCC) cytology also developed vaginal SCCs. Notably, samples from the MmuPV1-infected mice exhibited similar cellular abnormalities compared to the corresponding human samples at similar disease stages. Hence, Pap smear screening proves to be an effective tool for the longitudinal monitoring of disease progression in the MmuPV1 mouse model.
IMPORTANCE
Papanicolaou (Pap) smear has saved millions of women's lives as a valuable early screening tool for detecting human papillomavirus (HPV) cervical precancers and cancer. However, more than 200,000 women in the United States alone remain at risk for cervical cancer due to pre-existing HPV infection-induced precancers, as there are currently no effective treatments for HPV-associated precancers and cancers other than invasive procedures including a loop electrosurgical excision procedure (LEEP) to remove abnormal tissues. In the current study, we validated the use of Pap smears to monitor disease progression in our recently established mouse papillomavirus model. To the best of our knowledge, this is the first study that provides compelling evidence of applying Pap smears from cervicovaginal swabs to monitor disease progression in mice. This HPV-relevant cytology assay will enable us to develop and test novel antiviral and anti-tumor therapies using this model to eliminate HPV-associated diseases and cancers.
KEYWORDS: the mouse papillomavirus (MmuPV1), lower genital infection, Pap smear, LSIL, HSIL, squamous cell carcinoma, longitudinal, cytology, in situ hybridization, RNAscope, transmission electron microscope (TEM), 2014 Bethesda system, qPCR, mouse model, immunohistochemistry (IHC), viral copy number
INTRODUCTION
Human papillomavirus infection is one of the most common sexually transmitted diseases (1). HPV is the causative factor for almost all cervical cancers, 70% of vaginal and oropharyngeal cancers, and 90% of anal cancers (2–5). In 2020, as the fourth most common cancer, an estimated 604,127 cervical cancer cases and 341,831 deaths were reported globally (6). Worldwide, two in three women aged 30–49 years have never been screened for cervical cancer (7). More than three million cases of HPV-associated cervical intraepithelial neoplasia (CIN) were detected in the United States in 2019. Four prophylactic virus-like particle (VLP)-based vaccines have shown strong protection against up to nine high-risk HPV types included in the vaccines, even with the new single-dose regimen to counteract the limited vaccine supply (8, 9). However, these vaccines cannot protect against HPV infections from types not covered by the vaccine, and they do not exhibit therapeutic effects for pre-existing infections (10). Considering that the vaccine uptake rates for the full doses in eligible populations remain around 50% in the United States and even lower in low-income countries (11), HPV-associated infections and cancers will continue to be a significant public health problem (11, 12). Although excisional treatment of pre-invasive disease is effective, lesions often reoccur (2). Therefore, innovative therapeutic approaches to manage HPV-associated cancers are essential for the millions of people who contract HPV infections each year. A better understanding of the mechanisms behind HPV pathogenesis is essential for the development of these HPV-associated cancer treatments (13).
Early diagnosis is critical for the control of HPV-associated disease progression (14, 15). The Papanicolaou test, also called Pap smear or Pap test, has been used successfully for cervical screening since 1943 (15). This test includes collecting cells from the transition zone area of the cervix and detecting potentially precancerous and cancerous processes (16, 17). Detecting pre-invasive lesions has saved millions of lives (15). One critical challenge for studying HPV in vivo is that HPV infections are species-specific. Thus, no preclinical model system can be used to study HPV infection directly (18). Recently, we and others established a mouse papillomavirus (MmuPV1) model that displays relevance to HPV infections in the anogenital tract and oropharynx (19–31). Our published and unpublished data strongly support that the infection course of MmuPV1 mimics HPV infections in the human lower genital tract with increased detection of viral signals in the cervicovaginal specimens and disease progression from low-grade to high-grade dysplasia over time (21, 22, 32). Advanced squamous cell carcinomas (SCC) in the vaginal tissues have been demonstrated in different mouse strains, including immunocompetent mouse strains (FVB and NU/J heterozygotes), immunocompromised (athymic nude mice), and immunogenetically modified mice (19–21). We also developed noninvasive methods for longitudinally monitoring viral activities by quantitating viral DNA copies via qPCR analysis (21, 22, 32, 33). All these preliminary studies led us to postulate that a Pap smear method might be appropriate for monitoring disease progression in the mouse HPV model.
The current study took advantage of our longitudinal monitored MmuPV1 lower genital infection model (34), the well-established human Pap smear (16), and the 2014 Bethesda system (16, 17, 35) to demonstrate that Pap smear is a valuable method for monitoring disease progression in the mouse PV model. We included two clinically relevant immunogenetically modified mouse strains, NU/J heterozygous (Foxn1nu/+) and Rag1ko mice, to represent immunocompetent and immunocompromised backgrounds, respectively. Transcription factor forkhead box N1 (Foxn1) is the master regulatory gene of thymic epithelium development and function (36, 37). In heterozygous (Foxn1nu/+) human patients, the Foxn1 gene dosage is more critical during embryonic and early post-natal life, with an increased risk of complication due to increased susceptibility to pathogens. However, these patients showed normal T-lymphocyte function as adults (31). Likewise, after 3 weeks of age, NU/J heterozygous (Foxn1nu/+) mice also showed comparable thymocyte counts to wild type (Foxn1+/+) (31). Therefore, 4- to 6-week-old NU/J heterozygous (Foxn1nu/+) mice used in the current study are considered immunocompetent. These mice were tested in our previous study and proceeded to develop advanced diseases, including carcinoma in situ in the lower genital infected tissues (21). In addition to NU/J heterozygous (Foxn1nu/+) mice, a second mouse strain, recombination-activating genes 1 (Rag1) knockout mice, was included in the current study as the positive control for viral infection (27, 38–40). Rag1 and Rag2 proteins play an essential role in adaptive immunity by initiating V(D)J recombination (41–43). Patients without Rag1 or Rag2 display lymphocyte development arrested, resulting in T- and B-cell deficiency (41–43). Similarly, Rag1 and Rag2 knockout mice have no T- and B-cell responses and are used to study different diseases (41, 44–46). Rag1ko mice are susceptible to MmuPV1 infections in both cutaneous and mucosal tissues and have been used as the positive control for MmuPV1 infection in previous studies (27, 38, 40).
We followed vaginally infected mice longitudinally from benign to advanced disease stages, including squamous cell carcinoma in Rag1ko and NU/J heterozygous (Foxn1nu/+) mice for up to 32 weeks. Cervicovaginal swabs were periodically collected for viral DNA analysis via qPCR (32) and for cytological assays via Pap smears. At the termination of the experiments, infected tissues from these tested mice were further analyzed using additional in situ assays to confirm viral presence (21) and disease stages by histology. To further validate our observations and their relevance to HPV infections and associated cancer, we compared our results with corresponding human Pap smear samples and cervical interepithelial neoplasia tissues. The demonstrated surprising similarities in cytology and histology stages between diseased human and mouse cervicovaginal lavage samples will lay a solid foundation for us to study mechanisms of HPV disease progression and test novel anti-viral and anti-tumor compounds in our mouse PV model.
MATERIALS AND METHODS
Mice and MmuPV1 infection
All mouse work was approved by the Institutional Animal Care and Use Committee of Pennsylvania State University’s College of Medicine (COM), and all methods were performed in accordance with guidelines and regulations. The mice were housed (2–5 mice/cage) in sterile cages within sterile filter hoods in the COM BL2 animal core facility. Immunocompromised mouse strains, including Rag1ko (B6.129S7-Rag1tm1Mom/J, 002216, the Jackson laboratory), athymic nude mice (Hsd:NU, Envigo) (21), and immunocompetent mouse strain NU/J heterozygous mice (002019, the Jackson laboratory) (21) were used for the study.
Infectious MmuPV1 virions were isolated from lesions on the tails of athymic nude mice from our previous study (24). In brief, lesions scraped from the tails of the Hsd:NU athymic mice were homogenized in sterile phosphate-buffered saline (1 × PBS) using a polytron homogenizer (Brinkman PT10-35) at the highest speed for 3 minutes while chilling in an ice bath. The homogenate was spun at 10,000 rpm and the supernatant was mixed with glycerol (1:1) and decanted into Eppendorf tubes for long-term storage at −80°C. The viral DNA of the stock was quantitated by extracting the DNA from 5 µL of this stock using qPCR. For standard infection, 10 µL of MmuPV1 virus stock (1 × 108 viral DNA equivalents) was used for infection in the lower genital tract (47).
To increase the infectivity and generate robust results, all female mice were treated with 3 mg of medroxyprogesterone acetate (Depo-Provera, DMPA) subcutaneously 3 days before the viral infection as previously described (32). The day before infection, the mice were anesthetized, and the lower genital tracts were pre-wounded with Doctors’ Brush Picks coated with 4% nonoxynol-9 gel [Conceptrol(R)]. Twenty-four hours after pre-wounding, the mice were again anesthetized and challenged with infectious MmuPV1 virus (1 × 109 viral DNA equivalents) using a filtered pipette tip in the lower genital tract as described in our previous studies (47). For the titration study in NU/J heterozygous mice, four groups of mice (N = 5/group) were infected with the same volume of viral suspension between 1 × 105 and 1 × 108 viral DNA equivalents in sterile 1 × PBS.
Cervicovaginal swab for DNA extraction and qPCR analysis
Biweekly swabs from the lower genital tract were monitored for viral DNA by qPCR (32). Vaginal swabs (Puritan PurFlock Ultra; Puritan Diagnostics, LLC) soaked in 25 µL of sterile 0.9% NaCl were introduced into the vaginal canals and then stored at –20°C before processing for viral DNA quantification. Viral DNA was extracted (DNeasy Blood & Tissue Kit, 69506), and the qPCRs were run in an Agilent qPCR machine as described previously (24). The primer pairs (5′GGTTGCGTCGGAGAACATATAA 3′ and 5′ CTAAAGCTAACCTGCCACATATC 3′) for E2 amplification were used together with the probe (5′ 6-FAM-TGCCCTTTCA/ZEN/GTGGGTTGAGGACAG 3′-IBFQ). Each reaction contained 9 µL ultrapure water, 5 pmol of each primer, 9 µL Brilliant III qPCR Master Mix (Agilent), and 2 µL DNA template. The PCR conditions were initial denaturation at 95°C for 3 minutes, then 40 cycles at 95°C for 5 seconds, and 60°C for 10 seconds on an Agilent AriaMx. Viral copy numbers were converted into an equivalent DNA load: 1 ng viral DNA = 1.2 × 108 copies (http://cels.uri.edu/gsc/cndna.html). All samples were assessed in duplicates or triplicates (21). Viral titers were calculated according to the standard curve (32).
Viral RNA detection
RNA samples were extracted from mouse tissues that were flash-frozen in liquid nitrogen and stored at −80°C or liquid nitrogen. Approximately 20 mg of tissue was put into 1 mL TRIzol and processed for 5 minutes through a homogenizer (24). Following the standard Thermo Fisher protocols for TRIzol, the clear supernatant was collected, and 0.2 mL chloroform was added per 1 mL TRIzol solution. The mixtures were centrifuged, and the clear supernatant was retrieved. RNA was quantified using a nanodrop Spectrophotometer (NanoDrop Technologies; ND-1000), and 1 µg RNA was used for the reverse-transcriptase reaction using the SuperScript III RT Kit (Thermo Fisher Scientific; 18080044). The copy numbers were based on 200 ng RNA from each sample (Viral RNA E1^E4 transcripts were quantified using primers 5′-TAGCTTTGTCTGCCCGCACT-3′ and 5′-GTCAGTGGTGTCGGTGGGAA-3′ and probe 5’FAM-CGGCCCGAAGACAACACCGCCACG-3′TAMRA. RNA (0.2 µg) was mixed with 0.25 µg/µL random primers, 10 mM dNTP mix, and nuclease-free H2O for a total of 13 µL. This sample was then incubated at 65°C for 5 minutes. A mix of diluted 1 × first-strand buffer, 0.1 M dithiothreitol, 40 U/ µL RNase, and 200 U SuperScript III was added to the nucleotide mix. The sample was then incubated in a thermocycler. The program consisted of 5 minutes at 25°C, 1 hour at 50°C, and 15 minutes at 70°C. Following PCR, cDNA samples were diluted 1:9 using Ultra-Pure DNase/RNase-Free Distilled Water. Of the 1:9 dilution, 2 µL was used in the 20 µL quantitative PCR (qPCR) reaction purchased from Agilent.
Cytology and histology assessment and interpretation
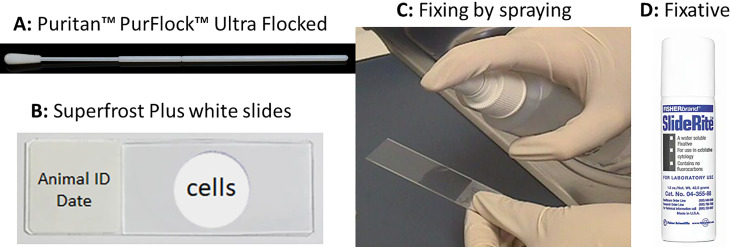
Cervicovaginal swab samples were collected from noninfected control and MmuPV1-infected animals at different times post-viral infection. Cells on the swabs were spread by rolling on labeled slides in a circle motion (22) and fixed by spraying the SlideRight cytology fixative (Fishersci) from 6 inches above the slide as shown in Fig. 1. The fixed slides were kept at 4°C before Pap smear staining. All cytology slides were automatically stained using an automatic stainer with the standard protocol Pap smear reagent used by human samples (Thermo Scientific/Richard Allen Scientific). In brief, the fixed slides were sequentially hydrated by being submerged in 100%, 95%, and 70% ethanol for 2 minutes each. After washing in ddH2O for 1 minute, the slides were stained with Gill’s hematoxylin for 3 minutes. After a 3-minute wash with ddH2O, the slides were placed in bluing for 1 minute, ddH2O for 1 minute, 70% ethanol for 1 minute, OG-6 for 3 minutes, two changes of 95% ethanol for 2 minutes, EA-50 for 3 minutes, and three changes of 95% ethanol for 2 minutes. Finally, slides were submerged in 100% ethanol for 3 minutes and xylene for 3 minutes before being mounted and cover-slipped with Cytoseal 60. The slides were scanned 7–10 days after staining to ensure the mounting media was completely dry.
Fig 1.
Pap smear slide preparation using a conventional swab. Single-used swabs (A, Puritan PurFlock Ultra Flocked Swabs) were dipped into sterile 0.9% NaCl and inserted into the vaginal tract of infected mice. The cells on the swabs were spread onto a superfrost white slide (B) by rotating the swabs on the slide in a circle manner. The cells will be fixed by spraying (C) the SlideRight cytology fixative (D, fishersci) from 6 inches above. The fixed slides were stored in a slide box at 4°C before staining with the standard Pap smear protocol.
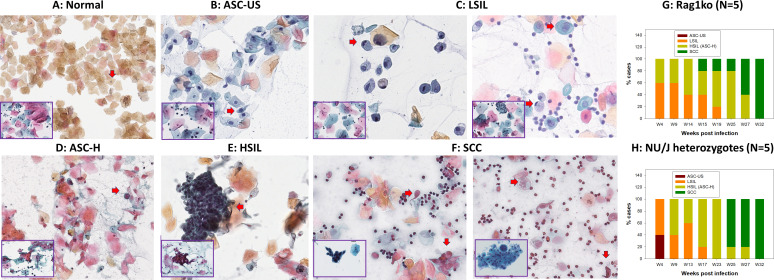
The stained slides were scanned digitally at 40× magnification using an Aperio AT2 digital slide scanner at the Aperio Slide Scanning Core at Penn State COM. The digital slides were independently evaluated by one veterinary pathologist and a physician pathologist for signs of epithelial cell abnormalities using the ImageScope x64 software (Table 1). Both pathologists (HMA and AU) were provided with sample IDs without knowledge of infection status or time post-infection before matching the assessments with the animal ID. Modified 2014 Bethesda system criteria comparable to that used for human cytological samples representing grades of intraepithelial neoplasia were used for these slides, considering normal mouse physiology (48): (i) Normal: normal squamous cells with oval-shaped and small nuclei. They are negative for intraepithelial lesion and malignancy; (ii) ASC-US: Atypical squamous cells of uncertain significance with nucleus size 2–2.5 times larger than a normal squamous cell; (iii) LSIL: Low-grade squamous intraepithelial lesion with hyperchromatic raisin shaped nuclei with a clear halo around it or binucleated cells; (iv) ASC-H: Atypical squamous cells with nucleus size 2–2.5 times larger than a normal squamous cell, cannot exclude HSIL; (E) HSIL: High-grade squamous intraepithelial lesion (HSIL, hyperchromatic cells with high N-C ratio and forming sheets), encompassing cervical intraepithelial neoplasia grades 2 and 3; CIN2 and CIN3; and F) SCC: squamous cell carcinoma (SCC, keratinizing with marked pleomorphism of cell size and shape, arrow; the presence of tadpole cells with dense orangeophilic cytoplasm, arrow; spindle-shaped cells, and inflammation and necrotic debris on the background).
TABLE 1.
The final diagnosis of Pap smear slides by the two pathologists (HMA and AU)
| Slide ID | HMA | Gul | Slide ID | HMA | Gul |
|---|---|---|---|---|---|
| 31592 | IELM Neg | IELM Neg | 31096 | HSIL | HSIL |
| 31593 | LSIL | LSIL | 31099 | HSIL | HSIL |
| 31594 | ASC-H | ASC-H | 31140 | SCC | SCC |
| 31595 | ASC-US | ASC-US | 31141 | SCC | SCC |
| 31596 | ASC-H | LSIL | 31142 | SCC | SCC |
| 31597 | ASC-US | ASC-US | 31144 | LSIL | LSIL |
| 31598 | IELM Neg | IELM Neg | 31145 | ASC-H | ASC-H |
| 31600 | IELM Neg | IELM Neg | 31146 | ASC-H | ASC-H |
| 31601 | IELM Neg | IELM Neg | 31147 | HSIL | HSIL |
| 31602 | ASC-US | ASC-US | 31149 | HSIL | HSIL |
| 31603 | HSIL | HSIL | 31151 | HSIL | HSIL |
| 31604 | LSIL | LSIL | 31152 | HSIL | HSIL |
| 31605 | ASC-US | ASC-US | 31157 | SCC | SCC |
| 31607 | IELM Neg | IELM Neg | 31158 | ASC-H | ASC-H |
| 31608 | LSIL | LSIL | 31159 | SCC | SCC |
| 31610 | LSIL | LSIL | 31160 | SCC | SCC |
| 31611 | IELM Neg | IELM Neg | 31161 | SCC | SCC |
| 31612 | LSIL | LSIL | 31162 | HSIL | HSIL |
| 31613 | LSIL | LSIL | 31163 | ASC-H | ASC-H |
| 31614 | LSIL | LSIL | 31164 | HSIL | HSIL |
| 31615 | ASC-US | ASC-US | 31165 | ASC-H | ASC-H |
| 31616 | ASC-US | ASC-US | 31167 | SCC | SCC |
| 31617 | ASC-H | ASC-H | 31168 | HSIL | HSIL |
| 31618 | SCC | SCC | 31169 | HSIL | HSIL |
Anti-MmuPV1 antibody detection by enzyme-linked immunosorbent assays (ELISA)
Mouse sera were collected at the termination of the experiment to assess for antibodies against E4. 0.1–0.5 µg of KLH-conjugated MmuPV1 E4 peptide (PKTTPPRRELFPPTPLTQPP, synthesized by China peptide)/well in 50 µL bicarbonate (pH 9.6) buffer was incubated overnight at 37°C. Coated plates were blocked with PBS/5% dry milk powder for 1 hour at room temperature. The ELISA was conducted as reported previously (24, 27). Each serum sample was diluted 1:100 to 1:100,000 in PBS/5% dry milk and then added to the 96-well plates for 1-hour incubation at RT. Plates underwent five washes with washing buffer [0.05% (vol/vol) Tween 20 in 1 × PBS], before the addition of secondary anti-Ig isotypes (IgG3)-alkaline phosphatase (AP) (Southern Biotech) diluted 1:2,000 in 5% milk/1 × PBS for 1 hour at RT. After five further washes, 100 µL of 1 mg/mL pNPP (para-nitrophenyl phosphate, Sigma), a substrate for AP, was added to each well for color development, and absorbance at 405 nm/450 nm was measured using a Fisher Microplate Reader.
Western blot analysis for detection of L1 protein
Proteins were extracted from paraffin-embedded vagina tissues using the Qproteome FFPE tissue kit (ID: 37623) according to the manufacturer’s instructions. Protein concentration was determined with a BCA Protein Assay kit (Thermo Fisher Pierce). 15 µg of proteins from vaginal tissues of either Rag1ko or NU/J heterozygous mice (four separate samples) was denatured at 95°C for 5 min, separated by gel electrophoresis using a Novex Wedge Well 4%–20% Tris-Glycine Gel (Thermo Fisher Invitrogen) and transferred to a PVDF membrane (Merck Millipore). MPV.B9 (1:200) or beta-actin (1:200) and Anti-mouse IgG1 HRP (1: 2,000, Thermo Fisher Pierce) were used to probe for L1 in western blot analysis (49). A 10 µg positive L1 VLP sample was used as a positive control. The samples were run and probed as described above. Protein loading controls were confirmed by Coomassie stain (data not shown).
Immunohistochemistry and in situ hybridization analyses of infected tissues
After the termination of the experiment, the animals were euthanized. Half of each reproductive tract, including vaginal tissue of interest, was frozen in liquid nitrogen and the other half was fixed in 10% buffered formalin then paraffin-embedded and sectioned as described previously (22). Regressive hematoxylin and eosin (H &E) staining, in situ hybridization (ISH), and immunohistochemistry (IHC) were conducted routinely in the laboratory as described in previous studies (22, 32, 47, 49–51). For IHC, an anti-MmuPV1 E4 rabbit polyclonal antibody (a generous gift from Dr. John Doorbar’s laboratory), anti-MmuPV1 L1 monoclonal antibody (MPV.B9), Ki67 (Invitrogen, rabbit monoclonal antibody, MA5-14520), and CD31 (PECAM-1) (D8V9E) XP Rabbit mAb #77699 (Cell Signaling) were used on formalin-fixed paraffin-embedded (FFPE) sections (52).
RNA ISH was performed on FFPE mouse tissue sections using RNAscope probes (the probe targeting MmPV1 E4 transcript was used in the current study), and protocols (Advanced Cell Diagnostics, ACD) as shown in a previous study (49). The HybEZ hybridization system of ACD was used to perform RNA-ISH. In brief, 5 µm sections were baked in the HybEZ Oven II (Advanced Cell Diagnostics; 321720) for 1 hour at 60°C and immediately deparaffinized in xylene, then rehydrated in an ethanol series. Epitope retrieval was performed by placing the slides in RNAscope 1 × Target Retrieval Reagent (Advanced Cell Diagnostics; 322000) at 100°C for 15 minutes and then washing it in 1 × washing buffer. Protease treatment was performed by adding RNAscope Protease Plus (Advanced Cell Diagnostics; 322331) to the section and incubating for 30 minutes at 40°C in a HybEZ Oven II (Advanced Cell Diagnostics; 321720). After probe hybridization with target probes, preamplifier, and amplifier, sections were stained with Fat Brown reagent (RNAscope 2.5 HD Detection Reagents—Brown; Advanced Cell Diagnostics; 322360). A counterstain of 50% hematoxylin and 0.02% ammonia water was used. Positive and negative probes were used in each assay to ensure proper controls. Slides from normal and infected tissues were used as negative and positive controls. Images of infected tissues with RNA ISH brown signals were captured through scanned slides using the ImageScope x64 software. Representative areas of each slide were imaged at 10× to 400× magnification and positive RNA ISH signals were counted with the specific infection status of the cells and tissues (51). E4 RNA signals should be absent in uninfected cells/tissues.
Transmission electron microscopy (TEM)
For TEM, the tissue or pellet of the swab sample was immersion fixed for 24 h in Karnovsky’s fixative (2.5% glutaraldehyde/2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.3) (32). Following fixation, the tissue was washed in 0.1 M sodium cacodylate buffer, post-fixed in buffered 1% osmium tetroxide/1.5% potassium ferrocyanide, rewashed with buffer, dehydrated in a graded series of ethanol, transferred to acetone, and embedded in Spurr low viscosity resin. Sections (70 nm) were cut with a diamond knife, mounted on copper grids, and stained with 2% aqueous uranyl acetate and lead citrate. The sections were examined in a JEOL JEM 1400 electron microscope and the pictures were recorded (32, 33).
RESULTS
Viral presence and anti-viral antibody determination
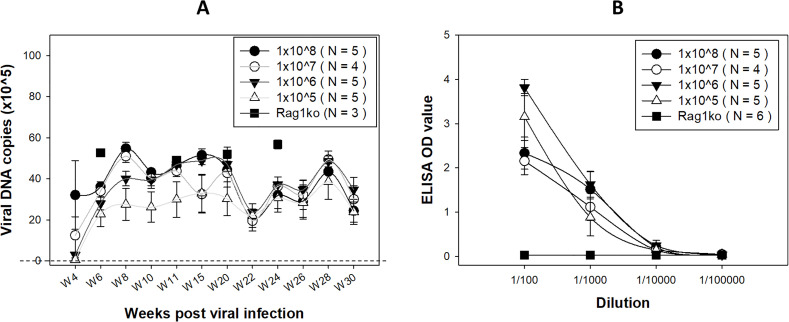
In addition to six Rag1ko mice that were monitored for viral activities biweekly using our previously described method, we also conducted a dose curve study to monitor viral DNA in the NU/J heterozygous mice that we demonstrated to develop carcinoma in situ in a previous study (21). Consistent with previous studies (21), viral DNA levels in cervicovaginal swabs of the NU/J heterozygous mice increased and peaked around 8–10 weeks and plateaued with a drop between weeks 20 and 22 (Fig. 2A). Our previous study also observed a fluctuation in viral DNA detection that corresponded to the estrous cycle in athymic nude mice (32). Significantly higher levels of viral DNA were detected in the infected Rag1ko mice at various representative time points (Fig. 2A, square). Mice with the lowest doses of viral infection (1 × 105 viral genomic copies) showed slightly lower levels of viral DNA detection by week 20 post-infection when compared to higher dose groups but no difference after week 22. Interestingly, a drop in viral DNA copies (down to 1 × 106 copies/swab) was detected in all groups at week 22 post-infection (53) although these levels are about the threshold of viral DNA detection (A, the dotted line). This decrease in viral DNA detection is inversely correlated with the progression of the disease to SCC. This may suggest a potential increase in viral integration, as previously reported in MmuPV1-infected tissues (53).
Fig 2.
Viral DNA (A) over the time course of papillomavirus infection and anti-MmuPV1 E4 antibodies (B) were detected in the NU/J heterozygous mice. In addition to the previously used high titers of virus for infection, we also assessed a viral titer as low as 1 × 105 genome equivalents. Despite a delayed detection of viral DNA copies in the group with the lower titer, no significant difference was observed among these different groups at week 22 post-infection (the dotted line indicates the threshold of viral detection). Higher levels of viral DNA were detected in Rag1ko mice at several representative time points (indicated by a square). Significantly higher levels of anti-MmuPV1 E4 IgG3 antibody were detected in all infected NU/J heterozygous mice initiated with different doses of viral infection, compared to those in Rag1ko mice (indicated by a square), at dilutions ranging from 1:100 to 1:1,000 (B, P < 0.05, one-way ANOVA).
NU/J heterozygous mice are immunocompetent (21). We therefore wanted to examine whether an anti-MmuPV1 antibody was generated after infection. Sera were harvested from all tested mice sacrificed at different time points up to week 30 post-viral infection. We recently reported that orally infected immunocompetent female mice develop antibodies against one of the most abundant viral proteins, E4, especially isotype IgG3 (27). Therefore, we tested these mice for E4 IgG3 isotype antibody. We ran a serial 1:10 dilution of serum samples for this assay. All infected NU/J heterozygous mice were positive for E4 isotype IgG3 despite the initial viral dose suggesting low dose such as 1 × 105 is sufficient for infection in the lower genital tract (Fig. 2B) (27). Significantly higher levels of anti-MmuPV1 E4 IgG3 antibody were detected in all infected NU/J heterozygous mice initiated with different doses of viral infection, compared to those in Rag1ko mice (indicated by a square), at dilutions ranging from 1:100 to 1:1,000 (B, P < 0.05, one-way ANOVA) (27, 40). This agrees with previous observations that a small amount of virus is needed to initiate a productive infection either via blood (47) or cutaneous (54) infections.
Virus-infected cells were identified from the cervicovaginal swabs
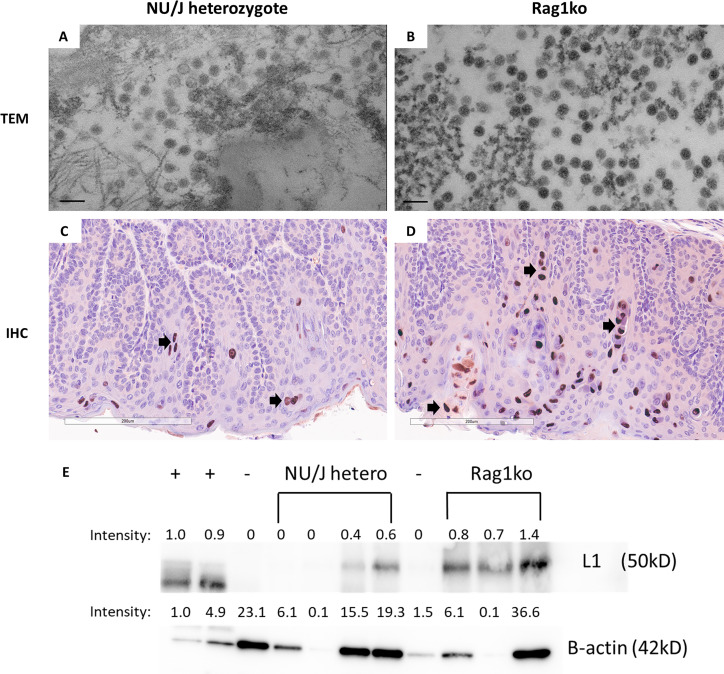
Our previous studies have demonstrated the presence of viral particles in virus-infected tissues by transmission electron microscope (TEM) in athymic nude mice (32). To test whether virions could be detected in the exfoliated cells from the lower genital tract from our tested animals, we pooled cervicovaginal swabs from either infected Rag1ko or NU/J heterozygous mice and pelleted cells for TEM-base viral particle evaluation (Fig. 3). Consistent with previous studies, viral particles (about 50 nm in diameter) were found in exfoliated cells in the cervicovaginal samples of NU/J heterozygous (Fig. 3A) and Rag1ko (Fig. 3B) mice. The infected tissues were further analyzed for viral capsid protein L1 by IHC. In agreement with the TEM findings, viral L1 capsid protein was also detected in the infected vaginal tissues of both NU/J heterozygous (Fig. 3C) and Rag1ko (Fig. 3D) mice. The levels of MmuPV1 L1 in the infected tissues were semi-quantified by Western blot, with increased levels of L1 detected in Rag1ko mice (Fig. 3E). These findings further demonstrated that immunodeficient mice (Rag1ko) produced higher levels of viral production compared to mice with more complete immunity (NU/J heterozygotes), as reported in our previous studies (21, 51).
Fig 3.
Viral particles were found in the cervicovaginal swabs of infected mice. Cervicovaginal swabs were collected from both NU/J heterozygous (A) and Rag1ko (B) infected mice and tested for viral particle presence by TEM (scale bar 100 nm). Consistent with previous reports, viral particles around 50 nm in diameter were found in both mouse strains. Rag1ko (B) mice produced more viral particles in the cervicovaginal swab samples when compared to those in NU/J heterozygous mice (A). Higher numbers of viral capsid protein L1 were detected in the infected vaginal tissues of Rag1ko (20×, D) mice than those in NU/J heterozygous (C) mice by IHC using in-house monoclonal antibody MPV.B9 (20×, Red dots, arrows). MmuPV1 L1 was detected using an in-house monoclonal antibody MPV.B9 via western blot (E).
Pap smear staining reveals pathogenic changes over the infection course
Our previous studies demonstrated that virally infected abnormal cells can be determined by cytological analysis from cervicovaginal lavage samples in vaginally infected mice using H&E staining (21, 22, 32). To further determine whether Pap smear staining could reveal similar cytological changes in the longitudinal cervicovaginal swabs as reported in humans and to determine the suitability of this approach as a model for human studies, we adapted the clinical Pap smear staining protocol for our collected slides. Slides were prepared according to the protocol described in Fig. 1 from five Rag1 and five NU/J heterozygous mice infected with 1 × 109 MmuPV1 viral DNA equivalent as shown in Fig. 4G and H, respectively. More than 200 Pap smear slides from infected and noninfected mice were evaluated without knowledge of infection status or time post-infection before matching the assessments with the animal ID (Table 1). In agreement with our previous cytological observations using H&E staining, we detected amphophilic cytoplasm (resembling inclusion) in atypical squamous cells with ribbon-like central chromatin (22, 32). Koilocytosis, a typic morphology of papillomavirus-infected cells, was easily identified in the slides harvested from these infected mice. The corresponding grades of the longitudinal samples were determined based on squamous cell morphology using the 2014 Bethesda system, the standard used to evaluate human Pap smears (Fig. 4A through F). Here, we showed an example of each category. We found a high convergence between mouse and human Pap smear slide diagnoses (insets of Fig. 4A through F). Increased HSIL cases were observed in both longitudinally Pap smear samples of Rag1ko (N = 5, Fig. 4G) and NU/J heterozygous (N = 5, Fig. 4H) mice after 6 months post-infection. All Rag1ko and NU/J heterozygous infected mice developed SCC according to cytological characterization after 8 months post-infection for both mouse strains.
Fig 4.
Papillomavirus infection induces stages of neoplastic progression identifiable by Papanicolaou-stained Pap smear in the form of cytologically characterized neoplasia in exfoliated cells of mouse and corresponding human Pap Smear samples (inserts at the left corner, 40×). Representative cytology images showing stages of progression of mucosal neoplasia after mouse papillomavirus infection in both immunocompromised (Rag1ko, N = 5) and immunocompetent (NU/J heterozygous, N = 5) mice infected with 1 × 109 MmuPV1 viral DNA equivalent and followed longitudinally. (A) Normal: normal squamous cells with oval-shaped and small nuclei are among a large number of anucleate cornified squamous cells. They are negative for intraepithelial lesion or malignancy (Fig. 4A); (B) ASC-US: Atypical squamous cells of uncertain significance with nucleus size 1.5–2 times larger than a normal squamous cell (Fig. 4B, arrow); (C) LSIL: Low-grade squamous intraepithelial lesion with koilocytes (hyperchromatic raisin-shaped nuclei with nucleus size 2.5–3 times larger than a normal squamous cell and a clear halo around it, arrow; or binucleated cells with a clear halo, arrow); (D) ASC-H: Atypical squamous cells with hyperchromatic nucleus sized 2–2.5 times larger than a normal squamous cell and increased N-C ratio (arrow), cannot exclude HSIL; (E) HSIL: High-grade squamous intraepithelial lesion (HSIL, hyperchromatic cells with high N-C ratio scattered individually or forming sheets, arrow), encompassing cervical intraepithelial neoplasia grades 2 and 3; CIN2 and CIN3; and F) SCC: squamous cell carcinoma (SCC, keratinizing with marked pleomorphism of cell size and shape, arrow; the presence of tadpole cells with dense orangeophilic cytoplasm and hyperchromatic nuclei, arrow; spindle-shaped cells, and inflammation and necrotic debris on the background). These stages of neoplastic progression are based on the 2014 Bethesda system and represent increased grades of intraepithelial neoplasia. Both immunocompromised (G, Rag1ko) and immunocompetent (H, NU/J heterozygous) mice infected with MmuPV1 at the lower genital tract developed LSIL as early as week four post-infection. An increased proportion of Pap smear samples became HSIL over time in both mouse strains. After 6 months, most infected animals developed HSIL and all of them progressed to SCC after 8 months post-infection in the vaginal tissues.
Validation of disease progression by in situ assays
To determine whether cytological results agreed with the histology of infected mice, we sacrificed Rag1ko and NU/J heterozygous mice at different time points post-viral infection. Viral signals, including viral DNA, RNA, and proteins, were detected as previously reported (21, 51). The histology of infected vaginal tissues of both mouse strains agreed with the Pap smear samples on the disease stages. We detected vaginal intraepithelial neoplasia (VAIN) 1 to squamous cell carcinoma (SCC) in the vaginal tissues of these infected mice over time (Fig. 5A). Surprisingly, we also detected SCC in NU/J heterozygous mice as early as 21 weeks post-virus infection in the current study. After week 28 post-infection, most tested mice (both Rag1ko and NU/J heterozygous mice) developed SCC at the infected vaginal tissues (Fig. 5B). The histological changes agree with HPV-induced cervical intraepithelial neoplasia shown in the corresponding insert of Fig. 5A.
Fig 5.
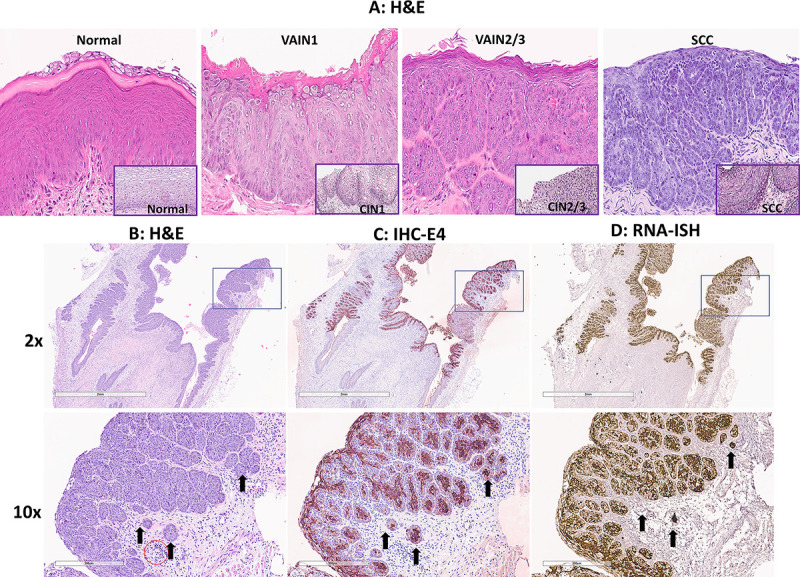
Papillomavirus infection induces identifiable stages from vaginal intraepithelial neoplasia (VAIN) to squamous cell carcinoma (A). Infected mice were sacrificed at different time points post-viral infection for histological validation. We observed the agreement between Pap smear LSIL and VAIN1; HSIL and VAIN2/3; and SCC in our tested animals. Overall, it may take around 5.5 months after mouse papillomavirus infection in the mucosa of infected mice to develop advanced diseases (HSIL to SCC). All mice developed SCC after 8 months post-infection. Corresponding human cervical intraepithelial neoplasia tissues (A, inserts at the right corner, 20×) share similar pathology with the mouse tissues. Histology of squamous cell carcinoma (2× and 10×, B) with a scirrhous response (B, arrows) typical of carcinomas was detected in SCC. Strong viral E4 protein (2× and 10×, C, arrows) and RNA transcripts (2× and 10×, D, arrows) were detected by IHC and RNA-ISH, respectively. Most inflammation was lymphoplasmacytic. There is a low level of immune cells in the stroma of the vagina (red circle), but this cluster was more than anticipated = score 1.
NU/J heterozygous mice were susceptible to MmuPV1 infection and developed VAIN3 in our previous study (21). In the current study, we further identified SCC in these infected mice. The histology of the infected tissues agreed with the Pap smear diagnoses collected simultaneously. In other words, the infected tissues of Pap smear SCC samples also displayed SCC by histological characterization. Figure 4B (2 × and 10×) shows an example of a scirrhous response typical of carcinomas of vaginal SCC tissues of a NU/J heterozygous mouse. The SCC tissues were further evaluated for viral presence using antibodies and probes targeting MmuPV1 E4 (Fig. 5C), an abundant protein produced after active viral infection (32). These SCC tissues are also positive for MmuPV1 E4 RNA transcripts (Fig. 5D). The infected cells invaded the underlying vaginal lamina propria and muscularis layers (Fig. 5B through D, arrows). Most inflammation was lymphoplasmacytic as shown in Fig. 5B. There is a low level of immune cells in the stroma of the vagina (Fig. 5B, red circle) but this cluster was more than anticipated = score 1.
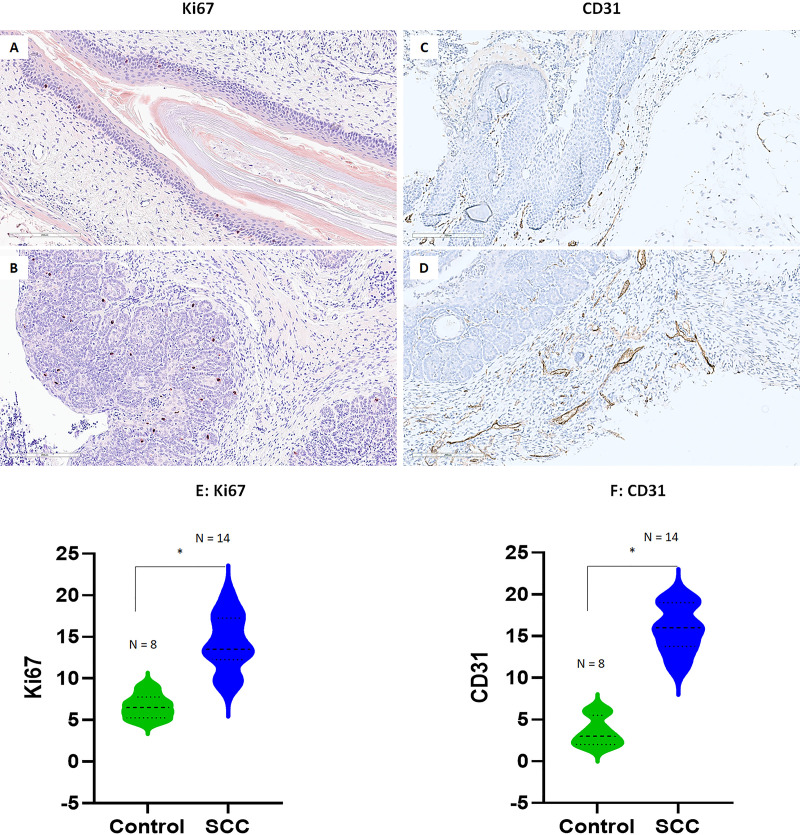
Increased Ki67 and CD31 expression in the infected vaginal tissue that developed SCC
Several biomarkers have been widely used for HPV pathogenesis and SCC (52, 55, 56). For example, cellular proliferation with increased numbers of mitotic figures in the nuclei could be found by Ki67 staining in tissues with more advanced diseases including SCC when compared to that of the normal tissues (20, 57). In addition, among many molecules involved in tumor angiogenesis, platelet-endothelial cellular adhesion molecule (PECAM-1), also known as CD31, has been widely used as a biomarker in highlighting tumor vascular invasion (52). Increased vascular growth has been found in HPV-infected tissues with advanced diseases and cancer and can be targeted for therapy (52, 58). We therefore conducted immunohistochemistry analysis to probe the changes in Ki67 (nuclear staining) and CD31 (vascular staining) expression in the vaginal tissue with normal (Fig. 6A) or low-grade disease (Fig. 6C) and SCC (Fig. 6B and D). For Ki67, we focused on the signals in the infected regions and compared them to corresponding control tissues. Ki67 was found in the nucleus of basal epithelial cells of both normal and infected cells that were actively growing and dividing. Based on the criteria used for Ki67 staining used in clinical evaluation (55), we counted Ki67-positive cell numbers of a 20× magnification view of both normal (N = 8) and infected tissues (N = 14). Although we detected Ki67-positive cells in both normal (Fig. 6A) and SCC (Fig. 6B) tissues, significantly higher numbers of Ki67-positive cells were found in SCC tissues (E, P < 0.01, unpaired Student’s t-test). Similarly, we counted CD31-positive signals of a 20× magnification view based on the size of vascular staining. Significantly higher levels of CD31-positive blood vessels were found in SCC (Fig. 6D) when compared to those in normal controls (Fig. 6F, P < 0.01, unpaired Student’s t-test) supporting the role of CD31 in tumor growth and malignancies.
Fig 6.
Increased mitotic signal (Ki67) and endothelial cellular adhesion molecule (CD31) were found in representative infected mucosa of mice. Representative images of either Ki67 or CD31 expression in the normal (20×, A, C) or infected vaginal tissues with advanced SCC (20×, B, D) of both tested Rag1ko and NU/J heterozygous mice. Significantly higher Ki67-positive cells (E, P < 0.01, unpaired Student’s t-test) and CD31-positive blood vessels (F, P < 0.01, unpaired Student’s t-test) were found in SCC tissues when compared to those in normal tissues.
DISCUSSION
To the best of our knowledge, this is the first report on longitudinally following viral infection to squamous cell carcinoma stages using the well-established mouse model to recapitulate human HPV lower genital tract infection and dysplasia. This study suggests that Pap smear together with viral DNA detection effectively monitored disease progression in the mouse model. These findings will allow us to better understand the dynamic changes during tumor progression in the lower genital tract and develop and test novel treatments to counteract disease progression at different disease stages.
As the fourth most common cancer, cervical cancer not only claims the lives of more than 300,000 annually but has become a heavy economic and health burden worldwide (6). In the United States, around 5.5 million new HPV infections are reported each year. Among them, 10% of HPV persistent infections progress to invasive cancers, which can take more than two decades (59). Treating HPV-associated low-grade and high-grade squamous intraepithelial lesions (SILs) costs billions of dollars each year in the United States (60). Therefore, novel strategies to prevent and treat HPV-associated diseases and cancers besides the HPV vaccines are highly desired.
Since the first report of the MmuPV1 as a cutaneous virus (61), this model has been studied extensively and helped to answer many questions relating to papillomavirus infection and pathogenesis (18, 34, 62, 63). For example, we use this model to understand infection at different anatomic sites and how host immunogenetic background contributes to different disease outcomes (19, 24, 40, 47). These studies will help us further answer some clinical questions including the role of both viral and host genes and pathways in infection control and discover novel biomarkers for diagnosis and therapies (34, 62, 63). MmuPV1 is a member of the pi papillomavirus genera and thus evolutionarily distinct from the alpha papillomaviruses most often associated with human cancers (64). However, MmuPV1 contains two functional oncogenes, E6 and E7, and a non-coding region with many of the same regulatory and transcription factor binding sites as shown in HPV genomes (34, 65) and most importantly, a tissue tropism that includes the anogenital tract (33, 39). The broad tissue tropism of the MmuPV1 has allowed us to model several types of prevalent HPV infections and cancers including oropharyngeal cancer in this amiable laboratory animal model (33, 66–68). Over the past decade, we have had a better understanding of viral-host interaction at both cutaneous and mucosal sites using this model system with the collective efforts of many researchers (19, 20, 23–31, 34, 38–40, 47, 49, 51, 53, 54, 62–65, 69–83). The most promising advancement is identifying additional immunocompetent mouse strains for studying viral persistence and clearance in different tissue types (19–22, 38, 70, 73). This tractable model is highly desirable especially for mucosal infections, as we can monitor disease progression longitudinally without sacrificing animals for the prospective study of HPV pathogenesis (22, 32).
Our previous cytological assay used H&E stain to determine abnormal cells in our infected animals (22, 32). To facilitate the clinical application of this model to discover mechanisms underlying disease progression and develop novel diagnostic and therapeutic markers, we adapted the well-established Pap smear protocol for human samples. To further determine whether Pap smear staining could reveal similar cytological changes in the longitudinal cervicovaginal swab samples as reported in humans, two pathologists independently evaluated more than 200 slides from infected and noninfected mice without knowledge of infection status or time post-infection before matching the assessments with the animal ID. There was high agreement between pathologists, suggesting the translated Bethesda scoring system and evaluation are consistent and robust for evaluating the Pap smear slides from mice. The discrepancies between the two diagnoses in some samples represent the natural spectrum of pathological evaluation in humans. We were able to monitor changes and correlate disease stages using Pap smear with corresponding human specimens. Therefore, the Pap smear is a valid and efficient method for monitoring MmuPV1-associated cervicovaginal disease progression longitudinally. We detected fluctuation in viral DNA detection according to the estrous state in the genital-infected athymic mice (32). In the current study, we attempted to correlate the estrus stage with Pap smear evaluation. However, it was impossible to identify the estrous state from virus-infected mouse Pap smear samples, especially later, due to increased signs of infection secondary to neoplastic invasion and cellular atypia. Therefore, we did not classify Pap smear samples by stage of estrous cycle in the infected slides.
We used Rag1ko mice and NU/J heterozygotes to represent both immunocompromised and immunocompetent backgrounds, respectively. Similar genetic deficiencies were found in humans (41, 84). Therefore, our mouse model can potentially be used to study the interaction between viral and host during papillomavirus infection in these patients which has not been reported. The primary and secondary immunocompromised people are more prone to HPV infections and associated malignancies (85). To generate robust data for Pap smear to cover all the cytological characterization stages, we followed both immunocompromised (Rag1ko) and immunocompetent (NU/J heterozygous) mice up to 32 weeks post-infection. Biweekly swabs were evaluated for both viral load and cytology. In addition, corresponding tissues were harvested periodically along with cytological samples for further validation. This design allowed us to gain time-sensitive data to closely monitor disease progression in these infected animals. Although Pap smears in human studies focus on cervical cytology, cervicovaginal swabs have been reported to screen for HPV infections in women using self-sampling for different populations and showed satisfactory results (86–88). The self-sampling strategy to collect cervix-vaginal swabs for HPV detection can overcome Pap smear challenges, including the requirement of professionals to collect Pap smear samples and the cost of conducting the procedure but cannot detect cytological changes (86). A recent study demonstrated that a combination of cytology and HPV testing would improve the detection of early cases of CIN2+ (89). In our study, we also combined viral detection, cytology, and tissue histology for more accurate evaluation of these infected tissues as used in clinical practice. The agreement between our mouse model and reports in humans further supports that this combination is essential for early diagnosis and development of novel stage-associated treatments for HPV-associated cancers.
Intriguingly, malignant conversion was detected in both immunocompromised (Rag1ko) and immunocompetent (NU/J heterozygous) mice as early as 22 weeks after viral infection in Pap smears and the corresponding tissues. Rag1ko mice are deficient in both T and B cells, key components of the adaptive immune system; therefore, MmuPV1 infection at both cutaneous and mucosal tissue persists and develops cancer over time. All tested immunocompetent NU/J heterozygous mice also developed SCC in the vaginal tissues in the current study. Our previous study also detected VAIN3 in NU/J heterozygous mice (21). Advanced disease including SCC in the infected genital tract was also reported in another immunocompetent mouse strain (FVB) after 6 months of infection (20) and some immune-deficient mouse strains by other groups (19). The mechanisms underlying SCC development in mice from different genetic backgrounds need further investigation. Ki67- (a cellular marker for proliferation) positive cells are found in high-risk HPV-associated cancer (55). Aligned with this, we also observed significantly higher numbers of Ki67-positive basal cells in infected epithelia of NU/J heterozygous mice compared to controls as shown in the infected FVB mice (20). However, more Ki67 signals were shown in the infected FVB vaginal tract in a previous study (20). The main difference was the type of the antibody for Ki67. We used a rabbit monoclonal antibody that usually shows a lower background compared to a mouse monoclonal antibody on mouse tissues. To confirm our observations, we tested infected FVB mice using our rabbit monoclonal antibody and detected a similar pattern as shown in NU/J heterozygous mice suggesting the different Ki67 antibodies contributed to these differences (Fig. S1A through C). Other factors were also reported to impact the detection of Ki67 in the vaginal tissues, these include hormone levels and estrus stages (90–92). The previous study treated FVB mice with 17-beta estradiol (20) while no hormone treatment was applied to either FVB or NU/J heterozygotes after the initial DMPA treatment for standard infection in our study. Whether 17-beta estradiol contributed to increased Ki67 detection in FVB mice needs further investigation. Nevertheless, we both observed the same increase in Ki67-positive cells in the infected basal layers compared to uninfected tissues. CD31 is a reliable marker of tumor vascular invasion and HPV malignancies. We further demonstrate the correlation between increased expression and advanced disease stages in both NU/J heterozygous and Rag1ko mice suggesting it can be a tumor invasion marker for cancer in the mouse model. Therefore, the mouse model recapitulates HPV-associated disease progression from early infection to SCC, making it a relevant model to study HPV pathogenesis. In our previous study, we observed increased Ly6B.2-positive cells in the lower genital tract of NU/J heterozygous mice with advanced disease (21). In the current Pap smear samples, we also detected increased numbers of neutrophils in advanced stages especially HSIL samples suggesting these immune cells might have played a role in accelerating cancer development. Infiltration of Ly6B.2-positive cells were found in tissues with high-grade dysplasia (21). We showed most heterozygous mice (9/10) showed increased neutrophil infiltration while only one of five Rag1ko mice showed neutrophil infiltration in the infected vaginal tissues (Fig. S1D through F) (21). Further investigation needs to be conducted to determine their function in HPV pathogenesis.
Both cytology and histology agree with each other in our tested animals, especially at the advanced disease stage such as HSIL and SCC. For some samples, HSIL was detected as early as 8 weeks after infection but fluctuated until three continuous time points of HSIL to correlate with SCC development in these mice. SCC was detected as early as 16 weeks post-infection in some mice. Tissues harvested at the corresponding time points also showed similar pathogenic stages, suggesting that Pap smear-based cytological evaluation is valid for diagnosing the disease progression. It is especially significant that we can use this traceable model to test anti-viral and anti-tumor compounds by intervening at different time points/disease stages and to differentiate their efficacy in clearing infections/tumors.
ACKNOWLEDGMENTS
We thank Marianne Klinger for her assistance with the CD31 immunohistochemistry analysis at the morphologic and molecular pathology research lab, Department of pathology and laboratory medicine.
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R21AI121822 (Christensen/Hu), National Institute of Dental and Craniofacial Research of the National Institutes of Health under Award Number 1R21DE028650 (Hu), National Cancer Institute of the National Institutes of Health under Award Number R21 CA271069 (J.H./T.T.M), Canadian Institutes of Health Research (CIHR) Team Grant (A.D.B/T.T.M./J.H.) (MRT-168046), National Institutes of Health Research (NIH) (R01CA266050) (A.D.B/T.T.M./J.H.), CIHR grant (University of Manitoba 52250/322744) (A.D.B/T.T.M./J.H.), and CIHR grant MRT-CDAA-159236 (T.T.M.), the pathology department research initiative fund (J.H.), and the Jake Gittlen Memorial Golf Tournament (N.D.C).
Conceptualization, J.H., H.A., A.A.U., and T.T.M.; methodology, J.L., D.A.S., K.B.B., H.C., P. L., M.K., D.N.C.A., S.A., and S.A.B. validation, J.H., H.A., P.L., D.N., J.C., M.K., H.C., and A.A.U.; formal analysis, J.L., D.A.S. K.B.B., S.A.B., H.A., D.N., H.C., S.A., and A.A.U and J.H.; resources, J.H., T.T.M., and N.D.C.; data curation, J.H., H.A., J.L., S.A.B., P.L., D.N., T.M., and A.A.U.; writing—original draft preparation, J.H.; writing—review and editing, J.L., S.A.B., J.H., M.K., H.A., D.N., A.A.U., T.T.M., K.C., C.A., A.D.B., S.A., Y.Z., and N.D.C.; funding acquisition, J.H., T.T.M., A.D.B., and N.D.C. All authors have read and agreed to the published version of the manuscript.
Contributor Information
Hannah M. Atkins, Email: hannah_atkins@med.unc.edu.
Aysegul Aksakal Uslu, Email: a.guluslu@hotmail.com.
Thomas T. Murooka, Email: thomas.murooka@umanitoba.ca.
Jiafen Hu, Email: Fjh4@psu.edu.
Mario Poljak, Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia.
DATA AVAILABILITY
The data supporting the findings of this study are available from the corresponding author upon request.
ETHICS APPROVAL
The animal study protocol was approved by the Institutional Animal Care and Use Committee of Penn State University, College of Medicine (PRAMS200747110, approved 05/06/2019, and PROTO202102089, approved 01/17/2022) for studies involving animals.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01420-24.
Increased mitotic signal (Ki67) and neutrophils (Ly6B.2) in red were found in vaginal tissues by immunohistochemistry.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. McBride AA. 2022. Human papillomaviruses: diversity, infection and host interactions. Nat Rev Microbiol 20:95–108. doi: 10.1038/s41579-021-00617-5 [DOI] [PubMed] [Google Scholar]
- 2. Guo L, Hua K. 2020. Cervical cancer: emerging immune landscape and treatment. Onco Targets Ther 13:8037–8047. doi: 10.2147/OTT.S264312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lechner M, Liu J, Masterson L, Fenton TR. 2022. HPV-associated oropharyngeal cancer: epidemiology, molecular biology and clinical management. Nat Rev Clin Oncol 19:306–327. doi: 10.1038/s41571-022-00603-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clarke MA, Deshmukh AA, Suk R, Roberts J, Gilson R, Jay N, Stier EA, Wentzensen N. 2022. A systematic review and meta-analysis of cytology and HPV-related biomarkers for anal cancer screening among different risk groups. Int J Cancer 151:1889–1901. doi: 10.1002/ijc.34199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo T, Kang SY, Cohen EEW. 2022. Current perspectives on recurrent HPV-mediated oropharyngeal cancer. Front Oncol 12:966899. doi: 10.3389/fonc.2022.966899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh D, Vignat J, Lorenzoni V, Eslahi M, Ginsburg O, Lauby-Secretan B, Arbyn M, Basu P, Bray F, Vaccarella S. 2023. Global estimates of incidence and mortality of cervical cancer in 2020: a baseline analysis of the WHO global cervical cancer elimination initiative. Lancet Glob Health 11:e197–e206. doi: 10.1016/S2214-109X(22)00501-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bruni L, Serrano B, Roura E, Alemany L, Cowan M, Herrero R, Poljak M, Murillo R, Broutet N, Riley LM, de Sanjose S. 2022. Cervical cancer screening programmes and age-specific coverage estimates for 202 countries and territories worldwide: a review and synthetic analysis. Lancet Glob Health 10:e1115–e1127. doi: 10.1016/S2214-109X(22)00241-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou L, Qiu Q, Zhou Q, Li J, Yu M, Li K, Xu L, Ke X, Xu H, Lu B, Wang H, Lu W, Liu P, Lu Y. 2022. Long-read sequencing unveils high-resolution HPV integration and its oncogenic progression in cervical cancer. Nat Commun 13:2563. doi: 10.1038/s41467-022-30190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kreimer AR, Cernuschi T, Rees H, Brotherton JML, Porras C, Schiller J. 2023. Public health opportunities resulting from sufficient HPV vaccine supply and a single-dose vaccination schedule. J Natl Cancer Inst 115:246–249. doi: 10.1093/jnci/djac189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei F, Alberts CJ, Albuquerque A, Clifford GM. 2023. Impact of human papillomavirus vaccine against anal human papillomavirus infection, anal intraepithelial neoplasia, and recurrence of anal intraepithelial neoplasia: a systematic review and meta-analysis. J Infect Dis 228:1496–1504. doi: 10.1093/infdis/jiad183 [DOI] [PubMed] [Google Scholar]
- 11. Ebrahimi N, Yousefi Z, Khosravi G, Malayeri FE, Golabi M, Askarzadeh M, Shams MH, Ghezelbash B, Eskandari N. 2023. Human papillomavirus vaccination in low- and middle-income countries: progression, barriers, and future prospective. Front Immunol 14:1150238. doi: 10.3389/fimmu.2023.1150238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Williamson AL. 2023. Recent developments in human papillomavirus (HPV) vaccinology. Viruses 15:1440. doi: 10.3390/v15071440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gravitt PE, Winer RL. 2017. Natural history of HPV infection across the lifespan: role of viral latency. Viruses 9:267. doi: 10.3390/v9100267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poljak M, Cuschieri K, Alemany L, Vorsters A. 2023. Testing for human papillomaviruses in urine, blood, and oral specimens: an update for the laboratory. J Clin Microbiol 61:e0140322. doi: 10.1128/jcm.01403-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ogilvie G, Nakisige C, Huh WK, Mehrotra R, Franco EL, Jeronimo J. 2017. Optimizing secondary prevention of cervical cancer: recent advances and future challenges. Int J Gynaecol Obstet 138:15–19. doi: 10.1002/ijgo.12187 [DOI] [PubMed] [Google Scholar]
- 16. Mayer C, Budh DP. 2021. Abnormal papanicolaou smear. StatPearls, Treasure Island (FL). [Google Scholar]
- 17. Kitchen FL, Cox CM. 2021. Papanicolaou smear. StatPearls, Treasure Island (FL). [PubMed] [Google Scholar]
- 18. Christensen ND, Budgeon LR, Cladel NM, Hu J. 2017. Recent advances in preclinical model systems for papillomaviruses. Virus Res 231:108–118. doi: 10.1016/j.virusres.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scagnolari C, Cannella F, Pierangeli A, Mellinger Pilgrim R, Antonelli G, Rowley D, Wong M, Best S, Xing D, Roden RBS, Viscidi R. 2020. Insights into the role of innate immunity in cervicovaginal papillomavirus infection from studies using gene-deficient mice. J Virol 94:e00087-20. doi: 10.1128/JVI.00087-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spurgeon ME, Uberoi A, McGregor SM, Wei T, Ward-Shaw E, Lambert PF. 2019. A novel in vivo infection model to study papillomavirus-mediated disease of the female reproductive tract. mBio 10:e00180-19. doi: 10.1128/mBio.00180-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cladel NM, Budgeon LR, Balogh KK, Cooper TK, Brendle SA, Christensen ND, Schell TD, Hu J. 2017. Mouse papillomavirus infection persists in mucosal tissues of an immunocompetent mouse strain and progresses to cancer. Sci Rep 7:16932. doi: 10.1038/s41598-017-17089-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cladel NM, Budgeon LR, Balogh KK, Cooper TK, Hu J, Christensen ND. 2015. A novel pre-clinical murine model to study the life cycle and progression of cervical and anal papillomavirus infections. PLoS One 10:e0120128. doi: 10.1371/journal.pone.0120128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spurgeon ME, Lambert PF. 2019. Sexual transmission of murine papillomavirus (MmuPV1) in Mus musculus. Elife 8:e50056. doi: 10.7554/eLife.50056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brendle SA, Li J, Cladel NM, Balogh KK, Booth J, Shearer DA, Walter V, Lu S, Christensen ND, Covington D, DeBroff J, Milici J, Zhu Y, Viscidi R, Hu J. 2022. Passive immunization with a single monoclonal neutralizing antibody protects against cutaneous and mucosal mouse papillomavirus infections. J Virol 96:e0070322. doi: 10.1128/jvi.00703-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. King RE, Rademacher J, Ward-Shaw ET, Hu R, Bilger A, Blaine-Sauer S, Spurgeon ME, Thibeault SL, Lambert PF. 2024. The larynx is protected from secondary and vertical papillomavirus infection in immunocompetent mice. Laryngoscope 134:2322–2330. doi: 10.1002/lary.31228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spurgeon ME, Townsend EC, Blaine-Sauer S, McGregor SM, Horswill M, den Boon JA, Ahlquist P, Kalan L, Lambert PF. 2024. Key aspects of papillomavirus infection influence the host cervicovaginal microbiome in a preclinical murine papillomavirus (MmuPV1) infection model. mBio 15:e0093324. doi: 10.1128/mbio.00933-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brendle SA, Li JJ, Walter V, Schell TD, Kozak M, Balogh KK, Lu S, Christensen ND, Zhu Y, El-Bayoumy K, Hu J. 2023. Immune responses in oral papillomavirus clearance in the MmuPV1 mouse model. Pathogens 12:1452. doi: 10.3390/pathogens12121452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang W, Spurgeon ME, Pope A, McGregor S, Ward-Shaw E, Gronski E, Lambert PF. 2023. Stress keratin 17 and estrogen support viral persistence and modulate the immune environment during cervicovaginal murine papillomavirus infection. Proc Natl Acad Sci U S A 120:e2214225120. doi: 10.1073/pnas.2214225120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. King RE, Ward-Shaw ET, Hu R, Lambert PF, Thibeault SL. 2022. Expanded basal compartment and disrupted barrier in vocal fold epithelium infected with mouse papillomavirus MmuPV1. Viruses 14:1059. doi: 10.3390/v14051059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. King RE, Bilger A, Rademacher J, Ward-Shaw ET, Hu R, Lambert PF, Thibeault SL. 2022. A novel in vivo model of laryngeal papillomavirus-associated disease using Mus musculus papillomavirus. Viruses 14:1000. doi: 10.3390/v14051000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blaine-Sauer S, Shin MK, Matkowskyj KA, Ward-Shaw E, Lambert PF. 2021. A novel model for papillomavirus-mediated anal disease and cancer using the mouse papillomavirus. mBio 12:e0161121. doi: 10.1128/mBio.01611-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu J, Budgeon LR, Cladel NM, Balogh K, Myers R, Cooper TK, Christensen ND. 2015. Tracking vaginal, anal and oral infection in a mouse papillomavirus infection model. J Gen Virol 96:3554–3565. doi: 10.1099/jgv.0.000295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cladel NM, Budgeon LR, Cooper TK, Balogh KK, Hu J, Christensen ND. 2013. Secondary infections, expanded tissue tropism, and evidence for malignant potential in immunocompromised mice infected with Mus musculus papillomavirus 1 DNA and virus. J Virol 87:9391–9395. doi: 10.1128/JVI.00777-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu J, Cladel NM, Budgeon LR, Balogh KK, Christensen ND. 2017. The mouse papillomavirus infection model. Viruses 9:246. doi: 10.3390/v9090246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Beyazit F, Sılan F, Gencer M, Aydin B, Paksoy B, Unsal MA, Ozdemir O. 2018. The prevelance of human papillomavirus (HPV) genotypes detected by PCR in women with normal and abnormal cervico-vaginal cytology. Ginekol Pol 89:62–67. doi: 10.5603/GP.a2018.0011 [DOI] [PubMed] [Google Scholar]
- 36. Du Q, Huynh LK, Coskun F, Molina E, King MA, Raj P, Khan S, Dozmorov I, Seroogy CM, Wysocki CA, Padron GT, Yates TR, Markert ML, de la Morena MT, van Oers NS. 2019. FOXN1 compound heterozygous mutations cause selective thymic hypoplasia in humans. J Clin Invest 129:4724–4738. doi: 10.1172/JCI127565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rota IA, Handel AE, Maio S, Klein F, Dhalla F, Deadman ME, Cheuk S, Newman JA, Michaels YS, Zuklys S, et al. 2021. FOXN1 forms higher-order nuclear condensates displaced by mutations causing immunodeficiency. Sci Adv 7:eabj9247. doi: 10.1126/sciadv.abj9247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang JW, Jiang R, Peng S, Chang Y-N, Hung C-F, Roden RBS. 2015. Immunologic control of Mus musculus papillomavirus type 1. PLoS Pathog 11:e1005243. doi: 10.1371/journal.ppat.1005243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sundberg JP, Stearns TM, Joh J, Proctor M, Ingle A, Silva KA, Dadras SS, Jenson AB, Ghim S. 2014. Immune status, strain background, and anatomic site of inoculation affect mouse papillomavirus (MmuPV1) induction of exophytic papillomas or endophytic trichoblastomas. PLoS One 9:e113582. doi: 10.1371/journal.pone.0113582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Béziat V, Rapaport F, Hu J, Titeux M, Bonnet des Claustres M, Bourgey M, Griffin H, Bandet É, Ma CS, Sherkat R, et al. 2021. Humans with inherited T cell CD28 deficiency are susceptible to skin papillomaviruses but are otherwise healthy. Cell 184:3812–3828. doi: 10.1016/j.cell.2021.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bosticardo M, Pala F, Notarangelo LD. 2021. RAG deficiencies: recent advances in disease pathogenesis and novel therapeutic approaches. Eur J Immunol 51:1028–1038. doi: 10.1002/eji.202048880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rios FJ, Montezano AC, Touyz RM. 2020. Lessons learned from RAG-1-deficient mice in hypertension. Hypertension 75:935–937. doi: 10.1161/HYPERTENSIONAHA.119.14406 [DOI] [PubMed] [Google Scholar]
- 43. Seniuk A, Thiele JL, Stubbe A, Oser P, Rosendahl A, Bode M, Meyer-Schwesinger C, Wenzel UO, Ehmke H. 2020. B6.Rag1 knockout mice generated at the Jackson laboratory in 2009 show a robust wild-type hypertensive phenotype in response to ANG II (angiotensin II). Hypertension 75:1110–1116. doi: 10.1161/HYPERTENSIONAHA.119.13773 [DOI] [PubMed] [Google Scholar]
- 44. Gramlich OW, Godwin CR, Heuss ND, Gregerson DS, Kuehn MH. 2020. T and B lymphocyte deficiency in Rag1-/- mice reduces retinal ganglion cell loss in experimental glaucoma. Invest Ophthalmol Vis Sci 61:18. doi: 10.1167/iovs.61.14.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Efremova M, Rieder D, Klepsch V, Charoentong P, Finotello F, Hackl H, Hermann-Kleiter N, Löwer M, Baier G, Krogsdam A, Trajanoski Z. 2018. Targeting immune checkpoints potentiates immunoediting and changes the dynamics of tumor evolution. Nat Commun 9:32. doi: 10.1038/s41467-017-02424-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Til NP, Sarwari R, Visser TP, Hauer J, Lagresle-Peyrou C, van der Velden G, Malshetty V, Cortes P, Jollet A, Danos O, Cassani B, Zhang F, Thrasher AJ, Fontana E, Poliani PL, Cavazzana M, Verstegen MMA, Villa A, Wagemaker G. 2014. Recombination-activating gene 1 (Rag1)-deficient mice with severe combined immunodeficiency treated with lentiviral gene therapy demonstrate autoimmune Omenn-like syndrome. J Allergy Clin Immunol 133:1116–1123. doi: 10.1016/j.jaci.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 47. Cladel NM, Jiang P, Li JJ, Peng X, Cooper TK, Majerciak V, Balogh KK, Meyer TJ, Brendle SA, Budgeon LR, Shearer DA, Munden R, Cam M, Vallur R, Christensen ND, Zheng ZM, Hu J. 2019. Papillomavirus can be transmitted through the blood and produce infections in blood recipients: evidence from two animal models. Emerg Microbes Infect 8:1108–1121. doi: 10.1080/22221751.2019.1637072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu YS, Feldman S, Leung SOA, Creer MH, Warrick J, Williams N, Mastorides S. 2023. AACC guidance document on cervical cancer detection: screening, surveillance, and diagnosis. J Appl Lab Med 8:382–406. doi: 10.1093/jalm/jfac142 [DOI] [PubMed] [Google Scholar]
- 49. Brendle S, Li JJ, Cladel NM, Shearer DA, Budgeon LR, Balogh KK, Atkins H, Costa-Fujishima M, Lopez P, Christensen ND, Doorbar J, Murooka TT, Hu J. 2021. Mouse papillomavirus L1 and L2 are dispensable for viral infection and persistence at both cutaneous and mucosal tissues. Viruses 13:1824. doi: 10.3390/v13091824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cladel NM, Budgeon LR, Cooper TK, Balogh KK, Christensen ND, Myers R, Majerciak V, Gotte D, Zheng ZM, Hu J. 2017. Mouse papillomavirus infections spread to cutaneous sites with progression to malignancy. J Gen Virol 98:2520–2529. doi: 10.1099/jgv.0.000926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu J, Brendle SA, Li JJ, Walter V, Cladel NM, Cooper T, Shearer DA, Balogh KK, Christensen ND. 2022. Depo medroxyprogesterone (DMPA) promotes papillomavirus infections but does not accelerate disease progression in the anogenital tract of a mouse model. Viruses 14:980. doi: 10.3390/v14050980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fei J, Hong A, Dobbins TA, Jones D, Lee CS, Loo C, Al-Ghamdi M, Harnett GB, Clark J, O’Brien CJ, Rose B. 2009. Prognostic significance of vascular endothelial growth factor in squamous cell carcinomas of the tonsil in relation to human papillomavirus status and epidermal growth factor receptor. Ann Surg Oncol 16:2908–2917. doi: 10.1245/s10434-009-0579-1 [DOI] [PubMed] [Google Scholar]
- 53. Yu L, Majerciak V, Xue XY, Uberoi A, Lobanov A, Chen X, Cam M, Hughes SH, Lambert PF, Zheng ZM. 2021. Mouse papillomavirus type 1 (MmuPV1) DNA is frequently integrated in benign tumors by microhomology-mediated end-joining. PLoS Pathog 17:e1009812. doi: 10.1371/journal.ppat.1009812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Egawa N, Shiraz A, Crawford R, Saunders-Wood T, Yarwood J, Rogers M, Sharma A, Eichenbaum G, Doorbar J. 2021. Dynamics of papillomavirus in vivo disease formation & susceptibility to high-level disinfection-Implications for transmission in clinical settings. EBioMedicine 63:103177. doi: 10.1016/j.ebiom.2020.103177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maebayashi T, Ishibashi N, Aizawa T, Sakaguchi M, Saito T, Kawamori J, Tanaka Y, Hirotani Y, Homma T. 2019. Roles of Ki-67 and p16 as biomarkers for unknown primary head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol 276:1221–1229. doi: 10.1007/s00405-019-05338-3 [DOI] [PubMed] [Google Scholar]
- 56. Suresh TN, Hemalatha A, Harendra Kumar ML, Azeem Mohiyuddin SM. 2015. Evaluation of histomorphological and immunohistochemical parameters as biomarkers of cervical lymph node metastasis in squamous cell carcinoma of oral cavity: a retrospective study. J Oral Maxillofac Pathol 19:18–24. doi: 10.4103/0973-029X.157195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dellas A, Schultheiss E, Oberholzer M, Torhorst J, Gudat F. 1996. Analysis of proliferative activity using Ki-67 on cervical precancerous lesions and the relationship to p53 expression. Anticancer Res 16:3403–3407. [PubMed] [Google Scholar]
- 58. Zhao X, Bose A, Komita H, Taylor JL, Chi N, Lowe DB, Okada H, Cao Y, Mukhopadhyay D, Cohen PA, Storkus WJ. 2012. Vaccines targeting tumor blood vessel antigens promote CD8+ T cell-dependent tumor eradication or dormancy in HLA-A2 transgenic mice. J Immunol 188:1782–1788. doi: 10.4049/jimmunol.1101644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shanmugasundaram S, You J. 2017. Targeting persistent human papillomavirus infection. Viruses 9:229. doi: 10.3390/v9080229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chesson HW, Ekwueme DU, Saraiya M, Watson M, Lowy DR, Markowitz LE. 2012. Estimates of the annual direct medical costs of the prevention and treatment of disease associated with human papillomavirus in the United States. Vaccine 30:6016–6019. doi: 10.1016/j.vaccine.2012.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ingle A, Ghim S, Joh J, Chepkoech I, Bennett Jenson A, Sundberg JP. 2011. Novel laboratory mouse papillomavirus (MusPV) infection. Vet Pathol 48:500–505. doi: 10.1177/0300985810377186 [DOI] [PubMed] [Google Scholar]
- 62. Uberoi A, Yoshida S, Lambert PF. 2018. Development of an in vivo infection model to study Mouse papillomavirus-1 (MmuPV1). J Virol Methods 253:11–17. doi: 10.1016/j.jviromet.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Uberoi A, Lambert PF. 2017. Rodent papillomaviruses. Viruses 9:362. doi: 10.3390/v9120362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Meyers JM, Uberoi A, Grace M, Lambert PF, Munger K. 2017. Cutaneous HPV8 and MmuPV1 E6 proteins target the NOTCH and TGF-beta tumor suppressors to inhibit differentiation and sustain keratinocyte proliferation. PLoS Pathog 13:e1006171. doi: 10.1371/journal.ppat.1006171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Joh J, Jenson AB, King W, Proctor M, Ingle A, Sundberg JP, Ghim S -j. 2011. Genomic analysis of the first laboratory-mouse papillomavirus. J Gen Virol 92:692–698. doi: 10.1099/vir.0.026138-0 [DOI] [PubMed] [Google Scholar]
- 66. Cladel NM, Budgeon LR, Balogh KK, Cooper TK, Hu J, Christensen ND. 2016. Mouse papillomavirus MmuPV1 infects oral mucosa and preferentially targets the base of the tongue. Virology 488:73–80. doi: 10.1016/j.virol.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Christensen ND, Chen KM, Hu J, Stairs DB, Sun YW, Aliaga C, Balogh KK, Atkins H, Shearer D, Li J, Brendle SA, Gowda K, Amin S, Walter V, Viscidi R, El-Bayoumy K. 2021. The environmental pollutant and tobacco smoke constituent dibenzo[def,p]chrysene is a co-factor for malignant progression of mouse oral papillomavirus infections. Chem Biol Interact 333:109321. doi: 10.1016/j.cbi.2020.109321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wei T, Buehler D, Ward-Shaw E, Lambert PF. 2020. An infection-based murine model for papillomavirus-associated head and neck cancer. mBio 11:e00908-20. doi: 10.1128/mBio.00908-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Saunders-Wood T, Egawa N, Zheng K, Giaretta A, Griffin HM, Doorbar J. 2022. Role of E6 in maintaining the basal cell reservoir during productive papillomavirus infection. J Virol 96:e0118121. doi: 10.1128/JVI.01181-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jiang RT, Wang JW, Peng S, Huang TC, Wang C, Cannella F, Chang YN, Viscidi RP, Best SRA, Hung CF, Roden RBS. 2017. Spontaneous and vaccine-induced clearance of Mus musculus papillomavirus 1 infection. J Virol 91:e00699-17. doi: 10.1128/JVI.00699-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xue XY, Majerciak V, Uberoi A, Kim BH, Gotte D, Chen X, Cam M, Lambert PF, Zheng ZM. 2017. The full transcription map of mouse papillomavirus type 1 (MmuPV1) in mouse wart tissues. PLoS Pathog 13:e1006715. doi: 10.1371/journal.ppat.1006715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Handisurya A, Day PM, Thompson CD, Buck CB, Pang Y-Y, Lowy DR, Schiller JT. 2013. Characterization of Mus musculus papillomavirus 1 infection in situ reveals an unusual pattern of late gene expression and capsid protein localization. J Virol 87:13214–13225. doi: 10.1128/JVI.02162-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Handisurya A, Day PM, Thompson CD, Buck CB, Kwak K, Roden RBS, Lowy DR, Schiller JT. 2012. Murine skin and vaginal mucosa are similarly susceptible to infection by pseudovirions of different papillomavirus classifications and species. Virology 433:385–394. doi: 10.1016/j.virol.2012.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Joh J, Jenson AB, Proctor M, Ingle A, Silva KA, Potter CS, Sundberg JP, Ghim S. 2012. Molecular diagnosis of a laboratory mouse papillomavirus (MusPV). Exp Mol Pathol 93:416–421. doi: 10.1016/j.yexmp.2012.07.001 [DOI] [PubMed] [Google Scholar]
- 75. Wei T, Grace M, Uberoi A, Romero-Masters JC, Lee D, Lambert PF, Munger K. 2021. The Mus musculus papillomavirus type 1 E7 protein binds to the retinoblastoma tumor suppressor: implications for viral pathogenesis. mBio 12:e0227721. doi: 10.1128/mBio.02277-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Joh J, Jenson AB, Ingle A, Sundberg JP, Ghim S. 2014. Searching for the initiating site of the major capsid protein to generate virus-like particles for a novel laboratory mouse papillomavirus. Exp Mol Pathol 96:155–161. doi: 10.1016/j.yexmp.2013.12.009 [DOI] [PubMed] [Google Scholar]
- 77. Kuehner F, Wong M, Straub E, Doorbar J, Iftner T, Roden RBS, Stubenrauch F. 2023. Mus musculus papillomavirus 1 E8;E2 represses expression of late protein E4 in basal-like keratinocytes via NCoR/SMRT-HDAC3 co-repressor complexes to enable wart formation in vivo. mBio 14:e0069623. doi: 10.1128/mbio.00696-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Strickley JD, Messerschmidt JL, Awad ME, Li T, Hasegawa T, Ha DT, Nabeta HW, Bevins PA, Ngo KH, Asgari MM, Nazarian RM, Neel VA, Jenson AB, Joh J, Demehri S. 2019. Immunity to commensal papillomaviruses protects against skin cancer. Nature 575:519–522. doi: 10.1038/s41586-019-1719-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Johnson LH, Son HG, Ha DT, Strickley JD, Joh J, Demehri S. 2023. Compromised T cell immunity links increased cutaneous papillomavirus activity to squamous cell carcinoma risk. JID Innov 3:100163. doi: 10.1016/j.xjidi.2022.100163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Romero-Masters JC, Grace M, Lee D, Lei J, DePamphilis M, Buehler D, Hu R, Ward-Shaw E, Blaine-Sauer S, Lavoie N, White EA, Munger K, Lambert PF. 2023. MmuPV1 E7's interaction with PTPN14 delays Epithelial differentiation and contributes to virus-induced skin disease. PLoS Pathog 19:e1011215. doi: 10.1371/journal.ppat.1011215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gunder LC, Johnson HR, Green HA, Bilger A, Moyer TH, Zhang W, Ziolkowski MR, Bertrang PA, Carchman EH. 2022. The use of a topical protease inhibitor, Saquinavir, to alleviate mouse papillomavirus-mediated anal disease. Virology 576:96–104. doi: 10.1016/j.virol.2022.09.012 [DOI] [PubMed] [Google Scholar]
- 82. Gunder LC, Blaine-Sauer S, Johnson HR, Shin MK, Auyeung AS, Zhang W, Leverson GE, Ward-Shaw ET, King RE, McGregor SM, Matkowskyj KA, Lambert PF, Carchman EH. 2022. Efficacy of topically administered dihydroartemisinin in treating papillomavirus-induced anogenital dysplasia in preclinical mouse models. Viruses 14:1632. doi: 10.3390/v14081632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Romero-Masters JC, Lambert PF, Munger K. 2022. Molecular mechanisms of MmuPV1 E6 and E7 and implications for human disease. Viruses 14:2138. doi: 10.3390/v14102138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Torres S, Marzullo M. 2022. FOXN1 gene considerations in severe combined immunodeficiency treatment in children. Cureus 14:e32040. doi: 10.7759/cureus.32040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Varada S, Posnick M, Alessa D, Ramírez-Fort MK. 2014. Management of cutaneous human papillomavirus infection in immunocompromised patients. Curr Probl Dermatol 45:197–215. doi: 10.1159/000357187 [DOI] [PubMed] [Google Scholar]
- 86. Haguenoer K, Sengchanh S, Gaudy-Graffin C, Boyard J, Fontenay R, Marret H, Goudeau A, Pigneaux de Laroche N, Rusch E, Giraudeau B. 2014. Vaginal self-sampling is a cost-effective way to increase participation in a cervical cancer screening programme: a randomised trial. Br J Cancer 111:2187–2196. doi: 10.1038/bjc.2014.510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bokan T, Ivanus U, Jerman T, Takac I, Arko D. 2021. Long term results of follow-up after HPV self-sampling with devices Qvintip and HerSwab in women non-attending cervical screening programme. Radiol Oncol 55:187–195. doi: 10.2478/raon-2021-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Forslund O, Miriam Elfström K, Lamin H, Dillner J. 2019. HPV-mRNA and HPV-DNA detection in samples taken up to seven years before severe dysplasia of cervix uteri. Int J Cancer 144:1073–1081. doi: 10.1002/ijc.31819 [DOI] [PubMed] [Google Scholar]
- 89. Koliopoulos G, Nyaga VN, Santesso N, Bryant A, Martin-Hirsch PP, Mustafa RA, Schünemann H, Paraskevaidis E, Arbyn M. 2017. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst Rev 8:CD008587. doi: 10.1002/14651858.CD008587.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chagas DC, Barros-Oliveira M da C, Lopes-Costa PV, Pereira R de O, Melo M de A, Costa-Silva DR, Borges CS, Viana JL, Santos ARD, Facina G, Silva B da. 2020. Effects of anastrozole on Ki-67 antigen expression in the vaginal epithelium of female rats in persistent estrus. Clinics (Sao Paulo) 75:e1643. doi: 10.6061/clinics/2020/e1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Marusak RA, Radi ZA, Obert L. 2007. Expression of Ki-67 in the uterus during various stages of the estrous cycle in rats. Exp Toxicol Pathol 59:151–155. doi: 10.1016/j.etp.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 92. Sukjumlong S, Kaeoket K, Dalin AM, Persson E. 2003. Immunohistochemical studies on oestrogen receptor alpha (ER alpha) and the proliferative marker Ki-67 in the sow uterus at different stages of the oestrous cycle. Reprod Domest Anim 38:5–12. doi: 10.1046/j.1439-0531.2003.00383.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Increased mitotic signal (Ki67) and neutrophils (Ly6B.2) in red were found in vaginal tissues by immunohistochemistry.
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author upon request.