Abstract
Intercellular transport of tobacco mosaic virus (TMV) RNA involves the accumulation of virus-encoded movement protein (MP) in plasmodesmata (Pd), in endoplasmic reticulum (ER)-derived inclusion bodies, and on microtubules. The functional significance of these interactions in viral RNA (vRNA) movement was tested in planta and in protoplasts with TMV derivatives expressing N- and C-terminal deletion mutants of MP fused to the green fluorescent protein. Deletion of 55 amino acids from the C terminus of MP did not interfere with the vRNA transport function of MP:GFP but abolished its accumulation in inclusion bodies, indicating that accumulation of MP at these ER-derived sites is not a requirement for function in vRNA intercellular movement. Deletion of 66 amino acids from the C terminus of MP inactivated the protein, and viral infection occurred only upon complementation in plants transgenic for MP. The functional deficiency of the mutant protein correlated with its inability to associate with microtubules and, independently, with its absence from Pd at the leading edge of infection. Inactivation of MP by N-terminal deletions was correlated with the inability of the protein to target Pd throughout the infection site, whereas its associations with microtubules and inclusion bodies were unaffected. The observations support a role of MP-interacting microtubules in TMV RNA movement and indicate that MP targets microtubules and Pd by independent mechanisms. Moreover, accumulation of MP in Pd late in infection is insufficient to support viral movement, confirming that intercellular transport of vRNA relies on the presence of MP in Pd at the leading edge of infection.
Intercellular communication in plants occurs through plasmodesmata (Pd), gatable channels in the cell wall that provide symplastic continuity between adjacent cells. Although the size exclusion limit (SEL) of Pd normally restricts diffusion for molecules larger than 1 kDa (22, 43, 45, 53), accumulating evidence indicates that Pd are dynamic and able to increase the SEL to allow the passage of macromolecules such as proteins, RNA, and protein-RNA complexes (13, 15, 18, 21, 29, 32, 33, 36, 47, 53). Most compelling evidence for macromolecular trafficking through Pd is provided by viruses which exploit Pd to move their genomes from cell to cell and to spread infection (18, 33). The spread of viruses represents an excellent model to study the mechanism by which nucleic acid molecules are targeted to Pd. Their trafficking depends on virus-encoded movement proteins (MPs) (12) which can be used to directly probe the intercellular communication machinery.
Much of what is known about MPs is derived from studies on the MP of tobacco mosaic virus (TMV). Consistent with its function in viral movement, this protein is targeted to Pd and increases their SEL (37, 53). The protein also binds single-stranded nucleic acids in vitro, which results in the formation of elongated and unfolded complexes (8, 9). Since the virus coat protein (CP) is not required for intercellular spread of TMV infection (10, 42) MP is believed to form a ribonucleoprotein complex (vRNP) with viral RNA (vRNA) and to facilitate its intercellular transport in nonencapsidated form (9).
To elucidate the mechanism by which MP and vRNA are targeted to Pd, we recently developed recombinant TMV derivatives which encode MP as a fusion with green fluorescent protein (GFP). Using these constructs, it was possible to perform studies in planta and to directly correlate the cell-to-cell spread of vRNA and the growth of the fluorescent infection site with the subcellular localization of the MP:GFP fusion protein (24, 25, 37). These studies demonstrated that during infection, the virus-encoded MP:GFP fusion protein associates with Pd, with irregularly shaped endoplasmic reticulum (ER)-derived inclusion bodies, as well as with microtubules (5, 24). The ER-derived inclusion bodies contain replicase (25) as well as vRNA (34) in addition to MP, thus providing evidence that these bodies represent sites of virus replication and protein synthesis. During the course of infection, MP:GFP produced in these sites is conveyed to microtubules (25), indicating a role of microtubules in the transport of vRNPs from membrane sites of synthesis toward Pd (25). Recent support for this hypothesis was provided by in situ hybridization experiments which localized TMV RNA to microtubules (34) as well as by in vivo observations which indicated that the amount of MP:GFP associated with microtubules at the leading edge of infection is positively correlated with the efficiency by which infection spreads from cell to cell (5). MP also appears to interact with actin (35). Since actin is a component of Pd (2, 14, 52), it may be possible that microfilaments and microtubules share synergistic functions in Pd targeting (6, 54).
This study was undertaken to further dissect the functional role of associations of MP with microtubules, ER inclusion bodies, and Pd. Based on the finding that the vRNA transport function of MP can tolerate large deletions at its C terminus but not at its N terminus (1), we generated a series of TMV derivatives in which we introduced N- and C-terminal deletions into MP fused to GFP. This generated a series of infectious and noninfectious virus derivatives which allowed us to correlate the activity of wild-type and mutant MP:GFP in viral movement with their subcellular distribution and to identify those interactions between MP and host components that are essential or dispensable in vRNA intercellular transport in planta.
MATERIALS AND METHODS
Constructs.
Plasmid pTf5-nx1, used as a template for in vitro mutagenesis, was constructed by substitution of the MluI-KpnI fragment of plasmid pLA41, a pUC119 derivative with an extended polylinker, with the BamHI-KpnI restriction fragment of pTMV-M:GfusBr (24, 25, 37). BalI-SpeI restriction sites were then used to replace most of the GFP coding sequence by a PCR fragment encoding an improved version of GFP (GFP5, without ER targeting sequences) which is characterized by increased thermostability and by two stabilized excitation absorbance maxima (395 and 473 nm) (41). Unique NruI and XhoI restriction sites were then introduced in front of and behind the MP:GFP5 open reading frame (ORF), resulting in plasmid pTf5-nx1. The BamHI-NotI fragment of pTf5-nx1 was then used to replace the BamHI-NotI fragment of pTMV-M:GfusBr to create plasmid pTf5-nx2, encoding infectious genomic RNA of TMV-MP:GFP5. It should be noted that “wild-type MP” refers to the MP as expressed from the original TMV-M:GfusBr construct (24). In this construct, fusion of the GFP ORF resulted in the deletion of 30 nucleotides from the 3′ end of the MP gene sequence. Thus, wild-type MP fused to GFP lacks amino acids 259 to 268 from its C terminus. Moreover, the MP is derived from a TMV variant that is distinguished from the published TMV U1 sequence by a replacement of glutamic acid in position 52 by glycine (27).
In vitro mutagenesis of the MP:GFP5 coding sequence was performed by PCR in the presence of pTf5-nx1 as the template and specific primer sets for mutagenesis to yield MP sequences encoding MP with specific N- or C-terminal deletions. The PCR fragments were blunt ended at the 5′ end of the corresponding plus strand and had an XmaI restriction site at their 3′ end, the latter located within the hinge region between MP and GFP (24). Following XmaI digestion, the sequences were cloned into plasmid pTf5-nx1 for in-frame fusion of the MP sequences with the GFP ORF, replacing the NruI-XmaI fragment. The resulting pTf5-nx1 derivative plasmids were cut with restriction enzymes BamHI and Asp718I to subclone the fragments in place of the respective fragment in pTf5-nx2. Viral infectious RNA was produced by in vitro transcription of plasmid pTf5-nx2 (wild-type MP:GFP) and its derivatives (dMP:GFP mutants) by using a MEGAscript T7 kit (Ambion).
Inoculation of plants and protoplasts.
Nicotiana tabacum cv. Xanthi nn, N. tabacum cv. Xanthi NN, and N. benthamiana plants (5 to 6 weeks old) were mechanically inoculated (in the presence of carborundum) with transcripts derived from in vitro reactions, and the plants were maintained in 70% humidity at 22°C during the 16-h photoperiod and at 20°C during the dark period.
Protoplasts of tobacco suspension cell line BY-2 were prepared and inoculated by electroporation with infectious transcripts as described elsewhere (51). Following inoculation, protoplasts were resuspended in 10 ml of medium and cultured as 1-ml aliquots in 35-mm-diameter petri dishes in the dark at 28°C. Actinomycin D (30 μg/ml) was added to the protoplasts to increase MP expression (3, 25).
Immunoblot analysis.
Protoplasts (5 × 105) were harvested at 20 h postinfection from culture medium by low-speed centrifugation, and pellets were immediately frozen on dry ice and stored at −80°C. Proteins from 125,000 protoplasts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride membranes (Amersham, Arlington Heights, Ill.). The membranes were probed with affinity-purified rabbit antibodies that were raised against synthetic peptides encompassing amino acid residues 6 to 22 (N-terminal anti-MP) or 209 to 222 (C-terminal anti-MP) of MP.
Immunofluorescent labeling and microscopy.
Fixation and immunostaining of protoplasts was performed as described elsewhere (24). Rhodamine-labeled secondary antibody was from Pierce (Rockford, Ill.). Fluorescence microscopy was performed with a Nikon Eclipse E800 microscope equipped with CFI Plan Apochromat objectives (Nikon Corp., Tokyo, Japan) and XF100 (Omega Optical, Inc., Brattleboro, Vt.) as well as G-2A (Nikon Corp.) filter sets for visualization of GFP and rhodamine fluorescence, respectively. Whole fluorescent infection sites on leaves were viewed with 2× or 4× lenses. Leaf disks placed on glass slides were used for high-magnification microscopy using 100× oil immersion objectives. Protoplasts were analyzed with 60× and 100× oil immersion lenses. Images were acquired and processed using an ORCA-100 progressive scan interline charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan) and Openlab 2 software (Improvision, Coventry, England). Images were prepared for publication using Adobe Photoshop software (Adobe Systems Inc., Mountain View, Calif.).
RESULTS
TMV derivatives replicate and produce ΔMP:GFP proteins of expected sizes.
The deletion mutations in MP used in this study (Fig. 1A) include two ΔMP:GFP mutants with 3 and 28 amino acids deleted from the N terminus (the first two amino acids, N5 and N30, were not deleted) and four ΔMP:GFP mutants with up to 81 amino acids deleted from the C terminus (C35, C55, C66, and C81) of MP. Except for encoding either wild-type MP:GFP or ΔMP:GFP, all viral constructs used in the analysis were identical and derived from TMV-MP:GFP (25). To test whether the viruses replicate and produce authentic ΔMP:GFP mutant proteins, we infected BY-2 protoplasts and confirmed expression of the ΔMP:GFP proteins by immunoblotting using antibodies raised against N- and C-subterminal peptides of the MP. As shown in Fig. 1B and C, the ΔMP:GFP fusion proteins were comparable to MP:GFP produced by the wild-type virus with respect to both amount and stability.
FIG. 1.
(A) Schematic representation of ΔMP:GFP constructs used in this study. MP and GFP sequences are represented by dark grey and white, dashed bars, respectively. MP:GFP (24) is shown at the top, and ΔMP:GFP derivatives are depicted below. The ΔMP:GFP derivatives include mutants with up to 81 amino acids deleted from the C terminus (C35, C55, C66, and C81) and 3 or 28 amino acids deleted from the N terminus (the first two amino acids, N5 and N30, were not deleted) of MP. (B and C) Immunoblot analysis of infected tobacco BY-2 protoplasts expressing wild-type MP:GFP and ΔMP:GFP mutants with antibody directed against amino acids 6 to 22 (B) and 209 to 222 (C) of MP. Mk, mock-infected protoplasts; M, protein size markers (positions are indicated in kilodaltons).
Activity of ΔMP:GFP mutant proteins in intercellular transport of vRNA.
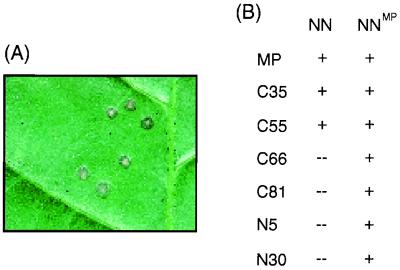
It was previously reported that MP can tolerate deletions of up to 55 amino acids at its C terminus, whereas even very small deletions within the N terminus destroy the function of MP in vRNA transport (1, 20). To test if this activity profile of MP deletion mutants is reflected by our ΔMP:GFP fusion proteins, we tested the infectivity of TMV variants encoding these mutant proteins by inoculation of N. tabacum cv. Xanthi NN plants. N. tabacum cv. Xanthi NN plants harbor the N gene, which triggers a hypersensitive cell death response in infected cells, resulting in the formation of local necrotic lesions (Fig. 2A). As summarized in Fig. 2B, local lesion formation on leaves of N. tabacum cv. Xanthi NN plants was observed when leaves were inoculated with virus expressing MP:GFP, C35:GFP, or C55:GFP. Thus, C35:GFP and C55:GFP are functional and, like MP:GFP, can transport vRNA from cell to cell. In contrast, no lesions were observed on N. tabacum cv. Xanthi NN plants when leaves were inoculated with virus encoding ΔMP:GFP with larger deletion mutations at the C terminus of MP (C66:GFP or C81:GFP). Lesions also did not develop when leaves were inoculated with virus expressing N5:GFP or N30:GFP. Thus, all mutant viruses except those that produced C35:GFP or C55:GFP were defective in local movement. Nevertheless, as summarized in Fig. 2B, the whole series of mutant and wild-type viruses was infectious on N. tabacum cv. Xanthi NN plants carrying a transgene for MP (line 2005 [11]). Thus, all mutant viruses were able to replicate and to move from cell to cell if complemented by wild-type MP. Collectively, these tests demonstrated that the ΔMP:GFP fusion proteins reflect the activity profiles reported for unfused ΔMP mutants (1, 20).
FIG. 2.
Infectivity of TMV derivatives encoding ΔMP:GFP fusion proteins. TMV variants encoding either wild-type MP:GFP or mutant ΔMP:GFP derivatives were inoculated onto leaves of N. tabacum cv. Xanthi NN plants to test for local lesion development (A) in response to local virus movement. (B) Results obtained following inoculation of nontransgenic (NN) and MP-transgenic (NNMP) plants, showing the presence (+) and absence (--) of local lesion formation. The ability of a virus mutant to induce the formation of lesions on MP-transgenic but not nontransgenic plants indicates that the virus replicates but encodes a ΔMP:GFP fusion protein deficient in TMV RNA transport.
Intracellular targeting of MP:GFP and mutant ΔMP:GFP derivatives.
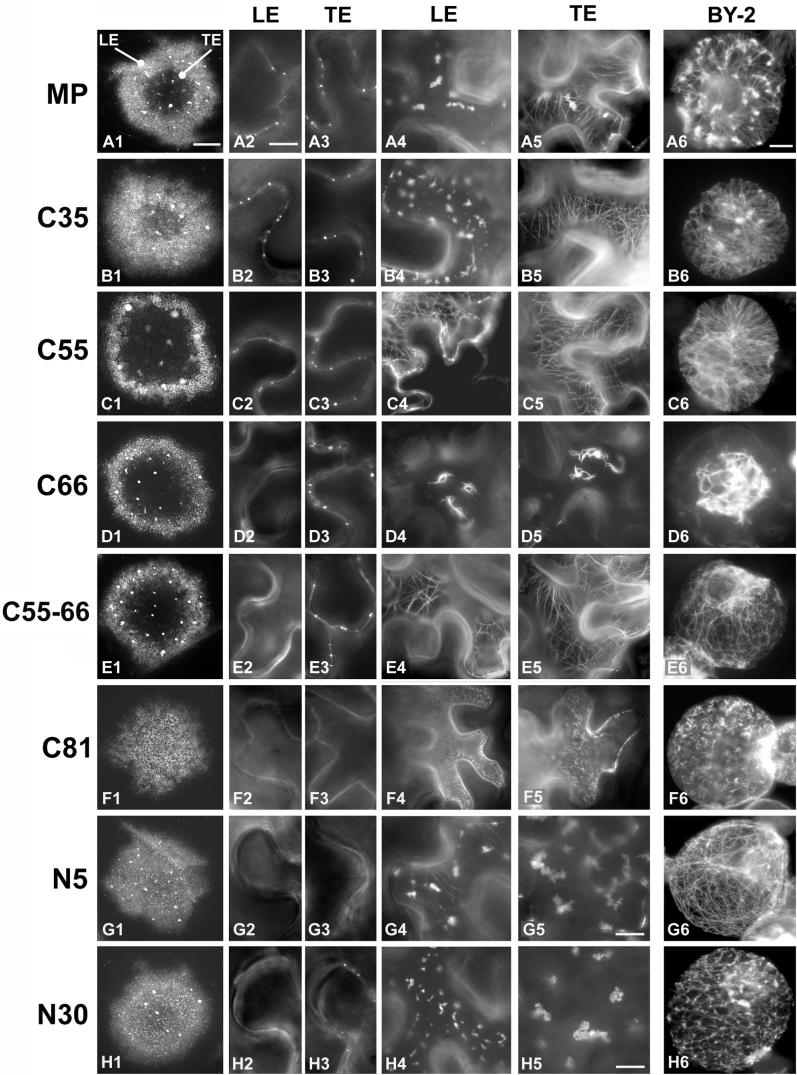
To correlate the activity profiles of mutant ΔMP:GFP proteins with their specific subcellular sites of accumulation, we analyzed infection sites formed at 22°C on leaves of N. benthamiana, a systemic host of TMV. As expected, viruses encoding C35:GFP or C55:GFP were infectious in N. benthamiana, whereas the other viral mutants were dependent on functional complementation by N. benthamiana transgenic for MP. Since we did not observe any difference in subcellular localization of MP:GFP, C35:GFP, and C55:GFP between infection sites on wild-type and MP-transgenic N. benthamiana plants, subcellular localization of each of the ΔMP:GFP mutants (Fig. 3) was examined in MP-transgenic plants. Nevertheless, to monitor possible effects of wild-type MP on the subcellular distribution of the ΔMP:GFP mutants, especially those that are impaired in facilitating vRNA movement, we compared the localization pattern of the proteins in N. benthamiana with their localization pattern in infected, nontransgenic BY-2 tobacco protoplasts. The time course of MP:GFP accumulation in epidermal cells and protoplasts has been described in detail elsewhere (25). Therefore, emphasis was given to the occurrence, absence, or modification of specific patterns, although a detailed time course analysis was performed for each of the viral constructs. Infected protoplasts were analyzed at 10, 14, 18, and 22 h postinfection; MP:GFP accumulation patterns in cells of infection sites on N. benthamiana leaves were carefully examined at high magnification, starting with cells at the outer leading edge and progressing toward cells within the center of the infection site.
FIG. 3.
Intracellular targeting of MP:GFP and of mutant ΔMP:GFP derivatives. Wild-type MP:GFP and mutant derivatives encoded by the virus used for infection are indicated on the left. Infection sites in MP-transgenic N. benthamiana are shown in column 1, and the subcellular distribution of the proteins in epidermal cells from these sites is shown in columns 2 to 5. Images in columns 2 and 3 show the presence or absence of the proteins in Pd of epidermal cell walls at the leading edge (LE) and trailing edge (TE) of infection; those in columns 4 and 5 show representative patterns of distribution of the proteins within the cytoplasm of cells at the LE and TE. Localization of the LE and TE of the ring-like infection site is shown in A1. In the cases of infection sites that appeared in the form of expanding disks (F1, G1, and H1), no TE could be defined and pictures were taken from cells close to the center of the infection site. Column 6 displays representative images of the subcellular distribution of the proteins in infected tobacco BY-2 protoplasts which are nontransgenic for MP. The size bar in A1 represents 1 mm and applies to all infection sites shown in column 1; the size bar in A2 represents 5 μm and applies to all images in columns 2 to 5 except for G5 and H5, where the size bar represents 2.5 μm; the size bar in A6 represents 10 μm and applies to all protoplasts shown in column 6.
Consistent with transient expression of MP early during infection (49), infection sites formed in N. benthamiana by virus producing fluorescent MP:GFP appear in the form of fluorescent rings (24, 25) (Fig. 3, A1). Like in previous studies (25), we observed that the MP:GFP localized to Pd in all cells of the infection site, including those at its center (Fig. 3, A2 and A3). Moreover, also as described previously, MP:GFP accumulated in inclusion bodies in cells at or near the leading edge of the infection site (Fig. 3, A4) and on microtubules in cells in the middle and at the trailing edge of the infection site (Fig. 3, A5). Association of MP:GFP with microtubules and inclusion bodies was also seen in infected protoplasts (Fig. 3, A6). Moreover, protoplasts displayed fluorescent puncta at or near the plasma membrane (25). However, because a clear relationship between these puncta in protoplasts and specific sites of MP:GFP accumulation in epidermal cells could not be established, we decided not to focus on the puncta.
Infection sites produced by virus encoding C35:GFP appeared as fluorescent rings (Fig. 3, B1) and thus were similar to those produced by virus encoding MP:GFP (Fig. 3, A1). The intracellular distribution of C35:GFP was also essentially like that of MP:GFP and indicated association of the protein with inclusion bodies at the leading edge of the ring (Fig. 3, B4), with microtubules at the trailing edge of the ring (Fig. 3, B5), as well as with Pd throughout the infection site (Fig. 3, B2 and B3). However, accumulation of C35:GFP in inclusion bodies was reduced (Fig. 3, B5) compared to MP:GFP (Fig. 3, A5). This was especially obvious in protoplasts, where the protein was mostly observed in association with microtubules (Fig. 3, B6). C55:GFP accumulated on microtubules (Fig. 3, C5) and in Pd (Fig. 3, C2 and C3) but not in inclusion bodies (Fig. 3, C4). Moreover, unlike MP:GFP and C35:GFP, C55:GFP was localized to microtubules throughout the infection site, even in cells at the very leading edge of infection (Fig. 3, C4 and C5). Similar findings were obtained in infected BY-2 protoplasts, where C55:GFP was predominantly found in association with microtubules throughout the time course of infection (Fig. 3, C6). Since C55:GFP is active in vRNA transport, these observations indicate that accumulation of MP:GFP in ER-derived inclusion bodies is not a requirement for its activity.
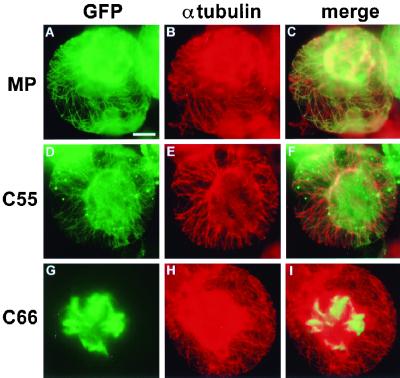
Virus encoding C66:GFP could form infection sites (Fig. 3, D1) only in N. benthamiana transgenic for MP. This was expected since this virus also was movement defective in N. tabacum cv. Xanthi NN and gave rise to infection and the formation of local lesions only in the presence of wild-type MP (Fig. 2). The functional deficiency of C66:GFP in vRNA movement was correlated with the lack of association of the protein with microtubules in epidermal cells of MP transgenic plants (Fig. 3, D4 and D5) as well as in nontransgenic protoplasts (Fig. 3, D6). Instead, the protein appeared in the form of thick bundles of short filaments that accumulated in the center of the cells (Fig. 3, D4 and D5; Fig. 4). To test if this aberrant localization of C66:GFP was caused by disruption of microtubules in infected cells, we stained infected protoplasts with antibodies directed against α-tubulin. As shown in Fig. 4, green fluorescence of MP:GFP (Fig. 4A) and C55:GFP (Fig. 4D) aligned rhodamine-labeled, red fluorescent microtubules (Fig. 4B and 4E) and gave rise to yellow-stained microtubules when images were merged (Fig. 4C and F). C66:GFP formed aberrant filaments (Fig. 4G) even though microtubules were intact (Fig. 4H and I), confirming that C66:GFP does not disrupt microtubules but is deficient in microtubule association. In addition to the formation of aberrant filamentous structures in the center of infected cells, C66:GFP was also characterized by its lack of accumulation in Pd of cells located at the leading front of infection (Fig. 3, D2). The protein accumulated normally in Pd of cells at the trailing edge and in the center of the infection site (Fig. 3, D3), showing that C66:GFP is not defective in Pd targeting per se. However, the deletion mutation in C66:GFP appears either to fully abolish Pd targeting of the protein early during infection or to reduce the rate of Pd targeting, thus prolonging the time required for accumulation and, as infection advances, visualization of the protein in Pd of cells located behind the infection front.
FIG. 4.
Intracellular distribution of MP:GFP (A to C), C55:GFP (D to F), and C66:GFP (G to I) in relation to the pattern of microtubules in infected BY-2 protoplasts. Microtubules were immunostained with antibody directed against α-tubulin followed by rhodamine-labeled secondary antibody. Bar = 10 μm. The strong red fluorescence in the center of protoplasts shown in panels B and H may be caused by insufficient fluorescence filtering and bleed-through from areas of very strong GFP fluorescence.
The finding that C55:GFP, but not C66:GFP, interacted with microtubules suggested that amino acids 203 to 213 may be involved in microtubule association of MP:GFP. To test this possibility, we constructed a virus in which only amino acids 203 to 213 were deleted from MP (C55-66). Like virus encoding C66:GFP, this virus was infectious only when inoculated onto plants expressing wild-type MP, where it formed normal ring-shaped infection sites (Fig. 3, E1). Unexpectedly, in epidermal cells as well as in protoplasts, we observed that C55-66:GFP, unlike C66:GFP, associated with microtubules (Fig. 3, E4, E5, and E6), indicating that amino acids 202 to 213 are not essential for microtubule association of MP. However, like C66:GFP, C55-C66:GFP did not accumulate in Pd at the leading edge of infection (Fig. 3, E2 and E3). We conclude that accumulation of MP in Pd behind the infection front is insufficient for intercellular transport of vRNA. In contrast, vRNA movement requires the presence of MP in Pd of cells at the leading front of infection.
Like C66:GFP, C81:GFP failed to associate with microtubules and formed short filamentous structures in epidermal cells (Fig. 3, F4 and F5) and protoplasts (Fig. 3, F6). However, in contrast to filaments formed by C66:GFP (Fig. 3, D4, D5, and D6), filaments formed by C81:GFP were more numerous, randomly distributed, and devoid of bundle formation (Fig. 3, F4, F5, and F6). More importantly, in contrast to C66:GFP, C81:GFP failed to accumulate in Pd anywhere within the infection site (Fig. 3, F2 and F3), suggesting that amino acids 188 to 202 contribute to Pd targeting of MP. Moreover, unlike virus expressing wild-type MP:GFP or virus expressing ΔMP:GFP with deletions of up to 66 amino acids from the C terminus, virus encoding C81:GFP produced disk-shaped rather than ring-shaped infection sites (Fig. 3, F1). This may suggest that a normal ring-shaped appearance of the infection site is linked to the ability of MP to target Pd.
Like C81:GFP, N5:GFP failed to accumulate in Pd throughout the infection site (Fig. 3, G2 and G3) and produced disk-shaped infection sites (Fig. 3, G1). However, in contrast to C81:GFP, this mutant had the ability to associate with microtubules (Fig. 3, G4 and G6) and inclusion bodies (Fig. 3, G5), indicating that these associations are not sufficient for Pd targeting of MP and for vRNA movement.
We noted that the intracellular distribution of N5:GFP differed between infected MP-transgenic epidermal cells and infected nontransgenic BY-2 protoplasts. In epidermal cells, N5:GFP tended to accumulate in the form of very large fluorescent inclusion bodies (Fig. 3, G5). Fluorescent filaments were seen only rarely and, if present, were aberrantly shaped (faintly visible in Fig. 3, G4). In contrast, in protoplasts N5:GFP was associated with microtubules and accumulated in inclusion bodies at much lower levels if at all (Fig. 3, G6). These findings suggest that N5:GFP may not be able to compete with the plant-encoded wild-type MP for binding sites on microtubules and therefore accumulates in inclusion bodies in epidermal cells. It may also be possible that N5:GFP and plant-encoded MP undergo an incompatible interaction in which wild-type MP and N5:GFP mutually interfere with microtubule association. Such a mechanism could contribute to MP-derived resistance developed by plants transgenic for a defective MP which lacks amino acids 3 to 5 (30; G. Kotlitzky, A. Katz, J. van der Laak, V. Boyko, M. Lapidot, R. N. Beachy, M. Heinlein, and B. L. Epel, submitted for publication) as is the case for N5:GFP.
N30:GFP was also impaired in efficient Pd accumulation (Fig. 3, H2 and H3) and produced disk-shaped infection sites (Fig. 3, H1). Like N5:GFP, this protein distributed differently between epidermal cells of MP-transgenic plants (Fig. 3, H4 and H5) and nontransgenic BY-2 protoplasts (Fig. 3, H6). However, the patterns observed in both systems indicate that N30:GFP has the capacity to accumulate in inclusion bodies as well as to interact with microtubules.
DISCUSSION
This study was undertaken to illuminate the mechanism by which TMV RNA is targeted to Pd for intercellular spread and to investigate the functional significance of the previously observed associations between TMV MP and subcellular components. We introduced N- and C-terminal deletion mutations into MP fused to GFP and correlated the activity of the resulting ΔMP:GFP mutants in mediating vRNA transport with subcellular localization. With regard to the effect of the deletion mutations on RNA transport, the MP:GFP fusion proteins behaved as expected from studies on unfused MP (1, 20). Thus, the GFP is unlikely to account for the subcellular distribution effects of the mutations reported here. ΔMP:GFP mutants with deletions of up to 55 amino acids from the C terminus of the MP were functional and allowed vRNA to move from cell to cell. In contrast, ΔMP:GFP mutants carrying either N-terminal deletions or deletions larger than 55 amino acids from the C terminus of MP were dysfunctional, and vRNAs produced neither lesions on NN tobacco plants nor fluorescent infection sites on leaves of N. benthamiana. The lack of infection sites was due to a defect in the vRNA transport function of ΔMP:GFP, since the movement-deficient viruses were able to replicate in protoplasts as well as in transgenic plants expressing functional MP. Using these TMV derivatives expressing functional and dysfunctional MP:GFP mutants together with immunolabeling of microtubules in protoplasts, this study shows that the vRNA transport function of the MP is tightly associated with specific targeting and accumulation of the protein to microtubules and to Pd at the leading edge of infection, thus supporting the involvement of these sites in intercellular transport of vRNA.
vRNA movement does not depend on accumulation of MP:GFP in ER inclusion bodies.
Deletion of the 35 C-terminal amino acids from MP had no effect on the function and subcellular localization of the protein. Deletion of 55 C-terminal amino acids also did not abolish the RNA transport function of the protein but led to a strong reduction, if not absence, of fluorescence in inclusion bodies. This finding indicates that accumulation of MP in ER-derived inclusion bodies proposed to represent sites of viral replication and protein synthesis (25, 34, 39) does not represent a functional requirement for intercellular movement of TMV RNA. The distribution of C55:GFP in infected cells was strikingly similar to the distribution of MP:GFP at elevated temperature (5). In both cases we observed that the lack of fluorescent bodies in cells at the leading edge of infection was complemented by the presence of highly fluorescent microtubules. In the case of MP:GFP, the association with microtubules in cells at the leading edge was correlated with a strong increase in the rate of intercellular spread of infection, supporting a role of microtubules in the trafficking of vRNA to Pd (5). These findings indicate that the distribution of MP:GFP between replication sites and microtubules is critical for the efficiency of viral movement. Since C55:GFP, in contrast to C35:GFP, accumulated predominantly on microtubules, this distribution appears to be controlled by determinants in MP, most likely by the domain encompassing amino acids 214 to 233. This domain is adjacent to C-terminal sequences that harbor phosphorylation sites (50) as well as host range determinants (17, 19). Moreover, in vivo phosphorylation in the C terminus of the MP was mapped to a peptide composed of amino acids 212 to 231 (23) as well as to Ser238 within the MP of the closely related tomato mosaic virus (28). The temperature-sensitive dynamics by which the MP associates with microtubules (5) following synthesis on the ER may thus be regulated by multiple phosphorylation and dephosphorylation events at the C terminus as well as by host-specific factors.
vRNA movement depends on accumulation of MP in Pd early in infection.
The observation that the inactivating N5 and N30 deletion mutations specifically interfered with the accumulation of the MP:GFP in Pd suggests that the N-terminal portion of the protein is involved in targeting or anchorage of MP to Pd and confirms that the presence of the protein in the cell wall channel is essential for intercellular movement of vRNA. The observations obtained by using these mutations also demonstrated that the ability of MP:GFP to accumulate in inclusion bodies and to interact with microtubules is insufficient for accumulation of the protein in Pd. In fact, microtubules appear not to be involved in the targeting of MP to Pd, as was shown by temperature-sensitive mutations in MP which interfere with microtubule association but not with Pd targeting at nonpermissive temperatures (4). In addition to the N5 and N30 mutations, accumulation of MP in Pd was also affected by the large C-terminal C66 and C81 deletion mutations, suggesting that almost all domains of MP, except the C-terminal amino acids 213 to 268, are required for Pd targeting or anchorage of the protein. It appears noteworthy that the C66 and C55-66 mutations interfered with the accumulation of MP:GFP in Pd at the infection front but not at the trailing edge and in the center of the infection site. It is possible that the mutations slow down the targeting of MP:GFP to Pd, thus requiring a longer time for detectable accumulation. However, based on this observation one could also speculate that targeting of MP to Pd is differentially regulated during early and late stages of infection and that the mutations specifically interfere with the targeting of MP:GFP to Pd early in infection. The results obtained with viruses encoding C55-C66:GFP and C66:GFP indicate that MP found in Pd only behind the infection front is unable to compensate for the lack of MP in Pd at the leading edge of infection. Thus, it is tempting to speculate that the protein may exist in two forms, an active form that is targeted to Pd early in infection and able to support viral movement and an inactive form which is targeted to Pd late in infection and not able to perform this function. This interpretation is consistent with microinjection studies which have shown that MP increases the SEL of Pd within the fluorescent halo of the infection site but not in its center (37). This view is also consistent with the proposal that MP which stays in Pd behind the infection front may be downregulated to reduce the level of interference of MP with normal intercellular communication (7). Regardless of whether active MP is replaced by inactive MP or active MP residing in Pd is downregulated, blocking Pd behind the infection front may represent an important strategy evolved by TMV to interfere with the spread of a virus-specific gene-silencing signal (40, 46) and therefore merits further investigation.
Involvement of microtubules in vRNA transport.
The mutants carrying N-terminal deletion mutations demonstrated that accumulation of MP:GFP in Pd is not a prerequisite for association of the protein with microtubules. Thus, the inability of C66:GFP to associate with microtubules is caused not by disruption of Pd accumulation at the leading edge of infection but rather by an independent effect of the mutation. In fact, the two effects could be separated when the C terminus of the MP was retained and only amino acids 203 to 213 of MP were deleted from MP:GFP (C55-66). It is intriguing that deletion of amino acids 203 to 268 interfered with microtubule association of the protein although neither the deletion of amino acids 203 to 213 nor the deletion of amino acids 214 to 268 had this effect. It thus appears that amino acids 203 to 268 do not provide a microtubule binding activity but that deletion of this stretch of the protein sequence affects a microtubule association domain located in another region of the MP. This conclusion is consistent with other studies indicating that microtubule association is mediated by the core region of the protein (4, 27). Nevertheless, the observation that the inactivating C66 mutation interferes with microtubule association of MP:GFP is consistent with a role of microtubules in intra-and intercellular transport of vRNA. A role of microtubules in vRNA movement is further supported by (i) temperature-sensitive mutations in MP that were used to directly correlate microtubule association of MP with its activity in intercellular movement of vRNA (4), (ii) a positive correlation between the amount of MP:GFP associated with microtubules and the efficiency of intercellular transport of vRNA (5), and (iii) the colocalization of viral genomic RNA with microtubules (34).
The appearance of the infection site is linked to the presence or absence of MP:GFP in Pd.
During infection, the tobamovirus MPs are transiently produced and then degraded (3, 16, 26, 31, 38, 44, 49), and a ring-shaped infection site develops because infection spreads radially, away from the origin of infection. We observed that several mutations in the MP gave rise to a change in the appearance of the infection site from the normal ring shape to a disk shape. Notably, disk-shaped infection sites are formed by viruses encoding defective MPs that have lost the ability to accumulate in Pd but still can associate with microtubules and accumulate in inclusion bodies (N5:GFP; N30:GFP, and C81:GFP). This suggests that the dynamics of MP production and degradation may be tightly linked to the successful targeting and anchorage of MP to Pd. Since the ability of MP to move by itself (48) likely contributes to its distribution within the infection site, it is also possible that MP:GFP mutants that have lost the ability to interact with Pd overaccumulate in infected cells, thus changing the appearance of the fluorescent infection site. Finally, ΔMP:GFP mutants that are not targeted to Pd may fail to downregulate Pd conductivity late in infection, thus perturbing the regulation of macromolecular trafficking, including that of vRNA.
ACKNOWLEDGMENTS
We thank Barbara Hohn, Witold Filipowicz, Ueli Grossniklaus, and Gertraud Orend for critically reading the manuscript before submission. We are also grateful to Sjoerd van Eeden and Markus Briker for continuous greenhouse support.
This work was supported by the Novartis Research Foundation.
REFERENCES
- 1.Berna A, Gafny R, Wolf S, Lucas W J, Holt C A, Beachy R N. The TMV movement protein: role of the C-terminal 73 amino acids in subcellular localization and function. Virology. 1991;182:682–689. doi: 10.1016/0042-6822(91)90609-f. [DOI] [PubMed] [Google Scholar]
- 2.Blackman L M, Overall R L. Immunolocalization of the cytoskeleton to plasmodesmata of Chara corallina. Plant J. 1998;14:733–741. [Google Scholar]
- 3.Blum H, Gross H J, Beier H. The expression of the TMV-specific 30-kDa protein in tobacco protoplasts is strongly and selectively enhanced by actinomycin. Virology. 1989;169:51–61. doi: 10.1016/0042-6822(89)90040-8. [DOI] [PubMed] [Google Scholar]
- 4.Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M. Function of microtubules in intercellular transport of plant virus RNA. Nat Cell Biol. 2000;2:826–832. doi: 10.1038/35041072. [DOI] [PubMed] [Google Scholar]
- 5.Boyko V, Ferralli J, Heinlein M. Cell-to-cell movement of TMV RNA is temperature-dependent and corresponds to the association of movement protein with microtubules. Plant J. 2000;22:315–325. doi: 10.1046/j.1365-313x.2000.00740.x. [DOI] [PubMed] [Google Scholar]
- 6.Carrington J C, Kasschau K D, Mahajan S K, Schaad M C. Cell-to-cell and long distance transport of viruses in plants. Plant Cell. 1996;8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Citovsky V. Tobacco mosaic virus: a pioneer of cell-to-cell movement. Philos Trans R Soc Lond B. 1999;354:637–643. doi: 10.1098/rstb.1999.0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citovsky V, Knorr D, Schuster G, Zambryski P. The P30 movement protein of tobacco mosaic virus is a single-stranded nucleic acid binding protein. Cell. 1990;60:637–647. doi: 10.1016/0092-8674(90)90667-4. [DOI] [PubMed] [Google Scholar]
- 9.Citovsky V, Wong M L, Shaw A L, Venkataram Prasad B V, Zambryski P. Visualization and characterization of tobacco mosaic virus movement protein binding to single-stranded nucleic acids. Plant Cell. 1992;4:397–411. doi: 10.1105/tpc.4.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson W O, Bubrick P, Grantham G L. Modifications of the tobacco mosaic virus coat protein gene affecting replication, movement, and symptomatology. Phytopathology. 1988;78:783–789. [Google Scholar]
- 11.Deom C M, Wolf S, Holt C A, Lucas W J, Beachy R N. Altered function of the tobacco mosaic virus movement protein in a hypersensitive host. Virology. 1991;180:251–256. doi: 10.1016/0042-6822(91)90029-b. [DOI] [PubMed] [Google Scholar]
- 12.Deom M C, Lapidot M, Beachy R N. Plant virus movement proteins. Cell. 1992;69:221–224. doi: 10.1016/0092-8674(92)90403-y. [DOI] [PubMed] [Google Scholar]
- 13.Ding B. Intercellular protein trafficking through plasmodesmata. Plant Mol Biol. 1998;38:279–310. [PubMed] [Google Scholar]
- 14.Ding B, Kwon M-O, Warnberg L. Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J. 1996;10:157–164. [Google Scholar]
- 15.Ding B, Kwon M O, Hammond R, Owens R. Cell-to-cell movement of potato spindle tuber viroid. Plant J. 1997;12:931–936. doi: 10.1046/j.1365-313x.1997.12040931.x. [DOI] [PubMed] [Google Scholar]
- 16.Epel B L, Padgett H S, Heinlein M, Beachy R N. Plant virus movement protein dynamics probed with a GFP-protein fusion. Gene. 1996;173:75–79. doi: 10.1016/0378-1119(95)00678-8. [DOI] [PubMed] [Google Scholar]
- 17.Fenczik C. Ph.D. thesis. St. Louis, Mo: Washington University; 1994. [Google Scholar]
- 18.Fenczik C A, Epel B L, Beachy R N. Role of plasmodesmata and virus movement proteins in spread of plant viruses. In: Verma D P S, editor. Signal transduction in plant development. New York, N.Y: Springer-Verlag; 1996. pp. 249–272. [Google Scholar]
- 19.Fenczik C A, Padgett H S, Holt C A, Casper S J, Beachy R N. Mutational analysis of the movement protein of odontoglossum ringspot virus to identify a host-range determinant. Mol Plant Microbe Interact. 1995;8:666–673. doi: 10.1094/mpmi-8-0666. [DOI] [PubMed] [Google Scholar]
- 20.Gafny R, Lapidot M, Berna A, Holt C A, Deom C M, Beachy R N. Effects of terminal deletion mutations on function of the movement protein of tobacco mosaic virus. Virology. 1992;187:499–507. doi: 10.1016/0042-6822(92)90452-u. [DOI] [PubMed] [Google Scholar]
- 21.Ghoshroy S, Lartley R, Sheng J, Citovsky V. Transport of proteins and nucleic acids through plasmodesmata. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:27–50. doi: 10.1146/annurev.arplant.48.1.27. [DOI] [PubMed] [Google Scholar]
- 22.Goodwin P B. Molecular size limit for movement in the symplast of the Eleodea leaf. Planta. 1983;157:124–130. doi: 10.1007/BF00393645. [DOI] [PubMed] [Google Scholar]
- 23.Haley A, Hunter T, Kiberstis P, Zimmern D. Multiple serine phosphorylation sites on the 30 kDa TMV cell-to-cell movement protein synthesized in tobacco protoplasts. Plant J. 1995;8:715–724. doi: 10.1046/j.1365-313x.1995.08050715.x. [DOI] [PubMed] [Google Scholar]
- 24.Heinlein M, Epel B L, Padgett H S, Beachy R N. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science. 1995;270:1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- 25.Heinlein M, Padgett H S, Gens J S, Pickard B G, Casper S J, Epel B L, Beachy R N. Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell. 1998;10:1107–1120. doi: 10.1105/tpc.10.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi S, Pleij C W A, Haenni A L, Chapeville F, Bosch L. Properties of the tobacco mosaic virus intermediate length RNA-2 and its translation. Virology. 1983;127:100–111. doi: 10.1016/0042-6822(83)90375-6. [DOI] [PubMed] [Google Scholar]
- 27.Kahn T W, Lapidot M, Heinlein M, Reichel C, Cooper B, Gafny R, Beachy R N. Domains of the TMV movement protein involved in subcellular localization. Plant J. 1998;15:15–25. doi: 10.1046/j.1365-313x.1998.00172.x. [DOI] [PubMed] [Google Scholar]
- 28.Kawakami S, Padgett H S, Hosokawa D, Okada Y, Beachy R N, Watanabe Y. Phosphorylation and/or presence of serine 37 in the movement protein of tomato mosaic tobamovirus is essential for intracellular localization and stability in vivo. J Virol. 1999;73:6831–6840. doi: 10.1128/jvi.73.8.6831-6840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kühn C, Franceschi V R, Schulz A, Lemoine R, Frommer W B. Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science. 1997;275:1298–1300. doi: 10.1126/science.275.5304.1298. [DOI] [PubMed] [Google Scholar]
- 30.Lapidot M, Gafny R, Ding B, Wolf S, Lucas W J, Beachy R N. A dysfunctional movement protein of tobacco mosaic virus that partially modifies the plasmodesmata and limits spread in transgenic plants. Plant J. 1993;4:959–970. [Google Scholar]
- 31.Lehto K, Bubrick P, Dawson W O. Time course of TMV 30k protein accumulation in intact leaves. Virology. 1990;174:290–293. doi: 10.1016/0042-6822(90)90077-5. [DOI] [PubMed] [Google Scholar]
- 32.Lucas W J, Bouche-Pillon S, Jackson D P, Nguyen L, Baker L, Ding B, Hake S. Selective trafficking of KNOTTED1 homeodomain protein and its RNA through plasmodesmata. Science. 1995;270:1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- 33.Lucas W J, Gilbertson R I. Plasmodesmata in relation to viral movement within leaf tissues. Annu Rev Phytopathol. 1994;32:387–411. [Google Scholar]
- 34.Mas P, Beachy R N. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement in intracellular distribution of viral RNA. J Cell Biol. 1999;147:945–958. doi: 10.1083/jcb.147.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean B G, Zupan J, Zambryski P C. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco plants. Plant Cell. 1995;7:2101–2114. doi: 10.1105/tpc.7.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mezitt L A, Lucas W J. Plasmodesmal cell-to-cell transport of proteins and nucleic acids. Plant Mol Biol. 1996;32:251–273. doi: 10.1007/BF00039385. [DOI] [PubMed] [Google Scholar]
- 37.Oparka K J, Prior D A M, Santa Cruz S, Padgett H S, Beachy R N. Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus. Plant J. 1997;12:781–789. doi: 10.1046/j.1365-313x.1997.12040781.x. [DOI] [PubMed] [Google Scholar]
- 38.Padgett H S, Epel B L, Kahn T W, Heinlein M, Watanabe Y, Beachy R N. Distribution of tobamovirus movement protein in infected cells and implications for cell-to-cell spread of infection. Plant J. 1996;10:1079–1088. doi: 10.1046/j.1365-313x.1996.10061079.x. [DOI] [PubMed] [Google Scholar]
- 39.Reichel C, Beachy R N. Tobacco mosaic virus infection induces severe morphological changes of the endoplasmatic reticulum. Proc Natl Acad Sci USA. 1998;95:11169–11174. doi: 10.1073/pnas.95.19.11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz M T, Voinnet O, Baulcombe D C. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siemering K R, Golbick R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- 42.Takamatsu K, Ishikawa M, Meshi T, Okada Y. Expression of bacterial chloramphenicol acetyltransferase gene in tobacco plants mediated by TMV-RNA. EMBO J. 1987;6:307–311. doi: 10.1002/j.1460-2075.1987.tb04755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terry B R, Robards A W. Hydrodynamic radius alone governs the mobility of molecules through plasmodesmata. Planta. 1987;171:145–157. doi: 10.1007/BF00391090. [DOI] [PubMed] [Google Scholar]
- 44.Tomenius K, Clapham D, Meshi T. Localization by immunogold cytochemistry of the virus-coded 30K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology. 1987;160:363–371. doi: 10.1016/0042-6822(87)90007-9. [DOI] [PubMed] [Google Scholar]
- 45.Tucker E B. Translocation in the staminal hairs of Setcreasea purpurea. I. Study of cell ultrastructure and cell-to-cell passage of molecular probes. Protoplasma. 1982;113:193–201. [Google Scholar]
- 46.Voinnet O, Baulcombe D C. Systemic signalling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- 47.Voinnet O, Vain P, Angell S, Baulcombe D C. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- 48.Waigmann E, Lucas W, Citovsky V, Zambryski P. Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc Natl Acad Sci USA. 1994;91:1433–1437. doi: 10.1073/pnas.91.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe Y, Emori Y, Ooshika I, Meshi T, Ohno T, Okada Y. Synthesis of TMV-specific RNAs and proteins at the early stage of infection in tobacco protoplasts: transient expression of 30k protein and its mRNA. Virology. 1984;133:18–24. doi: 10.1016/0042-6822(84)90421-5. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe Y, Meshi T, Okada Y. In vivo phosphorylation of the 30-kDa protein of tobacco mosaic virus. FEBS Lett. 1992;313:181–184. doi: 10.1016/0014-5793(92)81440-w. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe Y, Ohno T, Okada Y. Virus multiplication in tobacco protoplasts inoculated with tobacco mosaic virus RNA encapsulated in large unilamellar vesicle liposomes. Virology. 1982;120:478–480. doi: 10.1016/0042-6822(82)90048-4. [DOI] [PubMed] [Google Scholar]
- 52.White R G, Badelt K, Overall R L, Vesk M. Actin associated with plasmodesmata. Protoplasma. 1994;180:169–184. [Google Scholar]
- 53.Wolf S, Deom C M, Beachy R N, Lucas W J. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science. 1989;246:377–379. doi: 10.1126/science.246.4928.377. [DOI] [PubMed] [Google Scholar]
- 54.Zambryski P. Plasmodesmata: plant channels for molecules on the move. Science. 1995;270:1943–1944. doi: 10.1126/science.270.5244.1943. [DOI] [PubMed] [Google Scholar]