Abstract
Among a myriad of putative functions assigned to the hepatitis C virus (HCV) core protein, several studies suggest that it may modulate internal ribosome entry site (IRES)-mediated initiation of translation. We compared the translational activity of dicistronic reporter transcripts containing the HCV IRES within the intercistronic space fused to downstream sequence encoding either 22 amino acids (aa) or 173 aa of the core protein. The inclusion of the nearly full-length core protein-coding sequence significantly suppressed translation in vitro and in transfected HepG2 cells. However, this suppression was not eliminated by frameshift mutations introduced into the core sequence, suggesting that it occurred at the RNA level and not as a result of core protein expression in cis. Similarly, the expression of core protein (aa 1 to 191) in trans from a recombinant baculovirus did not suppress IRES-directed translation from any of these transcripts in transfected Huh-7 cells. While core protein expression did decrease IRES activity in HepG2 cells (up to 79% suppression), the expression of β-galactosidase from a control baculovirus also suppressed IRES activity (up to 56%), strongly suggesting that this suppression was nonspecific. Finally, the addition of purified recombinant core protein (aa 1 to 179) to in vitro translation reactions at concentrations up to a 10-fold molar excess over the RNA transcripts resulted in no significant reduction in IRES activity. Consistent with these results, a gel retention assay indicated no difference in the affinities of the recombinant HCV core protein and a recombinant Venezuelan equine encephalitis virus capsid protein for HCV IRES-containing RNA transcripts. We conclude that while the inclusion of core protein-coding sequence downstream of the IRES may reduce the efficiency of cap-independent translation on HCV RNA, the core protein itself has no biologically relevant activity in modulating HCV IRES activity.
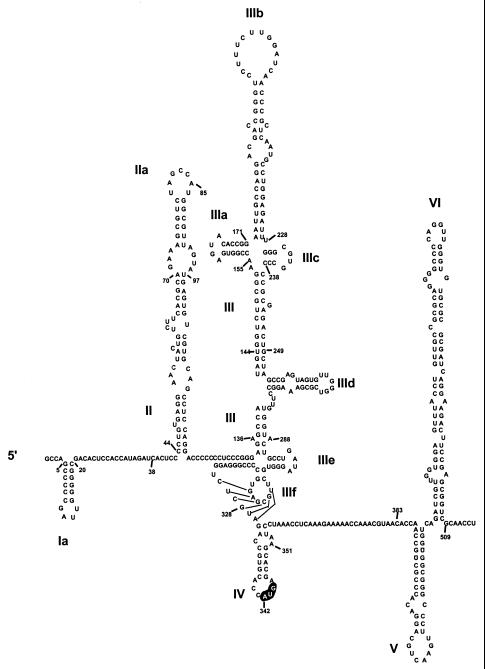
Hepatitis C virus (HCV), the pestiviruses, and GB virus B (GBV-B) are members of the family Flaviviridae that utilize a cap-independent mechanism to initiate translation on their genomic RNAs. This process involves an internal ribosome entry site (IRES) located in the 5′ nontranslated RNA (NTR), as well as both canonical and noncanonical translation initiation factors (12, 36, 45, 46, 59). The 5′ NTRs of these positive-strand RNA viruses range from 341 nucleotides (nt) in length in HCV to 445 nt in GBV-B and possess similar RNA secondary structures that can be divided into four major structural domains (Fig. 1). Domains II and III are complex stem-loop structures, the latter including an RNA pseudoknot, that are both required for efficient internal ribosome entry (17, 45, 46, 58). Domain IV consists of a stem-loop structure located immediately downstream of the pseudoknot in the genomic RNAs of HCV and GBV-B. It contains the polyprotein translation initiation codon within the loop sequence and has been proposed only for HCV and GBV-B (Fig. 1) (15, 56). A similar structure does not appear to be present in the genomic RNAs of the pestiviruses (15, 55), and this structure is not essential for translation of HCV or GBV-B RNA.
FIG. 1.
Proposed model of secondary and tertiary RNA structures within the 5′ NTR and the immediate downstream region of HCV-N strain, genotype 1b (18). The major structural domains are indicated by Roman numerals, while the authentic initiator AUG codon is highlighted at nt 342 in domain IV. The presence of domains V and VI was suggested by Smith and Simmonds (56).
Available evidence suggests that an early step in the internal initiation of translation on these viral RNAs involves a direct interaction between the 40S subunit and the viral RNA that is not dependent upon any canonical translation initiation factors (35). The 40S subunit interacts with HCV RNA at the site of the initiator AUG (domain IV) in the absence of any prior scanning of the ribosome along the RNA (15, 42), and mutations that enhance the predicted stability of the domain IV stem-loop adversely affect the rate of translation of both HCV and GBV-B RNAs (15, 44). Thus, it seems likely that the equilibrium between the folded and unfolded conformations of stem-loop IV is an important factor in controlling the interaction of the ribosome subunit with the RNA and thus in determining the efficiency of internal initiation of translation on HCV and GBV-B RNAs. It has been suggested that the cellular La autoantigen interacts with HCV RNA in the region of this stem-loop and that this interaction facilitates translation (1). However, a more attractive hypothesis is that the stem-loop IV RNA segment might specifically bind a product of viral translation, thereby down-regulating translation in a manner that would foster viral persistence (15).
The HCV core protein is the most N-terminal protein of the viral polyprotein, whose cleavage from the nascent polyprotein is mediated by host-signal peptidase (8, 13, 60). In addition to the mature full-length core protein, smaller C-terminally truncated products have been observed in transfected mammalian cells expressing this protein (49). Core protein is localized to both the cytoplasm and perinuclear regions of the cell (6, 25, 37). It is an RNA-binding protein, with RNA-binding domains localized to the N-terminal 75 amino acids (aa), a region rich in basic amino acids (49). It is a good candidate for a viral protein that might interact with the IRES. However, while Fan et al. (11) reported a specific interaction between the core protein and the HCV 5′ NTR, Santolini et al. (49) found that the RNA-binding activity of the HCV core protein is not specific for HCV RNA. Nonetheless, a specific interaction between the HCV core protein and the 5′ NTR was claimed recently by Shimoike et al. (52), who also suggested that there is specific suppression of HCV IRES-directed translation in cells expressing the core protein. If correct, this would add viral translational regulation to a lengthy list of putative core protein functions that include, among others, the regulation of cellular transcription, the modulation of cellular signal transduction pathways, cellular transformation, and immunosuppression (7, 21, 23, 30, 34, 38–41, 54, 57, 61), in addition to the role this protein seems certain to play in viral assembly and morphogenesis.
These observations have led us to investigate in greater detail whether the HCV core protein plays a role in IRES-directed translation. Our results indicate that the inclusion of the nearly full-length core protein-coding sequence downstream of the HCV IRES substantially reduces the efficiency of IRES-directed translation. However, in contrast to the recent report from Shimoike et al. (52), we show that the in vivo expression of core protein, either in cis or in trans, does not result in a specific inhibition of IRES-directed translation from dicistronic reporter RNAs. Moreover, we demonstrate that the core protein of HCV does not differ from the similarly basic capsid protein of Venezuelan equine encephalitis virus in terms of its ability either to interact with HCV RNA or to inhibit HCV IRES-dependent translation in a cell-free translation system. We conclude from these studies that translational regulation is not a biologically relevant property of the HCV core protein.
MATERIALS AND METHODS
Cells.
Huh-7 human hepatocellular carcinoma cells were maintained in Dulbecco's modified Eagle's medium. HepG2 cells were maintained in modified Eagle's minimum essential medium (Sigma) with l-glutamine, nonessential amino acids (Gibco-BRL), and sodium bicarbonate at concentrations recommended by the American Type Culture Collection. Sf9 insect cells were maintained in spinner flask culture with Grace's medium (Gibco-BRL) supplemented with yeastolate, insect hemolymph, and 0.2% Pluronic F-68 solution (Sigma). All media were supplemented with 10% fetal bovine serum, penicillin, and streptomycin (Gibco-BRL).
Plasmids.
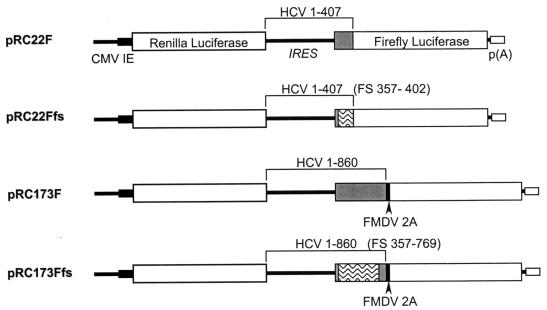
The dicistronic reporter construct pRC22F was previously referred to as pRL-HL (16). It contains the Renilla luciferase (RLuc) coding sequence under the control of a composite T7-cytomegalovirus (CMV) promoter, followed by the 5′ NTR of a genotype 1b HCV (HCV-N) and 66 nt of core protein-coding sequence fused in-frame directly with the firefly luciferase (FLuc) sequence. pRC22Ffs is identical to pRC22F except for two frameshift mutations, the removal of A at nt 357 and the addition of T after nt 402 of the HCV sequence, both inserted by PCR site-directed mutagenesis. These two frameshift mutations result in a nonsense protein sequence spanning from aa 5 to the end of the core-coding sequence. The plasmid pRC173F is similar to pRC22F but contains core sequence extending to HCV sequence nt 860, fused in-frame with sequence encoding the foot-and-mouth disease virus (FMDV) 2A proteinase, followed by FLuc. pRC173Ffs has two frameshift mutations: the removal of A at nt 357 (as in pRC22Ffs) and the addition of a C after nt 766, inserted by Quick-Change (Stratagene) site-directed mutagenesis. These mutations place approximately 80% of the core protein sequence out of frame and result in a protein that is largely nonsense sequence.
Recombinant baculovirus.
The recombinant baculoviruses AcCACore, AcCALacZ, and AcCAG express the HCV core protein (aa 1 to 191) or β-galactosidase protein under control of the composite mammalian CAG promoter or contain the CAG promoter only, respectively (53). These were a gift from Yoshiharu Matsuura of NIH, Japan. Recombinant baculoviruses were amplified in Sf9 cells using standard protocols (32) and were concentrated as follows. Seventy-two hours following inoculation, infected Sf9 cultures were collected and centrifuged at 6,000 × g for 15 min at 4°C to remove cell debris. Baculovirus contained in the supernatant was concentrated by ultracentrifugation through a 25% sucrose cushion and was resuspended in phosphate-buffered saline (PBS) at concentrations near 1010 PFU/ml. The titer of recombinant baculoviruses was determined by plaque assay on Sf9 cells. Briefly, 8 × 105 Sf9 cells per well were seeded into six-well plates. Serial dilutions of the concentrated baculovirus stocks were inoculated onto Sf9 cells in individual wells. The viral inoculum was removed after 1 h, and 2 ml of agarose overlay (0.5% SeaKem agarose and 2× complete Grace's medium; Gibco-BRL) was added. After the cultures were incubated for 3 to 5 days at 27°C, the cell monolayers were stained overnight with neutral red. Plaques were visible as clear areas surrounded by stained cells.
Assessment of HCV translation in vivo.
Huh-7 or HepG2 cells grown in six-well plates were transfected with plasmid DNAs when they were ∼70% confluent. Cells were washed with OptiMEM (Gibco-BRL) twice prior to transfection. Lipofectin-mediated transfection reactions were prepared according to manufacturer protocols (Gibco-BRL). For each well of a six-well plate, 15 μl of Lipofectin reagent (Gibco-BRL) was mixed with 1 μg of plasmid DNA in a total of 200 μl of OptiMEM and was incubated for 15 min at room temperature before the mixture was added to the cells. OptiMEM (800 μl) was added immediately after the addition of the Lipofectin-DNA mixture. For experiments involving protein expression from recombinant viruses, cells were subsequently incubated at 37°C for 24 h before infection. Huh-7 or HepG2 cells from a test well were trypsinized, removed, and counted to determine the quantity of baculovirus required for infection. The baculovirus was then added to the remaining wells at a multiplicity of infection (MOI) of 100. Following a 1-h virus adsorption period, 1.5 ml of complete medium was added to the cells without removal of the virus inoculum. Seventy-two hours posttransfection, cells were washed with 1× PBS and the media were replaced with 0.5 ml of PBS. Cells were then scraped, collected, and pelleted by centrifugation before being lysed with the passive lysis buffer provided with the dual luciferase assay (DLA; Promega). The luciferase reporter activities were determined by DLA and measured in a luminometer. All experiments were carried out with triplicate samples.
Antibodies and immunofluorescence staining of transfected cells.
Murine monoclonal antibody to the HCV core protein was generously provided by Johnson Lau of the Schering-Plough Research Institute, Kenilworth, N.J. Fluorescein isothiocyanate-labeled goat anti-mouse antibody (Sigma) was used as the secondary antibody for immunofluorescence. Cells were grown on glass coverslips in six-well plates and were air dried and fixed with 1:1 methanol-acetone solution. Coverslips were incubated with the murine monoclonal antibody at a 1:350 dilution for 1 h, washed three times with PBS, incubated with the goat anti-mouse antibody at a dilution of 1:64 for 30 min, washed, and then counterstained with DAPI (4′,6′-diamidino-2-phenylindole) (Molecular Probes) for 5 min to visualize nuclei. The coverslips were inverted onto glass slides with Vectashield immunofluorescence mounting fluid (Vector Laboratories, Inc.) and were examined under a Zeiss fluorescence microscope.
In vitro transcription and translation.
Dicistronic plasmids were linearized at the Acc65 I site located immediately downstream of the FLuc sequence. In vitro transcribed RNAs were synthesized using the T7 MEGAscript in vitro transcription kit (Ambion) and were quantitated by agarose gel electrophoresis. RNA transcripts were used to program the Flexi rabbit reticulocyte lysate system (Promega) at 12.5 μg/ml per 10 μl of translation reaction. Reactions were supplemented with HCV core protein (aa 1 to 179) or VEE capsid protein (aa 1 to 275), expressed in Escherichia coli, and purified to near homogeneity under denaturing conditions. These proteins were generously provided by Stanley Watowich of the University of Texas Medical Branch (unpublished data). The translation reactions were carried out at 30°C for 1.5 h, and products were separated by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS–12% PAGE). Reaction products were quantitated by PhosphorImager analysis (Molecular Dynamics).
Electrophoretic mobility shift assay.
An RNA probe (HCV nt 1 to 770) was synthesized by runoff transcription using as template the plasmid pMN2-1G (15), which was linearized by AvrII. Transcription was carried out with [α-32P]CTP (800 Ci/mmol) using the MEGAscript T7 in vitro transcription kit (Ambion) reagents. After 2 h of reaction time, the transcription mix was digested with RNase-free DNase I at 37°C for 15 min. At this point, trichloroacetic acid precipitation was carried out to assess the specific activity of the probe. Transcripts were then extracted with phenol-chloroform, precipitated with ammonium acetate and isopropanol, and resuspended in nuclease-free water. Binding reactions were adapted from the conditions described by Schultz et al. (50). Each reaction (10 μl) contained ∼0.5 pmol of radiolabeled probe (2.5 × 105 cpm), 2 μg of yeast tRNA, 1 U of RNasin (Promega), and the appropriate amount of purified recombinant core protein, VEE capsid protein, or bovine serum albumin (BSA) for the indicated RNA-to-protein molar ratios. The final binding reactions contained 20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10% (vol/vol) glycerol, and 0.4 mM dithiothreitol with 25 mM KCl (50). Binding reactions were incubated at room temperature for 10 min before electrophoresis through a nondenaturing 0.8% agarose gel containing 1× TBE (90 mM Tris borate, 2 mM EDTA) at 50 V until the bromophenol blue dye front reached the end of the gel. The gel was dried and subjected to PhosphorImager analysis.
RESULTS
Core protein-coding sequence suppresses HCV IRES-directed translation in vitro.
Previous reports have suggested that the inclusion of a minimal length of core protein-coding sequence downstream of the IRES is required either for efficient cap-independent translation of reporter proteins (43) or for the replication of a chimeric poliovirus in which the HCV IRES replaced the picornavirus translation element (26). In at least the latter case, it was suggested that this might reflect a specific transactivating effect of the core protein. On the other hand, a very recent report suggests that core protein may specifically suppress translation initiated by the HCV IRES (52). To assess the influence of core protein expressed in cis from RNAs that contain the HCV IRES, we used pRC22F and constructed plasmids pRC22Ffs, pRC173F, and pRC173Fts, all of which contain dicistronic transcriptional units, as shown in Fig. 2. Transcripts are produced from each of these plasmids under the control of a composite CMV-T7 promoter, and they contain the RLuc sequence as the upstream cistron and the FLuc sequence as the downstream cistron. The translation of FLuc from these transcripts is dependent on the HCV IRES sequence placed in the intercistronic space, while RLuc, translated from the upstream cistron, serves as an internal control for transcript abundance. These four plasmids differ with respect to the HCV core protein-coding sequence that is fused naturally to the 5′ NTR, just upstream of the FLuc sequence. These sequences encode either the amino-terminal 22 aa (66 nt) or 173 aa (519 nt) of the core protein in pRC22F and pRC173F, respectively, as indicated in Fig. 2. pRC22Ffs and pRC173Ffs contain similar lengths of the core protein-coding sequence, but each contains paired frameshift mutations that result in a significant length of the core protein being translated as a nonsense sequence out of the wild-type reading frame. For pRC173F and pRC173Ffs, a 20-aa-long FMDV 2A autoproteolytic sequence (10) was introduced between the core and FLuc sequences in order to release the FLuc product from the lengthier HCV core protein.
FIG. 2.
Schematic display of dicistronic reporter plasmids containing the RLuc and FLuc sequences as upstream and downstream reporters flanking various lengths of HCV sequences containing the 5′ NTR. The core sequences are represented as shaded boxes. The frameshifted regions are represented as wavy boxes. Numbers refer to the nucleotide sequence of HCV.
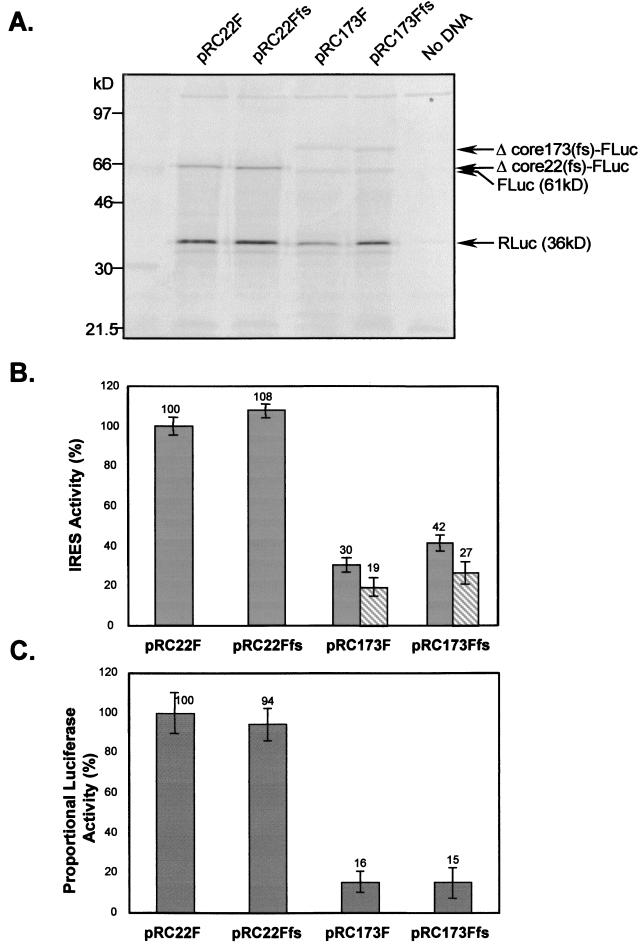
To assess the influence of the different downstream HCV protein-coding sequences on IRES-directed translation, we programmed in vitro coupled transcription-translation reactions with each of the four plasmids shown in Fig. 2. Figure 3A shows the SDS-PAGE analysis of the translation products, which clearly demonstrated the presence of each of the expected proteins. The pRC22F and pRC22Ffs plasmids resulted in the expression of both RLuc (36 kDa) and the fusion protein, Δcore22-FLuc (∼64 kDa). In addition to the expected RLuc protein, the products from pRC173F and pRC173Ffs included both the FLuc protein (61 kDa) produced by FMDV 2A-mediated cleavage and the cleavage precursor protein, Δcore173-FLuc (∼83 kDa). The nonsense core products produced from the frameshift mutants migrated slightly more rapidly than the native protein produced from both sets of plasmids. Cleavage of the Δcore173-FLuc and Δcore173(fs)-FLuc products at the FMDV 2A site was approximately 50% efficient. Thus, all four constructs expressed the expected products, indicating that they contain a functional HCV IRES. An interesting feature of the results shown in Fig. 3A, however, was the apparent lower abundance of the Δcore173-FLuc and Δcore173(fs)-FLuc products produced from pRC173F and pRC173Ffs compared with the Δcore22-FLuc and Δcore22(fs)-FLuc produced from pRC22F and pRC22Ffs, despite comparable amounts of RLuc produced from each. This indicates that the presence of the lengthier core-coding sequences in pRC173F and pRC173Ffs resulted in a significant attenuation of IRES-directed translation. These differences were not due to premature termination of transcription in the longer transcripts in these coupled transcription-translation reactions, as similar results were obtained in cell-free translation reactions programmed with RNAs prepared in separate in vitro transcription reactions and analyzed by SDS-PAGE (data not shown).
FIG. 3.
Products of in vitro coupled transcription-translation reactions programmed with the four dicistronic plasmids pRC22F, pRC22Ffs, pRC173F, and pRC173Ffs. (A) Equal amounts of DNA templates were used to program coupled transcription-translation reactions. Translation products were analyzed in an SDS–12% PAGE gel. Products are indicated by arrows on the right. (B) The in vitro products were quantitated by PhosphorImager analysis, and the IRES activity of each construct was determined by calculating the abundance of FLuc relative to RLuc, with that of pRC22F normalized to 100%. Shaded bars indicate IRES activity determined by measuring the abundance of both cleaved and uncleaved Fluc-containing products relative to RLuc. Hatched bars indicate IRES activity determined by measuring the abundance of only cleaved FLuc relative to RLuc. Each bar represents the average IRES activity of triplicate translation reactions. (C) IRES activity estimated from proportional luciferase activity (FLuc/RLuc) of in vitro transcription-translation products, with that of pRC22F normalized to 100%. Each bar represents the average IRES activity based on RLuc and FLuc activities from triplicate translation samples. (B and C) Error bars indicate the standard deviations of triplicate samples.
To better assess the extent to which the inclusion of the lengthier core-coding sequence affected translation, we quantified the translation products by PhosphorImager analysis (Fig. 3B). We estimated the relative activities of the IRES elements in pRC22F and pRC22Ffs by calculating the ratio of the Δcore22-FLuc to RLuc products produced in reactions programmed with these plasmids. The IRES activities of pRC173F and pRC173Ffs were similarly estimated by calculating the ratio of FLuc plus Δcore173-FLuc (or FLuc alone) to RLuc. To compare these constructs, the IRES activity of pRC22F was arbitrarily considered to be 100%, and adjustments were made for the number of Met residues in the translation products: 14 for pRC22F and pRC22Ffs and 18 for pRC173F and pRC173Ffs. This analysis indicated that the activity of the IRES in pRC173F was only 30% that in pRC22F (taking into account both the cleaved and uncleaved FLuc products from pRC173F) (Fig. 3B). Thus, the inclusion of the additional core-coding sequence in pRC173F substantially reduced IRES activity. The suppressive effect appeared to be related primarily to the inclusion of the additional RNA sequence and not to the expression of core protein, since there was little difference between the activity of pRC173F (30%) and pRC173Ffs (42%), which encodes a nonsense protein that shares only about 20% sequence identity with core protein (Fig. 2). Since pRC173F produces a nearly full-length core protein from the IRES-containing RNA, these results suggest that the expression of the core protein in cis neither transactivates nor down-regulates HCV IRES activity in this cell-free translation system.
The results of luciferase assays carried out on the products of these in vitro translation reactions are shown in Fig. 3C. Here, IRES activity is presented as the ratio of FLuc to RLuc activities, relative to that of pRC22F. This analysis confirmed that the transcripts of pRC22F (100%) and pRC22Ffs (94%) have comparable IRES activities (Fig. 3C), as shown by the PhosphorImager analysis (Fig. 3B). The enzymatic assays also confirmed a substantially lower IRES activity in transcripts derived from pRC173F (16%) or pRC173Ffs (15%) (Fig. 3C). However, these last two results were more consistent with the PhosphorImager quantitation of the cleaved FLuc product only (19 and 27%, respectively; Fig. 3B) than with the PhosphorImager analysis of the total products of translation (30 and 42%, respectively; Fig. 3B). This suggests that the cleavage precursors, Δcore173-FLuc and Δcore173fs-FLuc, have substantially reduced luciferase activities.
Expression of core protein in cis does not influence IRES-directed translation in transfected cells.
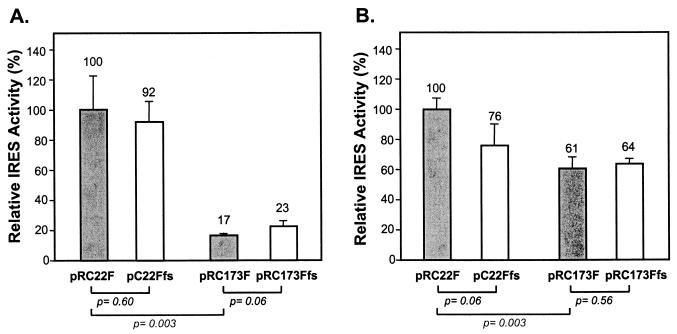
To determine whether the core protein-coding sequences included in the dicistronic constructs shown in Fig. 2 would similarly influence the activity of the IRES in vivo, we transfected two different human hepatocyte-derived cell lines, Huh-7 and HepG2, with these plasmids. The cells were harvested 72 h following transfection, and both reporter protein activities were determined by luciferase assay. The results in HepG2 cells (Fig. 4A) closely mimicked those obtained in the cell-free translation system (compare Fig. 4A and 3C). Compared with the relative luciferase activities expressed from pRC22F (100%), the translational activities of the pRC22Ffs, pRC173F, and pRC173Ffs transcripts were 92, 17, and 23%, respectively. Since the comparison of the quantitative PhosphorImager analysis and enzymatic assays carried out on in vitro translation products suggested that uncleaved Δcore173-FLuc fusion proteins have little luciferase activity (Fig. 3), these enzyme assays of cell lysates are likely to have overestimated the degree of suppression of IRES activity caused by the additional core protein-coding sequences in pRC173F and pRC173Ffs. Nonetheless, they show that the additional core sequences in pRC173F and pRC173Ffs reduce IRES activity in HepG2 cells to an extent similar to that observed in vitro. A statistical analysis indicated that these differences were highly significant (P < 0.005). As in the in vitro translation experiments, the suppression of translation appeared to be due to the inclusion of the additional RNA sequences and not to the expression of a specific protein. There was no significant difference between the IRES activities of pRC173F and pRC173Ffs (P > 0.05).
FIG. 4.
The relative IRES activities of dicistronic reporter transcripts produced in vivo following transfection of HepG2 (A) or Huh-7 (B) cells. Cells were transfected with equal amounts of the reporter plasmids and assayed for luciferase activity 72 h later. The IRES activity of each construct was determined as described in Results, with that of the pRC22F set at 100%. Error bars indicate the standard deviations of triplicate samples.
Qualitatively similar results were obtained in transfected Huh-7 cells (Fig. 4B). However, although the inclusion of additional core protein-coding sequence in pRC173F and pRC173Ffs resulted in a statistically significant reduction in the ratio of FLuc to RLuc activities produced relative to pRC22F (P < 0.005), this suppressive effect was quantitatively less than in the HepG2 cells. The relative luciferase activities expressed from the pRC173F and pRC173Ffs transcripts were 61 and 64% that of pRC22F, respectively. These differences may reflect only the reduced specific enzymatic activities of the uncleaved fusion proteins produced from these constructs. More importantly, however, there was no difference in the proportional RLuc and FLuc activities of Huh-7 cells transfected with pRC173F or pRC173Ffs, suggesting again that the expression of core protein in cis has no effect on translation in vivo.
Expression of core protein in trans in transfected cells.
The preceding experiments demonstrate that the inclusion of core-coding sequence significantly reduces the IRES activity of dicistronic reporter transcripts, at least in rabbit reticulocyte lysates and HepG2 cells, and suggest that the core protein does not have any IRES-modulating activity. To further investigate this possibility, we transfected the reporter plasmids into HepG2 and Huh-7 cells. Transfected cells were subsequently infected with AcCACore, a recombinant baculovirus that expresses the core protein under the transcriptional control of the composite CAG mammalian promoter. To determine whether any effects on translation were related specifically to expression of the core protein, we also transfected the reporter constructs into cells which were subsequently infected with AcCAG, a baculovirus containing the CAG promoter only, and AcCALacZ, a baculovirus that expresses β-galactosidase under the control of this promoter.
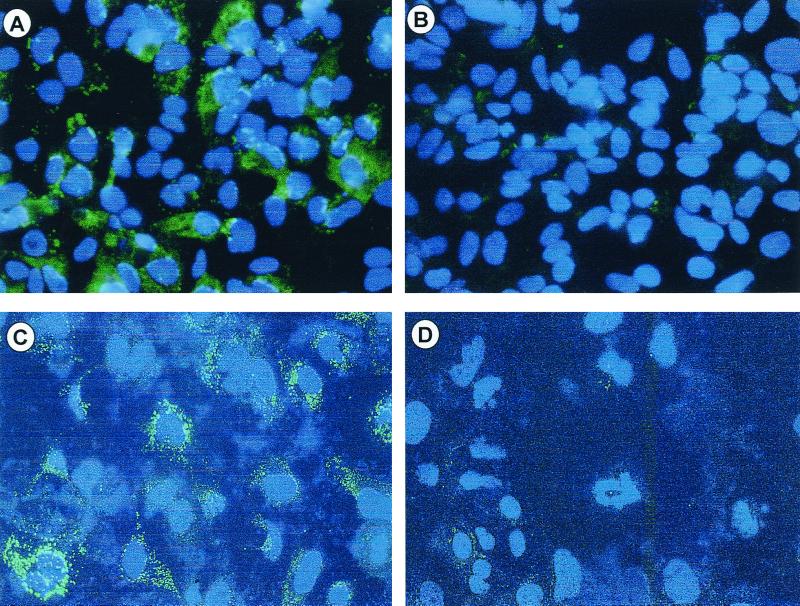
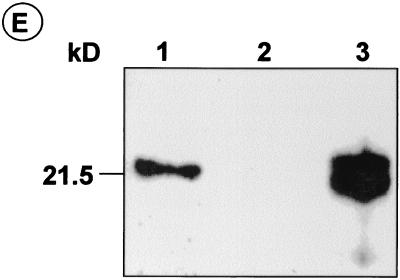
We confirmed the cytoplasmic expression of HCV core from the recombinant baculovirus AcCACore by immunofluorescence staining of Huh-7 and HepG2 cells 48 h after infection at an MOI of 100 (Fig. 5). Even at this high MOI, the infection of these cells resulted in no apparent cytopathology, consistent with the finding of Hofmann et al. (14). Infection with AcCACore at an MOI of 100 resulted in about 80 to 100% of HepG2 or Huh-7 cells staining positively for core protein (Fig. 5A and C), similar to the results reported by Shoji et al. (53). We found that AcCACore infection at an MOI lower than 100 resulted in a lower percentage of core-positive cells, while the level of core protein in the individual positively stained cells did not change (data not shown). No antigen was detectable in cells infected with the control baculovirus, AcCAG (Fig. 5B and D). In previous studies, core protein expressed from recombinant cDNA was localized to the cytoplasm and perinuclear regions (6, 22, 24, 25, 31, 48, 51). Although we demonstrated a similar lack of core within the nuclei of AcCACore-infected cells (Fig. 5), the cytoplasmic distribution of core in Huh-7 cells was different from that in HepG2 cells. In the former, core-specific fluorescence assumed a granular appearance, suggestive of localization of the protein within specific vacuoles or aggregates, while in the HepG2 cells the staining was diffuse throughout the cytoplasm (compare Fig. 5C and A). We also confirmed expression of the core protein by immunoblot analysis of S-100 fractions prepared from AcCACore-infected Huh-7 cells, as shown in Fig. 5E. These cells contained an immunoreactive 22-kDa core protein (Fig. 5E, lane 1) which was absent in AcCAG-infected Huh-7 cells (Fig. 5E, lane 2). For comparison, we also included core protein produced in an in vitro translation reaction (Fig. 5E, lane 3). These immunoblotting results confirmed the overexpression of a full-length, 22-kDa HCV core protein in AcCACore-infected Huh-7 cells that did not appear to be processed into smaller proteins as reported by Santolini et al. (49).
FIG. 5.
Core protein-specific immunofluorescence staining of HepG2 cells infected with AcCACore (A) or AcCAG (B) and of Huh-7 cells infected with AcCACore (C) or AcCAG (D). DAPI was used for nuclear staining. (E) Western blot analysis of HCV core protein with core-specific monoclonal antibody. S-100 lysates of Huh-7 cells were separated on an SDS–12% PAGE gel. Lane 1, lysate from AcCACore-infected Huh-7 cells; lane 2, lysate from AcCAG-infected Huh-7 cells; lane 3, in vitro translated core protein (aa 1 to 191).
To determine whether the core protein expressed in trans from the recombinant AcCACore baculovirus is capable of modulating HCV IRES activity, as suggested recently by Shimoike et al. (52), HepG2 or Huh-7 cells were transfected with the plasmids shown in Fig. 2 and infected with the recombinant baculoviruses 24 h later. The cells were harvested and assayed for luciferase activity following incubation for an additional 48 h. In general, infection of either HepG2 or Huh-7 cells with AcCAG, AcCACore, or AcCALacZ resulted in an increase in the expression of both the RLuc and FLuc reporter proteins compared with mock-infected cells (data not shown). This indicates that baculovirus infection of mammalian cells does not by itself impair cellular translation (consistent with the absence of a cytopathic effect) but suggests that baculovirus infection may enhance the efficiency with which transfected liposome-DNA complexes reach the nucleus and become transcriptionally active. Of much greater interest was the extent to which changes in the level of FLuc expression differed from changes in RLuc expression following infection with these recombinant baculoviruses, as such differences would be indicative of a specific effect on IRES-directed translation.
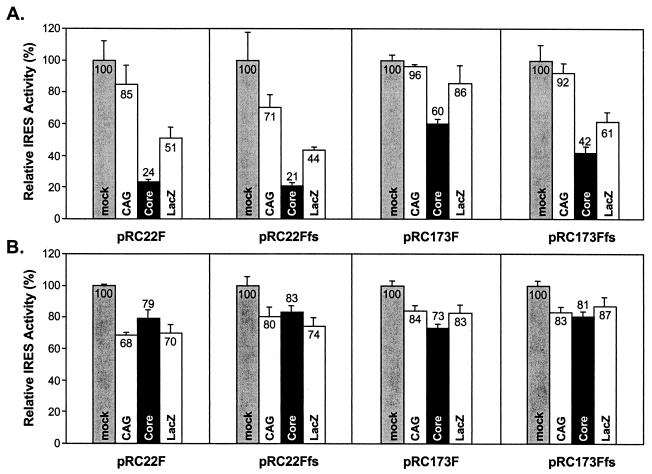
Figure 6 shows the IRES activities, estimated from the proportional RLuc and FLuc content, in HepG2 (Fig. 6A) or Huh-7 (Fig. 6B) cells transfected with each of the four dicistronic reporter constructs, with or without overexpression of the core protein or β-galactosidase. For each of the four dicistronic reporter plasmids, the activity of the IRES in mock-infected cells was arbitrarily set at 100% for this analysis. Compared with mock-infected HepG2 cells (Fig. 6A, lightly shaded columns), HCV IRES activities were significantly lower in cells overexpressing the core protein due to infection with AcCACore (Fig. 6A, solid columns), irregardless of the reporter construct tested. In contrast, infection with the AcCAG baculovirus, which contains only the CAG promoter and does not overexpress any protein, had no consistent effect on IRES activity, resulting in a significant lowering of relative IRES activity only with the pRC22Ffs reporter construct. Compared with AcCAG-infected cells, the estimated IRES activity ranged from 28 to 63% in core-expressing cells infected with AcCACore. These results are similar to those reported recently by Shimoike et al. (52). However, IRES activity was also substantially reduced in HepG2 cells infected with AcCALacZ, ranging from 60% (for pRC22Ffs) to 90% (for pRC173F) of the activity in AcCAG-infected cells (Fig. 6A). The substantial reduction in IRES activity observed with the overexpression of β-galactosidase from AcCALacZ, while always quantitatively less than that observed with overexpression of the core protein, substantially lessens the likelihood that the core-mediated translational suppression is biologically relevant. In support of this interpretation, the least degree of core-mediated translational suppression was observed with the pRC173F reporter construct, which contains the longest length of native HCV RNA sequence (Fig. 6A).
FIG. 6.
The relative IRES activities of dicistronic reporter transcripts in vivo in cells expressing the core protein from a recombinant baculovirus. HepG2 (A) and Huh-7 (B) cells were transfected with pRC22F, pRC22Ffs, pRC173F, or pRC173Ffs and then were infected with AcCAG (CAG), AcCACore (Core), or AcCALacZ (LacZ). mock, no baculovirus infection. The IRES activities of mock-infected cells are normalized to 100%. Error bars represent the standard deviations of triplicate samples.
The results obtained in baculovirus-infected Huh-7 cells provided further evidence that the core protein does not modulate IRES activity. As shown in Fig. 6B, infection with any of the baculoviruses slightly reduced IRES activity in Huh-7 cells. However, there were no significant differences between IRES activities in cells infected with AcCACore versus cells infected with either of the control baculoviruses, AcCAG and AcCALacZ. This lack of effect of the core protein on translation was evident with each of the four dicistronic reporter plasmids tested.
In the experiments shown in Fig. 6, HepG2 or Huh-7 cells were transfected with the reporter plasmids and then subsequently infected with the recombinant baculoviruses. To rule out the possibility that a minimal effect on IRES activity may have been missed due to the accumulation of FLuc reporter protein prior to effective baculovirus overexpression of the core, we reversed this sequence of events and transfected the reporter plasmids into cells that had been infected 24 h previously with baculovirus. As in the experiments shown in Fig. 6, we observed no specific changes in IRES activity related to expression of the core protein (data not shown). Thus, in contrast to the conclusion reached by Shimoike et al. (52), we conclude that the overexpression of core protein in trans in human hepatoma cells has no specific effect on HCV translation.
Recombinant HCV core protein does not influence IRES activity in vitro.
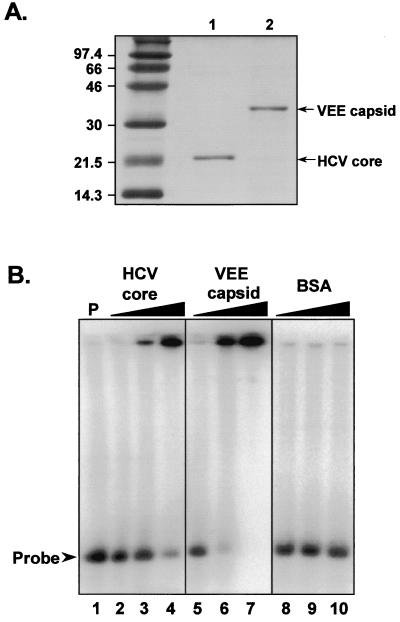
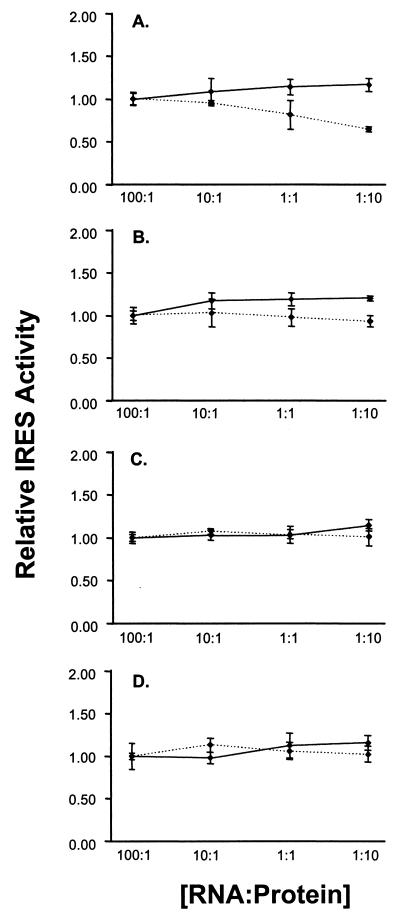
We considered the possibility that core protein expressed in trans from recombinant baculovirus might be sequestered in an intracellular compartment where it was not available for interaction with the RNA transcripts produced endogenously from the transfected reporter plasmids. To overcome this potential limitation to the experiments shown in Fig. 6, we determined whether core protein, added directly to an in vitro translation reaction, influences the efficiency of translation directed by the HCV IRES. Purified recombinant HCV core protein (aa 1 to 179) and VEE capsid protein were expressed in E. coli (S. Watowich, unpublished data). SDS-PAGE confirmed that these proteins were of the expected size and purity (Fig. 7A). The capsid proteins were added to reticulocyte lysates that were programmed for translation with RNAs transcribed in vitro from the plasmids shown in Fig. 2, with the amount of protein added calculated to produce an RNA/protein molar ratio ranging from 100:1 to 1:10. The reaction products were assayed for luciferase content by DLA to determine whether the addition of the core or VEE capsid protein altered IRES activity. The results of this experiment are shown in Fig. 8, in which the activity of the IRES at the lowest protein concentration (RNA/protein ratio = 100:1) is shown arbitrarily as 100%. The addition of purified core protein to the translation reactions had no effect on the activity of the IRES in any of the four dicistronic RNAs. In contrast, addition of the alphavirus capsid protein resulted in a slight suppression of IRES activity at the highest protein concentrations (RNA/protein = 1:10). This was most pronounced with the pRC22F reporter transcript (Fig. 8A) and was likely due to nonspecific RNA-protein interactions. The results strongly suggest that the purified core protein does not have a biologically relevant effect on IRES activity.
FIG. 7.
(A) SDS-PAGE analysis of purified HCV recombinant core (lane 1) and VEE capsid (lane 2) proteins. The gel was stained with Coomassie blue. (B) Electrophoretic mobility shift assays with 32P-labeled HCV RNA (nt 1 to 770). Lane 1, probe alone; lanes 2 to 4, probe with purified HCV core protein at RNA/protein molar ratios of 1:1, 1:10, and 1:20, respectively; lanes 5 to 7, probe with purified VEE capsid protein at RNA/protein ratios of 1:1, 1:10, and 1:100, respectively; lanes 8 to 10, probe with BSA at RNA/protein ratios of 1:1, 1:10, and 1:100, respectively.
FIG. 8.
Relative IRES activities of dicistronic reporter transcripts pRC22F (A), pRC22Ffs (B), pRC173F (C), and pRC173Ffs (D) in in vitro translation reactions supplemented with purified HCV core protein (solid line) or VEE capsid protein (dashed line) at the indicated RNA to protein molar ratios. RLuc and FLuc activities of the in vitro products were used to estimate the relative IRES activities. Each value represents the average IRES activity from triplicate translation samples. For each dicistronic RNA, the IRES activity of the 100:1 RNA/protein ratio sample was set arbitrarily at 1.00. Error bars indicate the standard deviations.
Core protein interaction with HCV RNA.
To determine whether the purified recombinant core protein was capable of a specific interaction with HCV RNA, we carried out electrophoretic mobility shift assays using as probe a 32P-labeled HCV RNA transcript extending from nt 1 to 770. Binding reactions contained the purified recombinant core protein at the indicated RNA/protein molar ratios, with control reactions containing indicated molar quantities of the recombinant VEE capsid protein or BSA (Fig. 7B). The recombinant HCV core protein bound to the RNA probe and retained the probe in the loading well of the gel when added to the RNA at a 10- (Fig. 7B, lane 3) or 20-fold (lane 4) molar excess. A similar phenomenon was observed with the addition of the VEE capsid protein at a 10- or 100-fold molar excess (Fig. 7B, lanes 6 and 7), while no retention of the probe was observed with BSA added at a 100-fold molar excess (Fig. 7B, lane 10). These data suggest a nonspecific interaction between the viral RNA probe and the HCV core protein at high protein concentrations that is shared with the VEE capsid protein. The retention of the protein-RNA complex in the loading well is consistent with the homotypic interaction of the core protein and its ability to form multimers under native conditions (29). Similar observations have been reported recently by Fan et al. (11).
DISCUSSION
In addition to its role as a structural protein in virus assembly, the HCV core protein has been suggested to have a wide range of biological activities, each of which could contribute to the pathogenesis of chronic hepatitis C. These include, among others, specific interactions with host cell proteins, including members of the tumor necrosis receptor family, the p53 tumor suppressor protein, apolipoprotein AII, DBX and other members of the DEAD box family of RNA helicases, and heterogeneous nuclear ribonuclear protein K (hnTNP K) (3, 19, 22, 27, 28, 33, 47, 62). However, despite an impressive number of in vitro experiments, evidence supporting the biological relevance of any these putative interactions has been limited by the absence of HCV-permissive cell cultures or small animal models of chronic hepatitis C.
Interactions of the core protein with the genomic RNA of HCV, and the potential effects of the core protein on cap-independent translation mediated by the HCV IRES, are similarly contentious. A possible role for core protein as a transactivator of HCV IRES activity was first suggested by studies of HCV/poliovirus chimeras in which the HCV IRES replaced the native picornavirus IRES (26). While the inclusion of a minimal length of core protein-coding sequence appeared to enhance translation from the chimeric RNA, subsequent work suggested that the inclusion of a lengthier HCV core sequence downstream of the IRES improved the replication capacity of these chimeric viruses by favorably influencing the processing of the chimeric polyprotein (62). These studies also demonstrated that the expression of the core protein was not necessary for HCV IRES-directed translation, nor for the replication of these chimeric viruses. On the other hand, early work by Reynolds et al. (43) suggested that as much as 33 nt of HCV core-coding sequence was required downstream of the IRES for efficient cap-independent translation. However, more recent evidence indicates that this requirement is conditional, dependent upon the specific reporter sequence, and not absolute (R. Rijnbrand, P. J. Bredenbeek, P. C. Haasnoot, W. J. Spaan, and S. M. Lemon, submitted for publication). Thus, there is no clear evidence that the core protein has any positive trans-activating effect on IRES activity.
More recently, Shimoike et al. (52) have suggested that the core protein is capable of a specific and profound down-regulation of HCV IRES activity. In large part, the conclusion that the core protein negatively modulates IRES activity was based on their finding of significant IRES suppression in HepG2 cells in which the core protein was expressed from a recombinant baculovirus, AcCACore, the same as that used by us in the experiment shown in Fig. 5 and 6. If correct, these data would be of substantial interest, since the structure of the HCV IRES and the immediately downstream core-coding sequence (Fig. 1) is very suggestive of the possibility that HCV translation might be tightly regulated by a viral or cellular protein binding to stem-loop IV. Minimal increases in the stability of this structure have been shown to profoundly reduce the efficiency of cap-independent translation (15). In some prokaryotic systems, the binding of translation products to RNA structures in the proximity of the initiation codon has been well documented as a mechanism of translational repression (2, 5, 9). The direct interaction of prokaryotic RNAs with the small ribosomal subunit is critically important to this mechanism of translational regulation and is a feature of prokaryotic translation that is shared by HCV (35).
Despite the attractive nature of this hypothesis, our data do not support a role for the core protein in regulating the initiation of translation on HCV RNA. Like Shimoike et al. (52), we observed significant repression of HCV IRES-directed translation when we expressed the full-length core protein from the recombinant baculovirus AcCACore in HepG2 cells (Fig. 6A). However, we also found substantial reductions in IRES activity in HepG2 cells expressing β-galactosidase, a finding that leads us to question the biological relevance of the repression observed with core protein. Importantly, our experimental approach differed from that of Shimoike et al. (52) in that we used a dicistronic IRES reporter system (Fig. 2) rather than monocistronic RNAs. The use of plasmid DNAs that express dicistronic transcripts in vivo allows a better discrimination between IRES-directed and cap-dependent translation, and it is possible that this technical difference contributes to the variance in the data presented by Shimoike et al. (52) and those presented here. We also infected cells with these baculoviruses at a higher MOI than Shimoike et al. (52) and may have achieved expression of the core and β-galactosidase protein in a greater proportion of cells. Whatever the basis is for the difference in our findings, the absence of biologically significant modulation of IRES-directed translation is strongly confirmed by the lack of any effect on translation from expression of either the core protein or β-galactosidase from recombinant baculoviruses in Huh-7 cells (Fig. 6B). The lack of even nonspecific suppression of translation in Huh-7 cells may be related to the different intracellular distribution of core protein, which was diffuse and cytoplasmic in HepG2 cells and discrete and granular in Huh-7 cells (Fig. 5). The basis for this difference is unknown, but a granular pattern of core expression has been noted previously in hepatocytes of HCV-infected chimpanzee (4).
Although it remains to be determined whether the core protein could specifically modulate viral translation in cell types other than those we have tested, in vitro evidence supports the conclusion that there is no biologically relevant repression of HCV translation by the core protein. Specifically, while we did observe a lower efficiency of translation in reticulocyte lysates programmed with dicistronic RNA transcripts encoding a nearly full-length core protein (pRC173F) than in transcripts containing only 66 nt of core protein-coding sequence (pRC22F), the analysis of related frameshift mutants indicated that this effect was due to the inclusion of the RNA sequence in the transcript and not specifically to the expression of the core protein from these RNAs in cis (Fig. 3). The suppressive effect of the additional core protein-coding sequence is likely to be related to the partial inclusion of a polypyrimidine-tract-binding protein binding domain near the 3′ end of the HCV sequence in these lengthier RNAs (20), a long-range RNA-RNA interaction between sequence in the core protein-coding region and that upstream of the IRES around nt 34 or 35 (18), and/or the presence of additional RNA secondary structures shown as stem-loops V and VI in Fig. 1 (56). Further studies are in progress to distinguish between these possibilities.
Finally, we observed no significant suppression of IRES-directed translation when purified recombinant core protein (aa 1 to 179) was added to reticulocyte lysates at a 10-fold molar excess over the HCV RNA used to program the translation reaction (Fig. 8). With each of the four dicistronic HCV transcripts tested, greater (but still only slight) repression of HCV translation was observed with the addition of recombinant VEE capsid protein. These results are consistent with the results of RNA gel retention experiments depicted in Fig. 7B, which show an interaction of both of these proteins with the viral RNA at high molar excess of the protein and which are consistent with the nonspecific RNA-binding activity of core protein described by Santolini et al. (49).
On the basis of these results, we are forced to conclude that the HCV core protein plays no specific role in regulating the translational activity of the viral IRES. The likelihood that this is so is strengthened by the probability that the molar concentration of the core protein in each of the experimental systems we used to assess this hypothesis is likely to significantly exceed that of the core protein present within the hepatocytes of persons with chronic hepatitis C. The discordance in the conclusions reached by Shimoike et al. (52) and those arrived at here attests to the difficulties inherent in working with a viral pathogen that cannot be propagated efficiently in any cell culture system and for which no small animal model is available. It also highlights the care that must be taken in claiming the biological relevance of intermolecular interactions observed in isolated in vitro or cell-based systems.
ACKNOWLEDGMENTS
We thank David Sangar and Kevin McKnight for their helpful discussions. We are grateful to Stanley Watowich for generously supplying us with the purified HCV core and VEE capsid proteins.
This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases (U19-AI40035) and the Advanced Technology Program of the Texas Higher Education Coordinating Board (004952-025). R.C.A.R. is the recipient of the Ridgeway-Blowitz Special Liver Scholar Fellowship of the American Liver Foundation.
REFERENCES
- 1.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrake M, Guild N, Hsu T, Gold L, Tuerk C, Karam J. DNA polymerase of bacteriophage T4 is an autogenous translational repressor. Proc Natl Acad Sci USA. 1988;85:7942–7946. doi: 10.1073/pnas.85.21.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki H, Hayashi J, Moriyama M, Arakawa Y, Hino O. Hepatitis C virus core protein interacts with 14-3-3 protein and activates the kinase raf-1. J Virol. 2000;74:1736–1741. doi: 10.1128/jvi.74.4.1736-1741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman M J, Miyamura T, Brechot C. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardi A, Spahr P F. Nucleotide sequence at the binding site for coat protein on RNA of bacteriophage R17. Proc Natl Acad Sci USA. 1972;69:3033–3037. doi: 10.1073/pnas.69.10.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buratti E, Baralle F E, Tisminetzky S G. Localization of the different hepatitis C virus core gene products expressed in COS-1 cells. Cell Mol Biol. 1998;44:505–512. [PubMed] [Google Scholar]
- 7.Chen C M, You L R, Hwang L H, Lee Y H. Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin-beta receptor modulates the signal pathway of the lymphotoxin-beta receptor. J Virol. 1997;71:9417–9426. doi: 10.1128/jvi.71.12.9417-9426.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke B. Molecular virology of hepatitis C virus. J Gen Virol. 1997;78:2397–2410. doi: 10.1099/0022-1317-78-10-2397. . (Review.) [DOI] [PubMed] [Google Scholar]
- 9.de Smit M H, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci USA. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnelly M L, Gani D, Flint M, Monaghan S, Ryan M D. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J Gen Virol. 1997;78(Pt. 1):13–21. doi: 10.1099/0022-1317-78-1-13. [DOI] [PubMed] [Google Scholar]
- 11.Fan Z, Yang Q R, Twu J S, Sherker A H. Specific in vitro association between the hepatitis C viral genome and core protein. J Med Virol. 1999;59:131–134. [PubMed] [Google Scholar]
- 12.Gosert R, Chang K H, Rijnbrand R, Yi M, Sangar D V, Lemon S M. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol Cell Biol. 2000;20:1583–1595. doi: 10.1128/mcb.20.5.1583-1595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P, Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc Natl Acad Sci USA. 1995;92:10099–10103. doi: 10.1073/pnas.92.22.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda M, Brown E A, Lemon S M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 16.Honda M, Kaneko S, Matsushita E, Kobayashi K, Abell G A, Lemon S M. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology. 2000;118:152–162. doi: 10.1016/s0016-5085(00)70424-0. [DOI] [PubMed] [Google Scholar]
- 17.Honda M, Ping L H, Rijnbrand R C, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 18.Honda M, Rijnbrand R, Abell G, Kim D, Lemon S M. Natural variation in translational activities of the 5′ nontranslated RNAs of hepatitis C virus genotypes 1a and 1b: evidence for a long-range RNA-RNA interaction outside of the internal ribosomal entry site. J Virol. 1999;73:4941–4951. doi: 10.1128/jvi.73.6.4941-4951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh T Y, Matsumoto M, Chou H C, Schneider R, Hwang S B, Lee A S, Lai M M. Hepatitis C virus core protein interacts with heterogeneous nuclear ribonucleoprotein K. J Biol Chem. 1998;273:17651–17659. doi: 10.1074/jbc.273.28.17651. [DOI] [PubMed] [Google Scholar]
- 20.Ito T, Lai M M. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology. 1999;254:288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- 21.Jin D Y, Wang H L, Zhou Y, Chun A C, Kibler K V, Hou Y D, Kung H, Jeang K T. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 2000;19:729–740. doi: 10.1093/emboj/19.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J E, Song W K, Chung K M, Back S H, Jang S K. Subcellular localization of hepatitis C viral proteins in mammalian cells. Arch Virol. 1999;144:329–343. doi: 10.1007/s007050050507. [DOI] [PubMed] [Google Scholar]
- 23.Large M K, Kittlesen D J, Hahn Y S. Suppression of host immune response by the core protein of hepatitis C virus: possible implications for hepatitis C virus persistence. J Immunol. 1999;162:931–938. [PubMed] [Google Scholar]
- 24.Liu Q, Tackney C, Bhat R A, Prince A M, Zhang P. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J Virol. 1997;71:657–662. doi: 10.1128/jvi.71.1.657-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo S Y, Masiarz F, Hwang S B, Lai M M, Ou J H. Differential subcellular localization of hepatitis C virus core gene products. Virology. 1995;213:455–461. doi: 10.1006/viro.1995.0018. [DOI] [PubMed] [Google Scholar]
- 26.Lu H H, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu W, Lo S Y, Chen M, Wu K, Fung Y K, Ou J H. Activation of p53 tumor suppressor by hepatitis C virus core protein. Virology. 1999;264:134–141. doi: 10.1006/viro.1999.9979. [DOI] [PubMed] [Google Scholar]
- 28.Mamiya N, Worman H J. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem. 1999;274:15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto M, Hwang S B, Jeng K S, Zhu N, Lai M M. Homotypic interaction and multimerization of hepatitis C virus core protein. Virology. 1996;218:43–51. doi: 10.1006/viro.1996.0164. [DOI] [PubMed] [Google Scholar]
- 30.Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065–1067. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- 31.Moriya K, Fujie H, Yotsuyanagi H, Shintani Y, Tsutsumi T, Matsuura Y, Miyamura T, Kimura S, Koike K. Subcellular localization of hepatitis C virus structural proteins in the liver of transgenic mice. Jpn J Med Sci Biol. 1997;50:169–177. doi: 10.7883/yoken1952.50.169. [DOI] [PubMed] [Google Scholar]
- 32.O'Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. Vol. 1. New York, N.Y: Oxford University Press; 1994. [Google Scholar]
- 33.Owsianka A M, Patel A H. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology. 1999;257:330–340. doi: 10.1006/viro.1999.9659. [DOI] [PubMed] [Google Scholar]
- 34.Park J S, Yang J M, Min M K. Hepatitis C virus nonstructural protein NS4B transforms NIH3T3 cells in cooperation with the Ha-ras oncogene. Biochem Biophys Res Commun. 2000;267:581–587. doi: 10.1006/bbrc.1999.1999. [DOI] [PubMed] [Google Scholar]
- 35.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole T L, Wang C, Popp R A, Potgieter L N, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 37.Ravaggi A, Natoli G, Primi D, Albertini A, Levrero M, Cariani E. Intracellular localization of full-length and truncated hepatitis C virus core protein expressed in mammalian cells. J Hepatol. 1994;20:833–836. doi: 10.1016/s0168-8278(05)80157-6. [DOI] [PubMed] [Google Scholar]
- 38.Ray R B, Lagging L M, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438–4443. doi: 10.1128/jvi.70.7.4438-4443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray R B, Lagging L M, Meyer K, Steele R, Ray R. Transcriptional regulation of cellular and viral promoters by the hepatitis C virus core protein. Virus Res. 1995;37:209–220. doi: 10.1016/0168-1702(95)00034-n. [DOI] [PubMed] [Google Scholar]
- 40.Ray R B, Steele R, Meyer K, Ray R. Hepatitis C virus core protein represses p21WAF1/Cip1/Sid1 promoter activity. Gene. 1998;208:331–336. doi: 10.1016/s0378-1119(98)00030-4. [DOI] [PubMed] [Google Scholar]
- 41.Ray R B, Steele R, Meyer K, Ray R. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J Biol Chem. 1997;272:10983–10986. doi: 10.1074/jbc.272.17.10983. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds J E, Kaminski A, Carroll A R, Clarke B E, Rowlands D J, Jackson R J. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 43.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rijnbrand R, Abell G, Lemon S M. Mutational analysis of the GB virus B internal ribosome entry site. J Virol. 2000;74:773–783. doi: 10.1128/jvi.74.2.773-783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rijnbrand R, van der Straaten T, van Rijn P A, Spaan W J, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rijnbrand R C, Abbink T E, Haasnoot P C, Spaan W J, Bredenbeek P J. The influence of AUG codons in the hepatitis C virus 5′ nontranslated region on translation and mapping of the translation initiation window. Virology. 1996;226:47–56. doi: 10.1006/viro.1996.0626. [DOI] [PubMed] [Google Scholar]
- 47.Sabile A, Perlemuter G, Bono F, Kohara K, Demaugre F, Kohara M, Matsuura Y, Miyamura T, Brechot C, Barba G. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology. 1999;30:1064–1076. doi: 10.1002/hep.510300429. [DOI] [PubMed] [Google Scholar]
- 48.Sansonno D, Cornacchiulo V, Racanelli V, Dammacco F. In situ simultaneous detection of hepatitis C virus RNA and hepatitis C virus-related antigens in hepatocellular carcinoma. Cancer. 1997;80:22–33. [PubMed] [Google Scholar]
- 49.Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schultz D E, Hardin C C, Lemon S M. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′-nontranslated RNA of hepatitis A virus. J Biol Chem. 1996;271:14134–14142. doi: 10.1074/jbc.271.24.14134. [DOI] [PubMed] [Google Scholar]
- 51.Selby M J, Choo Q L, Berger K, Kuo G, Glazer E, Eckart M, Lee C, Chien D, Kuo C, Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993;74:1103–1113. doi: 10.1099/0022-1317-74-6-1103. [DOI] [PubMed] [Google Scholar]
- 52.Shimoike T, Mimori S, Tani H, Matsuura Y, Miyamura T. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J Virol. 1999;73:9718–9725. doi: 10.1128/jvi.73.12.9718-9725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shoji I, Aizaki H, Tani H, Ishii K, Chiba T, Saito I, Miyamura T, Matsuura Y. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J Gen Virol. 1997;78:2657–2664. doi: 10.1099/0022-1317-78-10-2657. [DOI] [PubMed] [Google Scholar]
- 54.Shrivastava A, Manna S K, Ray R, Aggarwal B B. Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J Virol. 1998;72:9722–9728. doi: 10.1128/jvi.72.12.9722-9728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sizova D V, Kolupaeva V G, Pestova T V, Shatsky I N, Hellen C U. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith D B, Simmonds P. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J Mol Evol. 1997;45:238–246. doi: 10.1007/pl00006226. [DOI] [PubMed] [Google Scholar]
- 57.Tsuchihara K, Hijikata M, Fukuda K, Kuroki T, Yamamoto N, Shimotohno K. Hepatitis C virus core protein regulates cell growth and signal transduction pathway transmitting growth stimuli. Virology. 1999;258:100–107. doi: 10.1006/viro.1999.9694. [DOI] [PubMed] [Google Scholar]
- 58.Wang C, Le S Y, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasui K, Wakita T, Tsukiyama-Kohara K, Funahashi S I, Ichikawa M, Kajita T, Moradpour D, Wands J R, Kohara M. The native form and maturation process of hepatitis C virus core protein. J Virol. 1998;72:6048–6055. doi: 10.1128/jvi.72.7.6048-6055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.You L R, Chen C M, Lee Y H W. Hepatitis C virus core protein enhances NF-κB signal pathway triggering by lymphotoxin-beta receptor ligand and tumor necrosis factor alpha. J Virol. 1999;73:1672–1681. doi: 10.1128/jvi.73.2.1672-1681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.You L R, Chen C M, Yeh T S, Tsai T Y, Mai R T, Lin C H, Lee Y H. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol. 1999;73:2841–2853. doi: 10.1128/jvi.73.4.2841-2853.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao W D, Wimmer E, Lahser F C. Poliovirus/hepatitis C virus (internal ribosomal entry site-core) chimeric viruses: improved growth properties through modification of a proteolytic cleavage site and requirement for core RNA sequences but not for core-related polypeptides. J Virol. 1999;73:1546–1554. doi: 10.1128/jvi.73.2.1546-1554.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]