Abstract
Targeted epigenetic remodeling in the rat amygdala reverses the effects of adolescent alcohol consumption on excessive drinking and anxiety-like behavior in adulthood.
Alcohol consumption during adolescence is a risk factor for the development of alcohol use disorder, anxiety, and other neuropsychiatric conditions during adulthood (1). Alcohol consumption and binge drinking are common among adolescents, but how does such behavior impact the developing brain?
Adolescent alcohol consumption epigenetically suppresses the Arc gene
Persistent alcohol consumption during adolescence disrupts the normal development of the amygdala, a brain region nestled within the temporal lobe involved in associative learning and memory, and in processing emotionally salient stimuli. Corroborating evidence from studies in humans and rodents reveals that alcohol consumption during adolescence leads to a long-term reduction in ARC expression, a product of immediate early gene activation that is a key regulator of neuronal function and synaptic plasticity (2, 3), in the amygdala. The mechanism by which adolescent alcohol consumption decreases ARC expression is thought to be mediated by epigenetic suppression of an upstream Arc enhancer, known as synaptic activity response element (SARE). In the context of post-mitotic brain cells, the term “epigenetics” refers to post-translational modifications of chromatin without altering the base-pair sequence of DNA. DNA wraps around octamers of histone proteins known as nucleosomes to form higher order chromatin structures and enable compact packaging of DNA within the nucleus. Epigenetic modifications of chromatin are instrumental in controlling gene expression and cell function. Following adolescent alcohol consumption, increased repressive histone modifications and the removal of permissive histone modifications are observed at the Arc SARE site (4, 5). However, these measures of gene expression and histone modifications are correlative, falling short in defining a causal relationship.
Targeted epigenetic editing at Arc enhancer
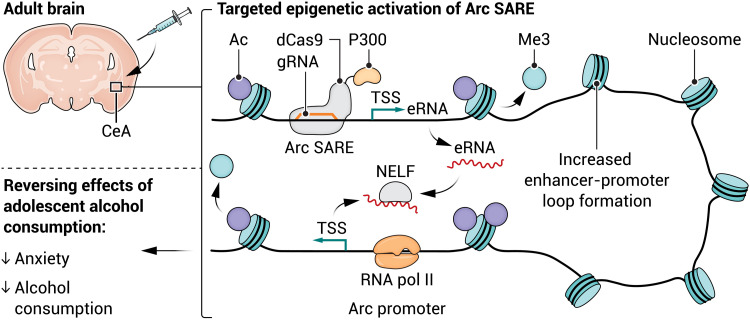
In this issue of Science Advances, Bohnsack et al. (6) demonstrate that targeted epigenetic remodeling of the Arc SARE site in the central nucleus of the amygdala (CeA) causally regulates Arc expression and the behavioral consequences of adolescent alcohol consumption. Targeted epigenetic remodeling is achieved using CRISPR-based epigenetic editing tools. These tools take advantage of a nuclease-dead Cas9 protein (dCas9) tethered to an epigenetic effector protein (e.g., histone-modifying enzyme, DNA methyltransferase, transcription factor) to alter the epigenetic landscape at a specific site in the genome. The dCas9-effector complex is directed toward a target gene using a guide RNA (gRNA) that associates with the dCas9 protein and undergoes complementary base-pairing with the target DNA sequence. These novel and rapidly evolving tools make it possible to selectively modify the epigenome at a single genomic locus and study the downstream effects at the molecular, cellular, circuit, and behavioral levels (7).
In this case, Bohnsack et al. (6) tether dCas9 to either P300, a histone acetyltransferase that promotes transcription, or Krüppel-associated box (KRAB), a zinc finger transcription factor moiety that suppresses transcription. While epigenetic editing tools are most often directed toward the promoter region of a target gene, Bohnsack et al. (6) opted to direct their tools to the Arc SARE site to determine whether epigenetic remodeling at this Arc enhancer causally regulates Arc expression.
Bohnsack et al. (6) found that targeting dCas9-P300 to the Arc SARE within the male rat CeA increased the levels of a permissive histone modification at this locus and increased Arc expression. By contrast, targeting dCas9-KRAB to the Arc SARE within the CeA decreased Arc expression, likely through numerous downstream epigenetic mechanisms. Together, these results support their hypothesis that epigenetic remodeling at the Arc enhancer is sufficient to bidirectionally regulate Arc expression.
Similar to findings in longitudinal human studies, rodents that consume alcohol during adolescence exhibit increased alcohol consumption and anxiety-like behavior during adulthood. Bohnsack et al. (6) demonstrated that rescuing Arc expression in the CeA using dCas9-P300 reversed the effects of adolescent alcohol consumption on excessive drinking and anxiety-like behavior. Conversely, suppressing Arc expression within the CeA using dCas9-KRAB increased alcohol consumption and anxiety-like behavior in naïve male rats, thus mimicking the behavioral consequences of adolescent alcohol consumption. While there are likely many other developing brain regions and molecular processes that are disrupted by adolescent alcohol consumption, Bohnsack et al. (6) present strong evidence that epigenetic remodeling of the Arc SARE site within the CeA is one key mechanism underlying the effects of adolescent alcohol consumption on excessive drinking and anxiety-like outcomes in adult males. Studies are now needed to determine if a similar mechanism operates in females.
Enhancer-promoter looping at Arc
How does epigenetic remodeling at an enhancer regulate the expression of a gene ~7 kb downstream? Distant regulatory elements and promoters come into proximity via three-dimensional chromatin looping. Over a decade of research emphasizes that the coordinated spatial arrangement of chromatin within the nucleus influences gene expression, cellular differentiation, and cell function (8). The observation that epigenetic remodeling at the Arc SARE site regulates Arc expression suggests that these genomic regions may interact directly via an enhancer-promoter loop. Bohnsack et al. (6) tested this hypothesis using a chromosome conformation capture (3C) assay and demonstrated that not only do the Arc SARE and Arc promoter interact via looping but also that targeting dCas9-P300 to the Arc SARE site increases the formation of this enhancer-promoter loop.
Enhancers are thought to regulate gene transcription in part by expressing enhancer RNAs (eRNAs) that prevent the negative elongation factor (NELF) complex from inhibiting RNA polymerase II function (9). Adolescent alcohol consumption decreases the expression of Arc eRNA, providing a potential mechanistic link between enhancer-promoter loop formation and changes in Arc expression (5). Building upon previous studies investigating epigenetic regulation of Arc (4, 5), the findings by Bohnsack et al. (6) point toward a detailed mechanism by which epigenetic remodeling at the Arc SARE site in the CeA regulates Arc gene expression (Fig. 1). A permissive epigenetic landscape at the Arc SARE enables Arc eRNA expression and formation of the Arc enhancer-promoter loop, the Arc eRNA binds to the NELF complex and releases it from the Arc promoter, and Arc mRNA production is increased.
Fig. 1. Theoretical epigenetic mechanisms regulating ARC expression.
Alcohol consumption during adolescence increases addiction- and anxiety-like behavior in adulthood through epigenetic suppression of the Arc gene. Targeting dCas9-P300 to the Arc SARE restores Arc mRNA expression and reverses these behavioral phenotypes. In the future, novel therapeutics aimed at reversing the epigenetic suppression of Arc may be an effective treatment for patients with anxiety or substance use disorders. gRNA, guide RNA; TSS, transcription start site. Credit: A. Mastin/Science Advances.
Implications and future directions
Despite the advantages offered by epigenetic editing tools, the diverse functions of the P300 and KRAB proteins make it difficult to draw conclusions about the specific type of epigenetic modification that leads to transcriptional regulation of Arc. The development of novel epigenetic editing tools that are selective for a single type of epigenetic modification is a high priority for future research. Another critical challenge in the field is to establish perfect single locus-specificity of any epigenome editing tool: It has not yet been feasible to demonstrate direct genomic binding sites of a dCas9 fusion protein in targeted populations of brain cells in vivo (7).
Bohnsack et al. (6) provide promising preclinical evidence indicating that treatments aimed at reversing the epigenetic suppression of the Arc SARE site within the CeA may alleviate addiction- and anxiety-like behaviors in adulthood. However, developing therapeutic interventions with this degree of specificity remains challenging and would require a multidisciplinary approach to overcome the barriers to clinical translation. Nonetheless, this study represents an important step forward in understanding how alcohol consumption during adolescence predisposes individuals to developing neuropsychiatric disorders later in life.
REFERENCES
- 1.Spear L., Effects of adolescent alcohol consumption on the brain and behaviour. Nat. Rev. Neurosci. 19, 197–214 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Shepherd J., Bear M., New views of Arc, a master regulator of synaptic plasticity. Nat. Neurosci. 14, 279–284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salery M., Trifilieff P., Caboche C., Vanhoutte P., From signaling molecules to circuits and behaviors: Cell-type-specific adaptations to psychostimulant exposure in the striatum. Biol. Psychiatry 87, 944–953 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Bohnsack J. P., Teppen T., Kyzar E. J., Dzitoyeva S., Pandey S. C., The lncRNA BDNF-AS is an epigenetic regulator in the human amygdala in early onset alcohol use disorders. Transl. Psychiatry 9, 34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyzar E. J., Zhang H., Pandey S. C., Adolescent alcohol exposure epigenetically suppresses amygdala Arc enhancer RNA expression to confer adult anxiety susceptibility. Biol. Psychiatry 85, 904–914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohnsack J. P., Zhang H., Wandling G., He D., Kyzar E. J., Lasek A., Pandey S. C., Targeted epigenomic editing ameliorates adult anxiety and excessive drinking after adolescent alcohol exposure. Sci. Adv. 8, abn2748 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yim Y. Y., Teague C. D., Nestler E. J., In vivo locus-specific editing of the neuroepigenome. Nat. Rev. Neurosci. 21, 471–484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vos S. M., Understanding transcription across scales: From base pairs to chromosomes. Mol. Cell 81, 1601–1616 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Schaukowitch K., Joo J.-Y., Liu X., Watts J. K., Martinez C., Kim T.-K., Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 56, 29–42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]