Abstract
A molecular epidemiology study was conducted among more than 100 human immunodeficiency virus type 1 (HIV-1) subtype C seropositive intravenous drug users (IDUs) from China. Genotyping based on the envelope C2V3 coding region revealed the highest homology of the most prevalent virus strains circulating throughout China to subtype C sequences of Indian origin. Based on these results, a virtually full-length genome representing the most prevalent class of clade C strains circulating throughout China was directly amplified from peripheral blood mononuclear cells of a selected HIV-infected IDU and subcloned. Sequence analysis identified a mosaic structure, suggesting extensive intersubtype recombination events between genomes of the prevalent clade C and (B′)-subtype Thai virus strains of that geographic region. Recombinant Identification Program analysis and phylogenetic bootstrapping suggested that there were 10 breakpoints (i) in the gag-pol coding region, (ii) in vpr and at the 3′ end of the vpu gene, and (iii) in the nef open reading frame. (B′)-sequences therefore include (i) several insertions in the gag-pol coding region; (ii) 3′-vpr, the complete vpu gene, and the first exons of tat and rev; and (iii) the 5′ half of the nef gene. Breakpoints located in the vpr/vpu coding region as well as in the nef gene of 97cn54 were found at almost identical positions of all subtype C strains isolated from IDUs living in different areas of China, suggesting a common ancestor for the C/B′ recombinant strains. More than 50% of well-defined subtype B-derived cytotoxic T-lymphocyte epitopes within Gag and Pol and 10% of the known epitopes in Env were found to exactly match sequences within in this clade C/B′ chimeric reference strain. These results may substantially facilitate a biological comparison of clade C-derived reference strains as well as the generation of useful reagents supporting vaccine-related efforts in China.
Human immunodeficiency virus (HIV) evolves by the rapid accumulation of mutations and intersubtype recombinations. Different subtypes cocirculating in the population of a geographical region represent the molecular basis for the generation and distribution of interclade mosaic viruses. Although the global HIV-1 variants have been studied intensively by means of serologic testing and heteroduplex DNA analysis, most phylogenetic studies are based on envelope sequences. Many of the prevalent subtypes and a variety of recombinant forms lack fully sequenced genomes. The increasing number of full-length HIV-1 genomes published recently in the databases indicate that full-length viral sequences are necessary for an optimal characterization of the phylogenetic relationship between a new isolate and the pre-existing HIV sequences, particularly in light of the potential for recombination (3, 4, 11, 12). A good example is provided by clade E viruses, which caused the major epidemics in Southeast Asia. Initially these viruses were classified as subtype E solely on the basis of envelope genotyping. Later they were shown to be members of an A/E recombinant strain by full-length genome sequences analysis (4, 12).
Each HIV epidemic in distinct geographical regions and population groups has its own specific characteristics and dynamics. In Asia, the HIV epidemic has spread extensively since the 1980s, with multiple, genetically divergent subtypes (38), complicating the development of effective vaccines for the affected countries (7, 8, 16). The experience in Thailand illustrates the potential for rapid HIV transmission in this area. Yunnan, a southwestern province of China bordering the drug triangle of Myanmar, Laos, and Thailand, was identified in the late 1980s as the first epidemic region in China, with prototype B strains circulating throughout the group of intravenous drug users (IDUs) (31, 42; Y. Ma, Z. Li, K. Zhang, et al., Abstract, Chin. J. Epidemiol. 11:184, 1990). With time a shift occurred toward B-Thai (B′) genotypes, and the former predominant prototype B has now been taken over by B-Thai variants (15, 36). The second epidemic was imported to the same area in the early 1990s, most probably by Indian IDUs carrying subtype C strains (30; C. C. Luo, C. Tian, D. J. Hu, M. Kai, T. Dondero, and X. Zhang, Letter, Lancet 345:1051–1052, 1995). Within a few years, subtype C viruses spread rapidly in southern, central, and even in northwestern China by drug trafficking and caused a widespread epidemic in China. According to a recent Chinese nationwide HIV molecular epidemiology survey, almost all the individuals infected with subtype C are IDUs and they include about 40% of HIV-infected IDUs in China, suggesting that subtype C is one of the major HIV-1 subtypes prevalent among IDUs in China (32; Y. Shao, L. Su, X. H. Sun, et al., Abstr. 12th World AIDS Conf., abstr. 13132, 1998). This suggests that the HIV epidemic among IDUs in China extended from a single predominant subtype (B) within a few years to at least two predominant subtypes, B-Thai and C, increasing the possibility of intersubtype recombination (2). All of the previous data on subtype C in China were limited to the genetic subtyping of the env gene (30, 40; Luo et al., Letter). Due to a lack of well-characterized molecular references, little information is available so far regarding the biological, immunogenic, and pathogenic properties of subtype C viruses in China. Accordingly, this study describes the identification and phylogenetic characterization of a clade C HIV isolate representing the most prevalent virus variants circulating throughout China.
MATERIALS AND METHODS
Blood samples.
All the blood samples used in this study were collected from HIV-1 subtype C-seropositive IDUs in several HIV-epidemic areas in China during the national molecular epidemiology survey in 1996 and 1997. Peripheral blood mononuclear cells (PBMCs) were separated on Ficoll gradients. Viruses were isolated by cocultivating the PBMCs from seropositive IDUs with phytohemagglutinin-stimulated donor PBMCs. Positive virus cultures were detected from cell culture supernatants by using the HIV-1 p24 Core Profile enzyme-linked immunosorbent assay kit (DuPont Inc., Boston, Mass.).
PCR amplifications and DNA sequencing.
Proviral DNA was extracted from productively infected PBMCs (Qiagen Inc., Valencia, Calif.). Nested PCR was used to amplify the envelope C2V3 coding region. PCR products were directly sequenced by Taq cycle sequencing using fluorescent dye-labeled terminators (no. 373A; Applied Biosystems, Foster City, Calif.) as previously described (1, 40). Multiple sequence alignments were performed by applying the Wisconsin software package from the Genetics Computer Group (GCG, version 9, 1997).
Virtually full-length HIV-1 genomes were amplified using the Expand Long Template PCR system (Boehringer, Mannheim, Germany) as described previously (15, 29). Primers were positioned in conserved regions within the HIV-1 long terminal repeats (LTR): TBS-A1 (5′- ATC TCT AGC AGT GGC GGC CGA A) and NP-6 (5′-GCA CTC AAG GCA AGC TTT ATT G). Purified PCR fragments were blunt-end ligated into a SrfI-digested pCR-Script vector (Stratagene, Heidelberg, Germany) and transformed into Escherichia coli strain DH5α. Several recombinant clones containing virtually full-length HIV-1 genomes were identified by restriction fragment length polymorphism analysis and sequencing of the V3-loop coding sequence. A provirus construct representing the vast majority of the positive clones was selected and sequenced as described above, using the primer-walking approach (primers were designed approximately every 300 bp along the genome for both strands).
Sequence analysis.
DNA sequences were assembled using Lasergene software (DNASTAR, Inc., Madison, Wis.) on Macintosh computers. All the reference subtype sequences in this study are from the Los Alamos HIV database. Nucleotide sequence similarities were calculated by the local-homology algorithm of Smith and Waterman (35). Multiple alignments of sequences with available sequence data of other subtypes were performed using the Wisconsin software package (version 9). Phylogenetic tree analyses were performed by using the PHYLIP software package (26). Evolutionary distances were calculated by the maximum-parsimony method and are indicated by cumulative horizontal branch lengths. The statistical robustness of the neighbor-joining tree was tested by bootstrap resampling as described previously (15).
Determination of intersubtype recombinations.
The Recombinant Identification Program (RIP) (version 1.3; http://hiv-web.lanl.gov/tools) was used to identify potential mosaic structures within the full-length sequence of this clone (window size, 200; threshold for statistical significance, 90%; gap handling, STRIP; informative mode, OFF). Gaps were introduced to create the alignment. The background subtypes sequences in this analysis were u455 (subtype A), RL42 [Chinese subtype B-Thai (B′)], eth2220 (subtype C), z2d2 (subtype D), and 93th2 (subtype A/E).
Nucleotide sequence accession number.
The full sequence of the cloned primary isolate 97cn54 has been submitted to the GenBank database.
RESULTS
Selection of a representative clade C HIV-1 isolate from Chinese IDUs.
Clade C HIV-1 C2V3 sequences were determined by direct sequencing from uncultured PBMCs of more than 100 preselected HIV-1-positive IDUs from the northwestern provinces of China. Based on the C2V3 sequences, a matrix of pairwise intra- and interisolate distances was generated using the Wisconsin Package (GCG 8.0 Unix) with the correction methods of Kimura. The calculated average intragroup distances ranged from 0.83 to 3.69 on the DNA level, indicating that the epidemic in this area is still very young. Intergroup differences between the Chinese clade C sequences and those of Indian, African, and South American origin were in the range of 7.36 to 11.98 (India) and 10.89 to 19.15 (Africa), respectively. This demonstrates a close phylogenetic relationship between Indian and Chinese clade C sequences (21) and a substantial genetic distance between these and the relatively heterogenous group of African clade C HIV-1 strains.
From the specimens analyzed, a representative isolate referred to as 97cn54 was identified as exhibiting the highest peptide homology (99.6%) to a calculated C2V3 consensus sequence, which has been established on the basis of the characterized local HIV sequences (Table 1). Multiple amino acid sequence alignments, including primary C-clade representative V3-loop sequences selected from different epidemic regions as well as consensus sequences of other clades (A to H, O, and CPZ), underlined the subtype C character of the selected primary isolate 97cn54 (Table 1). Compared with an overall V3 consensus sequence (consensus), 97cn54 and cn-con-c show amino acid alterations at positions 13 (H→R) and 19 (A→T), both of which are characteristic for subtype C isolates (C_consensus).
TABLE 1.
V3-loop amino acid sequence alignmenta
| Clade | Isolateb | Sequencec |
|---|---|---|
| 1 11 21 31 38 | ||
| Consensus | CTRPNNNTRK SIHIGPGQAF YA---TGDII GDIRQAHC | |
| C | 94IN11246 | ---------- --r-----t- --···--e-v -n------ |
| C | 93IN905 | ---------- --r-----t- --···----m -------- |
| C | 93IN999 | -vr------e --r-----t- --···--e-- -------- |
| C | Consensus | ---------- --r-----t- --···----- -------- |
| C | ind8 | ---------- -tr-----t- --···----- -------- |
| 97cn54-v3 | ----g----- --r-----t- --···----- -------- | |
| cn-con-v3 | ----g----- --r-----t- --···----- -------- | |
| C | bro025 | ---------- --r------- --···--e-- -------- |
| C | ind1024 | ---------- --r-----t- --···----- ----r-y- |
| C | nof | ---------- r-rv----tv --···-na-- -------- |
| C | zam20 | -a--g----- --r-----t- f-···--a-- -------- |
| C | sm145 | ---ya----- -vr-----t- -····-n--- -------- |
| A | Consensus | ---------- -vr------- --···----- -------- |
| B | Consensus | ---------- -------r-- -t···--e-- -------- |
| D | Consensus | ----y----q rt-------l -····-tr-- -------- |
| E | Consensus | ----s----t --t-----v- -r···----- ----k-y- |
| F | Consensus | ---------- ---l------ --···----- ----k--- |
| G | Consensus | ---------- --t------- --···----- -------- |
| H | Consensus | ---------- --s------- --···----- ----k-y- |
| O | Consensus | -e--gidiqe ·-r---·m-w -smglg-tng nss-a-y- |
The V3 amino acid alignment of consensus sequences from different HIV-1 clades (A to O) and selected subtype C isolates from different countries is given. The overall V3 consensus sequence was constructed by aligning consensus sequences from different clades (A to O).
cn-con-V3 represents the consensus sequence of HIV-1 subtype C strains prevalent in China. 97cn54 has been selected as the standard representative isolate of the most prevalent clade C HIV-1 strains circulating throughout China.
−, no exchange to the V3 consensus sequence; lowercase letters, an amino acid substitution; ·, gaps. All consensus and isolate sequences for multiple alignments were obtained from the Los Alamos database.
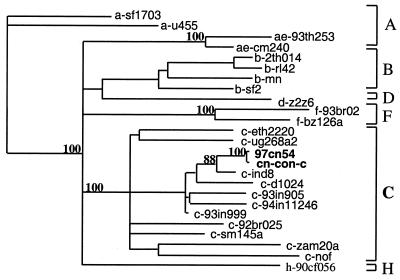
Phylogenetic tree analysis, initially based on the C2V3 sequences of the envelope gene, revealed that both 97cn54 and the consensus sequence of Chinese clade C isolates cluster with the subtype C strains from India (ind8, d1024, c-93in905, c-93in999, and c-93in11246), Africa (c-eth2220 and c-ug286a2), and South America (92br025, nof, cam20, and sm145). This suggests that the Indian clade C virus strains might be the source of the HIV-1 subtype C epidemic in China (Fig. 1). This hypothesis is also in agreement with our early epidemiology study confirming that the HIV-1 subtype C-infected individuals in Yunnan shared needles with the Indian jewellery businessmen in the boundary area (30).
FIG. 1.
Phylogenetic relationship of the env gene C2V3 coding region from clone 97cn54 with the representatives of the major HIV-1 (group M) subtypes. cn-con-c represents the env consensus sequence of HIV-1 subtype C strains prevalent in China. The phylogenetic tree was constructed using the neighbor-joining method. Values at the nodes indicate the percent bootstraps in which the cluster to the right was supported. Bootstraps of 70% and higher only are shown. Brackets on the right represent the major subtype sequences of HIV-1 group M.
Cloning and sequence analysis of 97cn54.
To obtain more complete information on the genetic structure of 97cn54, DNA was isolated from infected PBMCs and subjected to a long-template PCR analysis amplifying the complete coding proviral sequence. Several recombinant clones containing virtually full-length HIV genomes were identified by direct sequencing of PCR fragments generated by primer pairs located in the vector or at the very extreme ends in the conserved region of the LTRs. According to restriction fragment length polymorphism analysis, using different combinations of restriction endonucleases followed by sequencing of the V3-loop coding sequence, 77% of the positive full-length constructs were nearly identical. Based on this analysis, a provirus construct representing the vast majority of the cloned viral genomes was selected and fully sequenced.
The 9,078-bp genomic sequence derived from isolate 97cn54 contained all known structural and regulatory genes of an HIV-1 genome. No major deletions, insertions, or rearrangements were found. Nucleotide sequence similarities were examined by comparing all coding sequences of 97cn54 to consensus sequences of different genotypes and selected subtype isolates (Table 2). The highest homologies of the gag, pol, env, and vif reading frames to the corresponding clade C consensus sequences were within a range of 93.93 to 95.06%. This observation considerably extended the above C2V3-based sequence comparison and phylogenetic tree analysis (Table 1 and Fig. 1) and therefore clearly confirmed that the selected virus isolate belonged to the group of previously published clade C virus strains. However, the homology values determined by this kind of analysis for the tat, vpu, vpr, and nef genes (5, 6, 9) were not sufficient to allow a clear assignment of these reading frames to clade B or C virus strains (Table 2). For the vpu gene, the highest homologies were to clade B (94.24%), compared with only 78.23% to a clade C consensus sequence. Similar observations were made for the tat gene, for which the highest homology was to the B′-rl42 isolate (>91%), compared with 87.9% (C-92br025) and 85.5% (C-eth2220) for selected primary clade C representatives or 89.01% for the clade C consensus sequence. These data, together with the occurrence of B, C, and E genotypes throughout the epidemic area of Yunnan, suggested that the analyzed virus isolate might represent a mosaic virus strain that resulted from a B′/C interclade recombination event.
TABLE 2.
Comparison of 97cn54-derived coding sequences with the corresponding genes of reference strains and clade-specific consensus sequencesa
| Coding sequence | % Identity to 97cn54b in:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| gag | pol | vif | vpr | tat | rev | vpu | env | nef | |
| A | 87.68 | 91.80 | 86.81 | 83.66 | 84.90 | 83.97 | 79.82 | 85.75 | 84.19 |
| B | 90.43 | 91.93 | 88.04 | 90.31 | 86.56 | 82.08 | 94.24 | 84.52 | 88.13 |
| B-mn | 89.38 | 90.82 | 86.01 | 89.31 | 87.44 | 79.48 | 88.21 | 82.33 | 85.41 |
| B′-rl42 | 91.53 | 90.76 | 86.01 | 88.97 | 91.163 | 80.23 | 96.74 | 82.70 | 85.99 |
| C | 94.65 | 94.29 | 95.06 | 91.39 | 89.01 | 91.99 | 78.23 | 93.93 | 88.82 |
| C-92br025 | 92.19 | 92.91 | 88.51 | 90.03 | 87.91 | 89.70 | 76.13 | 88.51 | 86.20 |
| C-eth2220 | 91.4 | 92.06 | 87.15 | 90.77 | 85.57 | 88.08 | 80.09 | 87.15 | 87.08 |
| D | 89.80 | 91.08 | 87.74 | 87.94 | 83.93 | 84.39 | 87.30 | 85.26 | 86.88 |
| E/A | 86.324 | 89.07 | 86.59 | 83.39 | 81.44 | 81.74 | 77.31 | 82.09 | 84.18 |
| F | 88.02 | 88.99 | 86.36 | 86.25 | 80.65 | 86.25 | 82.33 | 84.02 | — |
| G | 88.08 | —c | — | — | — | — | — | 84.55 | — |
| H | 87.69 | 89.45 | 86.01 | 85.22 | — | — | — | 83.74 | — |
| O | 73.42 | 78.02 | 72.12 | 76.604 | 72.31 | 76.60 | 59.54 | 67.01 | 80.35 |
| CPZ | 74.14 | 78.80 | 93.75 | 75.44 | 76.00 | 75.44 | 64.41 | 72.42 | — |
Nucleotide sequence comparison of all coding sequences between 97cn54 and DNA sequences, representing either consensus sequences of distinct HIV-1 clades (obtained from the Las Alamos HIV database) or standard subtype C (92br025 and eth2220) and B (mn and rl42) isolates.
The data show the percent identity of a given sequence to 97cn54. Ambiguous nucleotide positions within consensus sequences were scored as a match. The highest degrees of homology are highlighted in boldface.
—, no consensus sequence was available from the Los Alamos database.
Recombination analysis.
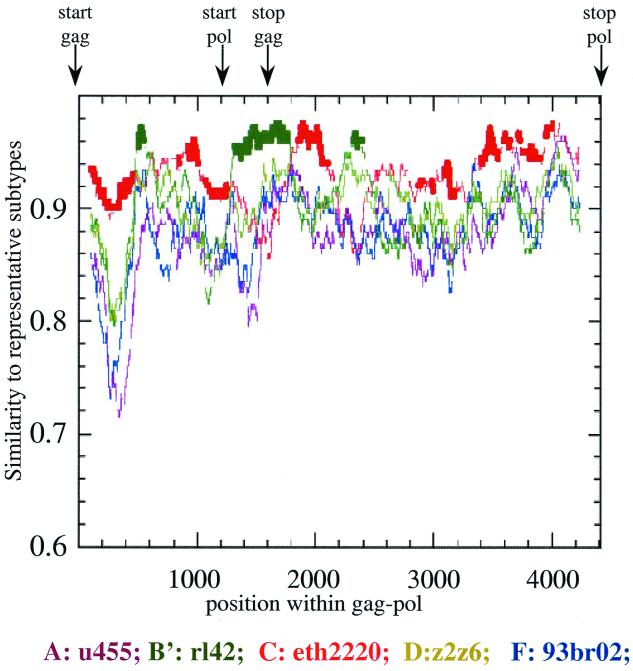
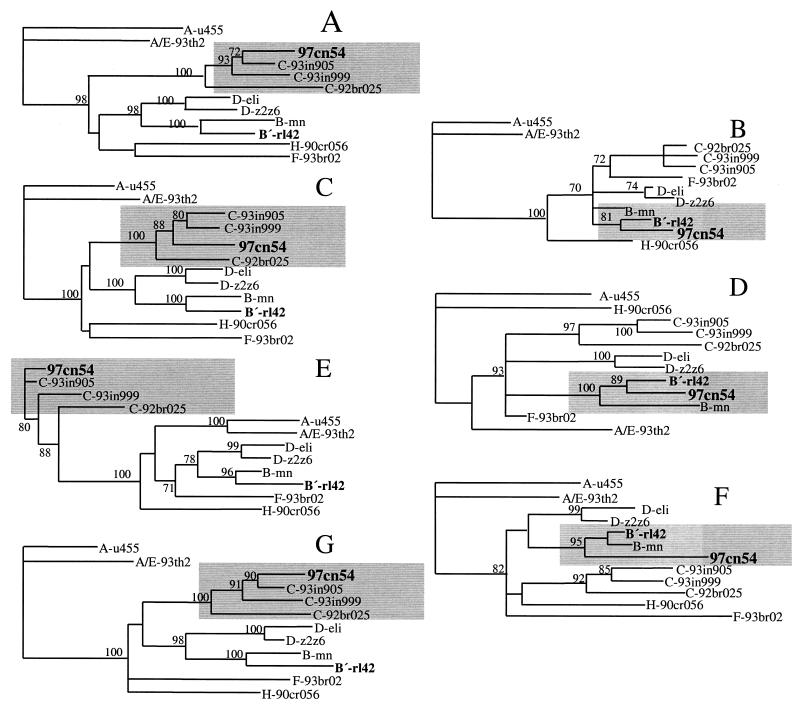
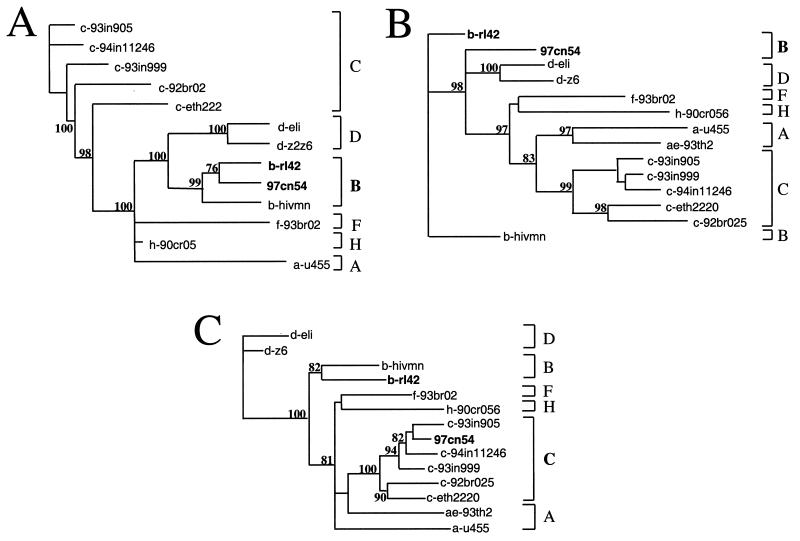
A more detailed sequence analysis program, RIP (28), was used to identify potential intersubtype mosaic structures within 97cn54. Although substantial homologies to clade C virus strains were observed within the highly conserved gag and pol reading frames, RIP analysis identified three areas of intraclade recombination within gag-pol around positions 478 to 620, 1290 to 1830, and 2221 to 2520 relative to the gag start codon. These dispersed stretches are located within gag and pol reading frames and encode (i) the C-terminal half of p17 (10, 41) including the amino-terminal 14 amino acids of the p24 capsid domain (amino acids 86 to 146 of Gag-Pol); (ii) the p7, p6 (14, 18, 25), and p6* moieties in the Gag and Gag-Pol polyproteins, respectively (amino acids 364 to 554); and (iii) amino acids 684 to 784 in the Gag-Pol precursor, extending into the active site of reverse transcriptase (RT) (17). These sequence stretches turned out to show the highest homology to prototype B strains (data not shown) and, in particular, the highest sequence similarity to a subtype B (B′) isolate originating from the Yunnan province, which we have described earlier (15) (Fig. 2). This observation clearly underlines the importance of RIP analysis, since simple homology alignments based on complete genes were not able to identify these small interpersed fragments of a different subtype. To confirm the data obtained by RIP analysis, several phylogenetic trees were generated using regions either flanking or spanning the stretches of proposed recombination (Fig. 3). Using various standard representatives of different subtypes and some selected clade C primary isolates, all proposed areas of recombination could be confirmed by differential clustering of 97cn54 with the respective clade C (Fig. 3A, C, E, and G) or clade B (Fig. 3B, D, and F) reference isolates.
FIG. 2.
RIP (version 1.3) analysis of the complete gag-pol coding region of 97cn54 (window size, 200; threshold for statistical significance, 90%; gap handling, STRIP). Positions of the gag and pol open reading frames are indicated by arrows above the diagram. RIP analysis was based on background alignments using reference sequences derived from selected virus strains representing the most relevant HIV-1 subtypes. The standard representatives are marked by different colors as indicated. The x axis indicates the nucleotide positions along the alignment. The y axis indicates the similarity of 97cn54 to the listed reference subtypes.
FIG. 3.
Phylogenetic relationship of different regions within the 97cn54-derived gag-pol reading frames to standard representatives of the major HIV-1 (group M) subtypes. Phylogenetic trees were constructed using the neighbor-joining method based on the following sequence stretches: nucleotides 1 to 478 (A), 479 to 620 (B), 621 to 1290 (C), 1291 to 1830 (D), 1831 to 2220 (E), 2221 to 2520 (F), and 2521 to 2971 (G). The indicated positions refer to the first nucleotide of the gag open reading frame. Grey areas highlight clustering of the analyzed sequences either with clade C-derived (A, C, E, and G) or with clade B-derived (B, D, and F) reference strains. Values at the nodes indicate the percent bootstraps in which the cluster to the right was supported. Bootstraps of 70% and higher only are shown.
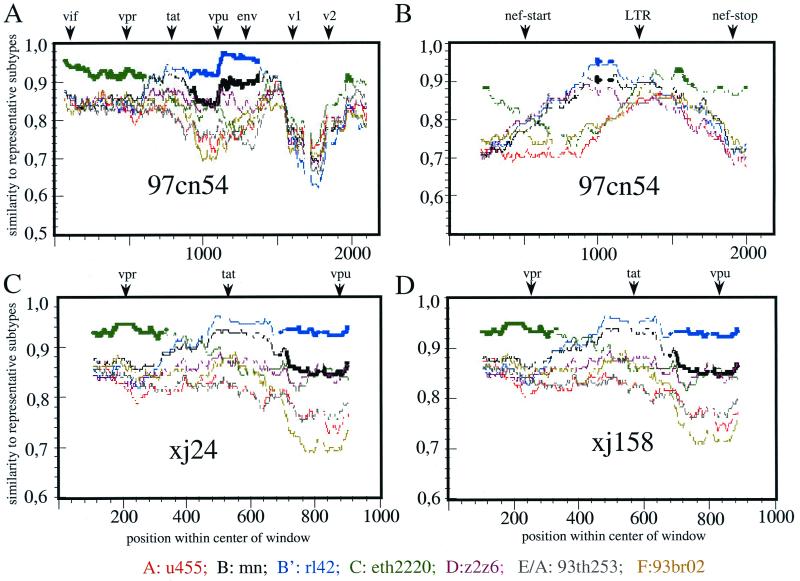
As expected from the sequence alignments summarized in Table 2, the RIP analysis clearly confirmed the intersubtype recombination between subtypes B-Thai (B′) and C (Fig. 4). A fragment of about 1,000 bp extending from 150 bp 3′ of vpr through exon 1 of tat and rev to vpu showed the highest degree of homology to the local subtype (B′) representative (rl42) (Fig. 4A). Furthermore, an about 300-bp sequence stretch overlapping the 5′ half of the nef gene showed highest homology to the B-Thai (B′) subtype whereas the remaining part, including a 300-bp fragment extending to the 3′ LTR, clustered with subtype C (Fig. 4B).
FIG. 4.
RIP analysis (version 1.3) of different regions of 97cn54 (window size, 200; threshold for statistical significance, 90%; Gap handling, STRIP). (A and B) The analysis included a sequence stretch of 1,500 bp from the start codon of the vif gene to the 5′ end of env, including vif, vpr, exon 1 of tat and rev, vpu, and the first 200 bp of env (A) and a ca. 700-bp fragment overlapping 300 bp from the 3′ end of env encompassing the complete nef gene and parts of the 3′ LTR (B). Positions of the start codons of vpr, tat, vpu, env, and nef, as well as the 5′ end of the 3′ LTR, are indicated by arrows above the diagrams. RIP analysis was based on background alignments using sequences derived from selected virus strains representing the most relevant HIV-1 subtypes. The indicated standard representatives are marked by different colors. The x axis indicates the nucleotide positions along the alignment. The y axis indicates the similarity of the 97cn54 to the listed reference subtypes. (C and D) RIP analysis of sequences from two independent clade C isolates (xj24 [C] and xj158 [D]) from China overlapping the vpr and vpu genes including the first exon of tat.
Extending the RIP analysis, phylogenetic trees showed the closest relationship of vpr/vpu and the 5′-portion of the nef gene to clade B isolates (Fig. 5A and B), whereas the 3′-nef fragment clearly clustered with subtype C representatives (Fig. 5C). Further analysis confirmed that the subtype B sequence within this mosaic was more closely related to a very recently described B′-Thai (B′) strain (rl42) isolated from a Chinese IDU (15) than to prototype B isolates (mn and sf2) (Table 2).
FIG. 5.
Phylogentic tree analysis. Phylogenetic trees were constructed using the neighbor-joining method based on a 380-bp fragment overlapping the 3′ 150 bp of the vpr gene to the end of the vpu reading frame (A), based on the first 290 bp of the nef coding region (B), and based on the 3′ 320 bp of the nef gene (C). Values at the nodes indicate the percent bootstraps in which the cluster to the right was supported. Bootstraps of 70% and higher only are shown. Brackets on the right represent the major subtype sequences of HIV-1 group M.
Similarity of breakpoints in independent isolates.
Breakpoints located in the vpr/vpu coding region as well as in the nef gene of 97cn54 were found at almost identical positions within all subtype C genomes isolated from 27 IDUs living along the drug-trafficking route from Yunnan via Sichuan up to the northwestern Xinjiang Province (Table 3). Whereas recombinations in the gag-pol gene seemed to be less frequent, the breakpoints in the vpr/vpu and nef coding regions were regularly found in all tested isolates. Figures 4C and D depict two representative examples selected from 27 investigated isolates indicating that the recombination points in the analyzed sequence stretches are close to identical. These data might suggest a common ancestor for the C/(B′) recombinant strains circulating throughout the northwestern drug trafficking road from Yunnan through Sichuan and Xinjiang and across the Chinese border to Kazakhstan.
TABLE 3.
Breakpoints shared by independent C/B′ chimeras isolated from various patients along the drug-trafficking route from Sichuan to Gansu, Ninxia, and Xinjiang Provinces in the west and far northwest of China
| Province where sample originatedb | No. of isolates | No. of sequences with breakpointsa found in:
|
||
|---|---|---|---|---|
| gag-pol | vpu-vpr | nef | ||
| Xinjiang | 20 | 4 | 20 | 20 |
| Sichuan | 5 | 2 | 5 | 5 |
| Ninxia | 1 | 0 | 1 | 1 |
| Gansu | 1 | 0 | 1 | 1 |
Location of breakpoints identical or closely related to 97cn54.
Independent isolates were grown on PBMCs as described in the text. Sequences of cloned isolates were obtained by automated Taq cycle sequencing.
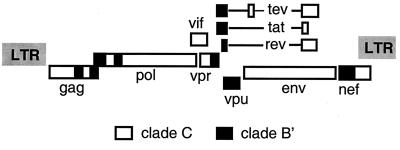
In conclusion, our results demonstrate that 97cn54 represents a C/(B′) interclade mosaic virus, with 10 breakpoints of intraclade recombination, that is most prevalent among the IDUs within the northwestern provinces of China. A schematic representation of the C/(B′) mosaic genome of isolate 97cn54 is given in Fig. 6.
FIG. 6.
Schematic representation of the mosaic genome organization of 97cn54.
Analysis of amino acid variation in known CTL epitopes.
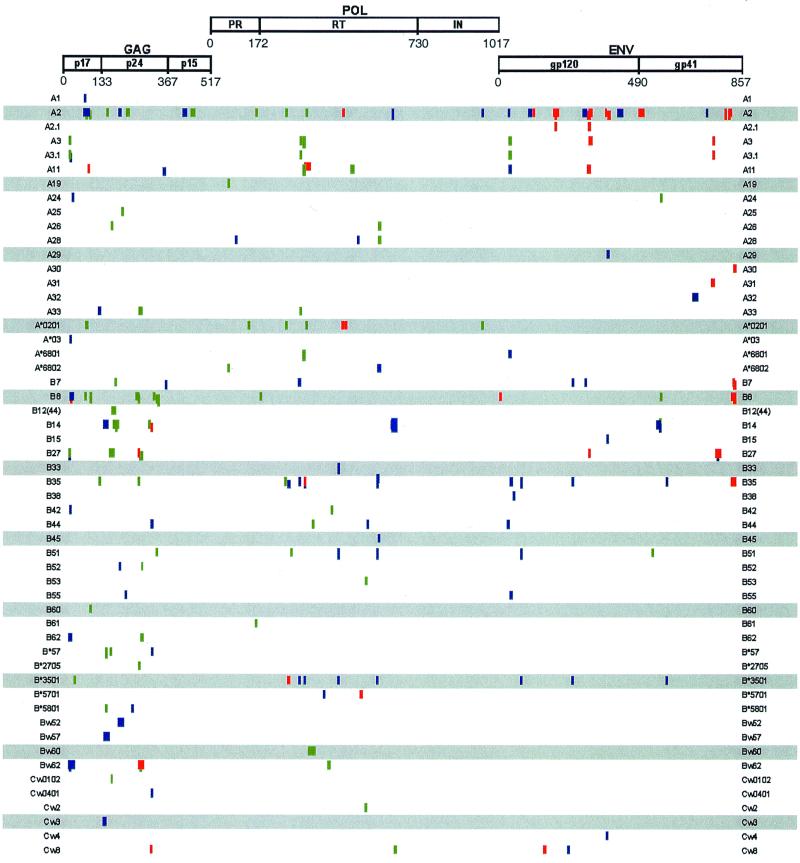
Genomic sequences offer the opportunity to assess the conservation of known cytotoxic T-lymphocyte (CTL) epitopes that may have an impact on the design of HIV-1 candidate vaccines. Most reagents for and data on CTL epitopes have been derived from clade B HIV-1LAI sequences. To provide an estimate of cross-clade CTL epitope conservation, the predicted protein sequences of 97cn54 were compared to the known and best-mapped LAI-specific CTL epitopes (obtained from the Los Alamos HIV Database) (Fig. 7). Of 194 reported HIV-1 CTL epitopes, 75, 55, 40, and 24 are located in Gag (p17, p24, and p15), in the RT, in gp120, and in gp41, respectively. Whereas only 5 and 17% of the gp120 and gp41 HIV-1LAI-derived CTL epitopes exactly matched the predicted amino acid sequences of 97cn54, about 50% of the epitopes in Gag and RT were completely identical among both virus strains. Taken together, these observations clearly predict a considerable cross-clade CTL reactivity, especially regarding the functionally and immunologically conserved HIV-1 proteins such as Gag and Pol. In addition, these data suggest that many of the reagents (peptides, vaccinia virus constructs) that have been synthesized and established for the mapping and characterization of clade B CTL epitopes may be also useful in determining CTL reactivities on the basis of clade C HIV sequences.
FIG. 7.
Comparison between known and experimentally proven prototype B (HIV-1LAI)-derived CTL epitopes and the corresponding amino acid sequences in the gag-, pol-, and env-encoded polypeptides of the clade C strain 97cn54. Functional domains in Gag (p17 matrix, p24 capsid, p15 nucleocapsid, and linker protein), Pol (protease [PR], RT, and integrase [IN]), and Env (gp120 external glycoprotein and gp41 transmembrane protein) are indicated. Numbers underneath the open reading frames indicate the amino acid position relative to the amino termini of the polypeptides. Haplotype restrictions of the known HIV-1LAI-derived CTL epitopes are indicated in the left and right margins. Green bars represent sequence identity between the known epitope and the corresponding clade C sequence; blue bars indicate two or fewer conservative missmatches; red bars represent clade C-derived sequence stretches with more than two conservative missmatches or any nonconservative substitution compared to the corresponding LAI-derived epitope.
DISCUSSION
Phylogenetic analyses of globally circulating HIV strains have identified a major group (M) of 10 different sequence subtypes (A to J) (13, 19, 20, 39) exhibiting sequence variations in the envelope protein of up to 24%, in addition to group O viruses, which differ from group M viruses by more than 40% in some reading frames (22, 23, 33, 34). Although the extent of global HIV-1 variation is well defined, little is known about the biological consequences of this genetic diversity and its impact on the design of candidate vaccines.
Due to a lack of well-characterized molecular references, little information is available so far regarding the biological, pathogenic, and immunological properties of subtype C viruses. Regarding the complex situation in developing countries, where multiple subtypes of HIV-1 are known to cocirculate, extensive molecular epidemiological studies are required to identify representative local virus strains. Particularly in the light of the potential for recombination (3, 4, 11, 12), full-length viral sequences are necessary for an optimal characterization of relevant virus strains. Accordingly, this study describes the identification and phylogenetic characterization of a clade C HIV isolate representing one of the most prevalent virus variants circulating throughout China.
Clade C HIV-1 strains play a leading role both in the total number of infected people and in the high incidence of new infections, especially in South America and Asia. Currently, there is an increasing number of nonrecombinant molecular clones and a few mosaic genomes available for viruses other than B. Regarding clade C HIV-1 viruses, only a few nonrecombinant representatives and four A/C recombinants have been published so far, all of them originating from Africa, South America, or India (11, 21, Luo et al., Letter).
With the exception of Thailand, limited information has been available until recently on the distribution and molecular characteristics of HIV-1 strains circulating throughout Asia. The World Health Organization estimates that South and Southeast Asia have the highest rate of HIV spread and will soon become the world's largest HIV epidemic region. China has very similar social and economic conditions and direct ethnic and economic connections to these regions. Since early 1995, a rapid increase in HIV infection was clearly seen in many provinces of China. Compared with the cumulative total of 1,774 cases of HIV infection and AIDS detected from 1985 to 1994, 1,421 cases were detected in 1995 and more than 4,000 cases were detected in 1997 alone. The World Health Organization estimated that there would be more than 400,000 HIV infections in China by the end of 1997, with an estimated 6,400 cumulative deaths and 4,000 people dying of AIDS in 1997 alone. In the recent national HIV molecular epidemiology survey, it was found that the prototype B and B′-subtype Thai strains in Dehong (15) were spread to central and eastern China by drug users and contaminated blood and plasma collection services. The subtype C strains of Yunnan were transmitted along the drug-trafficking routes to central western and northwestern China. Today, subtype C HIV-1 strains account for the majority of HIV-1 infections among IDUs in China.
In this report, we show for the first time that the prevalent HIV-1 strains transmitted among the IDUs in the northwestern provinces of China represent C/(B′) interclade mosaic strains. This study was based on genotyping the C2V3 envelope coding region amplified from proviral DNA isolated from PBMCs of more than 100 HIV-1 clade C-positive IDUs. The C2V3 nucleotide distances among the different virus isolates were in a range of 2 to 3%, indicating that the epidemic caused by the clade C HIV strains in this area is still very young (1, 30; Luo et al., Letter). Phylogenetic tree analysis based on the C2V3 region of a representative virus strain suggested that clade C HIV-1 strains circulating throughout China are closely related to those of Indian origin (21; Luo et al., Letter) and distinct from clade C viruses isolated in South America and Africa (11; Luo et al., Letter).
Detailed molecular characterization of a virtually full-length genome representing the most prevalent species of clade C HIV-1 strains circulating in China suggested several intersubtype recombination events between clade C and (B′)-Thai sequences. RIP analysis indicated a total of 10 breakpoints (i) in the gag-pol coding region, (ii) in vpr and at the 3′ end of the vpu gene, and (iii) in the nef open reading frame. This finding has been strongly supported by establishing distinct phylogenetic trees based on sequences flanking the recombination points.
The two parental (B′)-Thai and clade C HIV-1 subtypes had been reported earlier to cocirculate among IDUs in southwestern China, therefore clearly representing a potential reservoir for the observed interclade recombination (Luo et al., Letter; Ma et al., Abstract). The reason why Chinese B′ strains exclusively seem to exhibit homogeneous genome structures, whereas all so-called clade C virus strains identified in this area are interclade mosaics so far, remains unclear (1, 15, 37). However, the isolated appearance of C/(B′) chimeras in the northwestern provinces of China may be suggestive of a founder virus effect.
RIP analysis and phylogenetic bootstrapping of clade C sequences obtained from various independent IDUs living along the northwestern drug trafficking route from Yunnan to the northwestern Sichuan and Xinjiang Provinces revealed almost identical recombination points for all the analyzed subtype C/(B′) strains. This suggests that the observed recombination events had already occurred before this virus started to spread. Strikingly, only recombinant C/(B′) strains seem to travel along the drug-trafficking route to the far northwestern autonomous region, whereas the B′ parental strains are preferentially found in the Southwest (1, 32).
It is noteworthy that the C/(B′) recombinants found along the northwestern drug-trafficking route differ from the very recently reported C/(B′) recombinants isolated from IDUs living in the area of Guangxi neighboring the Yunnan Province and Myanmar (26a). The Guangxi-derived C/(B′) chimeras seem to share only part of the B′ sequence in the central portion of the RT gene whereas the breakpoints in the p17/p24 overlap region, in the p7/p6/p6* coding region, in the vpr/vpu genes, and in the 3′ portion of the nef gene are unique to those in the viruses found along the drug-trafficking route from Yunnan to Sichuan, Gansu, Ninxia, and Xinjiang Provinces. These data add clear evidence to previous observations that suggest two different routes of HIV subtype C/(B′) spread throughout China: one from Yunnan through the northwestern Sichuan, Gansu, Ninxia, and Xinjiang Provinces and across the border into Kasakhstan, and one spreading from Myanmar across the border into Yunnan and then through Guangxi and Hongkong to western countries. Taking these data into account, it seems as if each drug-trafficking route is associated with a different and relatively homogeneous HIV-1 recombinant, underlining the linkage between intravenous drug use, needle sharing, and HIV spread in China.
Based on these observations, it is tempting to speculate on whether interclade recombination events may confer selective advantages to the mosaic viruses. Each of the recombination events shown for 97cn54 might contribute to a more efficient transmission of the C/(B′) chimera compared to the proposed B′ parental virus. At least some of these questions may be answered by the availability of the B′ parental virus, the knowledge of breakpoints, the possibility of reconstituting replication-competent molecular clones (in process), and the existence of a wide variety of test systems allowing us to analyze distinct viral functions in vitro, in cell culture, and in appropriate small-animal models.
Finally, the high incidence of new infections in combination with the homogenous seed virus following a single and well-documented transmission route may suggest that IDUs from the northwestern and southwestern area form a potential high-risk population group for safety and efficacy vaccine trials in China. Initial analysis has demonstrated that probably about 50% of the CTL epitopes in Gag and RT are completely shared by prototype B variants and the C/(B′)-Thai interclade mosaics. These observations clearly predict a considerable cross-clade CTL reactivity, suggesting that the functionally and immunologically conserved HIV-1 proteins are strong potential candidates for future vaccine constructs.
In summary, this is to our knowledge the first report of a cloned virtual full-length C/(B′) chimeric HIV genome, which simultaneously represents one of the most prevalent subtype C virus strains from China. The reported data will be useful as a reference for future studies on the genetic diversity of HIV. Moreover, the established and carefully characterized clone may serve as a basis for the generation of subtype-specific immunological reagents and the development of candidate vaccines based on regional virus strains (27). Finally, the homogeneous seed virus with a single transmission route within a well-characterized population group suggests that this area is a good potential site for safety and efficacy vaccine trials.
ACKNOWLEDGMENTS
L.S. and M.G. contributed equally to this study.
We thank S. Piyasirisilp and X. Yu (Johns Hopkins University) and their colleagues at the Henry M. Jackson Foundation for helpful discussions and sharing of their data. We thank all the sample providers and the doctors from Health and Anti-Epidemic Stations in China for HIV serologic survey and blood sample collection.
This work was supported by European INCO-DC grant ERB 3514PL 962266.
REFERENCES
- 1.Bai X, Su L, Zhang Y, et al. Subtype and sequence analysis of the C2V3 region of gp120 gene among HIV-1 strains in Xinjiang. Chin J Virol. 1997;13:35–48. [Google Scholar]
- 2.Beyrer, C., M. H. Razak, K. Lisam, J. Chen, W. Lui, and X. F. Yu. Overland heroin trafficking routes and HIV-1 spread in south and south-east Asia. AIDS 14:75–83. [DOI] [PubMed]
- 3.Carr J K, Salminen M O, Albert J, Sanders Buell E, Gotte D, Birx D L, McCutchan F E. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- 4.Carr J K, Salminen M O, Koch C, Gotte D, Artenstein A W, Hegerich P A, St. Louis D, Burke D S, McCutchan F E. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–5943. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen B R. HIV-1 Nef protein: an invitation to a kill. Nat Med. 1999;5:985–986. doi: 10.1038/12417. [DOI] [PubMed] [Google Scholar]
- 6.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 7.Esparza J, Osmanov S, Heyward W L. HIV preventive vaccines. Progress to date. Drugs. 1995;50:792–804. doi: 10.2165/00003495-199550050-00003. [DOI] [PubMed] [Google Scholar]
- 8.Expert Group of Joint United Nations Programme on HIV/AIDS. Implications of HIV variability for transmission: scientific and policy issues. AIDS. 1999;11:UNAIDS 1–UNAIDS 15. [PubMed] [Google Scholar]
- 9.Frankel A D, Young J A. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao F, Robertson D L, Carruthers C D, Morrison S G, Jian B, Chen Y, Barré-Sinoussi F, Girard M, Srinivasan A, Abimiku A G, Shaw G M, Sharp P M, Hahn B H. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaywee J, Artenstein A W, VanCott T C, Trichavaroj R, Sukchamnong A, Amlee P, de Souza M, McCutchan F E, Carr J K, Markowitz L E, Michael R, Nittayaphan S. Correlation of genetic and serologic approaches to HIV-1 subtyping in Thailand. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:392–396. doi: 10.1097/00042560-199612010-00014. [DOI] [PubMed] [Google Scholar]
- 14.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graf M, Shao Y, Zhao Q, Seidl T, Kostler J, Wolf H, Wagner R. Cloning and characterization of a virtually full-length HIV type 1 genome from a subtype B′-Thai strain representing the most prevalent B-clade isolate in China. AIDS Res Hum Retroviruses. 1998;14:285–288. doi: 10.1089/aid.1998.14.285. [DOI] [PubMed] [Google Scholar]
- 16.Graham B S, Wright P F. Candidate AIDS vaccines. N Engl J Med. 1995;333:1331–1339. doi: 10.1056/NEJM199511163332007. [DOI] [PubMed] [Google Scholar]
- 17.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 18.Kondo E, Mammano F, Cohen E A, Göttlinger H G. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostrikis L G, Bagdades E, Cao Y, Zhang L, Dimitriou D, Ho D D. Genetic analysis of human immunodeficiency virus type 1 strains from patients in Cyprus: identification of a new subtype designated subtype I. J Virol. 1995;69:6122–6130. doi: 10.1128/jvi.69.10.6122-6130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leitner T, Albert J. A new genetic subtype of HIV-1. In: Myers G, Korber B, Wain-Hobson S, Jeang K T, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N, editors. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. pp. III147G–III150G. [Google Scholar]
- 21.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loussert Ajaka I, Chaix M L, Korber B, Letourneur F, Gomas E, Allen E, Ly T D, Brun Vezinet F, Simon F, Saragosti S. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J Virol. 1995;69:5640–5649. doi: 10.1128/jvi.69.9.5640-5649.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers G, Korber B, Foley B, Jeang K T, Mellors J W, Wain Hobson S. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1996. [Google Scholar]
- 24.Paulus C, Hellebrand S, Tessmer U, Wolf H, Krausslich H G, Wagner R. Competitive inhibition of human immunodeficiency virus type-1 protease by the Gag-Pol transframe protein. J Biol Chem. 1999;274:21539–21543. doi: 10.1074/jbc.274.31.21539. [DOI] [PubMed] [Google Scholar]
- 25.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philippe H. MUST, a computer package of Management Utilities for Sequences and Trees. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Piyasirisilp S, McCutchan F E, Carr J K, Sanders-Buell E, Wolf H, Liu W, Chen J, Shao Y, Lai S, Beyrer C, Yu X-F. A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J Virol. 2000;74:11286–11295. doi: 10.1128/jvi.74.23.11286-11295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruprecht R M. Live attenuated AIDS viruses as vaccines: promise or peril? Immunol Rev. 1999;170:135–149. doi: 10.1111/j.1600-065x.1999.tb01335.x. [DOI] [PubMed] [Google Scholar]
- 28.Salminen M O, Carr J K, Burke D S, McCutchan F E. Identification of breakpoints in intergenomic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 29.Salminen M O, Koch C, Sanders-Buell E, Ehrenberg P K, Michael N L, Carr J K, Burke D S, McCutchan F E. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology. 1995;213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 30.Shao Y, Guan Y, Zhao Q, et al. Genetic variation and molecular epidemiology of the Ruily HIV-1 strains of Yunnan in 1995. Chin J Virol. 1999;12:9. [Google Scholar]
- 31.Shao Y, Zeng Y, Chen Z, et al. Isolation of viruses from HIV infected individuals in Yunnan. Chin J Epidemiol. 1991;12:129. [Google Scholar]
- 32.Shao Y, Zhao Q B, Wang B, et al. Sequence analysis of HIV env gene among HIV infected IDUs in Yunnan epidemic area of China. Chin J Virol. 1994;10:291–299. [Google Scholar]
- 33.Sharp P M, Bailes E, Robertson D L, Gao F, Hahn B H. Origins and evolution of AIDS viruses. Biol Bull. 1999;196:338–342. doi: 10.2307/1542965. [DOI] [PubMed] [Google Scholar]
- 34.Sharp P M, Robertson D L, Hahn B H. Cross-species transmission and recombination of ‘AIDS’ viruses. Philos Trans R Soc London B Ser. 1995;349:41–47. doi: 10.1098/rstb.1995.0089. [DOI] [PubMed] [Google Scholar]
- 35.Smith T F, Waterman M S. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 36.Wagner R, Deml L, Notka F, Wolf H, Schirmbeck R, Reimann J, Teeuwsen V, Heeney J. Safety and immunogenicity of recombinant human immunodeficiency virus-like particles in rodents and rhesus macaques. Intervirology. 1996;39:93–103. doi: 10.1159/000150480. [DOI] [PubMed] [Google Scholar]
- 37.Wagner R, Deml L, Teeuwsen V, Heeney J, Yiming S, Wolf H. A recombinant HIV-1 virus-like particle vaccine: from concepts to a field study. Antibiot Chemother. 1996;48:68–83. doi: 10.1159/000425160. [DOI] [PubMed] [Google Scholar]
- 38.Weniger B G, Takebe Y, Ou C Y, Yamazaki S. The molecular epidemiology of HIV in Asia. AIDS. 1994;8(Suppl. 2):S13–28. [PubMed] [Google Scholar]
- 39.World Health Organization Network for HIV Isolation and Characterization. HIV-1 variation in WHO-sponsored vaccine-evaluation sites: genetic screening, sequence analysis and preliminary biological characterization of selected viral strains. AIDS Res Hum Retroviruses. 1994;10:1327–1344. doi: 10.1089/aid.1994.10.1327. [DOI] [PubMed] [Google Scholar]
- 40.Yu H, Su L, Shao Y. Identification of the HIV-1 subtypes by HMA and sequencing. Chin J Epidemiol. 1997;18:201–204. [Google Scholar]
- 41.Yu X, Yu Q C, Lee T H, Essex M. The C terminus of human immunodeficiency virus type 1 matrix protein is involved in early steps of the virus life cycle. J Virol. 1992;66:5667–5670. doi: 10.1128/jvi.66.9.5667-5670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Qu S, et al. A cohort study of HIV infection among intravenous drug users in Ruili and two countries in Yunnan Province, China. Chin J Epidemiol. 1997;18:1–4. [PubMed] [Google Scholar]