Abstract
Myelofibrosis and osteosclerosis are fibrotic diseases disrupting bone marrow function that occur in various leukemias but also in response to non-malignant alterations in hematopoietic cells. Here we show that endothelial cell–specific inactivation of the Lats2 gene, encoding Hippo kinase large tumor suppressor kinase 2, or overexpression of the downstream effector YAP1 induce myofibroblast formation and lead to extensive fibrosis and osteosclerosis, which impair bone marrow function and cause extramedullary hematopoiesis in the spleen. Mechanistically, loss of LATS2 induces endothelial-to-mesenchymal transition, resulting in increased expression of extracellular matrix and secreted signaling molecules. Changes in endothelial cells involve increased expression of serum response factor target genes, and, strikingly, major aspects of the LATS2 mutant phenotype are rescued by inactivation of the Srf gene. These findings identify the endothelium as a driver of bone marrow fibrosis, which improves understanding of myelofibrotic and osteosclerotic diseases, for which drug therapies are currently lacking.
Subject terms: Cardiovascular biology, Calcification
Sivaraj, Majev et al. demonstrate that the inactivation of Lats2 in endothelial cells triggers the upregulation of serum response factor and endothelial-to-mesenchymal transition, leading to myofibroblast formation, bone marrow fibrosis, osteosclerosis, impaired bone marrow function and extramedullary hematopoiesis.
Main
Fibrosis is associated with tissue injury, chronic inflammation and numerous other pathologies. In bone, fibrosis occurs in a variety of malignant and non-malignant disease conditions but also after acute or chronic exposure to radiation1. In certain groups of rare blood cancers, namely in primary myelofibrosis and in myelodysplastic syndromes, bone marrow (BM) fibrosis is a risk factor and most prevalent in patients with advanced disease and poor prognosis2. Excessive deposition of extracellular matrix (ECM) and tissue scarring contribute to impaired BM function, resulting in extramedullary hematopoiesis (EMH) and splenomegaly. Advanced bone fibrosis in human patients, but also in mouse models, can be accompanied by osteosclerosis, which manifests as irregular thickening of trabecular bone and abnormally elevated bone density3,4.
Bone marrow mesenchymal stromal cells (BMSCs), especially cells expressing Leptin receptor (LepR) or the zinc finger protein Gli1, were previously shown to contribute to osteogenesis5–7. These BMSC populations are also critical mediators of myelofibrosis and osteosclerosis8–10. Expression of tumor necrosis factor α and JAK-STAT signaling were shown to be increased in pre-fibrotic BMSCs, whereas strong upregulation of TGFβ signaling is predominant in overt fibrosis11. Elevated numbers and aberrant properties of megakaryocytes are also associated with myeloproliferation, BM fibrosis and destruction of the hematopoietic microenvironment12. Other alterations affecting hematopoietic cells, such as hypomorphic mutations in the transcription factor GATA1 or high expression of the glycoprotein hormone thrombopoietin (ThPO), cause myelofibrosis in mice13.
Although the role of endothelial cells (ECs) in myelofibrosis remains little understood, it was previously shown that ECs of small vessels in BM and spleen of the JAK2-V617F knock-in mouse model, but also in patients with primary myelofibrosis, acquire the expression of mesenchymal markers, which is a hallmark of a process known as endothelial-to-mesenchymal transition (EndMT)14. ECs play critical roles in normal osteogenesis and hematopoiesis15,16. In the metaphysis, specialized capillary ECs, termed type H because of high expression of the cell surface proteins CD31/Pecam1 and endomucin (EMCN), provide growth factor signals acting on perivascular osteoprogenitors and other osteoblast (OB) lineage cells expressing the transcription factors Runx2 and Osterix (OSX)16. The behavior and function of ECs is controlled by external growth factor signals but also by fluid shear stress, matrix stiffness and other mechanical signals17. The transcriptional coactivators YAP1 and TAZ, which are effectors in the Hippo signaling pathway, are capable of integrating a range of upstream signals to control cell proliferation, differentiation and tissue growth18. YAP1/TAZ interact in the nucleus with DNA-binding transcription factors, such as the TEA domain family members TEAD1–4, and thereby regulate gene expression19. Activation of the Hippo signaling cascade and large tumor suppressor homolog 1/2 (Lats1/2) kinase-mediated phosphorylation of YAP1/TAZ triggers exclusion of the transcriptional coactivators from the nucleus and promotes their proteolytic degradation20. In the endothelium of most organs, YAP1/TAZ activity promotes angiogenic vessel growth, but, surprisingly, the two coactivators suppress EC proliferation and postnatal angiogenesis in bone21.
Here we demonstrate that EC-specific loss of the Lats2 gene in ECs leads to extensive changes in the adult bone vasculature, EndMT and increased expression of ECM molecules and paracrine (also termed angiocrine) factors. These EC-specific changes promote the differentiation of metaphyseal BMSCs into myofibroblasts, resulting in bone fibrosis, osteosclerotic bone formation, impaired BM function and EMH. Loss of endothelial Lats2 also impairs the formation of cartilage-resorbing septoclasts and results in the accumulation of chondrocytes (CHOs) in the mutant metaphysis. Mutant defects appear to be confined to the skeletal system, presumably due to compensation by LATS1 in other organs. These findings and the analysis of mouse models of myelofibrosis support that ECs play an instrumental role in bone fibrosis and need to be considered in disease etiology and in the context of therapeutic approaches.
Results
Endothelial LATS2 prevents bone vessel defects and fibrosis
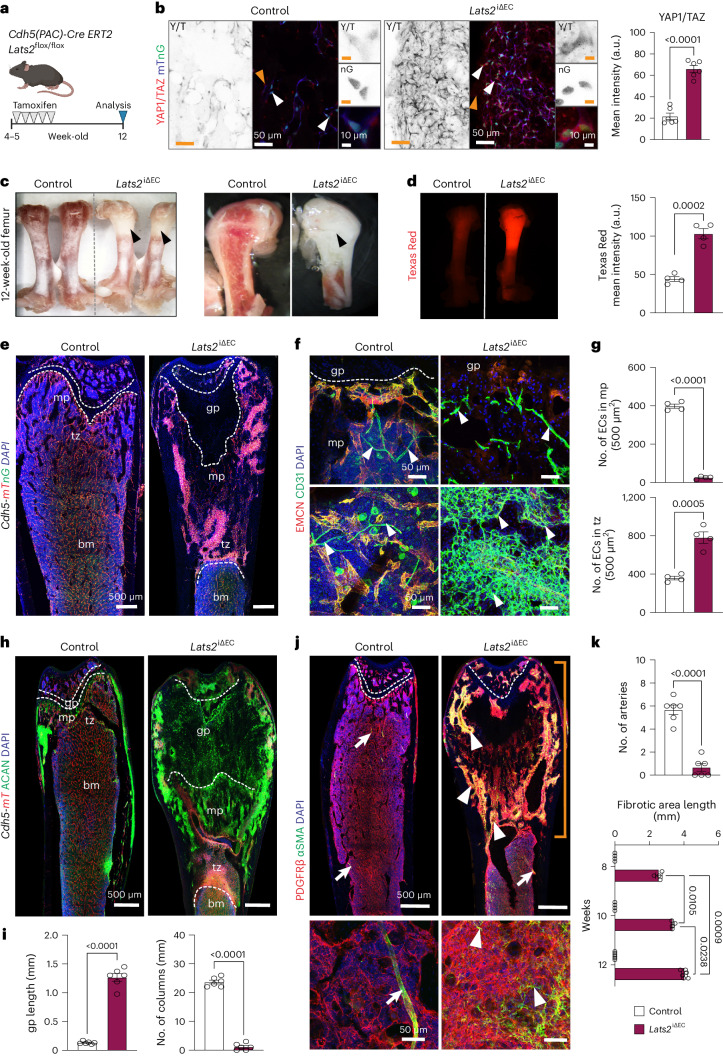
We increased YAP1/TAZ protein stability in ECs by deleting the upstream negative regulator LATS2 during bone remodeling. For this purpose, the tamoxifen-inducible, EC-specific Cdh5-CreERT2 transgene was introduced into animals carrying a loxP-flanked (floxed) version of the Lats2 gene. Tamoxifen administration for 5 d in a row at the age of 4–5 weeks generated Lats2 loss-of-function mutants (Lats2iΔEC), which were analyzed at the age of 12 weeks (Fig. 1a). Survival, body weight and the external appearance of Lats2iΔEC mutant mice is normal, and YAP1/TAZ immunostaining in the bone vasculature is increased relative to littermate controls (Fig. 1b and Extended Data Fig. 1a). Endothelial expression of Lats2, which is most prominent in arteries and metaphyseal (type H) vessels, is strongly reduced in Lats2iΔEC mutants both at the transcript and protein level (Extended Data Fig. 1b,c). The metaphysis of freshly isolated Lats2iΔEC femurs appears whiteish, indicating a decrease of red marrow, and shows enhanced vascular leakage (Fig. 1c,d). The Lats2iΔEC bone vasculature is substantially altered. Metaphyseal capillaries are very sparse, and an abnormally dense network of CD31+ ECs is visible in the transition zone connecting the metaphysis to the adjacent diaphysis (Fig. 1e–g and Extended Data Fig. 1d,e). Capillaries of the mutant metaphysis and transition zone show elevated CD31 immunostaining but decreased EMCN signal, have a small diameter or lack a lumen and express CAV1 (Caveolin 1), a caveolar protein that is highly abundant in control arterial ECs (Fig. 1f and Extended Data Fig. 1e). The Lats2iΔEC chondro-osseous junction next to the growth plate is strongly disorganized, and Aggrecan+ hypertrophic CHOs occupy a large region of the metaphysis, which is confirmed by both Safranin O histological staining and Collagen X immunostaining (Fig. 1h,i and Extended Data Fig. 2a,b).
Fig. 1. Endothelial Lats2 controls bone homeostasis and fibrosis.
a, Scheme showing tamoxifen-induced Lats2 inactivation in ECs with Cdh5-CreERT2 transgenic mice. b, Representative confocal images showing increased YAP1/TAZ immunostaining in Lats2iΔEC mutant bones. Endothelial YAP1/TAZ expression and nuclear localization were quantified (n = 6) by the mean intensity (a.u.) on the right side. Data are presented as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). c, Freshly dissected 12-week-old control and Lats2iΔEC mutant femurs (left) and longitudinal cross-section (right). Black arrowheads highlight pale areas. d, Microscopy images showing extravasation of Texas Red in control and Lats2iΔEC mutant femur. On the right, the mean intensity (a.u.) of Texas Red is quantified (n = 4). Data are presented as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). e, Confocal tile scan longitudinal view of Lats2iΔEC and control femur in Cdh5-mTnG background. ECs, red (dTomato) and green (nuclear H2B-GFP), DAPI (blue). f,g, Maximum intensity projections of EMCN+ (red) and CD31+ (green) vessels in the femoral metaphysis (mp) (top) and transition zone (tz) (bottom). Note the abundance of mutant CD31+ EMCN− ECs (f). Number of vessels and ECs is significantly reduced in the Lats2iΔEC mp and increased in the tz area compared to control (g). Data (n = 4) are presented as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). h,i, Tile scan confocal image showing avascular region filled with Aggrecan+ (ACAN; green) chondrocytes in Lats2iΔEC metaphysis (h). Quantification showing increased growth plate (gp) length and loss of type H vessel columns in Lats2iΔEC mp (n = 6). Presented as mean ± s.e.m. P values, Mann–Whitney test (two-tailed) (i). j, Tile scan confocal image and high-magnification image (below) showing widespread expression of PDGFRβ (red) and αSMA (green, arrowheads) in Lats2iΔEC mutant femur. αSMA signal in control is confined to arterial SMCs. k, Quantification of arteries (n = 6) and length of fibrotic area (8-week-old and 10-week-old mice, n = 4; 12-week-old mice, n = 6). Mean ± s.e.m. P values, Mann–Whitney test (two-tailed) and Tukey multiple comparison test (one-way ANOVA). n indicates the number of independent biological samples.
CHO resorption is mediated by vessel-associated FABP5+ septoclasts, which show high expression of metalloproteinases, including MMP9 (refs. 22,23). FABP5+ cells and MMP9 immunosignals are concentrated at the control chondro-osseous junction but are lost in Lats2iΔEC mutant samples (Extended Data Fig. 2c). Another striking alteration in the Lats2iΔEC mutant metaphysis is the strong upregulation of markers associated with tissue scarring and fibrosis, such as platelet-derived growth factor receptor β (PDGFRβ) and α-smooth muscle actin (αSMA+) (Fig. 1j,k and Extended Data Fig. 2d,e). Furthermore, AZAN trichrome and Reticulin histological staining, together with immunostaining for the ECM protein Collagen fibers, indicate widespread fibrosis in the Lats2iΔEC metaphysis (Extended Data Fig. 2f,g).
To gain insight into the emergence of vascular defects and fibrosis in Lats2iΔEC mutant bone, we analyzed samples at 8 weeks and 10 weeks of age. Increased perivascular αSMA expression is already visible at 8 weeks, and the disorganized area is further increased by 10 weeks (Extended Data Fig. 3a,b). Changes in the vasculature of the metaphysis and the transition zone bordering the diaphysis follow a similar timecourse (Extended Data Fig. 3c,d). Remarkably, fibrosis and vascular alterations in 12-week-old mutants are confined to bone, whereas no overt defects can be seen in kidney, lung, liver and heart sections (Extended Data Fig. 3e). Thus, loss of endothelial LATS2 leads to organ-specific defects in bone.
Osteosclerotic defects after loss of endothelial LATS2
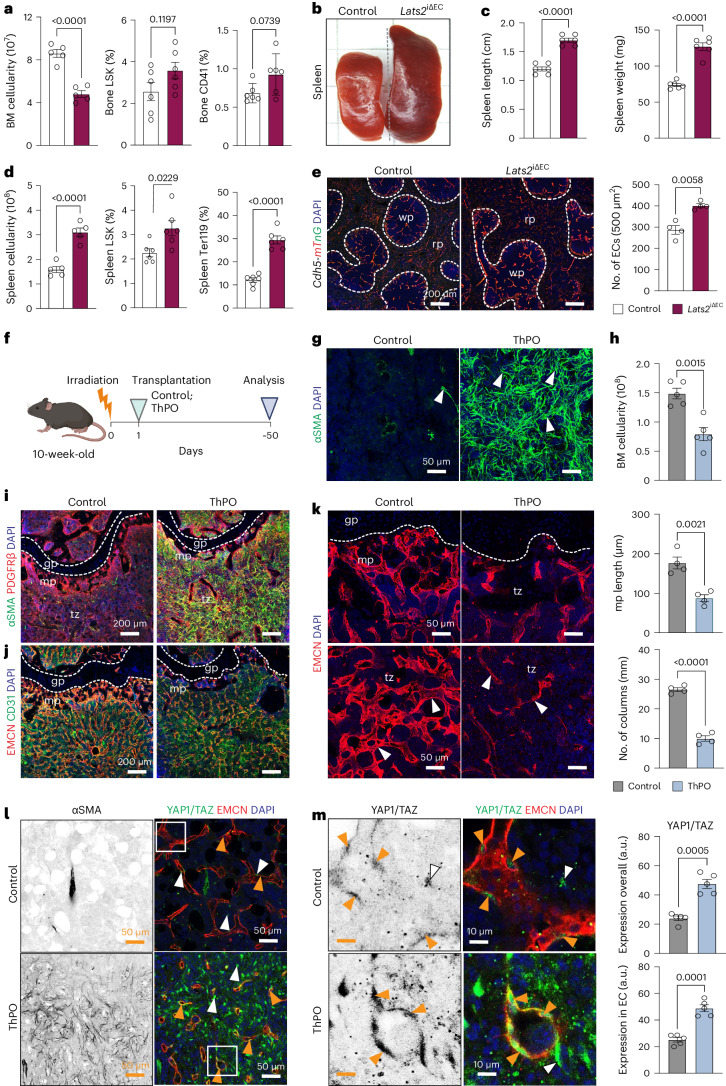
Bone fibrosis is known to cause the displacement of hematopoietic cells from BM, which leads to progressive splenomegaly due to EMH24. Analysis of Lats2iΔEC peripheral blood shows an elevation of white blood cells, whereas red blood cells and platelets are similar to control (Extended Data Fig. 4a). In Lats2iΔEC BM, total cellularity and Ter119+ erythrocytes and B220+ B lymphocytes are decreased, whereas Lin−Sca-1+c-Kit+ (LSK) hematopoietic stem and progenitor cells (HSPCs) are not significantly altered (Fig. 2a and Extended Data Fig. 4b). CD41+ megakaryocytes are known to act as main drivers of myelofibrosis24, but these cells are absent from the Lats2iΔEC fibrotic area, and their total number is not significantly altered in mutant femurs due to their presence in the diaphyseal BM (Fig. 2a and Extended Data Fig. 4c,d). Consistent with EMH, Lats2iΔEC mutants show increased spleen volume and cellularity together with an increase in splenic HSPCs and Ter119+ cells, whereas the fractions of CD45+ leukocytes and B220+ cells are reduced (Fig. 2b–d and Extended Data Fig. 4e). In spleen sections, the area of red pulp is increased relative to white pulp, a feature of EMH (Fig. 2e and Extended Data Fig. 4f,g). These alterations are likely to be secondary to the defects in Lats2iΔEC BM, as YAP1/TAZ immunostaining in splenic vessels is not increased (Extended Data Fig. 4h). Thus, Lats2iΔEC mutants display splenomegaly, BM fibrosis and impaired BM function, resembling key features of myelofibrosis.
Fig. 2. Loss of LATS2 promotes EMH.
a, Quantification of BM cellularity and FACS analysis of LSK and CD41+ cell frequency (n = 6). Mean ± s.e.m. P values, Mann–Whitney test (two-tailed). b,c, Images showing freshly dissected Lats2iΔEC and control spleen (b) and quantification of spleen size and weight (c) (n = 6). Data are presented as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). d, Quantification of spleen cellularity and frequency of LSK and Ter119 cells (n = 6). Data are presented as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). e, Sections of control and Lats2iΔEC spleens in Cdh5-mTnG reporter background. Red pulp (rp) and white pulp (wp) are indicated. Nuclei, DAPI (blue). Quantification showing increased EC number in Lats2iΔEC mutant (n = 4). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). f, Scheme showing ThPO-induced BM fibrosis. g, Representative confocal images showing αSMA (green) immunostaining in ThPO and control bone. Nuclei, DAPI (blue). h, Quantification of BM cellularity of control and ThPO mice (n = 5). Data are presented as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). i,j, Representative confocal images showing increase of metaphyseal αSMA signal (i) and reduction of EMCN+ CD31+ vessels (j) in ThPO femur relative to control. k, EMCN+ ECs (red) in metaphysis (mp) (top) and diaphysis (dp) (bottom). Nuclei, DAPI (blue). Graphs on the right show decreased mp length and vessel columns after ThPO (n = 4). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). l,m, Representative single optical confocal images showing increased expression of αSMA (gray) and YAP1/TAZ (green) in EMCN (red) ECs (orange arrowheads) in ThPO bone (l). Cropped zoomed-in image displays YAP1/TAZ nuclear (DAPI, blue) localization in ECs (m). Quantification shows increase of overall and EC-specific Yap1/TAZ immunosignal in ThPO bones (n = 5). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). n indicates the number of independent biological samples.
The etiology of myelofibrosis in human subjects is poorly understood, but previous work established that BM fibrosis in mice can be induced by overexpressing ThPO from hematopoietic cells, which results in the expansion of megakaryocytes, elevated expression of platelet-derived growth factor and transforming growth factor β and the activation of mesenchymal stromal cells10,25. ThPO overexpression in HSPCs first leads to leukocytosis and thrombocytosis and, over time, with onset of fibrosis, reduced hemoglobin and erythrocyte counts (Fig. 2f and Extended Data Fig. 5a). The ThPO overexpression model mirrors aspects of the Lats2iΔEC phenotype: increased and widespread αSMA immunostaining reflecting fibrosis, a general loss of cellularity in bone as well as an increase in spleen weight (Fig. 2g–i and Extended Data Fig. 5b,d). Remarkably, ThPO overexpression also induces a marked reduction of metaphyseal type H vessels and lowers endothelial EMCN expression (Fig. 2j,k and Extended Data Fig. 5c). Consistent with the loss of type H vessels, perivascular OSX+ OB lineage cells are strongly reduced in long bones of ThPO-overexpressing mice (Extended Data Fig. 5e). Strikingly, ThPO induces a strong increase in YAP1/TAZ immunostaining in fibrotic BM (Extended Data Fig. 5f), but it also significantly enhances YAP1/TAZ in ECs relative to control (Fig. 2l,m). Somatic activating mutations (such as W515L) in the transmembrane domain of the ThPO receptor, encoded by the MPL gene, are a driver of primary myelofibrosis in humans. Transplantation of MPLW515L-expressing hematopoietic cells can reproduce major features of the disease, including splenomegaly and extramedullary hematopoiesis, in mice26. In the MPLW515L model, fibrosis is accompanied by widespread upregulation of αSMA (Extended Data Fig. 5g,h). Strikingly, the EMCN+ BM vasculature of these mice is significantly reduced, and YAP1/TAZ immunostaining in vessels is upregulated (Extended Data Fig. 5h,i). These data show that vascular alterations and upregulation of YAP1/TAZ are a common feature in two established models of myelofibrosis.
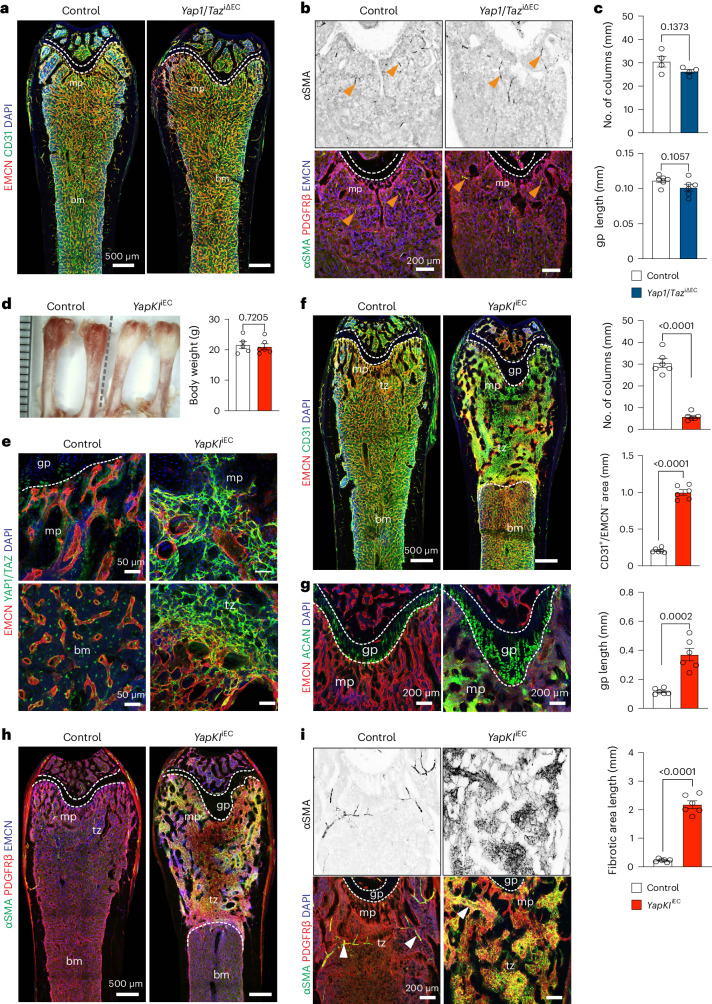
LATS2 and the related LATS1 kinase phosphorylate YAP1 and TAZ, which limits the activity of the two transcriptional coregulators due to cytoplasmic retention and proteasomal degradation. To directly address the role of YAP1 and TAZ, we generated tamoxifen-inducible, EC-specific loss-of-function double mutant mice (Yap1/TaziΔEC), which were given tamoxifen at the age of 4–5 weeks before analysis at 12 weeks. In contrast to the enhanced developmental angiogenesis in postnatal Yap1/TaziΔEC bone21, adult mutants display a normal organization of the bone vasculature and also lack alterations in PDGFRβ and αSMA expression (Fig. 3a–c). Overexpression of stabilized YAP1 (YAP1S112A) in ECs, however, causes the formation of sclerotic lesions, vascular impairment and the appearance of an EMCN-deficient endothelial network in the metaphysis of the resulting YapKIiEC gain-of-function mutants (Fig. 3d–f). Key defects observed in Lats2iΔEC mutants, such as expansion of chondrocytes and widespread expression of PDGFRβ and αSMA, are also observed in YapKIiEC animals (Fig. 3g–i).
Fig. 3. Overexpression of a stabilized Yap1 in ECs promotes BM fibrosis.
a, Tile scan confocal overview images of EMCN (red) and CD31 (green) immunostained vasculature in control and Yap1/TaziΔEC compound mutant femur. Metaphysis (mp), bone marrow (bm) and growth plate (gp) (dashed lines) are indicated. b,c, Representative confocal image of femoral αSMA+ (green) smooth muscle cells covering arteries (arrows) (above). EMCN+ (blue) blood vessels and PDGFRβ+ cells (red) in Yap1/TaziΔEC mutant and littermate control (below). Metaphyseal vessel columns and gp are not altered in Yap1/TaziΔEC bone (c) (n = 4, 6). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). d, Freshly dissected 12-week-old YapKIiEC gain-of-function and control femurs. Quantitative analysis of YapKIiEC and littermate control body weight (n = 5). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). e, Representative confocal images showing increased YAP1/TAZ (green) expression in YapKIiEC blood vessels (EMCN+, red) relative to control. f. Tile scan longitudinal view of EMCN and CD31 immunostaining in femur sections. Quantification shows decrease in number of vessel columns and increase in CD31+/EMCN− vascular area in YapKIiEC mutant (n = 6). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). g, Representative confocal images showing increase in Aggrecan+ (ACAN+) chondrocytes in YapKIiEC femur. Graph shows increased size of gp relative to control (n = 6). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). h,i. Tile scan confocal longitudinal view of PDGFRβ+ (red) and αSMA+ (green) cells. αSMA in control sections is largely confined to arterial smooth muscle cells (h). High-magnification images of αSMA+ (gray) fibrotic area is shown on the top right. gp (dashed line), metaphysis (mp) and transition zone (tz) to BM are indicated (i) (n = 6). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). n indicates the number of independent biological samples.
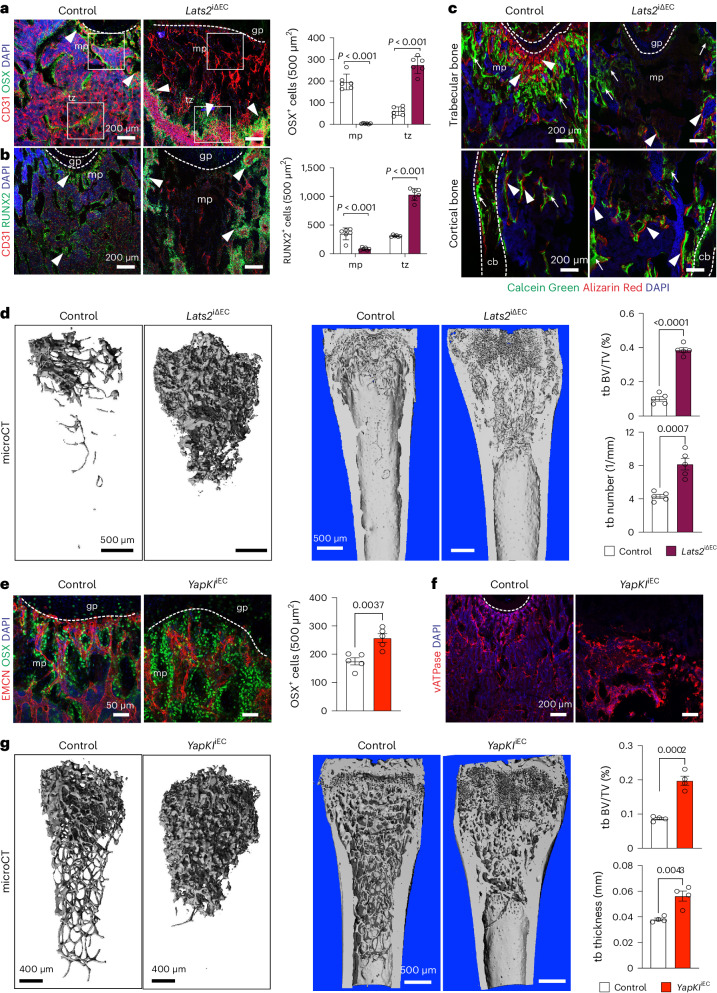
Previous work showed that metaphyseal type H capillaries are associated with osteoprogenitor cells and actively promote osteogenesis16. OSX+ cells and RUNX2+ osteoprogenitors are strongly reduced in the Lats2iΔEC metaphysis in regions that have lost capillaries. In contrast, OB lineage cells are enriched around the dense but disorganized endothelial network in the mutant transition zone (Fig. 4a,b). Analysis of bone remodeling using the Calcein Green and Alzarin Red double labeling approach (Methods) shows decreased osteogenesis and resorption in Lats2iΔEC long bone (Fig. 4c). Vacuolar ATPase (vATPase)-expressing osteoclasts are decreased in the avascular metaphysis and elevated in the mutant transition zone (Extended Data Fig. 6a). Microcomputed tomography (µCT) shows highly increased trabecular bone volume, quantity and thickness in Lats2iΔEC mutant femurs, whereas cortical bone thickness is not changed (Fig. 4d and Extended Data Fig. 6b,c). Supporting that the role of LATS2 in bone endothelium involves the modulation of YAP1/TAZ, YapKIiEC gain-of-function increases the number of osteoprogenitors, alters the distribution of osteoclasts and leads to sclerotic mineralization (Fig. 4e–g). In contrast, adult Yap1/TaziΔEC loss-of-function mutants show a modest decrease of OSX+ osteoprogenitors, whereas bone mineralization is unaffected (Extended Data Fig. 6d,e). These findings indicate that YAP1/TAZ levels are normally kept low in the adolescent/adult bone endothelium so that EC-specific overexpression of YAP1 or loss of LATS2 severely impair bone remodeling and homeostasis.
Fig. 4. Endothelial Hippo signaling controls osteosclerosis.
a,b, Representative confocal images showing OSX+ cells (a) and RUNX2+ osteoprogenitors (b) (arrowheads) in 12-week-old Lats2iΔEC femur and littermate control (n = 6). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). c, Representative confocal image showing double labeling with Calcein Green and Alizarin Red in metaphysis (mp) (top) and border to diaphysis (dp) (bottom) in control and Lats2iΔEC femur. Arrow highlights Calcein Green deposition, and arrowheads show double labeling of deposition/resorption. d, Representative µCT images of Lats2iΔEC and control femoral trabecular bone (tb). Quantitative analysis of tb volume (bone volume/total volume (BV/TV)) and number are shown on the right (n = 5). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). e,f, Representative confocal image showing OSX+ cells in control and YapKIiEC femoral metaphysis (n = 5). Data are shown as mean ± s.e.m. P values, Mann–Whitney test (two-tailed) (e). vATPase+ osteoclasts (red) are reduced in the YapKIiEC fibrotic area (f). g, Representative µCT images of tb in control and YapKIiΔEC femur. Quantitative analysis of tb volume (BV/TV) and number (n = 4). Data are presented as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). n indicates the number of independent biological samples.
LATS2 deletion induces EndMT
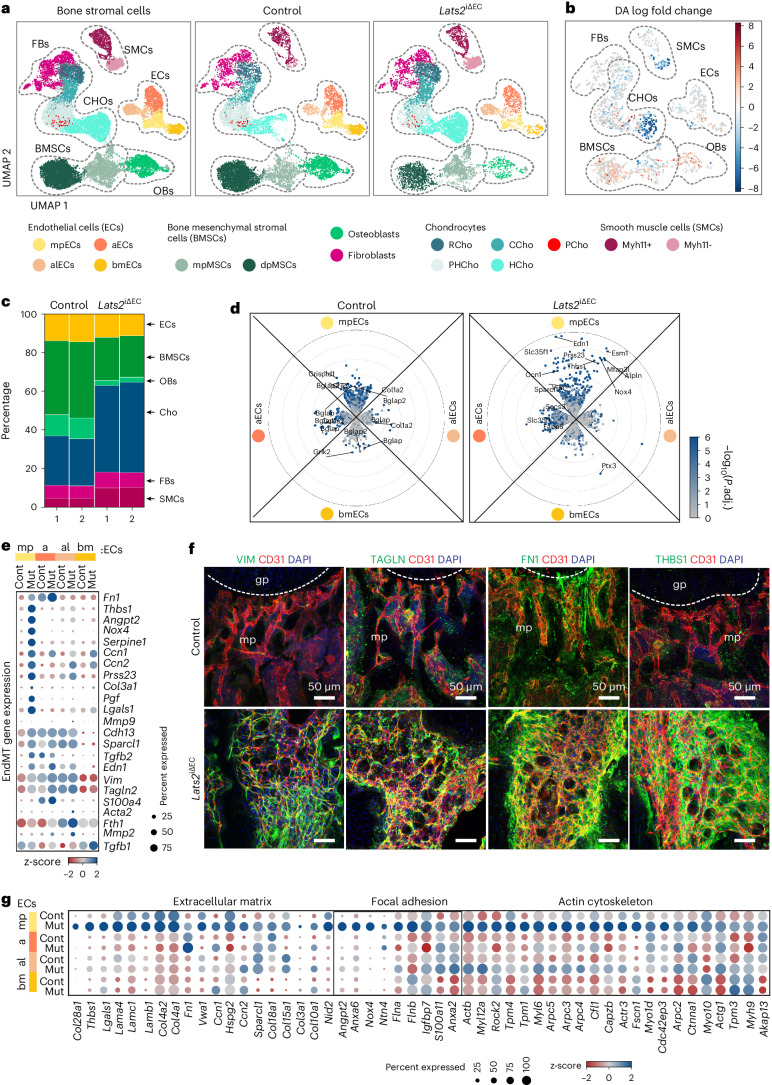
To determine stromal cell heterogeneity, fate specification and cellular crosstalk in Lats2iΔEC mutant and littermate control long bone, we performed droplet-based single-cell RNA sequencing (scRNA-seq) of non-hematopoietic bone stromal cells with the 10x Chromium platform. We obtained a total of 20,184 cells after removing doublets and residual hematopoietic cells from scRNA-seq experiments of control and Lats2iΔEC mutants with two biological replicates each. Based on the expression of known marker genes, we identified six primary cell types: ECs, BMSCs, OBs, smooth muscle cells (SMCs), CHOs and fibroblasts as well as their subpopulations. BMSC numbers are decreased and CHO and SMC numbers are increased in Lats2iΔEC mutants (Fig. 5a–c and Extended Data Fig. 7a–c). Within the EC cluster, metaphyseal (type H) ECs (mpECs, Ramp3+), bone marrow (type L) ECs (bmECs, Stab2+), arteriole-like ECs (alECs, Fmo2+) and arterial ECs (aECs, Gja4+) can be readily identified (Extended Data Fig. 7d–f). When compared to controls, Lats2iΔEC ECs exhibit numerous downregulated but also strongly upregulated differentially expressed genes (DEGs) (Extended Data Fig. 7g). Lats2iΔEC mutant mpEC and aEC subpopulations show particularly large numbers of upregulated genes (Fig. 5d). Expression of the YAP1/TAZ-controlled target genes Ccn1 (encoding cysteine-rich angiogenic inducer 61 (CYR61)), Ccn2 (connective tissue growth factor (CTGF)), Angpt1 (angiopoietin 1) and Thbs1 (thrombospondin 1) is strongly increased (Extended Data Fig. 7h). Surprisingly, genes typical for mesenchymal cells and fibroblasts are also strongly upregulated in Lats2iΔEC mpECs. The differential expression of a subset of these genes, including Vim (encoding the intermediate filament protein vimentin), Tagln (smooth muscle 22α, SM22α), Fn1 (fibronectin 1), Thbs1 and Angpt2 is confirmed by immunostaining of the corresponding gene products in bone sections (Fig. 5e,f and Extended Data Fig. 8a). Additionally, we noticed that Lats2iΔEC mutant type H ECs (mpECs) show expression of gene signatures associated with ECM, cell–matrix adhesion and actin cytoskeleton (Fig. 5g and Extended Data Fig. 8b). Furthermore, immunostaining of two critical downstream regulators controlling cell–matrix interactions, active integrin β1 and phosphorylated focal adhesion kinase (FAK), is strongly increased (Extended Data Fig. 8b). Ultrastructural analysis by transmission electron microscopy confirms the profoundly increased deposition of matrix components in the Lats2iΔEC metaphysis and pronounced actin fibers in mutant ECs (Extended Data Fig. 8c). Genetic lineage tracing supports that Lats2iΔEC ECs are not fully converted into mesenchymal stromal cells (Extended Data Fig. 8d) but acquire a mesenchymal cell-like phenotype through EndMT, including the expression of typical mesenchymal markers and elevated cell–matrix interactions.
Fig. 5. EndMT after loss of LATS2.
a,b, UMAP plots showing merged (left) and separated control and Lats2iΔEC mutant (right) bone stromal cells color-coded by cell type (a). Changes in cellular abundance (differential abundance (DA)) as calculated with milopy. Negative values indicate increased abundance in Lats2iΔEC mutant (b). Metaphyseal ECs (mpECs), arteriole-like ECs (alECs), arterial ECs (aECs), metaphyseal MSCs (mpMSCs, Osteo-CAR), diaphyseal MSCs (dpMSCs, Adipo-CAR), resting chondrocytes (RCHOs), columnar chondrocytes (CCHOs), proliferating chondrocytes (PCHOs), pre-hypertrophic chondrocytes (PHCHOs), hypertrophic chondrocytes (HCHOs) as well as Myh11+ and Myh11− SMCs are indicated. c, Bar graph displaying percentage of each main bone stromal cell type. d, Radial expression plot (Methods) showing DGE of control and Lats2iΔEC EC subclusters. y axis shows log2 fold expression changes; color scale represents the adjusted P values. e, Dot plot for normalized and averaged expression indicates high expression of mesenchymal markers and fibroblast genes in Lats2iΔEC mpECs. z-scaled per row. f, Immunostaining showing high levels of Vimentin (VIM), Transgelin (TAGLN), Fibronectin 1 (FN1) and Thrombospondin 1 (THBS1) in Lats2iΔEC metaphysis (mp). Control growth plate (gp) is marked by the dashed line. ECs (CD31, red) and nuclei (DAPI, blue) are labeled. g, Row-wise z-scaled normalized expression values of ECM, focal adhesion and actin cytoskeleton genes in Lats2iΔEC mutant and control EC populations. FB, fibroblast.
Endothelial signals control myofibroblast generation
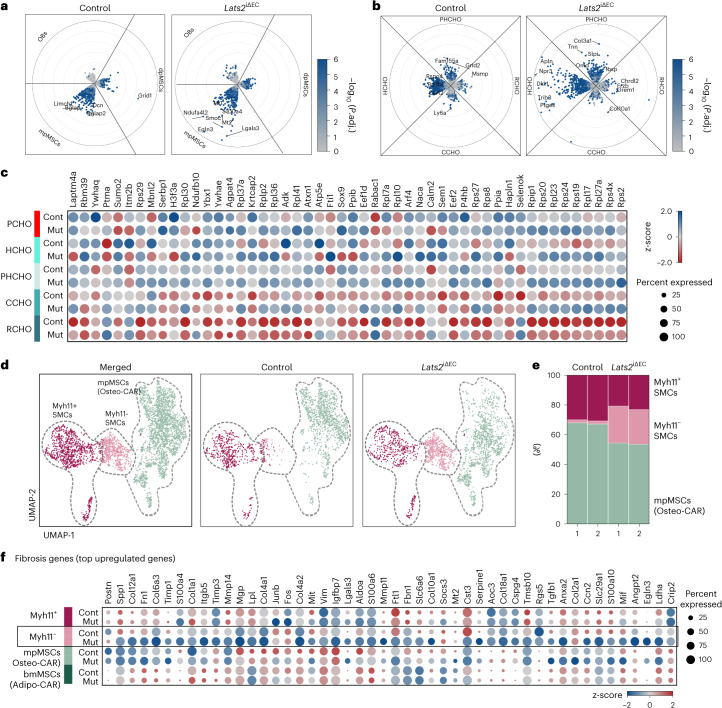
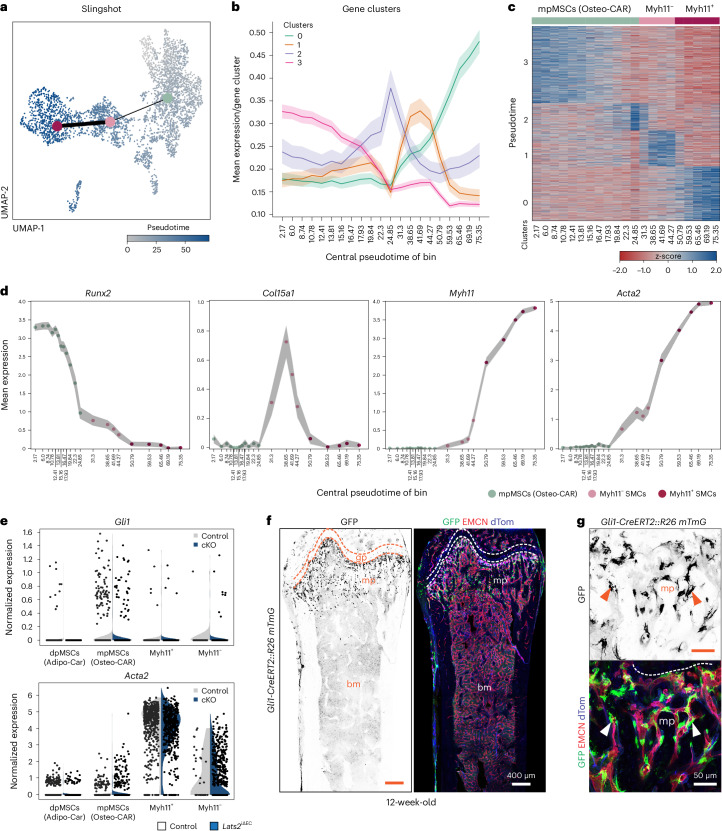
Previous research revealed that Lepr-Cre+ and Gli1+ mesenchymal stromal cells give rise to myofibroblasts during hematopoietic cell-induced myelofibrosis9,10. As the exact nature of the processes leading to bone fibrosis is incompletely understood, we analyzed gene expression changes in different cell populations. We noted that transcriptional alterations in Lats2iΔEC mpMSCs (Osteo-CAR) cells are more extensive than those seen in diaphyseal mesenchymal stromal cells (dpMSCs; Adipo-CAR cells) or OBs (Fig. 6a). In the CHO cluster, pre-hypertrophic chondrocytes (PHCHOs) and hypertrophic chondrocytes (HCHOs) show the largest number of DEGs (Fig. 6b,c). We also noted a strong enrichment of Myh11− cells in the Lats2iΔEC SMC cluster, which show increased expression of markers associated with fibrosis and are likely to represent myofibroblasts (Fig. 6d–f). Analysis of the same scRNA-seq data also shows a pronounced shift within the Lats2iΔEC mpMSC subcluster, indicative of altered gene expression relative to control (Fig. 6d). Pseudotime27 and partition-based graph abstraction (PAGA)28 analysis of the Lats2iΔEC scRNA-seq data suggest a potential differentiation trajectory from mpMSCs to Myh11− cells in the SMC cluster (Fig. 7a–d). Although Gli1 expression is most prominent in the mpMSC/Osteo-CAR subcluster, Acta2 transcripts (encoding αSMA) are abundant in Myh11+ SMCs and strongly upregulated in Lats2iΔEC Myh11− SMCs/myofibroblasts (Fig. 7e). Consistent with previous studies highlighting the role of Gli1+ cells as a source of myofibroblasts in bone fibrosis10, Gli1-CreERT2-mediated recombination of a Cre reporter allele labels vessel-associated BMSCs in the 12-week-old metaphysis, namely mpMSC/Osteo-CAR cells (Fig. 7f,g).
Fig. 6. EC-derived signals control mpMSCs and CHOs.
a–c, Radial expression plot (Methods) of BMSC/OB subsets, showing that most significant differences in regulation between conditions occur in mpMSCs (a). Within the CHO subset, most differences between conditions are confined to HCHOs. Proliferating CHOs are not shown, as number was less than 50 (b). Genes significantly differentially expressed (P.adj ≤ 0.01, log2FC < 0, frac. expressed ≥ 0.2) between control (Cont) and mutant (Mut) in at least one CHO sub-identity (c). d,e, UMAP visualization of mpMSC and SMC subclusters showing all cells (left) or split by genotype (right) (d). Percentage of cells in each cell type per sample is shown. Note increase in Lats2iΔEC Myh11− SMCs representing myofibroblasts (e). f, Row-wise z-scaled normalized expression values of fibrosis genes in Myh11− cells relative to mpMSCs, dpMSCs and Myh11+ SMCs.
Fig. 7. Conversion of mpMSCs into myofibroblasts.
a–c, UMAP plot colored by slingshot pseudotime. Centroid dots show cell type identity, and connecting lines indicate cell type connectivities calculated by PAGA (a). Visualization of mean expression in gene clusters as determined by Leiden clustering genes along the slingshot pseudotime trajectory; filled areas show 95% confidence interval (b). Heatmap of these gene clusters showing gene expression changes across pseudotime (c). d, Selected marker genes showing continuous gene expression changes across the pseudotime trajectory from mpMSC to Myh11+ SMC. Normalized expression counts averaged per pseudotime bin are shown; filled areas show 95% confidence interval. e, Violin plots showing Gli1 and Acta2 expression in dpMSCs/Adipo-CAR cells, mpMSCs/Osteo-CAR cells as well as Myh11+ and Myh11− SMCs of Lats2iΔEC mutants relative to control. f,g, Longitudinal tile scan confocal images showing Gli1-CreERT2-labeled cells (green) in the metaphysis (f). High-magnification images confirm perivascular location of GFP+ cells (g). EMCN (red) and dTomato (blue).
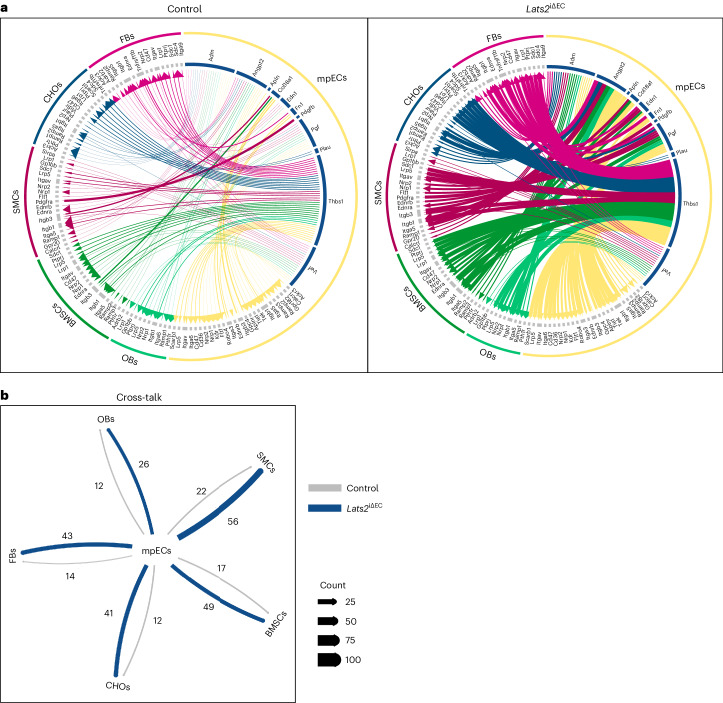
Next, we sought to understand which EC-derived signals might lead to the conversion of mpMSC/Osteo-CAR cells into myofibroblasts and the emergence of fibrosis in the Lats2iΔEC metaphysis. To identify potential ligand–receptor interactions (LRIs), we employed a newly developed approach (Methods) similar to CellPhoneDB29. Lats2iΔEC mutant mpECs show substantially increased paracrine signaling capability compared to other EC subtypes, reflected by strongly elevated expression of secreted molecules relative to control (Extended Data Fig. 9a). In turn, many of the respective receptors for these EC-derived ligands are expressed or even elevated in other cell types in Lats2iΔEC bone (Fig. 8a,b and Extended Data Fig. 9b). Specifically, genes involved in cell–matrix interactions (Thbs1, Fn1, Col4a1, Col18a1 and Plau), the glycoprotein von Willebrand factor (Vwf) and secreted signaling molecules, such as adrenomedullin (Adm), apelin (Apn), angiopoietin 2 (Angpt2) and endothelin 1 (Edn1), are upregulated in Lats2iΔEC mutant mpECs (Extended Data Fig. 9c). The corresponding receptors are expressed by multiple cell populations but are frequently found in mpMSCs and the Myh11− SMC subcluster (Extended Data Fig. 9c,d). Treatment of cultured BMSCs with recombinant endothelin 1 can increase the expression of fibrosis markers similar to TGFβ1, whereas adrenomedullin has no significant effect under the same conditions (Extended Data Fig. 9e). Based on these results, we propose that loss of LATS2 in ECs is sufficient to initiate bone fibrosis through the upregulation of EC-derived secreted molecules.
Fig. 8. Loss of Lats2 increases endothelial signaling interactions.
a, Ligand-receptor pairings indicating increased mpECs signaling to stromal cells in Lats2iΔEC mutants. Relative (min-max normalized) changes in normalized expression counts of ligand and receptor for selected interactions (LRI) (scaled between 0 and 1) originating from mpECs. Interactions are significant at P.adj of LRI score in at least one condition and LRIDiffScore ≤ 0.05 and LRIDiff ≥ 1. b, Number of significant LRIs between cell types of interest (P.adj. LRIScore ≤ 0.05; P.adj. LRIDiffScore ≤ 0.05; |LRIDiffScore| ≥ 1). FB, fibroblast.
YAP1/TAZ control EC plasticity through serum response factor
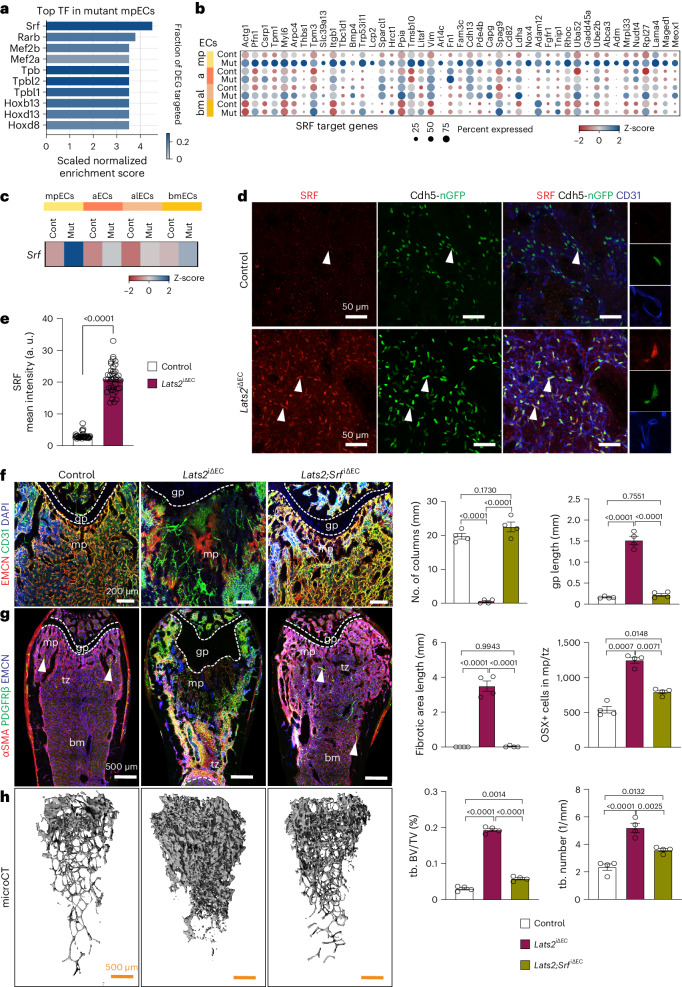
To gain insight into the mechanism controlling EndMT in Lats2iΔEC mutants, we used iRegulon30 to examine which transcription factors are likely regulators of the gene expression changes in mpECs. Serum response factor (SRF) is the top hit in this analysis, and SRF target genes are upregulated in Lats2iΔEC mpECs (Fig. 9a,b). Further arguing for a potential role of SRF, transcript and protein are highly increased in mutant mpECs relative to control (Fig. 9c–e). Srf inactivation in ECs (Srf iΔEC) alone has no significant effect on the bone vasculature, tissue organization, markers of fibrosis, growth plate size or mineralized bone, but CD31+/EMCN− vessels, representing arteries/arterioles, are slightly reduced, and OSX+ cells are increased (Extended Data Fig. 10a–e). In EC-specific Lats2 and Srf compound mutants (Lats2 Srf iΔEC) generated with the same tamoxifen administration regime described earlier, the lack of SRF largely restores the normal organization of metaphyseal vasculature and EMCN expression in the Lats2iΔEC background (Fig. 9f and Extended Data Fig. 10f–h). Vascular normalization is accompanied by the restoration of normal growth plate size and bone mineralization as well as reduction of PDGFRβ and αSMA immunostaining in Lats2 Srf iΔEC compound mutants (Fig. 9g,h and Extended Data Fig. 10i,j). The sum of these data shows that LATS2 limits YAP1/TAZ expression in bone ECs to prevent EndMT and SRF-mediated processes, leading to fibrosis and osteosclerotic defects (Extended Data Fig. 10k).
Fig. 9. YAP1/TAZ regulate EC plasticity through SRF.
a,b, Top transcription factors (TFs), predicted by iRegulon using genes significantly upregulated in Lats2iΔEC mpECs (325 total genes; log2FC ≥ 1, P.adj ≤ 0.05, frac. expressed ≥ 0.2) compared to control (a). Row-wise z-scaled normalized expression values of known SRF target genes (b). c–e, Srf transcript levels in row-wise z-scaled normalized expression counts (c) and immunostaining signals (d) are strongly increased in Lats2iΔEC mutant mpECs. Bar graph shows SRF mean intensity (a.u.) in ECs (e). Data (n = 4; 40 cells) are presented as mean ± s.e.m. P values, Mann–Whitney test (two-tailed). f, Maximum intensity projections of EMCN (red) and CD31 (green) immunostaining in control, Lats2iΔEC and Lats2 Srf iΔEC double mutant femur. Quantification shows number of type H vessel columns and growth plate (gp) length. g, Tile scan confocal longitudinal view showing PDGFRβ and αSMA immunostaining. Arrowheads indicate arterial αSMA+ SMCs. Graphs show length of fibrotic area and number of OSX+ cells. h, Representative µCT images of femoral trabecular bone (tb). Quantitative analysis of tb volume (bone volume/total volume (BV/TV)) and number. Data for f–h (n = 4 each) are shown as mean ± s.e.m. P values, Tukey multiple comparison test (one-way ANOVA). n indicates the number of independent biological samples.
Discussion
Our results demonstrate that loss of the LATS2 kinase endows bone ECs with disease-promoting properties, resulting in osteosclerotic defects, bone fibrosis, EMH and splenomegaly. This involves the upregulation of YAP1/TAZ, and, accordingly, EC-specific overexpression of stabilized YAP1S112A protein phenocopies major aspects of the defects seen in Lats2iΔEC loss-of-function bone. YAP1 and TAZ are well known for their prominent role in the regulation of cell proliferation and tissue growth. In the endothelium of most organs, combined inactivation of Yap1 and Taz impairs angiogenic blood vessel growth31,32, whereas the opposite phenotype, namely vascular overgrowth, occurs in the postnatal bone vasculature21. Our findings presented here indicate that YAP1 and TAZ are no longer required in the adult bone vasculature, and, instead, it is critical for healthy homeostasis that the activity of the two transcriptional coregulators is actively suppressed by LATS2.
Remarkably, loss of LATS2 is not sufficient to cause defects in the endothelium of multiple other organs, which might reflect redundant activity of the related kinase LATS1. It has been shown that LATS1 and LATS2 act non-redundantly in some biological contexts but can often compensate for each other33,34. In ECs of the growing retina and brain, combined inactivation of Lats1 and Lats2 results in hyperproliferation and metabolic activation through upregulation of the proto-oncogene c-Myc, which is rescued by the simultaneous inactivation of Yap1/Taz31. In the bone endothelium, however, LATS1 is unlikely to be a major player because it cannot prevent major defects in Lats2iΔEC mutants.
At the mechanistic level, our results link LATS2 to EndMT, SRF and the regulation of exocytosis. EndMT is a physiological process during embryonic heart development where it mediates the conversion of ECs into valve interstitial cells but also into cardiac pericytes and vascular SMCs35. By contrast, EndMT in the adult organism refers typically to the pathogenic acquisition of a mesenchymal gene signature by ECs in atherosclerosis, hypertension and some other cardiovascular diseases36. SRF is a critical regulator of actin polymerization, which, together with myocardin family cofactors, controls the expression of effectors of actin dynamics and, thereby, myofibroblast differentiation and tissue fibrosis in multiple organs37,38. Cooperation between YAP1/TAZ and SRF was reported for the differentiation of vascular SMCs but also in certain cancer cells and cancer-associated fibroblasts39–41. However, it was also shown that the crosstalk between MRTF-SRF and YAP1/TAZ is often indirect and mediated by cytoskeletal dynamics and cell shape changes40,42. Although the exact nature of the link between SRF and Hippo pathway components in the bone endothelium requires further investigation, our findings show that SRF is required for YAP1/TAZ-induced EndMT, which involves marked changes in EC morphology, gene expression and vascular architecture. These alterations in ECs, through increased expression of matrix molecules and other secreted factors, affect other cell types, most likely mesenchymal mpMSCs/Osteo-CAR cells, resulting in fibrosis and osteosclerotic lesions. Furthermore, the identification of vascular defects in the ThPO and MPL models of myelofibrosis raises the possibility that EndMT, increased biological activity of YAP1/TAZ and SRF-mediated changes in gene expression, but also EC cytoskeletal function, might contribute to fibrosis. These findings are consistent with an earlier study reporting EndMT in ECs of small vessels in the BM and spleen of the JAK2-V617F knock-in mouse model but also in patients with primary myelofibrosis14. Although the molecular interactions linking diseased hematopoietic cells and blood vessels remain to be unraveled, TGFβ family transforming growth factors were shown to be upregulated in megakaryocytes but also in monocytes/macrophages from patients with myelofibrosis. Inhibition of TGFβ signaling ameliorates fibrosis in vitro and in mouse models of myelofibrosis43,44. TGFβ signaling is also a potent inducer of EndMT, has been linked to the loss (rarefaction) of microvessels and can potentiate MRTF-SRF signaling38,45,46. Future studies will have to address the role of TGFβ and other molecular regulators and provide further insight into the role of blood vessels in animal models of myelofibrosis and in human patients.
Methods
Animal models
For the inducible and EC-specific inactivation of Lats2 in mice, we bred Lats2lox/lox conditional mutants to Cdh5(PAC)-CreERT2 transgenic mice21 to generate Cre-positive EC-specific Lats2iΔEC mutants. Cdh5(PAC)-mT/nG transgenic reporter mice21 were introduced into the Lats2iΔEC mutant background and corresponding controls. Mice carrying loxP-flanked Yap1 and Taz (Wwtr1) alleles47 were also bred to the Cdh5(PAC)-CreERT2 line to generate EC-specific Yap1lox/lox and/or Tazlox/lox conditional mutants21. To generate EC-specific constitutive-active Yap1 knock-in (Yap1-KI), mutated Yap1 amino acid 112 serine to alanine (Yap1S112A) mice21 were bred with Cdh5(PAC)-CreERT2. Using Srflox/lox mice with a similar breeding strategy, we generated inducible and EC-specific SrfiΔEC mutant48 and Lats2 Srf double loss-of-function mutants (Lats2 SrfiΔEC). In all these experiments, Cre-negative littermates subjected to the same tamoxifen administration regime were used as controls. For inducible Cre-mediated recombination, mice received daily intraperitoneal injections of 1 mg of tamoxifen for 5 d at the age of 4–5 weeks. For the characterization of BMSCs in the metaphysis, Gli1-CreERT2 (ref. 49) transgenic mice were interbred with Gt(Rosa26) ACTB-tdTomato-EGFP reporter mice50. These mice received 1 mg of tamoxifen for 3 d at 8 weeks of age and were analyzed 4 weeks later. Tamoxifen stocks were prepared as described previously21. Mice were backcrossed and maintained in a C57/BL6 genetic background, and both sexes were used for analysis.
For the ThPO model of bone fibrosis, wild-type (WT) cKit+-enriched cells were transduced with ThPO or empty vector (EV) control lentivirus and transplanted into lethally irradiated (split-dose) B6.Cg-Tg(Ly6a-EGFP)G5Dzk/J (n = 5 animals per group) and B6.SJL recipient mice (n = 3–4 mice per group). Blood was periodically collected via submandibular bleeds into microtainer tubes (Becton Dickinson), and blood counts were performed on a Horiba SciI Vet abc Plus hematology system. Transplantation of MPLW515L-expressing and matching control hematopoietic cells into sub-lethally irradiated mice was conducted as described previously51. Mice were euthanized for analysis when they displayed signs of fibrosis, as indicated by the dropping of hemoglobin levels or weight loss according to defined humane endpoints. Femurs were collected for imaging.
All animal experiments were performed according to institutional guidelines and laws, approved by local animal ethical committees and conducted at the Max Planck Institute for Molecular Biomedicine with necessary permissions (02.04.2015.A185, 84-02.04.2016.A160, 81-02.04.2017.A238 and 81-02.04.2019.A164) granted by the Landesamt für Natur, Umwelt und Verbraucherschutz of North Rhine-Westphalia, Germany. ThPO and MPL bone fibrosis experiments were conducted according to protocols approved by the Central Animal Committee (Centrale Commissie Dierproeven (CCD), The Netherlands) in accordance with legislation in The Netherlands (approval no. AVD1010020173387).
Sample processing and immunostaining
Mice were euthanized, and long bones (femur and tibia) were harvested and fixed immediately in ice-cold 2% paraformaldehyde (PFA) for 6–8 h under gentle agitation. Bones were decalcified in 0.5 M EDTA for 48–72 h at 4 °C under gentle shaking agitation, which was followed by overnight incubation in cryopreservation solution (20% sucrose, 2% PVP) and embedding in bone embedding medium (8% gelatine, 20% sucrose, 2% PVP). Samples were stored overnight at −80 °C. Then, 80–100-μm-thick cryosections were prepared for immunofluorescence staining.
Bone immunostaining was performed as described previously. Bone sections were washed in ice-cold PBS and permeabilized with ice-cold 0.3% Triton X-100 in PBS for 10 min at room temperature (RT). Samples were incubated in blocking solution (5% heat-inactivated donkey serum in 0.3% Triton X-100) for 30 min at RT. Primary antibodies: rat monoclonal anti-endomucin (V.7C7) (Santa Cruz Biotechnology, sc-65495, 1:100 dilution), goat polyclonal anti-CD31 (R&D Systems, AF3628, 1:100 dilution), rabbit polyclonal anti-Fabp5 (Lifespan Biosciences, C312991, 1:100 dilution), goat anti-PDGFRβ (R&D Systems, AF1042, 1:100 dilution), mouse monoclonal alpha-smooth muscle actin-Cy3 (Sigma-Aldrich, C6198, 1:200 dilution), mouse monoclonal alpha-smooth muscle actin-eFluor660 (eBioscience, 50-9760-82, 1:200 dilution), rabbit monoclonal anti-Yap1/Taz (D24E4, Cell Signaling Technology, 8418, 1:100 dilution), rabbit monoclonal anti-vimentin (D21H3, Cell Signaling Technology, 5741, 1:100 dilution), rabbit polyclonal anti-Tagln (Sm22a, Abcam, ab14106, 1:100 dilution), rabbit polyclonal anti-fibronectin (Fn1, Sigma-Aldrich, F3648, 1:100 dilution), rabbit polyclonal anti-thrombospondin 1 (Thbs1, Abcam, ab85762, 1:100 dilution), rabbit polyclonal anti-Angpt2 (Abcam, ab8452, 1:100 dilution), rabbit polyclonal anti-Cav1 (Cell Signaling Technology, 3238, 1:100 dilution), rabbit polyclonal anti-Col4 (AbD Serotec, 2150-1470, 1:100 dilution), rabbit-polyclonal anti-Osterix (Abcam, ab22552, 1:300 dilution), goat polyclonal anti-Mmp9 (R&D Systems, AF909, 1:200 dilution), rabbit polyclonal anti-Col-I (Millipore, AB765P, 1:100 dilution), rabbit polyclonal anti-Col-X (Abcam, ab58632, 1:100 dilution), rabbit polyclonal anti-LATS2 (GeneTex, GTX87529, 1:50 dilution), rabbit polyclonal anti-Aggrecan (Milipore, AB1031, 1:100 dilution), rabbit anti-vATPaseB1/B2 (Abcam, 200839, 1:100 dilution), rat anti-mouse Ter119 (BD Biosciences, 553673, 1:200 dilution), rat anti-mouse B220 (BD Biosciences, 553090, 1:200 dilution), rat-monoclonal anti-active Itgb1 (BD Biosciences, 553715, 1:500 dilution), rabbit polyclonal anti-FAK (pY397) (Thermo Fisher Scientific, 44-624G, 1:50 dilution), rabbit monoclonal anti-Runx2 (Abcam, ab192256, 1:200 dilution) or rat monoclonal anti-Srf (from Alfred Nordheim, 1:50 dilution) were diluted in 5% donkey serum mixed PBS and incubated overnight at 4 °C. Next, slides were washed 3–5 times in PBS in 5–10-min intervals. Species-specific Alexa Fluor secondary antibodies Alexa Fluor 488 (Thermo Fisher Scientific, A21208), Alexa Fluor 546 (Thermo Fisher Scientific, A11056), Alexa Fluor 594 (Thermo Fisher Scientific, A21209), Alexa Fluor 647 (Thermo Fisher Scientific, A31573 or A21447) or Phalloidin-488 (Invitrogen, A12379) diluted 1:100 in PBS were added and incubated for 2–3 h at RT. Immunostained bone sections were imaged with a Leica SP8 confocal microscope.
Vascular permeability assay
To conduct in vivo vascular permeability tests on adult mice, 100 µl of 1 mg of Dextran Texas Red (Thermo Fisher Scientific, D1830, 70,000 MW) was injected into the tail vein and left to circulate for 15 min. Femurs were removed from euthanized mice, immediately dropped in ice-cold 4% PFA and fixed overnight. Fluorescence microscopy was used to capture images of the femur without any signal enhancement.
Histological staining methods
For Safranin O staining, long bones from Lats2iΔEC mutants and control animals were harvested, fixed and decalcified. The bones were dehydrated and embedded using paraffin wax by stranded histology methods. Bone paraffin blocks were cut at 5 µm, and sections were deparaffinized and hydrated. Then, stain with 0.1% Safranin O for 8 min and wash with water. Next, slides were dehydrated and dried and mounted with non-aqueous mounting medium (Entellan, Sigma-Aldrich).
AZAN trichrome staining after deparaffinization and hydration of the slides. After 5 min at 55 °C in the azocarmin solution, the slides were washed with distilled water. The sections were then immersed in an aniline-ethanol solution for 18 minutes, before they were exposed to 1% acetic acid in ethanol solution for 45 s. Next, rinse with distilled water and then immerse in 5% phosphotungstic acid for 2 h. After that, place in a mixture of Aniline Blue–Orange G for 8 min. Use distilled water to rinse. Next, sections were dehydrated and mounted.
For Reticulin staining, bones were hydrated with distilled water. After a 5-min addition of potassium permanganate solution, distilled water was used to rinse. The next solutions—oxalic acid, silver nitrate, gold chloride and sodium thiosulfate—were added for 2 min and then individually washed with distilled water. Sections were dehydrated and mounted.
Calcein Green and Alizarin Red double labeling assay
For in vivo double labeling with Calcein Green and Alizarin Red, 10-week-old control and Lats2iΔEC mutant mice were injected intraperitoneally with 100 µl of Calcein Green (40 mg kg−1; Sigma-Aldrich, C0875) and 100 µl of Alizarin Red (40 mg kg−1; Sigma-Aldrich, A3882) 1 week later. Mice were euthanized at 12 weeks, and femurs were dissected and immediately placed in ice-cold 4% PFA for overnight fixation. Next, samples were cryopreserved, embedded in bone embedding media and kept at −80 °C overnight. Bones were cryosectioned at 30-µm thickness, stained for 30 min with DAPI and then washed twice with washing buffer before imaging by confocal fluorescence microscopy.
µCT analysis
Femurs were fixed in 4% PFA overnight at 4 °C and subjected to µCT analysis by Scanco Medical AG. In the whole three-dimensional evaluation of the trabecular structure, distal section of femur, a voxel size of 5 mm was used. We evaluated 833 slices covering 4.988 mm in height for each sample.
Fluorescence-activated cell sorting
Single-cell suspensions were prepared from bone and spleen as described above. Primary antibody lineage cell detection cocktail-biotin (Miltenyi Biotec, 130-092-613), rat monoclonal anti-Ly6A-PE-cy7 (BD Pharmingen, clone D7, 558162), rat monoclonal anti-CD117-APC (BD Pharmingen, clone 2B8, 553356), hamster monoclonal anti-CD3e-FITC (eBioscience, clone 145-2C11, 11-0031), rat monoclonal anti-B220-APC (Invitrogen, RA3-6B2, RM2605), rat monoclonal anti-CD11b-APC (BioLegend, M1/70, 101212), rat monoclonal anti-Ly-6G(Gr-1)-BV605 (BioLegend, RB6-8C5, 108440), rat monoclonal anti-CD45-PE (BD Pharmingen, 30-F11, 553081), rat monoclonal anti-TER-119-APC (eBioscience, 17-5921) or rat monoclonal anti-CD41-BV605 (BioLegend, MWReg30, 133921) were diluted in fluorescence-activated cell sorting (FACS) buffer (2% FCS, 0.5% BSA in PBS solution) and incubated with cells on ice for 40 min. Cells were washed 2–3 times in FACS buffer and incubated with secondary antibodies for 40 min. Cells were washed two further times and used for flow cytometry. Cells were resuspended in FACS buffer supplemented with 1 mg ml−1 DAPI to allow exclusion of non-viable cells when required. Cell sorting was performed on a FACSAriaIIu cell sorter (BD Biosciences). Cell analysis was performed using FlowJo software.
BMSC in vitro culture and quantitative PCR
BMSCs were isolated from metaphysis of WT mice and cultured as described previously52. For agonist-induced BMSC differentiation assay, cells were serum starved for overnight and then stimulated with agonist TGF-β1, Endothelin-1 (ET-1) or Adrenomedullin (ADM) for 24 h and analyzed for quantitative PCR. Next, total RNA was isolated from these cells using an RNA Plus Mini Kit (Qiagen, 74134) according to the manufacturer’s instructions. RNA was reverse transcribed using the an iScript cDNA Synthesis Kit (Bio-Rad, 1708890). Quantitative PCR was carried out using gene-specific TaqMan probes Eukaryotic 18S rRNA (4319413E), Acta2 (Mm00725412_s1), Tagln (Mm00441661_g1) and Fn1 (Mm01256744_m1) (Thermo Fisher Scientific) using a C1000 Touch thermal cycler (Bio-Rad).
Electron microscopy
Femurs were removed and directly cut into half in fixative. For ultrastructural analysis, fixative comprised 2% glutaraldehyde, 2% PFA, 20 mM CaCl2, 20 mM MgCl2 in 0.1 M cacodylate buffer, pH 7.4. Fixed material was post-fixed in 1% osmium tetroxide containing 1.5% potassium ferrocyanide, dehydrated, including uranyl-en-bloc staining, and embedded stepwise in epon. Ultramicrotomy was performed until reaching the area of interest, where 60-nm ultra-thin sections were collected on formvar-coated 1 slot copper grids. Sections were further stained with lead citrate and finally analyzed with a transmission electron microscope (Tecnai-12-biotwin, Thermo Fisher Scientific).
Bone stromal cells preparation for scRNA-seq
For scRNA-seq analysis, femur and tibia were harvested from Lats2iΔEC mutant and control mice and cleaned from attached surrounding tissue before the metaphysis region was dissected and collected in digestion enzyme solution (Collagenase type I and type IV, 2 mg ml−1). Next, bones were crushed using mortar and pestle. Samples were digested for 45 min at 37 °C under gentle agitation. Digested samples were transferred to 70-μm strainers in 50-ml tubes to obtain a single-cell suspension, which was resuspended in blocking solution (1% BSA, 1 mM EDTA in PBS without Ca2+/Mg2+), centrifuged at 300g for 5 min, washed 2–3 times with ice-cold blocking solution and filtered through 50-μm strainers. Pellets were resuspended in respective volume of blocking solution. Single-cell suspensions were subjected to lineage depletion using a lineage cell depletion kit (MACS, 130-090-858) following the manufacturer’s instructions. Next, lineage-negative (Lin−) cells were depleted by CD45 and CD117 using microbeads (MACS, 130-052-301 and 130-091-224) from Lin− bone cells to enrich bone stromal cells. The remaining cells were resuspended to final concentration of 106 cells per milliliter in 0.05% BSA in PBS, examined by microscopy and used for scRNA-seq.
Single-cell suspensions were subjected to droplet-based scRNA-seq. Single cells were encapsulated into emulsion droplets using the Chromium controller (10x Genomics). scRNA-seq libraries were prepared using a Chromium Single Cell 3′ Reagent Kit (V3) (10x Genomics, PN-10000075) according to the manufacturer’s protocol. scRNA-seq libraries were evaluated and quantified by an Agilent Bioanalyzer using a High Sensitivity DNA Kit (cat. no. 5067-4626) and a Qubit (Thermo Fisher Scientific, Q32851). Individual libraries were diluted to 4 nM and pooled for sequencing. Pooled libraries were sequenced by using a High Output Kit (150 cycles) (Illumina, TG-160-2002) with a NextSeq 500 sequencer (Illumina).
scRNA-seq data analysis
Detailed settings for the analysis steps mentioned below can be found in the accompanying GitLab repository (see ‘Data availability’ section) and in Supplementary Table 1. Default parameters were used, if not detailed otherwise, there or in the text below.
Read data quality control and mapping
Illumina sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software (version 2.20.0.422). Raw reads were processed using cutadapt53 (version 3.5). Pre-processed read data were aligned to the mouse reference genome (version GRCm39) with GENCODE M26 annotation54 extended by adding sequences for CreERT2 and Rosa26-mTmG and counted with STARsolo55 (version 2.7.10a) using its electron microscopy implementation and GeneFull_Ex50pAS read counting. The official 10x Genomics barcode whitelist56 was used.
Data filtering and visualization
Pre-processing of the feature-count-matrix output by STARsolo was largely performed within the Python (version 3.8.10)-based scanpy (version 1.9.1) ecosystem57. Exceptions are functionalities that are only available for R (ref. 58) (version 4.2.1). These include: sequencing depth normalization using scran (version 1.24.1)59, doublet detection using scDblFinder (version 1.11.4)60 and calculation of DEGs using DESeq2 (version 1.36)61. All visualization was done using matplotlib (version 3.5.3)62 functions. Barcodes with fewer than 1,000 unique molecular identifiers (UMIs) or fewer than 800 features (>0 counts per feature) detected were discarded. Similarly, only barcodes with less than 15% of counts stemming from mitotic genes or 5% from hemoglobin genes were retained. Furthermore, barcodes were filtered for a complexity value above 0.8, where complexity is defined as the log10-transformed number of expressed genes divided by the log10-transformed number of total counts per barcode. Additionally, only genes with at least 10 counts total from at least 10 distinct barcodes were kept. scran normalization was applied using min.size = 100 and min.mean = 0.1 in the quickCluster and computeSumFactors functions, respectively. The separate samples were merged into one AnnData (version 0.8.0) object; 4,000 highly variable genes were calculated from the normalized matrix; and the matrix was scaled to unit variance and zero mean, capping values at 10. The 50 first principal components were used for integration using Harmony-Py63 (version 0.0.9, Python implementation by Kamil Slowikowski) with max_iter_harmony = 1,000, max_iter_kmeans = 1,000, epsilon_cluster = 1 × 10−6 and epsilon_harmony = 1 × 10−5. Uniform manifold approximation and projection (UMAP) dimensionality reduction64 and Leiden clustering65 at a resolution of 1 were performed. The resulting 30 clusters, the uncorrected principal component analysis (PCA) embedding and the raw count and normalized matrices were used for scDblFinder doublet prediction. Barcode clusters were annotated with the broad cell identity that they most likely represent based on marker genes provided in Supplementary Table 1. Undesired populations, such as myeloid or lymphoid cells; clusters of barcodes that, either manually based on gene expression or by scDblFinder, could be classified as consisting of more than 20% doublets; and very small clusters (<100 cells) were removed. All remaining 20,383 barcodes will hereafter be referred to as cells.
For each of the four main groups of broad cell identities defined previously, we created subsets and performed scaling, PCA calculation, harmony-py integration, UMAP visualization and Leiden clustering anew as before, to detect more fine-grained sub-cell-type identities. Details on the settings used in each subclustering and marker genes used for annotation can be found in Supplementary Table 1. Some subclustering revealed small cell clusters that could not be identified based on their expression and were, therefore, removed. The subset was then visualized in a second passthrough (indicated in Supplementary Table 1) with the same settings as the first one. Afterwards, either all subsets (20,184 cells) or only the mp-MSC and SMC subsets were merged again for comprehensive visualization and analysis.
Pseudotime analysis was performed using slingshot66 (version 2.6.0). We defined the mpMSC cluster as starting and the Myh11 SMC cluster as the end node of the trajectory. Cells were binned into 21 bins of equal cell numbers along the pseudotime trajectory. Gene-to-gene Pearson correlation of normalized and z-scaled gene expression counts across the ordered pseudotime bins was calculated for the top 8,000 highly variable genes. Correlations with a correlation coefficient above 0.5 were kept, and the data were transformed into an igraph (Python igraph 0.10.4)67 network with genes representing nodes and their mutual correlation coefficients representing the edge weights. Leiden clustering was applied with a resolution parameter of 1.
Differential gene expression (DGE) analysis was performed using DESeq2 on pseudobulk expression matrices. When the expression of genes in one cluster was to be compared to several other clusters, all pairwise comparisons were calculated, and then the mean log2fold change (FC) of these comparisons was output as log2FC, and the minimum P value (alpha = 0.01) calculated from the respective adjusted P values using scipy.stats’s combine_pvalues function (version 1.9.1)68 was used as ‘adjusted P value’ of the upregulation of gene expression in the cluster of interest. It was also counted in how many comparisons the overexpression in the cell cluster of interest compared to each other one was above a log2FC threshold of 2. If not mentioned otherwise, only genes expressed in at least 20% of cells of the respective group in either condition were kept.
Radial expression plots were created by calculating DGE (using DESeq2) between the control and conditional knockout (cKO) conditions for each cell identity presented in the respective graph. Only genes expressed in at least 20% of cells of the respective group in either condition were kept. Each quadrant represents genes with upregulated expression in the respective condition and cell type. For each quadrant, all log2FC values below 0 were set to 0, and the accompanying adjusted P values were set to 1. Then, genes were plotted in a scatterplot by jittering points on the x axis, representing their log2FC on the y axis and the adjusted P value of the expression difference as the point color. This means that, per cell population, all genes present in the analysis are represented in each quadrant, showing which of these are upregulated in the respective condition and cell type. The main goal of this plot is to show in which condition and cell types most genes are differentially expressed.
Additional computational analyses
Differential abundance of cell neighborhoods was quantified using milopy (version 0.1.0)69 with alpha = 0.01. Potentially regulating transcription factors for selected gene sets were calculated using the iRegulon70 plugin to Cytoscape (version 3.9.1)71 with the 6K Motif collection, looking 500 bp upstream of provided genes in seven species. PAGA28 was applied to the data with default settings via scanpy’s implementation.
Predicting LRIs
To determine possible LRIs, a permutation-based approach similar to the one introduced in CellPhoneDB72 was used. The normalized count matrix was used as a basis for 10,000 cell type label permutations where cell type labels were randomly re-assigned. Ligand–receptor (LR) gene identity and interaction pairs were taken from the consensus database of LIANA73, excluding bidirectional annotations and genes expressed in less than 10% of cells. For each LR interaction, first the mean expression of the ligand (Lmean) and receptor (Rmean) were calculated for each cell type in each condition, and then an interaction score (LRIScore) was calculated for all possible cell type interactions per condition separately as the product of Lmean and Rmean. This approach differs from the methodology employed by CellPhoneDB in that our approach uses the product instead of the mean value of ligand and receptor mean expression. This is intended to implicitly penalize interactions where one or both partners are lowly expressed. Based on the number of permutation results matching or exceeding the observed value, values of P were calculated for each interaction. Values of P calculated in this test are approximations of P but will be referred to as P values nonetheless. Multiple testing correction was performed using the Benjamini–Hochberg74 implementation in the statsmodels (version 0.13.1)75 function multipletests on all P values. These corrected P values will be referenced as adjusted P values. Additionally, using the created background distribution, the z-score of the original LRIScore was determined.
In the same fashion as for cell type labels, condition labels (control/cKO) were permuted an equal number of times as cell type labels and a difference score between the LRI scores of the two conditions were calculated for each cell type interaction as:
This scoring is similar to a previously proposed perturbation score76, but the mean and not the product of the absolute log2FC was used. This was done to emphasize coordinated upregulation or downregulation of ligand and receptor instead of just the overall magnitude of change. P values and adjusted P values were calculated from these scores in a manner sensitive to the direction of change and used to assess differences in LRI scores between conditions. This is similar to a very recently published approach77.
This procedure was done once for a dataset, where, of all ECs, only mpECs were kept, and major cell type labels were used for all other cells. Another analysis focused on mpECs, mpMSCs, bmMSCs, Myh11+ and Myh11− SMCs compared to all other major cell types as background, but only the results for the mentioned subtypes were visualized.
Statistics and reproducibility
Images were analyzed, quantified and processed using Volocity (PerkinElmer), Fiji (ImageJ) and Adobe Photoshop and Illustrator software 2020.
Statistical analysis was performed using GraphPad Prism 9 software or the R statistical environment (http://r-project.org). All data are presented as mean values ± s.e.m. unless indicated otherwise. Comparisons between two groups were performed with unpaired two-tailed Mann–Whitney test. Comparisons between more than two groups were made by Tukey multiple comparison test to determine statistical significance. P < 0.05 was considered significant unless stated otherwise. For DGE (Figs. 5d and 6a,b and Extended Data Fig. 7g): DESeq2 (internally uses Wald test statistics) yielding false discovery rate (FDR)-corrected P values (P.adj). Extended Data Fig. 9a: genes filtered based on pairwise comparisons between mp-ECs and all other EC subtypes using DESeq2 yielding FDR-corrected P values (P.adj) combined across comparisons using the Fisher method implemented in scipy’s combine_p_values function. LR pairings and the significant interactions between cell types of interest (Fig. 6g,h) were analyzed with a permutation test (one-sided) with Benjamini–Hochberg-corrected P values (P.adj). Extended Data Fig. 9a,c,d used a permutation test (one-sided) with Benjamini–Hochberg-corrected P values (P.adj). Four biological independent samples were used in Figs. 1e,f, 2i,j and 5f, and three biological independent samples were used in Figs. 3a,e and 4c and Extended Data Figs. 1c–e, 2b–g, 3b, 4f–h, 5f,g,i, 6a and 8b–d. Sample numbers are indicated in the figure legends and were chosen based on experience from previous experiments. Reproducibility was ensured by more than three independent experiments (aside from scRNA-seq). No statistical method was used to predetermine sample size, and no animals were excluded from analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Integrated supplementary information
a. Lats2iΔEC mutant mice look normal and body weight is comparable to control littermates (n = 6). Data shown as mean ± SEM. P values, Mann–Whitney test (two-tailed). b. Dot plot for normalised and averaged expression indicates of Lats1 and Lats2 expression in the indicated control (Cont) and Lats2iΔEC mutant (Mut) EC subpopulations. Z-scaled per row. Note reduction of Lats2 transcripts and absence of compensatory Lats1 upregulation. c. Reduced LATS2 immunostaining (gray/green) in Lats2iΔEC mutant bone endothelium (EMCN, red) (arrowheads). Nuclei, DAPI (blue). d. Representative high magnification images of control and Lats2iΔEC mutant in Cdh5-mTnG reporter background. Femoral metaphysis (mp), bone marrow (bm), growth plate (gp) and transition zone between metaphysis and diaphysis (tz) are indicated. e. Representative confocal images showing increase CAV1+ (red), CD31+ (green) and EMCN+ (red) ECs in Lats2iΔEC femoral transition zone (tz). Note that CAV1 labels artery in control but microvascular structures in mutant (arrowheads). n = independent biological samples.
a. Tile scan confocal longitudinal views of EMCN and CD31 stained femoral blood vessels. Note enlargement of Lats2iΔEC growth plate and metaphysis. Type H capillaries are visible in control but are replaced by CD31+/EMCN- ECs in mutants (arrowheads). Graphs on the right show quantification of ECs in bone marrow (bm) (top) and CD31+/EMCN- capillaries (bottom) (n = 6). Data presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). b. Representative images showing expansion of hypertrophic chondrocytes in Lats2iΔEC mutants by Safranin O staining and collagen type X (COL-X) immunostaining. c. FABP5+ septoclasts and MMP9 expression (arrowheads) at chondro-osseous border near growth plate (gp) are lost in Lats2iΔEC mutants. d, e. Lats2iΔEC femurs show increase in PDGFRβ+ and αSMA+ immunosignals (arrows). αSMA+ in control is largely confined to arterial SMCs (arrowheads) (d). Higher magnification images are shown in (e). f. AZAN trichrome staining of femoral sections showing expansion of the Lats2iΔEC growth plate (gp) (left) and ectopic bone (center). Insets on the right show deposition of collagen fibers (arrowheads), which is visible at high magnification. g. Representative images showing increased collagen type I (COL-I) immunosignal and reticulin stained fibers in Lats2iΔEC mutant sections. n = independent biological samples.
a, b. Tamoxifen injection scheme (a). Tile scan confocal longitudinal view of femurs stained for EMCN (red) and CD31 (green) in 8 and 10-week-old control or Lats2iΔEC mutant mice (b). Nuclei, DAPI (blue). c. Confocal images of αSMA+ (green) cells in 8-week-old and 10-week-old control or Lats2iΔEC femoral metaphysis (mp). EMCN (red), DAPI (blue). d. Quantification of number and length of vessels columns as well as CD31+/EMCN- area (n = 4). Data presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). e. Representative images of control and Lats2iΔEC organs in the Cdh5-mTnG reporter background. Sections are stained for αSMA, EMCN and YAP1/TAZ, as indicated. Quantification shows that EC number is unchanged in these organs (n = 4). Data shown as mean ± SEM. P values, Mann–Whitney test (two-tailed). n = independent biological samples.
a, b. Quantification of white blood cells (WBC), red blood cells (RBC), and platelets (PLT) in peripheral blood (a). FACS analysis of Ter119+, B220+, and CD45+ cell frequencies in bone (b). c, d. Representative confocal images of CD41+ megakaryocytes (green) in control and Lats2iΔEC femoral sections (c). Quantification shows significant reduction of CD41+ cells in the Lats2iΔEC metaphysis fibrotic area but not in diaphysis of non-fibrotic area (d). e. Quantification of B220+ and CD45+ cell frequencies in spleen. f. Confocal tile scans of control and Lats2iΔEC spleen vasculature labelled with the Cdh5-mTnG reporter (left). Images at high magnification show αSMA (green), ECs (Cdh5-mTnG, red), and DAPI (blue). White pulp (wp) and red pulp (rp) are indicated. g, h. Representative confocal images of the spleen showing B220 (green) and Ter119 (red) positive white pulp (wp) and red pulp (rp), respectively. Blood vessels (Cdh5-mT, blue) (g). Immunostaining of YAP1/TAZ in Cdh5-mTnG reporter of control and Lats2iΔEC spleen (h). Sample number is n = 6 (a, b), n = 4 (d), n = 6 (e). Data are presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). n = independent biological samples.
a-c. Quantification of white blood cells (WBC), red blood cells (RBC) and platelets (PLT) in peripheral blood (a), spleen weight (b), and number of type H vessel buds (c) in control and ThPO gain-of-function bone. (n = 5). Data are presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). d, e. Representative confocal images showing increase of αSMA immunosignals in diaphysis (d) and reduction of OSX+ cells (e) in ThPO femur relative to control. (n = 4). Data are presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). f. Representative confocal images showing strong and widespread increase in αSMA (gray) immunostaining together with increased YAP1/TAZ expression ThPO-treated BM. g. Experimental scheme of MPLW515L-induced BM fibrosis. Representative confocal images (bottom) show αSMA immunostaining in control and MPLW515L (MPL) bone sections. h. Confocal images showing reduced EMCN+ (red) vessels and increase of αSMA (green) signal in MPL bone relative to control. Nuclei, DAPI (blue). Graphs on the right show decreased EMCN+ vessels area and expression in MPL bones. (n = 3-4). Data shown as mean ± SEM. P values, Mann–Whitney test (two-tailed). i. Immunostaining of YAP1/TAZ (green), EMCN (red), and DAPI (blue) as indicated, in control and MPL bone sections. n = independent biological samples.
a. Confocal images showing vATPase+ osteoclasts (red, arrowheads) in Lats2iΔEC femur. b, c. Representative microCT images of cortical bone (cb) in Lats2iΔEC and control femur (b). Graphs show quantitative analysis of trabecular bone (tb) thickness, tb separation, and cortical bone thickness (c). d. Representative confocal images and quantification showing slight reduction of femoral OSX+ cells in Yap1/TaziΔEC double mutant relative to littermate control. e. Representative µCT images of trabecular bone in control and Yap1/TaziΔEC femur. Quantitative analysis of trabecular (tb) volume (BV/TV, bone volume/total volume) and tb number. Sample numbers n = 5 (b, c), n = 4 (d), n = 5 (e). Data are presented as mean ± SEM. P values, statistical analysis performed using the Mann–Whitney test (two-tailed). n = independent biological samples.
a-c. UMAP plot showing color-coded cell clusters of control and Lats2iΔEC bone stromal cells (a). Markers for main bone stromal cell populations (b) and their subclusters (c) are shown in dot plots. d- f. UMAP plot of EC subsets, split by condition (right) (d). Bar graph displaying percentage of endothelial cells subtypes in control and Lats2iΔEC mutants (e). UMAP plot colored by normalized expression of EC subset identity marker genes: Ramp3 (metaphyseal ECs, mpECs), Gja4 (arterial ECs, aEC), Fmo (arteriole-like ECs, alECs) and Stab2 (bone marrow ECs, bm) (f). g, h. Volcano plot of differentially expressed genes (frac. expressed > 0.2 in either condition) between control and mutant condition across all ECs (blue color: log2FC >=1,p.adj <=0.001,) (g). Visualisation of selected known Hippo pathway target genes: Ccn1 (Cyr61), Ccn2 (Ctgf), Angpt2 and Thbs1 in Lats2iΔEC mutant and control (h).
a. Confocal images showing strongly elevated Angiopoietin 2 (ANGPT2), Thrombospondin 1 (THBS1), Fibronectin 1 (FN1), Transgelin (TAGLN), and Vimentin (VIM) expression in Lats2iΔEC bone sections. (n = 4). Data in graphs are presented as mean ± SEM. P values, statistical analysis performed using the Mann–Whitney test (two-tailed)). b. Confocal images showing increased staining for collagen IV (COL IV), the actin cytoskeleton (with Phalloidin), active-integrin β1 and phosphorylated focal adhesion kinase (FAK pY397) in Lats2iΔEC mutant relative to control. c. Transmission electron micrographs showing increased subendothelial basement membrane and accumulation of ECM (yellow arrowheads) in Lats2iΔEC femur. Mutant ECs are rich in actin filaments (red arrowheads). d. Confocal images of Lats2iΔEC mutant in R26-dTomato reporter background showing αSMA staining in dTomato+ ECs. In control samples, αSMA signal is confined to SMCs covering arteries. n = independent biological samples.
a. Ligands showing on average significantly (log2FC >=1,p.adj <=0.05, frac. expressed >=0.1) larger counts in mutant mpECs than in any other EC subidentity and are significantly elevated (see Methods) relative to at least one other EC subpopulation. b. Selected ligand-receptor parings of main bone stromal cells, which are increased in Lats2iΔEC mutants. c. Mean normalized expression counts of selected, significant ligand-receptor interactions (p.adj of LRI score in either condition and LRIDiff Scores <= 0.05, |LRIDiff| >= 1) in mpECS and all other major celltypes. Ligand (left, dark blue bar) and receptor (right, red bar). d. Mean normalized expression counts of selected, significant ligand-receptor interactions (p.adj of LRI score in either condition and LRIDiff Scores <= 0.05, LRIDiff >= 1) in mpECS, mpMSCs and SMCs. Ligand (left, dark blue bar) and receptor (right, red bar). e. Quantitative PCR (qPCR) shows expression of myofibroblasts markers Acta2, Tagln and Fn1 in cultured BMSCs upon treatment with recombinant TGFβ1, endothelin 1 (ET1) or adrenomedullin (ADM). (n = 3). Data in graphs are presented as mean ± SEM. P values, statistical analysis performed using the Mann–Whitney test (two-tailed)). n = independent biological samples.
a, b. Representative confocal images of control and SrfiΔEC femur show type H (EMCNhigh; CD31high) capillaries, Nuclei, DAPI (blue) (a) as well as PDGFRβ+ cells (red) and αSMA+ arterial SMCs (green) and EMCN (blue) in the femoral metaphysis (b). Graphs in (b) show quantification of vessel columns and arteries (n = 6). Data presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). c-e. Confocal images showing ACAN+ (green) chondrocytes in femoral growth plate (gp) (c) and OSX+ cells (green) (d). µCT images of trabecular bone of SrfiΔEC mutant and control(e) and quantitation of trabecular (tb) volume (BV/TV, bone volume/total volume) tb number and gp length (n = 6). Data presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). f, g. Longitudinal tile scan confocal images showing EMCN+ (red) CD31+ (green) vessels in control, Lats2iΔEC and Lats2 SrfiΔEC compound mutant femur (f). Metaphysis (mp), growth plate (gp, dashed lines) and bone marrow (bm) are indicated. Quantification shows increase in CD31+ EMCN- area in Lats2iΔEC metaphysis and rescue by inactivation of Srf (g) (n = 4). Data presented as mean ± SEM. P values, Tukey multiple comparison test (one-way Anova). h. Representative images showing OSX+ cells in control, Lats2iΔEC, and Lats2 SrfiΔEC compound mutant femur. i, j. Representative μCT images of femoral trabecular bone (i). Quantitative analysis of trabecular bone (tb) thickness and separation (j) (n = 4). Data presented as mean ± SEM. P values, Tukey multiple comparison test (one-way Anova). k. Proposed model of YAP1/TAZ-induced endothelial-to-mesenchymal transition (EndMT) as a driver of myelofibrosis and osteosclerosis. n = independent biological samples. Created with BioRender.com.
Supplementary information
Supplementary Fig. 1 and Supplementary Tables 1 and 2.
Source data
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Acknowledgements
We thank K. Kruse for providing feedback on the computational methods used in this work as well as for support and fruitful scientific discussions. We thank K. Mildner for helping with electron microscopy sample preparation. This work was supported by the Max Planck Society (R.H.A.), the Deutsche Forschungsgemeinschaft (CRC 1366; RHA), the Leducq Foundation (R.H.A.) and the European Research Council (AdG 786672, PROVEC; AdG 101139772, PROTECT) (R.H.A.).
Author contributions
K.K.S., P.-G.M. and R.H.A. designed experiments and interpreted results. K.K.S. generated and characterized mouse mutant lines and conducted all experiments, including FACS, bone sectioning and staining, confocal imaging, quantifications and scRNA-seq. P.-G.M. and K.K.S. analyzed the scRNA-seq data. B.D. performed staining and analysis. M.S. performed FACS. D.Z. performed electron microscopy. S.S. provided technical assistance. A.N. generated and contributed reagents and mouse strains. B.B. and R.K.S. performed the ThPO fibrosis experiment. K.K.S., P.-G.M. and R.H.A. wrote the manuscript.
Peer review
Peer review information
Nature Cardiovascular Research thanks Mei Xin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Funding
Open access funding provided by Max Planck Society.
Data availability
The scRNA-seq data generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE225429. The mouse reference genome GRCm39 with GENCODE M26 anotation (https://www.gencodegenes.org/mouse/release_M26.html) was used for mapping the reads in this study. All other relevant data supporting the key findings of this study are available within the article and its Supplementary Information files.
Code availability
All code used to produce the computational analysis results and figures in this publication are available at https://gitlab.gwdg.de/pmajev/sivaraj_majev_lats2_ecs. The assigned DOI is 10.5281/zenodo.11456485.
Competing interests
R.K.S. is a co-founder and shareholder of Sequantrix GmbH and a consultant for Active Biotech. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kishor K. Sivaraj, Paul-Georg Majev.
Contributor Information
Kishor K. Sivaraj, Email: kishor.kumar-sivaraj@mpi-muenster.mpg.de
Rebekka K. Schneider, Email: reschneider@ukaachen.de
Ralf H. Adams, Email: ralf.adams@mpi-muenster.mpg.de
Extended data
is available for this paper at 10.1038/s44161-024-00508-x.
Supplementary information
The online version contains supplementary material available at 10.1038/s44161-024-00508-x.
References
- 1.Zahr, A. A. et al. Bone marrow fibrosis in myelofibrosis: pathogenesis, prognosis and targeted strategies. Haematologica101, 660–671 (2016). 10.3324/haematol.2015.141283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li, B. et al. Bone marrow fibrosis grade is an independent risk factor for overall survival in patients with primary myelofibrosis. Blood Cancer J.6, e505 (2016). 10.1038/bcj.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boudin, E. & Van Hul, W. Sclerosing bone dysplasias. Best Pract. Res. Clin. Endocrinol. Metab.32, 707–723 (2018). 10.1016/j.beem.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 4.Spampinato, M. et al. Focus on osteosclerotic progression in primary myelofibrosis. Biomolecules11, 122 (2021). [DOI] [PMC free article] [PubMed]
- 5.Mizoguchi, T. et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev. Cell29, 340–349 (2014). 10.1016/j.devcel.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou, B. O., Yue, R., Murphy, M. M., Peyer, J. G. & Morrison, S. J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell15, 154–168 (2014). 10.1016/j.stem.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi, Y. et al. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun.8, 2043 (2017). 10.1038/s41467-017-02171-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kramann, R. et al. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell16, 51–66 (2015). 10.1016/j.stem.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decker, M. et al. Leptin-receptor-expressing bone marrow stromal cells are myofibroblasts in primary myelofibrosis. Nat. Cell Biol.19, 677–688 (2017). 10.1038/ncb3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider, R. K. et al. Gli1+ mesenchymal stromal cells are a key driver of bone marrow fibrosis and an important cellular therapeutic target. Cell Stem Cell20, 785–800 (2017). 10.1016/j.stem.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leimkuhler, N. B. et al. Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell28, 637–652 (2021). 10.1016/j.stem.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Psaila, B. et al. Single-cell analyses reveal megakaryocyte-biased hematopoiesis in myelofibrosis and identify mutant clone-specific targets. Mol. Cell78, 477–492 (2020). 10.1016/j.molcel.2020.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vannucchi, A. M. et al. A pathobiologic pathway linking thrombopoietin, GATA-1, and TGF-β1 in the development of myelofibrosis. Blood105, 3493–3501 (2005). 10.1182/blood-2004-04-1320 [DOI] [PubMed] [Google Scholar]
- 14.Erba, B. G. et al. Endothelial-to-mesenchymal transition in bone marrow and spleen of primary myelofibrosis. Am. J. Pathol.187, 1879–1892 (2017). 10.1016/j.ajpath.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 15.Ding, L., Saunders, T. L., Enikolopov, G. & Morrison, S. J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature481, 457–462 (2012). 10.1038/nature10783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusumbe, A. P., Ramasamy, S. K. & Adams, R. H. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature507, 323–328 (2014). 10.1038/nature13145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kretschmer, M., Rudiger, D. & Zahler, S. Mechanical aspects of angiogenesis. Cancers13, 4987 (2021). [DOI] [PMC free article] [PubMed]
- 18.Moya, I. M. & Halder, G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol.20, 211–226 (2019). 10.1038/s41580-018-0086-y [DOI] [PubMed] [Google Scholar]
- 19.Yu, F. X. & Guan, K. L. The Hippo pathway: regulators and regulations. Genes Dev.27, 355–371 (2013). 10.1101/gad.210773.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piccolo, S., Dupont, S. & Cordenonsi, M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev.94, 1287–1312 (2014). 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- 21.Sivaraj, K. K. et al. YAP1 and TAZ negatively control bone angiogenesis by limiting hypoxia-inducible factor signaling in endothelial cells. eLife9, e50770 (2020). [DOI] [PMC free article] [PubMed]
- 22.Bando, Y. et al. Expression of epidermal fatty acid binding protein (E-FABP) in septoclasts in the growth plate cartilage of mice. J. Mol. Histol.45, 507–518 (2014). 10.1007/s10735-014-9576-1 [DOI] [PubMed] [Google Scholar]
- 23.Sivaraj, K. K. et al. Mesenchymal stromal cell-derived septoclasts resorb cartilage during developmental ossification and fracture healing. Nat. Commun.13, 571 (2022). 10.1038/s41467-022-28142-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine, R. L., Pardanani, A., Tefferi, A. & Gilliland, D. G. Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat. Rev. Cancer7, 673–683 (2007). 10.1038/nrc2210 [DOI] [PubMed] [Google Scholar]
- 25.Yan, X. Q. et al. A model of myelofibrosis and osteosclerosis in mice induced by overexpressing thrombopoietin (mpl ligand): reversal of disease by bone marrow transplantation. Blood88, 402–409 (1996). 10.1182/blood.V88.2.402.bloodjournal882402 [DOI] [PubMed] [Google Scholar]
- 26.Pikman, Y. et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med.3, e270 (2006). 10.1371/journal.pmed.0030270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haghverdi, L., Buttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods13, 845–848 (2016). 10.1038/nmeth.3971 [DOI] [PubMed] [Google Scholar]
- 28.Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol.20, 59 (2019). 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protocols15, 1484–1506 (2020). 10.1038/s41596-020-0292-x [DOI] [PubMed] [Google Scholar]
- 30.Janky, R. et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol.10, e1003731 (2014). 10.1371/journal.pcbi.1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, J. et al. YAP/TAZ regulates sprouting angiogenesis and vascular barrier maturation. J. Clin. Invest.127, 3441–3461 (2017). [DOI] [PMC free article] [PubMed]
- 32.Wang, X. et al. YAP/TAZ orchestrate VEGF signaling during developmental angiogenesis. Dev. Cell42, 462–478 (2017). 10.1016/j.devcel.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 33.McPherson, J. P. et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J.23, 3677–3688 (2004). 10.1038/sj.emboj.7600371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furth, N. & Aylon, Y. The LATS1 and LATS2 tumor suppressors: beyond the Hippo pathway. Cell Death Differ.24, 1488–1501 (2017). 10.1038/cdd.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, Q. et al. Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat. Commun.7, 12422 (2016). 10.1038/ncomms12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovacic, J. C. et al. Endothelial to mesenchymal transition in cardiovascular disease: JACC State-of-the-Art Review. J. Am. Coll. Cardiol.73, 190–209 (2019). 10.1016/j.jacc.2018.09.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olson, E. N. & Nordheim, A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol.11, 353–365 (2010). 10.1038/nrm2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Small, E. M. The actin–MRTF–SRF gene regulatory axis and myofibroblast differentiation. J. Cardiovasc. Transl. Res.5, 794–804 (2012). 10.1007/s12265-012-9397-0 [DOI] [PubMed] [Google Scholar]
- 39.Liu, C. Y. et al. MRTF/SRF dependent transcriptional regulation of TAZ in breast cancer cells. Oncotarget7, 13706–13716 (2016). 10.18632/oncotarget.7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foster, C. T., Gualdrini, F. & Treisman, R. Mutual dependence of the MRTF–SRF and YAP–TEAD pathways in cancer-associated fibroblasts is indirect and mediated by cytoskeletal dynamics. Genes Dev.31, 2361–2375 (2017). 10.1101/gad.304501.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pagiatakis, C., Sun, D., Tobin, S. W., Miyake, T. & McDermott, J. C. TGFβ-TAZ/SRF signalling regulates vascular smooth muscle cell differentiation. FEBS J.284, 1644–1656 (2017). 10.1111/febs.14070 [DOI] [PubMed] [Google Scholar]
- 42.Fearing, B. V. et al. Mechanosensitive transcriptional coactivators MRTF-A and YAP/TAZ regulate nucleus pulposus cell phenotype through cell shape. FASEB J.33, 14022–14035 (2019). 10.1096/fj.201802725RRR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varricchio, L. et al. TGF-β1 protein trap AVID200 beneficially affects hematopoiesis and bone marrow fibrosis in myelofibrosis. JCI Insight6, e145651 (2021). [DOI] [PMC free article] [PubMed]
- 44.Lecomte, S. et al. Therapeutic activity of GARP:TGF-β1 blockade in murine primary myelofibrosis. Blood141, 490–502 (2023). 10.1182/blood.2022017097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu, P., Feng, X. H. & Li, L. Interaction of Smad3 and SRF-associated complex mediates TGF-β1 signals to regulate SM22 transcription during myofibroblast differentiation. J. Mol. Cell. Cardiol.35, 1407–1420 (2003). 10.1016/j.yjmcc.2003.09.002 [DOI] [PubMed] [Google Scholar]
- 46.Goumans, M. J., van Zonneveld, A. J. & ten Dijke, P. Transforming growth factor β-induced endothelial-to-mesenchymal transition: a switch to cardiac fibrosis? Trends Cardiovasc. Med.18, 293–298 (2008). 10.1016/j.tcm.2009.01.001 [DOI] [PubMed] [Google Scholar]
- 47.Reginensi, A. et al. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet.9, e1003380 (2013). 10.1371/journal.pgen.1003380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiebel, F. F., Rennekampff, V., Vintersten, K. & Nordheim, A. Generation of mice carrying conditional knockout alleles for the transcription factor SRF. Genesis32, 124–126 (2002). 10.1002/gene.10049 [DOI] [PubMed] [Google Scholar]
- 49.Ahn, S. & Joyner, A. L. Dynamic changes in the response of cells to positive Hedgehog signaling during mouse limb patterning. Cell118, 505–516 (2004). 10.1016/j.cell.2004.07.023 [DOI] [PubMed] [Google Scholar]
- 50.Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis45, 593–605 (2007). 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- 51.Gleitz, H. F. E. et al. Increased CXCL4 expression in hematopoietic cells links inflammation and progression of bone marrow fibrosis in MPN. Blood136, 2051–2064 (2020). 10.1182/blood.2019004095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sivaraj, K. K. et al. Regional specialization and fate specification of bone stromal cells in skeletal development. Cell Rep.36, 109352 (2021). 10.1016/j.celrep.2021.109352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal17, 10–12 (2011). 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 54.Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids. Res.47, D766–D773 (2019). 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun.8, 14049 (2017). 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol.19, 15 (2018). 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Core Team. R: a language and environment for statistical computing. https://www.R-project.org/ (2017).
- 59.Lun, A. T. L., Bach, K. & Marioni, J. C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol.17, 75 (2016). 10.1186/s13059-016-0947-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Germain, P.-L., Lun, A., Meixide, C. G., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res. 10, 979 (2022). [DOI] [PMC free article] [PubMed]
- 61.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng.9, 90–95 (2007). 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- 63.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: uniform manifold approximation and projection. J. Open Source Softw.3, 861 (2018). 10.21105/joss.00861 [DOI] [Google Scholar]
- 65.Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep.9, 5233 (2019). 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics19, 477 (2018). 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Csárdi, G. & Nepusz, T. The Igraph software package for complex network research. InterJournalComplex Systems, 1695 (2006).
- 68.Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods17, 261–272 (2020). 10.1038/s41592-019-0686-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dann, E., Henderson, N. C., Teichmann, S. A., Morgan, M. D. & Marioni, J. C. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol.40, 245–253 (2022). 10.1038/s41587-021-01033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Janky, R. S. et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol.10, e1003731 (2014). 10.1371/journal.pcbi.1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vento-Tormo, R. et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature563, 347–353 (2018). 10.1038/s41586-018-0698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dimitrov, D. et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat. Commun.13, 3224 (2022). 10.1038/s41467-022-30755-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Methodol.57, 289–300 (1995). 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 75.Seabold, S. & Perktold, J. Satsmodel: econometric and statistical modeling with Python. In Proc. of the 9th Python in Science Conference. 92–96 (SciPy, 2010).
- 76.Raredon, M. S. B. et al. Single-cell connectomic analysis of adult mammalian lungs. Sci. Adv.5, eaaw3851 (2019). 10.1126/sciadv.aaw3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lagger, C. et al. scDiffCom: a tool for differential analysis of cell-cell interactions provides a mouse atlas of aging changes in intercellular communication. Nat. Aging3, 1446–1461 (2023). 10.1038/s43587-023-00514-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a. Lats2iΔEC mutant mice look normal and body weight is comparable to control littermates (n = 6). Data shown as mean ± SEM. P values, Mann–Whitney test (two-tailed). b. Dot plot for normalised and averaged expression indicates of Lats1 and Lats2 expression in the indicated control (Cont) and Lats2iΔEC mutant (Mut) EC subpopulations. Z-scaled per row. Note reduction of Lats2 transcripts and absence of compensatory Lats1 upregulation. c. Reduced LATS2 immunostaining (gray/green) in Lats2iΔEC mutant bone endothelium (EMCN, red) (arrowheads). Nuclei, DAPI (blue). d. Representative high magnification images of control and Lats2iΔEC mutant in Cdh5-mTnG reporter background. Femoral metaphysis (mp), bone marrow (bm), growth plate (gp) and transition zone between metaphysis and diaphysis (tz) are indicated. e. Representative confocal images showing increase CAV1+ (red), CD31+ (green) and EMCN+ (red) ECs in Lats2iΔEC femoral transition zone (tz). Note that CAV1 labels artery in control but microvascular structures in mutant (arrowheads). n = independent biological samples.
a. Tile scan confocal longitudinal views of EMCN and CD31 stained femoral blood vessels. Note enlargement of Lats2iΔEC growth plate and metaphysis. Type H capillaries are visible in control but are replaced by CD31+/EMCN- ECs in mutants (arrowheads). Graphs on the right show quantification of ECs in bone marrow (bm) (top) and CD31+/EMCN- capillaries (bottom) (n = 6). Data presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). b. Representative images showing expansion of hypertrophic chondrocytes in Lats2iΔEC mutants by Safranin O staining and collagen type X (COL-X) immunostaining. c. FABP5+ septoclasts and MMP9 expression (arrowheads) at chondro-osseous border near growth plate (gp) are lost in Lats2iΔEC mutants. d, e. Lats2iΔEC femurs show increase in PDGFRβ+ and αSMA+ immunosignals (arrows). αSMA+ in control is largely confined to arterial SMCs (arrowheads) (d). Higher magnification images are shown in (e). f. AZAN trichrome staining of femoral sections showing expansion of the Lats2iΔEC growth plate (gp) (left) and ectopic bone (center). Insets on the right show deposition of collagen fibers (arrowheads), which is visible at high magnification. g. Representative images showing increased collagen type I (COL-I) immunosignal and reticulin stained fibers in Lats2iΔEC mutant sections. n = independent biological samples.
a, b. Tamoxifen injection scheme (a). Tile scan confocal longitudinal view of femurs stained for EMCN (red) and CD31 (green) in 8 and 10-week-old control or Lats2iΔEC mutant mice (b). Nuclei, DAPI (blue). c. Confocal images of αSMA+ (green) cells in 8-week-old and 10-week-old control or Lats2iΔEC femoral metaphysis (mp). EMCN (red), DAPI (blue). d. Quantification of number and length of vessels columns as well as CD31+/EMCN- area (n = 4). Data presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). e. Representative images of control and Lats2iΔEC organs in the Cdh5-mTnG reporter background. Sections are stained for αSMA, EMCN and YAP1/TAZ, as indicated. Quantification shows that EC number is unchanged in these organs (n = 4). Data shown as mean ± SEM. P values, Mann–Whitney test (two-tailed). n = independent biological samples.
a, b. Quantification of white blood cells (WBC), red blood cells (RBC), and platelets (PLT) in peripheral blood (a). FACS analysis of Ter119+, B220+, and CD45+ cell frequencies in bone (b). c, d. Representative confocal images of CD41+ megakaryocytes (green) in control and Lats2iΔEC femoral sections (c). Quantification shows significant reduction of CD41+ cells in the Lats2iΔEC metaphysis fibrotic area but not in diaphysis of non-fibrotic area (d). e. Quantification of B220+ and CD45+ cell frequencies in spleen. f. Confocal tile scans of control and Lats2iΔEC spleen vasculature labelled with the Cdh5-mTnG reporter (left). Images at high magnification show αSMA (green), ECs (Cdh5-mTnG, red), and DAPI (blue). White pulp (wp) and red pulp (rp) are indicated. g, h. Representative confocal images of the spleen showing B220 (green) and Ter119 (red) positive white pulp (wp) and red pulp (rp), respectively. Blood vessels (Cdh5-mT, blue) (g). Immunostaining of YAP1/TAZ in Cdh5-mTnG reporter of control and Lats2iΔEC spleen (h). Sample number is n = 6 (a, b), n = 4 (d), n = 6 (e). Data are presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). n = independent biological samples.
a-c. Quantification of white blood cells (WBC), red blood cells (RBC) and platelets (PLT) in peripheral blood (a), spleen weight (b), and number of type H vessel buds (c) in control and ThPO gain-of-function bone. (n = 5). Data are presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). d, e. Representative confocal images showing increase of αSMA immunosignals in diaphysis (d) and reduction of OSX+ cells (e) in ThPO femur relative to control. (n = 4). Data are presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). f. Representative confocal images showing strong and widespread increase in αSMA (gray) immunostaining together with increased YAP1/TAZ expression ThPO-treated BM. g. Experimental scheme of MPLW515L-induced BM fibrosis. Representative confocal images (bottom) show αSMA immunostaining in control and MPLW515L (MPL) bone sections. h. Confocal images showing reduced EMCN+ (red) vessels and increase of αSMA (green) signal in MPL bone relative to control. Nuclei, DAPI (blue). Graphs on the right show decreased EMCN+ vessels area and expression in MPL bones. (n = 3-4). Data shown as mean ± SEM. P values, Mann–Whitney test (two-tailed). i. Immunostaining of YAP1/TAZ (green), EMCN (red), and DAPI (blue) as indicated, in control and MPL bone sections. n = independent biological samples.
a. Confocal images showing vATPase+ osteoclasts (red, arrowheads) in Lats2iΔEC femur. b, c. Representative microCT images of cortical bone (cb) in Lats2iΔEC and control femur (b). Graphs show quantitative analysis of trabecular bone (tb) thickness, tb separation, and cortical bone thickness (c). d. Representative confocal images and quantification showing slight reduction of femoral OSX+ cells in Yap1/TaziΔEC double mutant relative to littermate control. e. Representative µCT images of trabecular bone in control and Yap1/TaziΔEC femur. Quantitative analysis of trabecular (tb) volume (BV/TV, bone volume/total volume) and tb number. Sample numbers n = 5 (b, c), n = 4 (d), n = 5 (e). Data are presented as mean ± SEM. P values, statistical analysis performed using the Mann–Whitney test (two-tailed). n = independent biological samples.
a-c. UMAP plot showing color-coded cell clusters of control and Lats2iΔEC bone stromal cells (a). Markers for main bone stromal cell populations (b) and their subclusters (c) are shown in dot plots. d- f. UMAP plot of EC subsets, split by condition (right) (d). Bar graph displaying percentage of endothelial cells subtypes in control and Lats2iΔEC mutants (e). UMAP plot colored by normalized expression of EC subset identity marker genes: Ramp3 (metaphyseal ECs, mpECs), Gja4 (arterial ECs, aEC), Fmo (arteriole-like ECs, alECs) and Stab2 (bone marrow ECs, bm) (f). g, h. Volcano plot of differentially expressed genes (frac. expressed > 0.2 in either condition) between control and mutant condition across all ECs (blue color: log2FC >=1,p.adj <=0.001,) (g). Visualisation of selected known Hippo pathway target genes: Ccn1 (Cyr61), Ccn2 (Ctgf), Angpt2 and Thbs1 in Lats2iΔEC mutant and control (h).
a. Confocal images showing strongly elevated Angiopoietin 2 (ANGPT2), Thrombospondin 1 (THBS1), Fibronectin 1 (FN1), Transgelin (TAGLN), and Vimentin (VIM) expression in Lats2iΔEC bone sections. (n = 4). Data in graphs are presented as mean ± SEM. P values, statistical analysis performed using the Mann–Whitney test (two-tailed)). b. Confocal images showing increased staining for collagen IV (COL IV), the actin cytoskeleton (with Phalloidin), active-integrin β1 and phosphorylated focal adhesion kinase (FAK pY397) in Lats2iΔEC mutant relative to control. c. Transmission electron micrographs showing increased subendothelial basement membrane and accumulation of ECM (yellow arrowheads) in Lats2iΔEC femur. Mutant ECs are rich in actin filaments (red arrowheads). d. Confocal images of Lats2iΔEC mutant in R26-dTomato reporter background showing αSMA staining in dTomato+ ECs. In control samples, αSMA signal is confined to SMCs covering arteries. n = independent biological samples.
a. Ligands showing on average significantly (log2FC >=1,p.adj <=0.05, frac. expressed >=0.1) larger counts in mutant mpECs than in any other EC subidentity and are significantly elevated (see Methods) relative to at least one other EC subpopulation. b. Selected ligand-receptor parings of main bone stromal cells, which are increased in Lats2iΔEC mutants. c. Mean normalized expression counts of selected, significant ligand-receptor interactions (p.adj of LRI score in either condition and LRIDiff Scores <= 0.05, |LRIDiff| >= 1) in mpECS and all other major celltypes. Ligand (left, dark blue bar) and receptor (right, red bar). d. Mean normalized expression counts of selected, significant ligand-receptor interactions (p.adj of LRI score in either condition and LRIDiff Scores <= 0.05, LRIDiff >= 1) in mpECS, mpMSCs and SMCs. Ligand (left, dark blue bar) and receptor (right, red bar). e. Quantitative PCR (qPCR) shows expression of myofibroblasts markers Acta2, Tagln and Fn1 in cultured BMSCs upon treatment with recombinant TGFβ1, endothelin 1 (ET1) or adrenomedullin (ADM). (n = 3). Data in graphs are presented as mean ± SEM. P values, statistical analysis performed using the Mann–Whitney test (two-tailed)). n = independent biological samples.
a, b. Representative confocal images of control and SrfiΔEC femur show type H (EMCNhigh; CD31high) capillaries, Nuclei, DAPI (blue) (a) as well as PDGFRβ+ cells (red) and αSMA+ arterial SMCs (green) and EMCN (blue) in the femoral metaphysis (b). Graphs in (b) show quantification of vessel columns and arteries (n = 6). Data presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). c-e. Confocal images showing ACAN+ (green) chondrocytes in femoral growth plate (gp) (c) and OSX+ cells (green) (d). µCT images of trabecular bone of SrfiΔEC mutant and control(e) and quantitation of trabecular (tb) volume (BV/TV, bone volume/total volume) tb number and gp length (n = 6). Data presented as mean ± SEM. P values, Mann–Whitney test (two-tailed). f, g. Longitudinal tile scan confocal images showing EMCN+ (red) CD31+ (green) vessels in control, Lats2iΔEC and Lats2 SrfiΔEC compound mutant femur (f). Metaphysis (mp), growth plate (gp, dashed lines) and bone marrow (bm) are indicated. Quantification shows increase in CD31+ EMCN- area in Lats2iΔEC metaphysis and rescue by inactivation of Srf (g) (n = 4). Data presented as mean ± SEM. P values, Tukey multiple comparison test (one-way Anova). h. Representative images showing OSX+ cells in control, Lats2iΔEC, and Lats2 SrfiΔEC compound mutant femur. i, j. Representative μCT images of femoral trabecular bone (i). Quantitative analysis of trabecular bone (tb) thickness and separation (j) (n = 4). Data presented as mean ± SEM. P values, Tukey multiple comparison test (one-way Anova). k. Proposed model of YAP1/TAZ-induced endothelial-to-mesenchymal transition (EndMT) as a driver of myelofibrosis and osteosclerosis. n = independent biological samples. Created with BioRender.com.
Supplementary Fig. 1 and Supplementary Tables 1 and 2.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Data Availability Statement
The scRNA-seq data generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE225429. The mouse reference genome GRCm39 with GENCODE M26 anotation (https://www.gencodegenes.org/mouse/release_M26.html) was used for mapping the reads in this study. All other relevant data supporting the key findings of this study are available within the article and its Supplementary Information files.
All code used to produce the computational analysis results and figures in this publication are available at https://gitlab.gwdg.de/pmajev/sivaraj_majev_lats2_ecs. The assigned DOI is 10.5281/zenodo.11456485.