Abstract
Quadruple-hit lymphomas are extremely rare non-Hodgkin lymphomas with a reported dismal prognosis in the few reported cases. A “quadruple hit” has been defined by the presence of concurrent MYC, BCL2, BCL6, and CCND1 chromosomal rearrangements. We report a new case of a quadruple hit lymphoma in a 73-year-old Hispanic man who presented with an enlarging left-sided neck mass. Computed tomography showed a 1.9-cm mass in left the tonsil with bulky cervical lymphadenopathy. The presence of all four chromosomal rearrangements can reportedly occur with disease progression in both diffuse large B-cell lymphomas and mantle cell lymphomas. Further characterization of the tumor by next-generation sequencing may be of benefit to delineate between these two possibilities. Immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), and next-generation sequencing were used to confirm and classify the diagnosis. Histologic sections of the cervical lymph node demonstrated an atypical lymphoid infiltrate with large and pleomorphic cells, which were positive for CD20, CD10, BCL1 (Cyclin D1), BCL2, BCL6, and cMYC and negative for CD5 and SOX11 on immunohistochemistry with a Ki-67 proliferative index of 70%. FISH demonstrated MYC, BCL2, BCL6, and CCND1 rearrangements and the diagnosis of high-grade B-cell lymphoma with MYC, BCL2, BCL6, and CCND1 was rendered. Our patient was treated with dose adjusted etoposide, doxorubicin, cyclophosphamide, prednisone, and rituximab chemotherapy and has been in remission for 20 months.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12308-024-00593-8.
Keywords: Quadruple hit lymphoma, MYC-BCL2 rearrangements, MYC-BCL6 rearrangements, Lymphadenopathy, KMT2D
Introduction
Diffuse large B-cell lymphoma (DLBCL) is an aggressive and biologically heterogeneous non-Hodgkin lymphoma group of B-cell lymphomas. High-grade B cell lymphoma (HGBL) with translocations involving MYC and BCL2 is retained as a so-called double hit (DH) lymphoma, by the 5th edition World Health Organization (WHO) classification (2022) [1]. DH lymphomas are rare entities with poor prognosis. In the revised 4th edition of the WHO Classification (2017), these lymphomas were categorized as HGBL with MYC, BCL2, and/or BCL6 rearrangements. However, the 5th edition WHO classification has excluded MYC and BCL6 rearranged cases from the DH category. B-cell lymphomas with MYC and BCL6 rearrangements are now reclassified as a subtype of DLBCL, NOS or HGBL, NOS according to their cytomorphological features [1]. In comparison, the International Consensus Classification (ICC) has retained these cases as a DH sub-category on the basis that some studies have recorded poor outcomes [2]. “Quadruple-hit” lymphomas, while not a defined entity in either the WHO or ICC classification schema, have been characterized by the concurrent presence of MYC, BCL2, BCL6, and CCND1 rearrangements, and are extremely rare with an apparent dismal prognosis. Currently, only 10 such cases have been reported in the literature [3–10]. Herein, we report a case of HGBL with MYC, BCL2, BCL6, and CCND1 rearrangements, a so-called quadruple hit lymphoma and describe its molecular and cytogenetic features.
Case presentation
A 73-year-old male with no significant past medical history presented with a left-sided neck swelling for a few days’ duration. Initially, he was treated with antibiotics for presumed underlying infection. Three months later, he returned to clinic with further enlargement of swelling in the left neck. Laryngoscopy revealed an enlarged, firm, and erythematous left tonsil with an exophytic mass. The right tonsil was unremarkable. CT scan of the neck demonstrated a 1.9-cm mass in the left tonsil with bulky cervical lymphadenopathy; a subsequent PET scan showed the mass and lymph nodes to be hypermetabolic; no other hypermetabolic lesions were present.
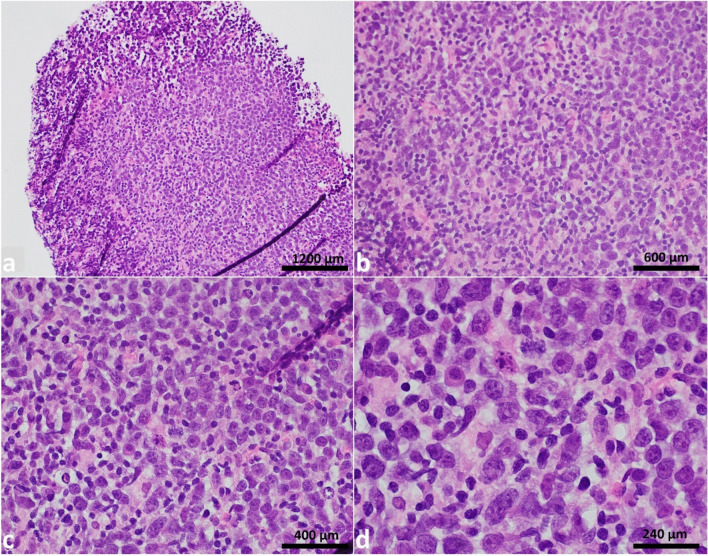
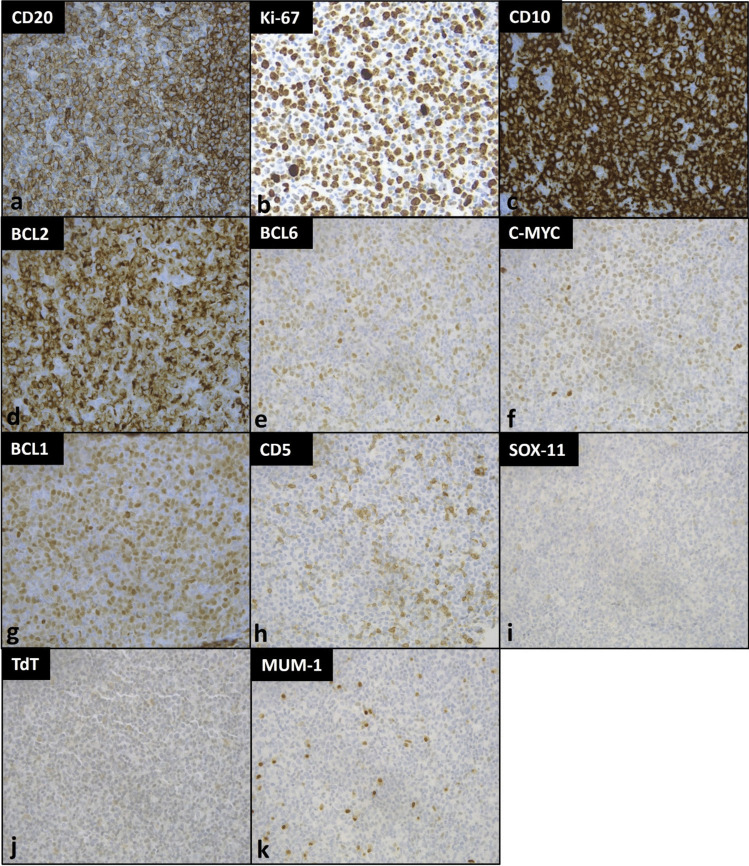
A core biopsy of the left cervical lymph node was obtained. Histology showed complete architectural effacement by large and pleomorphic cells (Fig. 1) which were positive for CD20, CD10, BCL1, BCL2, BCL6 (subset, weak), and cMYC. The Ki-67 proliferative rate was estimated at 70% (Fig. 2). Tumor cells were negative for CD5, CD23, SOX11, MUM1, TdT, CD30, and EBER by in situ hybridization. Given the overall findings, a preliminary diagnosis of DLBCL of germinal center origin was rendered, pending fluorescence in situ hybridization (FISH) studies to rule out a possible HGBL. In addition to MYC, BCL2, and BCL6 probes, testing for CCND1 rearrangement was also performed due to diffuse expression of BCL1.
Fig. 1.
Hematoxylin & eosin (H&E) stain of left cervical lymph node. Complete architectural effacement of lymph node core tissue by large pleomorphic nuclei with irregular contour, blastoid chromatin, and prominent eosinophilic nucleoli. The magnifications in figures a to d in order are 200 × , 400 × , 600 × , and 1000 ×
Fig. 2.
Immunohistochemical stains of left cervical lymph node. The immunohistochemical stains at 400 × magnification show tumor cells positive for a CD20, b Ki-67, c CD10, d BCL-2 (> 50%), e BCL-6 (weak, > 30%), f c-MYC (weak, > 40%), and g BCL1, and negative for h CD5, i SOX-11, j TdT, and k MUM-1
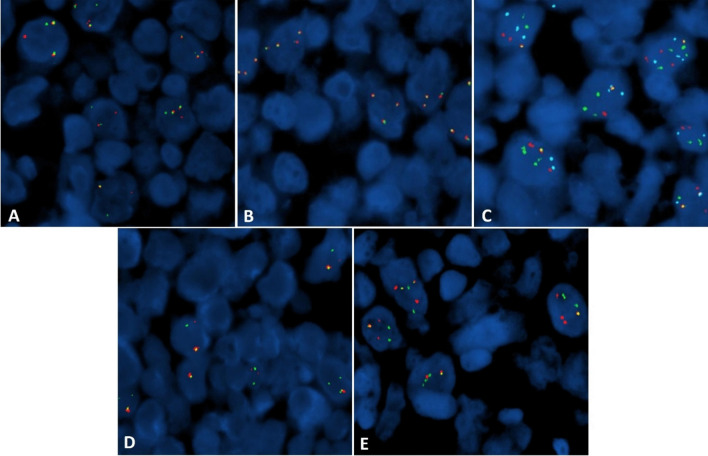
FISH studies (Fig. 3) revealed variant MYC (84.5%), CCND1 (77.5%), and BCL2 (95%) rearrangements, all involving the IGH gene locus and showing the presence of only a single fusion signal on dual color FISH. The translocations were specifically identified as t(8;14)(q24;q32), t(11;14)(q13;q32), and t(14;18)(q32;q21), respectively. Rearrangement of BCL6 (3q27) (72.5%) was also detected on break-apart probe testing. Studies were initially negative for rearrangement of the MYC (8q24) locus using a dual color break-apart probe; however, 3–4 intact (non-rearranged) copies of 8q24 were observed in 55% of the cells analyzed, prompting the MYC::IGH fusion testing with dual fusion probes. FISH studies also demonstrated the presence of trisomy 8 and additional signals for IGH, BCL6, and CCND1. No fusion partner testing was performed for BCL6, or for potential partners from the variant rearrangements of MYC, BCL2, and CCND1.
Fig. 3.
Fluorescence in situ hybridization studies of lymph node (FISH). Break-apart FISH studies showed breaks in BCL6 (A), but no breaks in MYC (B). Dual fusion FISH studies showed the presence of only a single fusion signal and variable additional probe signals for: IGH::MYC {green: IGH, red: MYC (8q24), blue: CEP8}(C), IGH::BCL2 {green: IGH, red: BCL2}(D), and IGH::CCND1 {green: IGH, red: CCND1}(E)
Based on the FISH results, a modified diagnosis of HGBL with MYC, BCL2, BCL6, and CCND1 rearrangements was made. Next-generation-sequencing (NGS) was performed for further characterization, with results detailed in Table 1. No abnormalities were seen in the TP53 gene. Tumor mutation burden is calculated as 7.55 mutations/Mb.
Table 1.
Gene mutations in the present case
| Gene name | Chr | Exon (E)/intron (I) | Nucleotide | Amino acid change | VAF | Mutation type | Variant classification |
|---|---|---|---|---|---|---|---|
| ARID1A | 1 | E: 1/20 | c.837_862del | p.Ser280AlafsTer111 | 11% | Frameshift variant | Likely pathogenic |
| BCL2 | 18 | I: 2/2 | c.585 + 4G > C | - | 5% | Splice region variant and intron variant | Uncertain significance |
| CCND3 | 6 | E: 5/5 | c.774_775delinsTG | p.Ser259Ala | 42% | Missense variant | Uncertain significance |
| FANCA | 16 | E: 31/43 | c.3032G > A | p.Arg1011His | 7% | Missense variant | Uncertain significance |
| KMT2D | 12 | E: 32/54 | c.8200C > T | p.Arg2734Ter | 43% | Stop gained | Pathogenic |
| MYC | 8 | E: 2/3 | c.475C > T | p.Leu159Phe | 14% | Missense variant | Uncertain significance |
| MYC | 8 | E: 2/3 | c.339G > C | p.Gln113His | 13% | Missense variant | Uncertain significance |
| MYC | 8 | E: 2/3 | c.265 T > A | p.Tyr89Asn | 12% | Missense variant | Uncertain significance |
| PIM1 | 6 | E: 2/6 | c.113A > T | p.Tyr38Phe | 9% | Missense variant | Uncertain significance |
| PIM1 | 6 | E: 2/6 | c.87G > C | p.Lys29Asn | 8% | Missense variant | Uncertain significance |
| PIM1 | 6 | E: 1/6 | c.68C > T | p.Thr23Ile | 7% | Missense variant | Uncertain significance |
| PIM1 | 6 | E: 4/6 | c.286G > C | p.Val96Leu | 7% | Missense variant | Uncertain significance |
| PXDNL | 8 | E: 11/23 | c.1357A > T | p.Thr453Ser | 8% | Missense variant | Uncertain significance |
| SF3B1 | 2 | E: 14/25 | c.1998G > C | p.Lys666Asn | 9% | Missense variant | Likely pathogenic |
| SOCS1 | 16 | E: 2/2 | c.46G > A | p.Ala16Thr | 7% | Missense variant | Uncertain significance |
| TNFRSF14 | 1 | E: 5/8 | c.472C > T | p.Gln158Ter | 8% | Stop gained | Likely pathogenic |
| TSC2 | 16 | E: 2/42 | c.58G > T | p.Gly20Ter | 8% | Stop gained | Likely pathogenic |
Chro chromosome, VAF variant allele frequency
A staging bone marrow biopsy was negative for lymphoma (clinical stage I). The patient was treated with 6 cycles of dose adjusted etoposide, doxorubicin, cyclophosphamide, prednisone, and rituximab (DA-R-EPOCH). This regimen was completed and restaging PET scan at 2 months showed complete resolution of the hypermetabolic lesions. He has been in complete remission for approximately 20 months.
Material and methods
Immunohistochemistry
IHC was performed per routine hospital procedures at CHI Health Bergan Mercy Hospital, Omaha, NE, according to the manufacturer’s protocols.
Fluorescence in situ hybridization
FISH was performed at the Warren G. Sanger Human Genetics Laboratory at Nebraska Medicine, Omaha, NE, utilizing the Locus Specific Identifier (LSI) IGH::MYC t(8;14) Dual Fusion Translocation Probe with CEP 8, the LSI IGH::CCND1 t(11;14) Dual Fusion Translocation Probe, the LSI IGH::BCL2 t(14;18) Dual Fusion Translocation Probe, the LSI BCL6 (3q27) Major and Alternate Breakpoint Dual Color Break-apart Probe, and the LSI MYC (8q24) Dual Color Break-apart Probe. Standard FISH protocol (co-denaturation of the probe and target at 74 °C for 4 min, hybridization overnight at 37 °C, washing at 72 °C for 2 min) was followed and images were captured using ASI software (Applied Spectral Imaging, Chicago, IL).
Molecular pathology
NGS was performed at Cedars-Sinai Medical Center, Los Angeles, CA, with further details provided in supplementary material.
Discussion
DLBCL is an aggressive and heterogeneous grouping of non-Hodgkin lymphomas encompassing a spectrum of different immunophenotypic and molecular variants. The definition and categorization of DLBCL with multiple gene rearrangements (so-called hits) have been evolving in recent years. The 2022-ICC retains a subgrouping for cases with MYC and BCL6 rearrangement; this is recognized as a heterogeneous category with variable gene expression profiles and mutational spectra [2]. Neither the WHO nor ICC classification schema recognize HGBL with MYC, BCL2, BCL6, and CCND1 rearrangements (a so-called quadruple hit) as a unique entity.
The tumor cells in our case showed mild pleomorphism with eosinophilic nucleoli and some blastoid chromatin. Tumor cells were positive for BCL1, raising the possibility of a blastoid/pleomorphic variant of mantle cell lymphoma (MCL). However, CD5 and SOX11 were negative, arguing against a diagnosis of MCL. TdT was also negative. Tumor cells were also positive for CD10 and BCL6 (subset), further supporting germinal center derivation. Following the currently accepted classification schema, our case is best classified as a HGBL with MYC and BCL2 rearrangement.
B-cell lymphomas with MYC, BCL2, BCL6, and CCND1 rearrangements are rare entities. Available outcomes data from previous publications suggest this entity has a poor prognosis [3–10]. Among the reported cases, there were four males and three females with median age 74 years (range 51–81 years); lymphadenopathy was seen in four of the seven cases. Two cases reported staging information, and both were stage III. Most cases were negative for CD5 and SOX11 with Ki-67 in the range of 60–90%. Demographic data was not provided for the three remaining cases. There is no consensus on the optimal treatment and the outcomes have been dismal, despite aggressive initial therapies [7]. The clinical features of all reported cases are summarized in Supplementary Table 1 and the histopathological features, diagnosis, ancillary studies, treatment, and outcome/overall survival are summarized in Supplementary Table 2.
CCND1 gene rearrangement or other genetic alterations involving the gene can lead to aberrant BCL1 protein expression. Among hematolymphoid tumors, BCL1 is found to be overexpressed in > 90% of MCL cases and about 40% of plasma cell myelomas, both of which are caused by a translocation which juxtaposes the immunoglobulin heavy chain (IGH) gene to the CCND1 gene. BCL1 protein expression is rarely seen in DLBCL, which has been linked to copy number gains of CCND1 or via mRNA dysregulation [11, 12]. FISH studies in our case demonstrated additional signals of CCND1, a likely mechanism for the observed protein expression by IHC.
In DLBCL, the data suggests that the CCND1 rearrangement is a secondary event during lymphoma evolution [12]. Cheng et al. included a quadruple hit lymphoma in their report of DLBCL with CCND1 rearrangements considered to represent secondary genetic events [9]. This is in contrast to MCL, where CCND1 rearrangement is considered to be a primary genetic event [12]. MCL may also gain secondary BCL2, BCL6, and MYC rearrangements, as proposed in three [3, 9, 10] of the previously reported quadruple hit lymphomas.
In our case, it is difficult to confidently determine the sequence of genetic alterations. All translocations were found in relatively (and similarly) high proportions of tumor cells. However, sequencing results were more typical of DLBCL than MCL. The combination of mutations best fits in the EZB-DLBCL molecular cluster according to Morin et al. [13]; similar reported molecular clusters include C3, BCL2, and MYC/BCL2-DH [14]. Interestingly, a previously sequenced quadruple hit lymphoma, designated as a pleomorphic MCL, shows limited overlapping mutations with our case [10]. The PIM1 gene was the sole shared mutation with our case. Mutations in the PIM1 gene are frequently seen in lymphomas and have been implicated in DLBCL pathogenesis [15]. Although PIM1 mutations identified in our case are variants of uncertain significance (VUS), three out of the four mutations (c.87G > C, c.68C > T, and c.286G > C) have been reported multiple times in DLBCL [16–18] respectively; the other PIM1 mutation (c.113A > T) has not been reported to our knowledge. NGS findings from the present case and the two previously sequenced quadruple hit lymphomas are summarized in Supplementary Table 3.
MYC mutations in DLBCL are more frequently seen in cases with MYC and BCL6 rearrangement, which was present in our case [19]. This is thought to be due to aberrant somatic hypermutation from activation-induced cytidine deaminase, also implicated in the genesis of MYC rearrangement [20]. All three MYC mutations in our case are identified as VUS and have not been previously reported. Cases exhibiting intact (non-rearranged) copies of MYC on break apart probe testing, including some cases with additional intact MYC signals, as in our case, have revealed the presence of MYC gene fusions when followed by an IGH::MYC dual fusion probe [21].
FISH studies also showed additional copies of BCL6. This finding has been seen in nearly half of MYC-rearranged DLBCLs according to one study [22].
KMT2D is frequently mutated in DLBCL (~ 30% of de novo cases, including both germinal center B-cell and activated B-cell subtypes and follicular lymphoma (~ 90%)) [23]. Recent studies have suggested KMT2D mutations represent early events in a common progenitor before divergent evolution of follicular lymphoma or DLBCL, the latter occurs through acquisition of additional genetic lesions and clonal expansion [23, 24]
Among lymphomas, TNFRSF14 mutation has been reported in follicular lymphoma as well as DLBCL, NOS (EZB) [25] TNFRSF14 mutations have not been reported in mantle cell lymphoma [9]. The specific TNFRSF14 mutation found in our case (c.472C > T) has been previously reported in follicular lymphoma and DLBCL [26].
In addition to BCL2 translocation, our case also harbored BCL2 gene mutation (c.585 + 4G > C). This was categorized as a VUS for our case but been previously reported in follicular lymphoma and DLBCL [27].
B-cell lymphomas with concurrent MYC, BCL2, BCL6, and CCND1 rearrangements appear to be a rare occurrence; however, current standard approaches to DLBCL/HGBL classification do not require routine testing for CCND1 rearrangement. With the current classification schemes de-emphasizing the importance of BCL6 rearrangement, this may no longer be routinely assessed as well. The addition of CCND1 rearrangement in the workup for a DLBCL/HGBL might only be sought in cases with BCL1 protein expression, as seen in our case. Sequencing may be of benefit for delineating DLBCL from MCL in the quadruple hit setting, although current data is limited.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank late Dr. Bhavana J. Dave for her contributions and the staff at the Warren G. Sanger Human Genetics Laboratory at Nebraska Medicine, and the Molecular Pathology Laboratory at Cedars-Sinai Medical Center for their contributions in performing and evaluating the ancillary studies.
Funding
This study was not supported by any funding.
Declarations
Ethical approval
Informed consent was obtained from the patient.
Informed consent
Informed consent was obtained from the patient.
Consent for publication
A written informed consent was obtained from the patient.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alaggio R et al (2022) The 5th edition of the World Health Organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia 36(7):1720–1748. 10.1038/s41375-022-01620-2 10.1038/s41375-022-01620-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falini B, Martino G, Lazzi S (2023) A comparison of the International Consensus and 5th World Health Organization classifications of mature B-cell lymphomas. Leukemia 37(1):18–34. 10.1038/s41375-022-01764-1 10.1038/s41375-022-01764-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawakami K et al (2004) Case of B-cell lymphoma with rearrangement of the BCL1, BCL2, BCL6, and c-MYC genes. Int J Hematol 79(5):474–479. 10.1532/IJH97.03105 10.1532/IJH97.03105 [DOI] [PubMed] [Google Scholar]
- 4.Bacher U, Haferlach T, Alpermann T, Kern W, Schnittger S, Haferlach C (2011) Several lymphoma-specific genetic events in parallel can be found in mature B-cell neoplasms. Genes Chromosomes Cancer 50(1):43–50. 10.1002/gcc.20831 10.1002/gcc.20831 [DOI] [PubMed] [Google Scholar]
- 5.Yoshida M et al (2015) Clinicopathological features of double-hit <scp>B</scp> -cell lymphomas with <scp>MYC</scp> and <scp>BCL</scp> 2, <scp>BCL</scp> 6 or <scp>CCND</scp> 1 rearrangements. Pathol Int 65(10):519–527. 10.1111/pin.12335 10.1111/pin.12335 [DOI] [PubMed] [Google Scholar]
- 6.Ittel A et al (2015) Four genetic lymphoma-specific events (MYC, BCL2, BCL6 and CCND1) identified in a high grade B lymphoma case. Blood Cancer J 5(12):e374–e374. 10.1038/bcj.2015.99 10.1038/bcj.2015.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proulx J et al (2018) Quadruple hit lymphoma: a rare entity with dismal prognosis. Blood 132(Supplement 1):5305–5305. 10.1182/blood-2018-99-120147 10.1182/blood-2018-99-120147 [DOI] [Google Scholar]
- 8.Meloni-Ehrig A et al (2020) 36. Quadruple-hit B-cell lymphoma with simultaneous BCL2, BCL6, CCND1, and MYC rearrangements: two new cases. Cancer Genet 244:14. 10.1016/j.cancergen.2020.04.040 10.1016/j.cancergen.2020.04.040 [DOI] [Google Scholar]
- 9.Cheng J et al (2021) CCND1 genomic rearrangement as a secondary event in high grade B-cell lymphoma. Hemasphere 5(1):e505. 10.1097/HS9.0000000000000505 10.1097/HS9.0000000000000505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W et al (2021) Quadruple-hit pleomorphic mantle cell lymphoma with MYC, BCL2, BCL6, and CCND1 gene rearrangements. Br J Haematol 195(4):634–637. 10.1111/bjh.17729 10.1111/bjh.17729 [DOI] [PubMed] [Google Scholar]
- 11.Vela-ChÁvez T et al (2011) Cyclin D1 positive diffuse large B-cell lymphoma is a post-germinal center-type lymphoma without alterations in the CCND1 gene locus. Leuk Lymphoma 52(3):458–466. 10.3109/10428194.2010.540361 10.3109/10428194.2010.540361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juskevicius D, Ruiz C, Dirnhofer S, Tzankov A (2014) Clinical, morphologic, phenotypic, and genetic evidence of cyclin D1-positive diffuse large B-cell lymphomas with CYCLIN D1 gene rearrangements. Am J Surg Pathol 38(5):719–727. 10.1097/PAS.0000000000000120 10.1097/PAS.0000000000000120 [DOI] [PubMed] [Google Scholar]
- 13.Morin RD, Arthur SE, Hodson DJ (2022) Molecular profiling in diffuse large B-cell lymphoma: why so many types of subtypes? Br J Haematol 196(4):814–829. 10.1111/bjh.17811 10.1111/bjh.17811 [DOI] [PubMed] [Google Scholar]
- 14.Zhuang Y et al (2022) Altered pathways and targeted therapy in double hit lymphoma. J Hematol Oncol 15(1):26. 10.1186/s13045-022-01249-9 10.1186/s13045-022-01249-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H et al (2022) PIM1 genetic alterations associated with distinct molecular profiles, phenotypes and drug responses in diffuse large B‐cell lymphoma. Clin Transl Med 12(4). 10.1002/ctm2.808 [DOI] [PMC free article] [PubMed]
- 16.COSMIC (n.d.) Catalogue Of Somatic Mutations In Cancer, COSV65165568 PIM1 gene. Wellcome Sanger Institute. https://cancer.sanger.ac.uk/cosmic/mutation/overview?id=119825033. Accessed 22 Apr 2024
- 17.COSMIC (n.d.) Catalogue Of Somatic Mutations In Cancer, COSV65165412 PIM1 gene. Wellcome Sanger Institute. https://cancer.sanger.ac.uk/cosmic/mutation/overview?id=119827076. Accessed 22 Apr 2024
- 18.COSMIC (n.d.) Catalogue Of Somatic Mutations In Cancer, COSV65165396 PIM1 gene. Wellcome Sanger Institute. https://cancer.sanger.ac.uk/cosmic/mutation/overview?id=119824464. Accessed 22 Apr 2024
- 19.Xu-Monette ZY et al (2016) Clinical and biologic significance of MYC genetic mutations in denovo diffuse large B-cell lymphoma. Clin Cancer Res 22(14):3593–3605. 10.1158/1078-0432.CCR-15-2296 10.1158/1078-0432.CCR-15-2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenz G et al (2007) Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell–like diffuse large B cell lymphoma. J Exp Med 204(3):633–643. 10.1084/jem.20062041 10.1084/jem.20062041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King RL et al (2019) False-negative rates for MYC fluorescence insitu hybridization probes in B-cell neoplasms. Haematologica 104(6):e248–e251. 10.3324/haematol.2018.207290 10.3324/haematol.2018.207290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krull JE et al (2020) Somatic copy number gains in MYC, BCL2, and BCL6 identifies a subset of aggressive alternative-DH/TH DLBCL patients. Blood Cancer J 10(11):117. 10.1038/s41408-020-00382-3 10.1038/s41408-020-00382-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J et al (2015) Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 21(10):1190–1198. 10.1038/nm.3940 10.1038/nm.3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okosun J et al (2014) Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 46(2):176–181. 10.1038/ng.2856 10.1038/ng.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitz R et al (2018) Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med 378(15):1396–1407. 10.1056/NEJMoa1801445 10.1056/NEJMoa1801445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.COSMIC (n.d.) Catalogue Of Somatic Mutations In Cancer, COSV63185584 TNFRSF14 gene. Wellcome Sanger Institute. https://cancer.sanger.ac.uk/cosmic/mutation/overview?id=111831892. Accessed 22 Apr 2024
- 27.COSMIC (n.d.) Catalogue Of Somatic Mutations In Cancer, COSV61374190 BCL2 gene. Wellcome Sanger Institute. https://cancer.sanger.ac.uk/cosmic/mutation/overview?id=122082405. Accessed 22 Apr 2024
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.