Abstract
Genomic RNA sorting between translation and packaging was examined for human immunodeficiency virus type 1 (HIV-1) and HIV-2 using actinomycin D and leptomycin B treatment. Both viruses behaved differently from a simple retrovirus under actinomycin D treatment. With leptomycin B, the lack of apparent functional separation between translation and packaging functions in lentiviruses was confirmed. HIV-2 RNA levels were more stable, but reverse transcriptase production declined similarly to HIV-1.
Assembly of infectious retroviral particles requires encapsidation of the RNA genome by the structural protein Gag (2). Since Gag is translated from the genomic transcript, this RNA species provides both assembly components. It is of interest to determine whether an individual genomic RNA can fulfill both destinies or whether a functional separation exists between viral RNA molecules committed to different roles.
In simple retroviruses this question has been addressed by time course studies using Rauscher and AKR murine leukemia viruses (MLV) (10–12). Treatment of chronically infected cells with the transcription inhibitor actinomycin D revealed two nonequilibrating pools of genomic RNA: encapsidated RNA is not detectable at late time points of treatment, though message capable of protein production persists in the cytoplasm.
No studies of RNA sorting have been reported for complex retroviruses. Because of the relevance to vector and antiviral design, we applied this analysis to human immunodeficiency virus type 1 (HIV-1). We studied HIV-2 due to evidence of a close association of translation and packaging (7). We included the simple retrovirus Moloney murine leukemia virus (MMLV) to validate our system and as a basis for comparison.
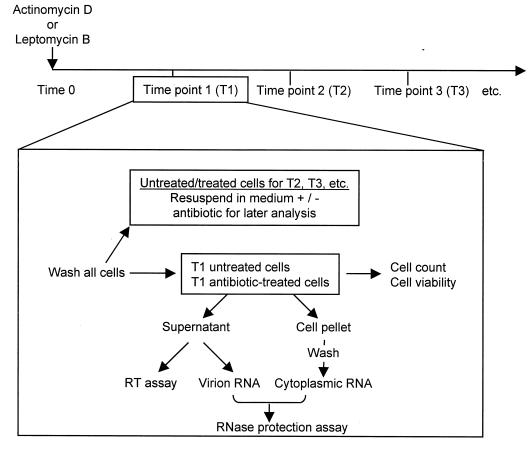
Chronically infected cultures of MMLV in NIH 3T3 cells and of HIV-1 and HIV-2 in Jurkat T cells were used in time course assays. Figure 1 shows the manipulations conducted at each time point. The time course length was longer for HIV-1 and HIV-2 because of the longer half-life observed for the HIV RNA (data not shown). This difference also necessitated a fivefold-higher concentration (5 μg/ml) of actinomycin D. These differing concentrations inhibit transcription comparably in the two cell types (1, 10).
FIG. 1.
Organization of the time course assays. MMLV infection was initiated by transfection of the proviral clone pNCA (a gift of S. Russell), and the Jurkat cells were infected with stocks of HIV-1 IIIB or HIV-2 ROD. After 2 to 3 weeks of culture, cells were used in time course assays. Chronically infected cells were split into an appropriate number of flasks or dishes, half of which were treated with antibiotic. At each time point, all cells were washed and either harvested or resuspended with or without drug treatment. Since all cells were washed whether or not they were harvested at the next time point, later-time-point cells undergo more washes. The practice of expressing values relative to control cells washed in parallel compensates for this disparity.
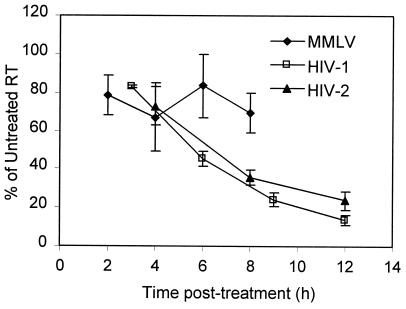
As previously (11), supernatant reverse transcriptase (RT) activity acted as a surrogate for translation of genomic RNA; levels were measured as previously described (8, 13). Figure 2 shows RT levels plotted as a percentage of the untreated control RT reading and represents the average of two to three experiments. Plots of RT decline from independent experiments were superimposable for HIV-1 and HIV-2. Although MMLV values varied more, the absence of a marked decline in MLV RT activity is consistent with published activities of 30 to 80% of control values at 6 h posttreatment (10, 11). RT activity reductions are not an artifact of actinomycin D concentration, since time courses performed at 1 μg/ml for HIV-1 and HIV-2 showed decreases of 47 and 70% from 2 to 8 h, respectively, while the MMLV decrease was just 12% (data not shown). Actinomycin D interferes with strong-stop and DNA-dependent DNA synthesis by HIV-1 RT (5, 6). However, the concentration used our assays is 17- to 25-fold lower than the 50% inhibitory concentrations (IC50) given for inhibition of these processes. Strand transfer is inhibited at concentrations lower than those used here (5, 6), but a decrease in new infections would not be apparent in chronically infected cultures over the time period examined.
FIG. 2.
Actinomycin D (5 μg/ml)-treated RT levels as percentages of those in untreated controls for MMLV, HIV-1, and HIV-2. Values reflect averages of PEG-precipitated supernatant RT measurements for two to three independent time course experiments for each virus. Error bars represent standard errors of the means.
To investigate viral RNA, cytoplasmic and virion RNA samples were extracted for use in RNase protection assays (RPA) as previously described (8). Riboprobes distinguished between viral DNA, full-length RNA, and spliced RNA. HIV-1 and HIV-2 probes were produced from KSIIΨCS (9) and KS2HIV-2 (7), respectively. MMLV riboprobes were generated from Bluescript KS II (Stratagene) containing SacI (position 414) to SpeI (position 731) (provirus numbering). Prior titrations established the quantity of cytoplasmic and virion RNA, ensuring a molar excess of probe (data not shown). RPA contained constant input amounts of cytoplasmic RNA, and virion RNA was normalized for RT activity. The HIV experiments included controls containing twice the usual input of RNA to confirm probe excess. Jurkat RNA was spiked into the virion samples so that β-actin mRNA or 28S rRNA probes (Ambion) could be used to detect loading variation; the latter probe also permitted normalization of cytoplasmic RNA.
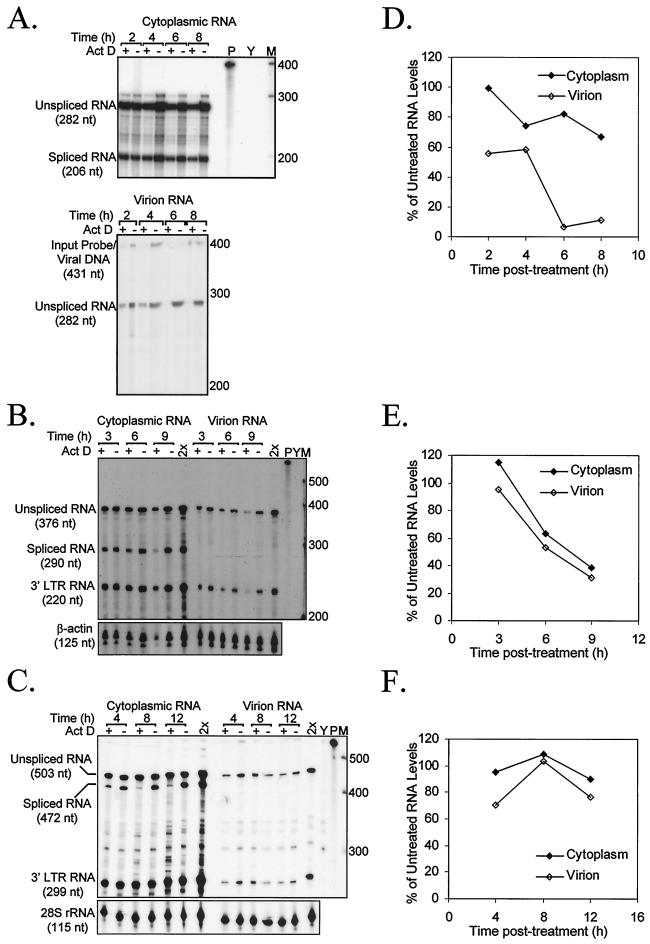
A representative RPA of two to three independent time course experiments is shown (Fig. 3A to C), with plots of RNA level as a percentage of that in an untreated sample (Fig. 3D to F). As previously described (10, 11), MMLV showed a differential decline in cytoplasmic and virion RNA levels. By 6 h posttreatment, virtually no encapsidated RNA was detectable, while only a moderate decline in cytoplasmic RNA was evident. By contrast, although HIV-1 virion RNA declines, it is in direct proportion to the level of cytoplasmic RNA. HIV-2 RNA levels were more stable but maintained the same virion/cytoplasmic ratios. Even in a time course performed at 10 μg of actinomycin D/ml, levels did not fall (data not shown).
FIG. 3.
RNase protection analysis of actinomycin D (5 μg/ml) time course RNA levels. A representative RPA of at least two independent experiments is shown for MMLV (A), HIV-1 (B), and HIV-2 (C). RPA used a constant cytoplasmic RNA amount (2.5 μg for MMLV; 0.5 μg for HIV-1 and HIV-2) and virion RNA corresponding to a constant level of RT activity (475 cpm of RT activity for MMLV; 25,000 cpm of RT activity for HIV-1 and HIV-2). Act D, actinomycin D; 2×, double input control; Y, torula yeast RNA and probe; P, undigested probe; M, RNA Century markers (Ambion; numbers adjacent to bands indicate nucleotide length of marker RNA). Levels of genomic-length RNA relative to those in untreated controls are plotted for MMLV (D), HIV-1 (E), and HIV-2 (F). MMLV gels were analyzed with NIH Image, version 1.62, and HIV gels were quantified with an Instant Imager (Packard).
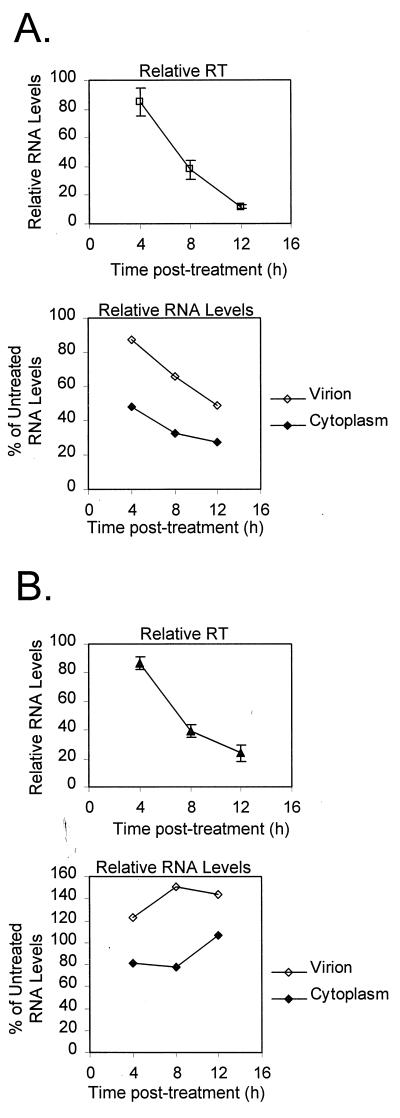
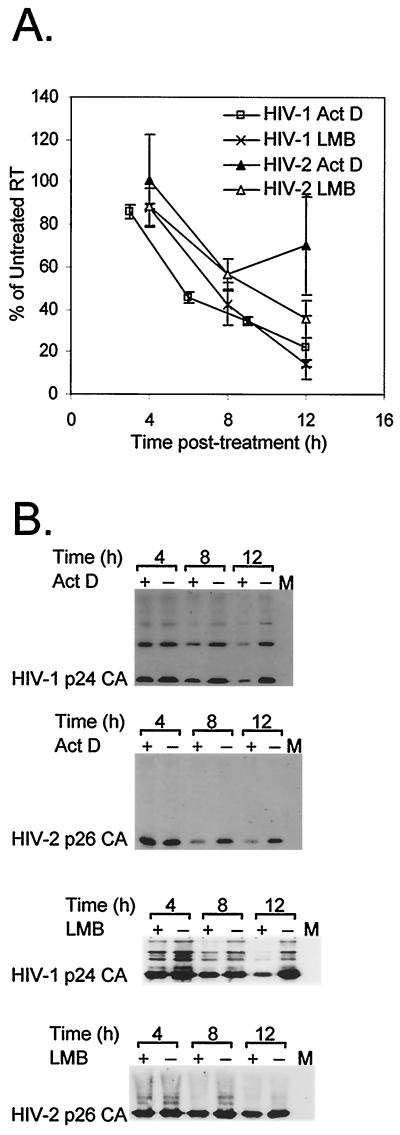
Actinomycin D can affect ribosomal loading (14), so to control for such secondary effects, the HIV experiments were repeated with leptomycin B, an inhibitor of Rev-mediated CRM1-dependent nucleocytoplasmic mRNA transport (3, 15). Leptomycin B-induced changes in RT activity and RNA levels are plotted in Fig. 4. As with actinomycin D treatment, RT activity and both viral and cytoplasmic RNA levels declined in HIV-1-infected Jurkat cell cultures. HIV-2 was treated with leptomycin B at double the concentration used with HIV-1 in an attempt to limit cytoplasmic RNA levels. However, as before, RNA levels remained more or less constant, while RT values declined similarly to those for HIV-1.
FIG. 4.
Relative RT and RNA levels of HIV-1 (A) and HIV-2 (B) in the leptomycin B time courses. HIV-1-infected cells were treated with 10 nM leptomycin B; HIV-2-infected cells were treated with 20 nM leptomycin B. Three independent experiments were performed. Average RT activities are plotted, and levels of genomic-length RNA from representative RPA are shown. Error bars reflect standard errors of the means.
The consistency of results obtained with the two antibiotics suggests that the findings reflect drug-induced changes in cytoplasmic RNA levels, not reagent-specific side effects. However, both agents are cytotoxic, resulting in viabilities 63 to 75% of control levels by the 12-h time point (data not shown). Nevertheless, normalizing RT activity for cell viability demonstrates that the RT reduction is not merely the result of nonspecific cytotoxicity (Fig. 5A). Furthermore, the measured RT activity is consistent with a real decline in Gag protein amount. Supernatant fractions of time course experiments were assayed by Western blotting with anticapsid monoclonal antibodies. In each case, there is a decline in Gag levels in treated cells (Fig. 5B). Direct effects on RT incorporation or release are unlikely and were not observed in a simple retrovirus (4, 11).
FIG. 5.
Detailed examination of Gag-Pol and Gag release from actinomycin D- and leptomycin B-treated cells. Relative RT measurements were normalized for cell viability to correct for drug-induced cytotoxicity (A). Aliquots of HIV-1- and HIV-2-infected cells were examined at each time point to assess fraction of viable cells. Levels of Gag were assayed by Western blotting (B). Time courses were performed with actinomycin D (5 μg/ml) or leptomycin B (10 nM for HIV-1; 20 nM for HIV-2). The HIV-1 antibody is reactive against p24CA, and the HIV-2 antibody (Chemicon) is reactive against p26CA; both also react with capsid-containing Gag and Gag-Pol products. Act D, actinomycin D; LMB, leptomycin B; M, mock-infected cells.
These data indicate major differences in virion assembly between the simple murine retroviruses and HIV-1. Although a subset of cytoplasmic RNA in MLV appears to be inaccessible to packaging, there is apparently no functional separation of HIV-1 transcripts. When the supply of cytoplasmic RNA is cut off during transcription (actinomycin D) or nucleocytoplasmic transport (leptomycin B), the amount of RNA packaged drops off at the same rate as the amount of viral RNA remaining in the cytoplasm. The level of RT activity declines analogously, reflecting the diminishing amount of RNA available for Gag-Pol translation. Although it is possible that translation and packaging pools exist in HIV-1 but are the same size or decay at the same rate, the simplest explanation is that there is no functional separation of HIV-1 RNA.
The differences between HIV-1 and HIV-2 are less pronounced, but it is apparent that HIV-2 cytoplasmic RNA levels are more stable under actinomycin D or leptomycin B treatment. Although leptomycin B sensitivity has not been examined for HIV-2 Rev-dependent transport, both viruses should be equally sensitive to actinomycin D-mediated transcription inhibition. HIV-2 transcripts may be more stable, the pool of cytoplasmic viral transcripts may be relatively larger, or the virus may have a lower rate of particle release. Although the relationship between particle number and RT activity has not been analyzed here, untreated supernatant RT activities were similar in the HIV-1 and HIV-2 time courses (data not shown), suggesting that a lower rate of virion production is not a likely explanation for the differences seen.
The rates of RT activity decline are similar in HIV-1 and HIV-2 despite the relative stability of HIV-2 RNA, suggesting that a factor needed for translation of HIV-2 Gag-Pol is limited. This agent may be translation-competent RNA or a cofactor necessary for RT production or release. The possible involvement of a labile cellular cofactor was not addressed in earlier MLV studies. Even if HIV-2 and MLV both rely upon a nonviral factor for virion production, their behavior is strikingly different. It is RT production that decays disproportionately in HIV-2, and it is packaging that diminishes in MMLV. Intriguingly, HIV-2 packages RNA predominantly in cis (7), such that the newly synthesized Gag preferentially encapsidates the RNA from which it was produced. This mechanism might sequester RNA in the packaging pathway and, without new RNA appearing in the cytoplasm, cause a decline in RT levels.
Understanding virion assembly better in lentiviruses such as HIV-1 may unveil further targets for antiviral chemotherapy and will aid in the design of efficient and safe lentiviral vectors for gene therapy. This study suggests that, in a lentivirus, genomic RNA is interchangeable between translation and packaging. Additionally, translation of structural proteins in HIV-2 is exquisitely sensitive to agents blocking the appearance of nascent transcripts in the cytoplasm.
Acknowledgments
MMLV proviral plasmid pNCA was a kind gift of S. Russell. We are grateful to M. Yoshida for the gift of leptomycin B. HIV-1 IIIB, HIV-2 ROD, and the p24CA monoclonal antibody were supplied by the NIBSC AIDS Reagent Programme. S. Griffin is thanked for useful discussions.
N.D. is a recipient of a National Science Foundation Graduate Student Fellowship and acknowledges support from the Marshall Aid Commemoration Commission. This work was supported by the Medical Research Council and the Sykes Trust.
REFERENCES
- 1.Arya S K, Gallo R C. Transcriptional modulation of human T-cell growth factor gene by phorbol ester and interleukin 1. Biochemistry. 1984;23:6685–6690. doi: 10.1021/bi00321a062. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 3.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 4.Gerwin B I, Levin J G. Interactions of murine leukemia virus core components: characterization of reverse transcriptase packaged in the absence of 70S genomic RNA. J Virol. 1977;24:478–488. doi: 10.1128/jvi.24.2.478-488.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo J, Wu T, Bess J, Henderson L E, Levin J G. Actinomycin D inhibits human immunodeficiency virus type 1 minus-strand transfer in in vitro and endogenous reverse transcriptase assays. J Virol. 1998;72:6716–6724. doi: 10.1128/jvi.72.8.6716-6724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeeninga R E, Huthoff H T, Gultyaev A P, Berkhout B. The mechanism of actinomycin D-mediated inhibition of HIV-1 reverse transcription. Nucleic Acids Res. 1998;26:5472–5479. doi: 10.1093/nar/26.23.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaye J F, Lever A M. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J Virol. 1999;73:3023–3031. doi: 10.1128/jvi.73.4.3023-3031.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaye J F, Lever A M. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J Virol. 1998;72:5877–5885. doi: 10.1128/jvi.72.7.5877-5885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaye J F, Richardson J H, Lever A M. cis-acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J Virol. 1995;69:6588–6592. doi: 10.1128/jvi.69.10.6588-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin J G, Grimley P M, Ramseur J M, Berezesky I K. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levin J G, Rosenak M J. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc Natl Acad Sci USA. 1976;73:1154–1158. doi: 10.1073/pnas.73.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messer L I, Levin J G, Chattopadhyay S K. Metabolism of viral RNA in murine leukemia virus-infected cells; evidence for differential stability of viral message and virion precursor RNA. J Virol. 1981;40:683–690. doi: 10.1128/jvi.40.3.683-690.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potts B J. “Mini” reverse transcriptase (RT) assay. In: Aldovini A, Walker B D, editors. Techniques in HIV research. New York, N.Y: Stockton Press; 1990. pp. 103–106. [Google Scholar]
- 14.Singer R H, Penman S. Stability of HeLa cell mRNA in actinomycin. Nature. 1972;240:100–102. doi: 10.1038/240100a0. [DOI] [PubMed] [Google Scholar]
- 15.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]