Abstract
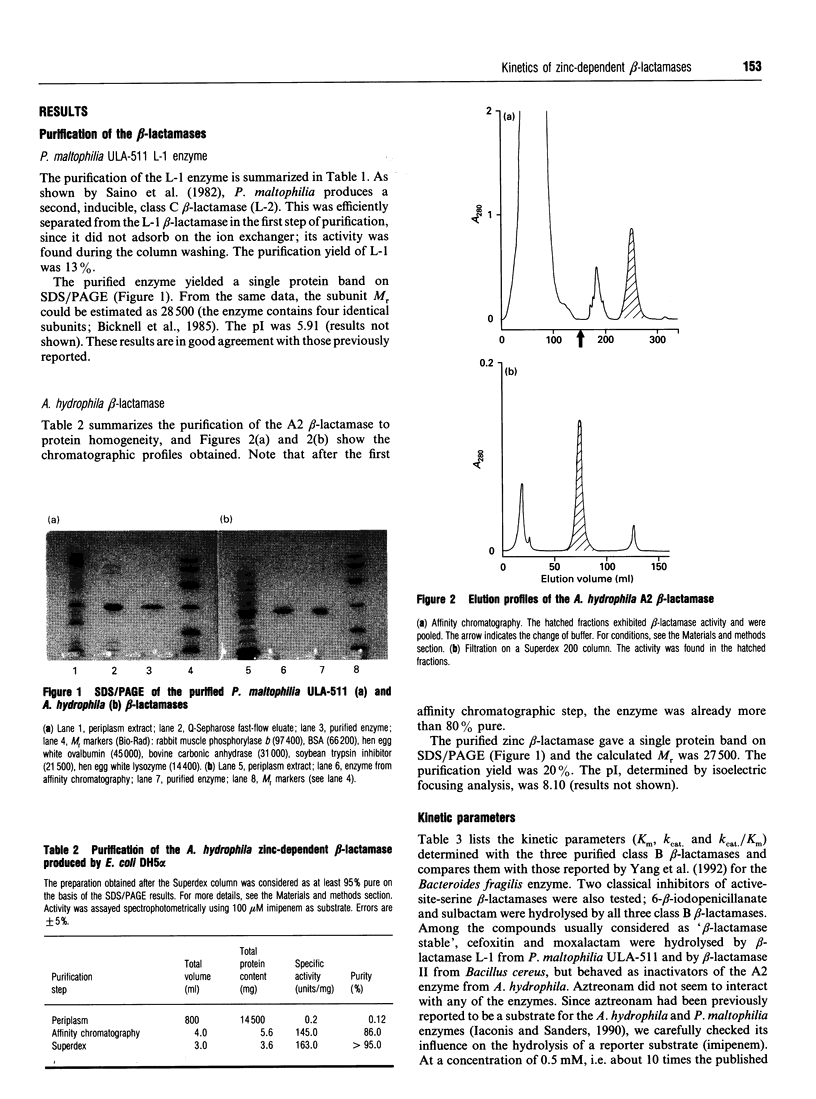
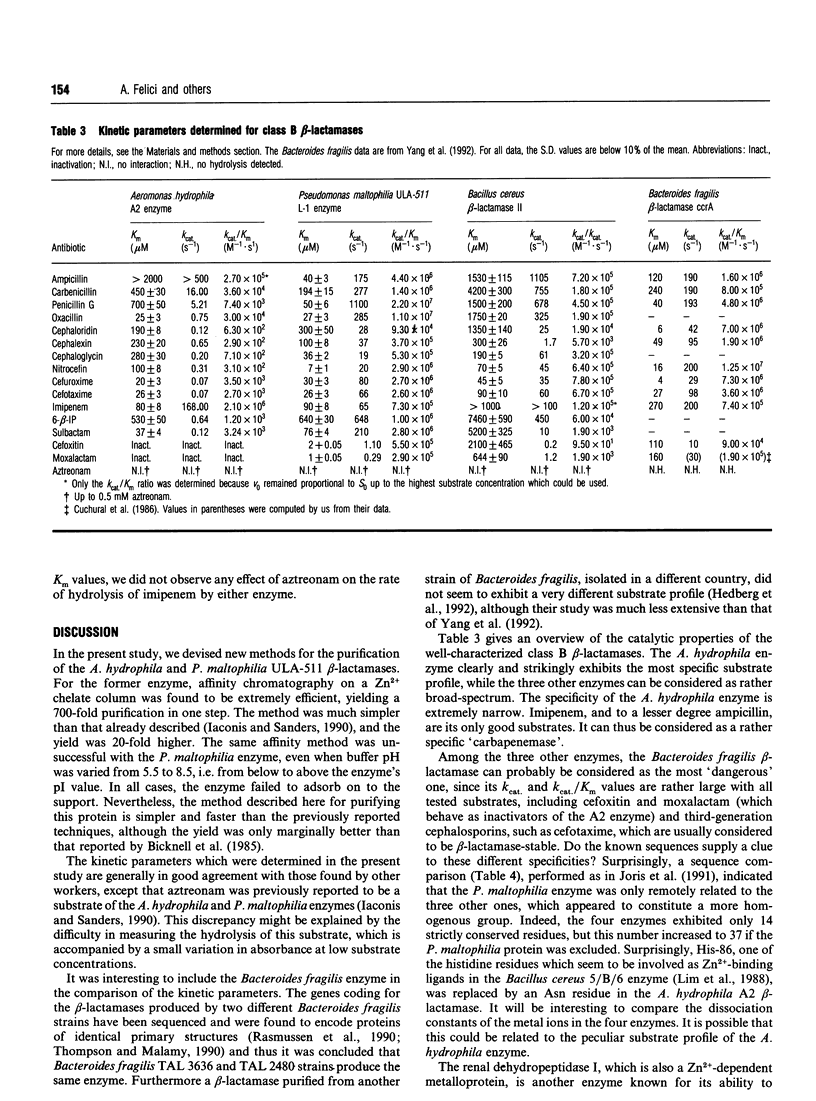
The catalytic properties of three class B beta-lactamases (from Pseudomonas maltophilia, Aeromonas hydrophila and Bacillus cereus) were studied and compared with those of the Bacteroides fragilis enzyme. The A. hydrophila beta-lactamase exhibited a unique specificity profile and could be considered as a rather specific 'carbapenemase'. No relationships were found between sequence similarities and catalytic properties. The problem of the repartition of class B beta-lactamases into sub-classes is discussed. Improved purification methods were devised for the P. maltophilia and A. hydrophila beta-lactamases including, for the latter enzyme, a very efficient affinity chromatography step on a Zn(2+)-chelate column.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi H., Tawaragi Y., Inuzuka C., Kubota I., Tsujimoto M., Nishihara T., Nakazato H. Primary structure of human microsomal dipeptidase deduced from molecular cloning. J Biol Chem. 1990 Mar 5;265(7):3992–3995. [PubMed] [Google Scholar]
- Ambler R. P., Coulson A. F., Frère J. M., Ghuysen J. M., Joris B., Forsman M., Levesque R. C., Tiraby G., Waley S. G. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991 May 15;276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandoh K., Muto Y., Watanabe K., Katoh N., Ueno K. Biochemical properties and purification of metallo-beta-lactamase from Bacteroides fragilis. Antimicrob Agents Chemother. 1991 Feb;35(2):371–372. doi: 10.1128/aac.35.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell R., Emanuel E. L., Gagnon J., Waley S. G. The production and molecular properties of the zinc beta-lactamase of Pseudomonas maltophilia IID 1275. Biochem J. 1985 Aug 1;229(3):791–797. doi: 10.1042/bj2290791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly A. K., Waley S. G. Characterization of the membrane beta-lactamase in Bacillus cereus 569/H/9. Biochemistry. 1983 Sep 27;22(20):4647–4651. doi: 10.1021/bi00289a006. [DOI] [PubMed] [Google Scholar]
- Cuchural G. J., Jr, Malamy M. H., Tally F. P. Beta-lactamase-mediated imipenem resistance in Bacteroides fragilis. Antimicrob Agents Chemother. 1986 Nov;30(5):645–648. doi: 10.1128/aac.30.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale J. W., Godwin D., Mossakowska D., Stephenson P., Wall S. Sequence of the OXA2 beta-lactamase: comparison with other penicillin-reactive enzymes. FEBS Lett. 1985 Oct 21;191(1):39–44. doi: 10.1016/0014-5793(85)80989-3. [DOI] [PubMed] [Google Scholar]
- Davies R. B., Abraham E. P. Separation, purification and properties of beta-lactamase I and beta-lactamase II from Bacillus cereus 569/H/9. Biochem J. 1974 Oct;143(1):115–127. doi: 10.1042/bj1430115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester F., Joris B., Reckinger G., Bellefroid-Bourguignon C., Frère J. M., Waley S. G. Automated analysis of enzyme inactivation phenomena. Application to beta-lactamases and DD-peptidases. Biochem Pharmacol. 1987 Jul 15;36(14):2393–2403. doi: 10.1016/0006-2952(87)90609-5. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg M., Edlund C., Lindqvist L., Rylander M., Nord C. E. Purification and characterization of an imipenem hydrolysing metallo-beta-lactamase from Bacteroides fragilis. J Antimicrob Chemother. 1992 Feb;29(2):105–113. doi: 10.1093/jac/29.2.105. [DOI] [PubMed] [Google Scholar]
- Hussain M., Carlino A., Madonna M. J., Lampen J. O. Cloning and sequencing of the metallothioprotein beta-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J Bacteriol. 1985 Oct;164(1):223–229. doi: 10.1128/jb.164.1.223-229.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaconis J. P., Sanders C. C. Purification and characterization of inducible beta-lactamases in Aeromonas spp. Antimicrob Agents Chemother. 1990 Jan;34(1):44–51. doi: 10.1128/aac.34.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaurin B., Grundström T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Ledent P., Dideberg O., Fonzé E., Lamotte-Brasseur J., Kelly J. A., Ghuysen J. M., Frère J. M. Comparison of the sequences of class A beta-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991 Nov;35(11):2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropp H., Sundelof J. G., Hajdu R., Kahan F. M. Metabolism of thienamycin and related carbapenem antibiotics by the renal dipeptidase, dehydropeptidase. Antimicrob Agents Chemother. 1982 Jul;22(1):62–70. doi: 10.1128/aac.22.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lim H. M., Pène J. J., Shaw R. W. Cloning, nucleotide sequence, and expression of the Bacillus cereus 5/B/6 beta-lactamase II structural gene. J Bacteriol. 1988 Jun;170(6):2873–2878. doi: 10.1128/jb.170.6.2873-2878.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linström E. B., Boman H. G., Steele B. B. Resistance of Escherichia coli to penicillins. VI. Purification and characterization of the chromosomally mediated penicillinase present in ampA-containing strains. J Bacteriol. 1970 Jan;101(1):218–231. doi: 10.1128/jb.101.1.218-231.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massidda O., Rossolini G. M., Satta G. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-beta-lactamases. J Bacteriol. 1991 Aug;173(15):4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne A., Misselyn-Bauduin A. M., Joris B., Erpicum T., Granier B., Frère J. M. The diversity of the catalytic properties of class A beta-lactamases. Biochem J. 1990 Jan 1;265(1):131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. B., Lampen J. O. Beta-lactamase III of Bacillus cereus 569: membrane lipoprotein and secreted protein. Biochemistry. 1983 Sep 27;22(20):4652–4656. doi: 10.1021/bi00289a007. [DOI] [PubMed] [Google Scholar]
- POLLOCK M. R. The cell-bound penicillinase of Bacillus cereus. J Gen Microbiol. 1956 Aug;15(1):154–169. doi: 10.1099/00221287-15-1-154. [DOI] [PubMed] [Google Scholar]
- Porath J., Carlsson J., Olsson I., Belfrage G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature. 1975 Dec 18;258(5536):598–599. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- Porath J., Olin B. Immobilized metal ion affinity adsorption and immobilized metal ion affinity chromatography of biomaterials. Serum protein affinities for gel-immobilized iron and nickel ions. Biochemistry. 1983 Mar 29;22(7):1621–1630. doi: 10.1021/bi00276a015. [DOI] [PubMed] [Google Scholar]
- Rasmussen B. A., Gluzman Y., Tally F. P. Cloning and sequencing of the class B beta-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1990 Aug;34(8):1590–1592. doi: 10.1128/aac.34.8.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saino Y., Kobayashi F., Inoue M., Mitsuhashi S. Purification and properties of inducible penicillin beta-lactamase isolated from Pseudomonas maltophilia. Antimicrob Agents Chemother. 1982 Oct;22(4):564–570. doi: 10.1128/aac.22.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes R. B., Matthew M. The beta-lactamases of gram-negative bacteria and their role in resistance to beta-lactam antibiotics. J Antimicrob Chemother. 1976 Jun;2(2):115–157. doi: 10.1093/jac/2.2.115. [DOI] [PubMed] [Google Scholar]
- Thatcher D. R. Beta-lactamase (Bacillus cereus). Methods Enzymol. 1975;43:640–652. doi: 10.1016/0076-6879(75)43129-9. [DOI] [PubMed] [Google Scholar]
- Thompson J. S., Malamy M. H. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J Bacteriol. 1990 May;172(5):2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Rasmussen B. A., Bush K. Biochemical characterization of the metallo-beta-lactamase CcrA from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1992 May;36(5):1155–1157. doi: 10.1128/aac.36.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]



