Highlights
-
•
A new critical role of hsa_circ_0008035 in gastric cancer progression.
-
•
The functional partners of hsa_circ_0008035 were identified in gastric cancer cells.
-
•
The mechanisms underlying hsa_circ_0008035 effects were revealed in gastric cancer progression.
-
•
The results provide new potential targets for gastric cancer progression and treatment.
Keywords: Circ_0008035, EXT1, PKM2, Immune evasion, Gastric cancer
Abstract
Circular RNAs (circRNAs) have been reported to be associated with the malignant phenotypes of cancer. However, the role and underlying mechanism of hsa_Circ_0008035 in colorectal cancer (CRC) remains unclear. In this study, we elucidated the pivotal role of hsa_circ_0008035 in gastric cancer progression and immune evasion. Elevated hsa_circ_0008035 levels in gastric cancer patient serum correlated positively with disease advancement, including tumor stages and lymph node metastasis. Functional analyses revealed a negative association between hsa_circ_0008035 and CD8+ T cell number and function. Mechanistically, hsa_circ_0008035 encoded the novel protein EXT1–219aa, suppressing EXT1 phosphorylation and expression. Additionally, hsa_circ_0008035 regulated pyruvate metabolism by influencing the nucleus localization of PKM2. The identified EXT1/PKM2 axis further underscored the intricate regulatory mechanisms orchestrated by hsa_circ_0008035 in gastric cancer, offering potential diagnostic and therapeutic implications in the ongoing pursuit of targeted therapies for gastric cancer patients.
Graphical abstract
Introduction
Circular RNAs (circRNAs) have emerged as pivotal players in the intricate landscape of gene expression regulation, wielding significant influence in various physiological and pathological processes, including cancer [1,2]. Amongst these malignancies, gastric cancer stands out as a formidable adversary, marked by its high morbidity and mortality rates worldwide [3]. In the context of gastric cancer, circRNAs have garnered attention for their multifaceted roles in tumorigenesis, progression, and metastasis [4]. The unique circular structure of circRNAs endows them with remarkable stability and resistance to exonucleases, enhancing their potential as stable biomarkers and functional effectors. Specifically, in gastric cancer, certain circRNAs have been identified as key modulators of crucial signaling pathways implicated in cancer development [4]. These pathways include those governing cell proliferation, apoptosis, migration, and invasion. Mechanistically, circRNAs have been implicated in the intricate regulatory networks involving microRNAs (miRNAs) and messenger RNAs (mRNAs), commonly known as competing endogenous RNA (ceRNA) networks [5], and encoding novel proteins [4]. Through their ability to sequester miRNAs and thereby modulate the expression of downstream target genes, circRNAs contribute to the dysregulation of critical cellular processes in gastric cancer cells. Therefore, understanding the intricate interplay between circRNAs and the molecular mechanisms underlying gastric cancer holds promise for the identification of novel diagnostic biomarkers and therapeutic targets.
EXT1 (Exostosin 1), a member of the Exostosin family, plays a pivotal role in glycoprotein synthesis [6]. This enzyme, primarily located in the endoplasmic reticulum and Golgi apparatus, catalyzes the addition of sugar chains during glycosylation, which is essential for maintaining the structural and functional integrity of proteins [7]. Beyond its canonical function, EXT1 has been implicated in various cellular processes, including cell adhesion [8], signaling, and tumor suppression [9]. In the context of cancer, dysregulation of EXT1 has been associated with aberrant glycosylation patterns, impacting cell adhesion and motility [10,11]. Notably, the loss of EXT1 function has been linked to the pathogenesis of hereditary multiple exostoses (HME), a genetic disorder characterized by the development of benign bone tumors [12,13]. However, the intricate interplay between EXT1 and the progression of gastric cancer, particularly in the context of immune regulation and metabolism, remains largely unexplored.
PKM2 (Pyruvate Kinase M2), a pivotal enzyme in glycolysis, catalyzes the conversion of phosphoenolpyruvate (PEP) to pyruvate, generating ATP and pyruvate in the final steps of glycolysis [14]. While PKM2 is a well-known regulator of cellular energy metabolism, its diverse functions extend beyond glycolysis. In cancer, PKM2 has emerged as a key player in orchestrating metabolic reprogramming to meet the energy demands of rapidly proliferating cells [15]. One distinctive feature of PKM2 is its existence in two isoforms: a highly active tetrameric form (PKM1) and a less active dimeric form (PKM2). Cancer cells often favor the expression of PKM2, conferring a proliferative advantage by diverting glycolytic intermediates into biosynthetic pathways [16]. Moreover, PKM2′s non-metabolic roles include the regulation of gene transcription and involvement in signaling pathways associated with cell proliferation and survival [17]. In the intricate landscape of cancer biology, PKM2 has been implicated in tumor initiation, progression, and immune evasion. Its nuclear localization, in particular, has been associated with transcriptional regulation, contributing to the Warburg effect and facilitating immune escape mechanisms [15]. However, the regulatory networks governing PKM2 in the context of gastric cancer, especially its interaction with circRNAs, remain largely unexplored.
CircRNA hsa_circ_0008035 was firstly identified in the previous study [18]. Subsequently, numerous studies have revealed its roles in numerous tumors, including bladder cancer [19], osteosarcoma [20], and gastric cancer [18,21]. In these above-mentioned studies, hsa_circ_0008035 functions through miRNA sponging activity. However, it is still unclear whether hsa_circ_0008035 can encode a novel protein and its roles in the immunotherapy of gastric cancer are still confusing. Here, it was identified that hsa_circ_0008035 could encode a novel protein, named EXT1–219aa. And this study unveiled the interconnected roles of hsa_circ_0008035, EXT1, and PKM2 in orchestrating gastric cancer progression, immune evasion, and metabolic reprogramming.
Materials and methods
Clinical sample collection and bioinformatic analysis
GSE78092 was used for analyze the expression of circRNAs based on gastric cancer and normal tissues. 43 pairs of gastric cancer paraffin-embedded tissue and serum samples from healthy controls and gastric cancer patients were randomly selected from the Department of Oncology, the First Affiliated Hospital of Nanjing Medical University between October 2019 and June 2023. Written informed consent from all patients and approval of the Hospital Ethic Review Committees was obtained (2023-SRFA-079).
Real-time quantitative PCR (RT-qPCR)
The extraction of total RNA from gastric cancer cells was carried out using RNAiso™ Plus reagent (Cat # T9108, TaKaRa, Dalian, China). For hsa_circ_0008035 and its linear ETX1 mRNA, a High-Capacity cDNA Reverse Transcription Kit (Cat # 436,881, Applied Biosystems, Foster City, CA) was then applied to reversely transcribe 0.2 μg of purified total RNA into complementary DNA (cDNA). cDNA was then amplified by qRT-PCR analysis using SYBR Premix Ex Taq II (Cat # RR820A, TaKaRa). All PCR reactions were performed on an ABI viiA7 Real-Time PCR System (Applied Biosystems, Foster City, CA). The relative expression of hsa_circ_0008035 and its linear mRNA was normalized to GADPH expression. The relative gene expression was computed by the 2−ΔΔCt method.
Cell culture
Human gastric epithelial cell line GES-1, gastric cancer cell lines (SGC7901, MKN-45, AGS, BGC-823, and MGC803), HEK293T cells, and Murine gastric cancer YTN16 cells were bought from Chinese Academy of Sciences Cell Resource Center (Shanghai, China) and maintained in DMEM (Cat # 10,564,011, Thermo Fisher Scientific, Inc., Waltham, MA) with the existence of 10 % heated inactivated fetal bovine serum (Cat # SH30071, HyClone, Beijing, China), 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were kept at 37 °C in a humidified incubator with 5 % CO2.
Western blot
Total protein extracts were obtained using pre-cold RIPA buffer containing 1 mM phenylmethylsulfonyl fluoride (Cat # 329–98–6, PMSF; Sigma). Bradford protein assay (Cat # A55866, Thermo Fisher Scientific, Inc.) was applied to quantify the protein concentration. Protein samples were subjected to SDS-PAGE and then shifted to nitrocellulose membranes. Following non-specific binding with 5 % nonfat milk for 1 h, membranes were probed with specific primary antibodies overnight at 4 °C and then with horseradish peroxidase conjugated anti-rabbit IgG secondary antibody (Cat # sc-2004, Santa Cruz Biotechnology) at 37 °C for 2 h. At last, the EasySee Western Blot Kit (Cat # DW101, Transgen Biotech, Beijing, China) was utilized to visualize the immune blot signals.
Lentivirus package and infection
HEK293T cells were cultured to 70–80 % confluency and transfected with a lentiviral vector plasmid containing hsa_circ_0008035, hsa_circ_0008035 shRNA, EXT1, EXT1 shRNA, PKM2 shRNA, respectively, along with packaging plasmids (psPAX2 and pMD2.G) using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA). Lentivirus-containing supernatant was collected 72 h post-transfection, processed to remove debris, and stored at −80 °C. For stable cell line selection, target cells were plated, and lentivirus containing the gene of interest was added with Polybrene. Following a 24-h incubation, cells were replenished with fresh growth medium and allowed to recover for 72 h. Selection was initiated with puromycin, and resistant colonies were expanded and characterized for the expression of the gene of interest.
RNA stability assay
To detect the existence of hsa_circ_0008035, total RNA (10 μg) was digested twice with or without 4 U RNase R (Cat # RNR07250, Epicentre Technologies, Madison, WI) at 37 °C for 15 min, followed by the determination of expression of hsa_circ_0008035 and linear EXT1 mRNA (control) by qRT-PCR. Additionally, Cells were exposed to DMSO or 2 µg/mL actinomycin D (Cat # A4262, Sigma-Aldrich, USA) to block transcription for 2, 4, and 6 h. Then the expression of hsa_circ_0008035 and its linear transcript EXT1 mRNA (control) were detected using qRT-PCR.
Cell viability assay
CD8+ T cells were isolated from the spleen using the MojoSort™ Mouse CD8+ T Cell Isolation Kit (Cat # 480,007, BioLegend). Subsequently, these isolated CD8+ T cells were activated for 3 days using Ultra-LEAF™ Purified anti-mouse CD3ε/CD28 (Cat # 100,339/102,115, BioLegend), following the manufacturer's protocol. The experiments were conducted in RPMI-1640 supplemented with IL-2 (Cat # xyA073Gu, 10 ng/mL, BioLegend). Lentivirally-transfected cancer cells were allowed to adhere to the plate overnight, followed by a 48-hour incubation with activated CD8+ T cells. The ratio between cancer cells and CD8+ TILs was 5:1. After the incubation period, T cells and cell debris were removed through PBS wash, and the viable cancer cells were quantified using a spectrometer at OD 595 nm, followed by crystal violet staining.
Flowcytometry analysis
Mice were humanely sacrificed using the spinal dislocation method. Tumors were carefully excised in their entirety with surgical instruments that had been sterilized with 75 % ethanol. Subsequently, the tumors were sectioned into small pieces using sterile ophthalmic surgical scissors and subjected to digestion using 0.25 % trypsin for a period of 10 - 20 min. To generate single-cell suspensions, the digested material was filtered through a 300-mesh nylon mesh two or three times. The resulting cell concentration was adjusted to approximately 5 × 105/ml. Following this adjustment, the single-cell suspensions were subjected to antibody staining, with incubation lasting a minimum of 20 min. Stained cells were analyzed using a BD FACS Celesta (BD Biosciences), and the acquired data were processed using the FlowJo software program.
Hsa_circ_0008035 localization analysis and fluorescence in situ hybridization (FISH)
For hsa_circ_0008035 localization, the cytoplasmic and nuclear RNAs of cells were separated by a Cytoplasmic and Nuclear RNA Purification Kit (Cat # NGB-21,000, Norgen Biotek, Canada), and hsa_circ_0008035 expression in the cytoplasm and nucleus was measured by qRT-PCR. 18S rRNA and U6 snRNA were used as the positive control in the cytoplasm and nucleus, respectively. Additionally, Cy3-labeled oligonucleotide probe for hsa_circ_0008035 was purchased (Geneseed, Guangzhou, China). The hybridization was then performed in gastric cancer cells. Briefly, cell slides fixed with 4 % paraformaldehyde were treated with several buffers in RNA-FISH Kit (Cat # F11201, GenePharma). After denaturing at 73 °C for 5 min, probe mixture was hybridized in cell slide overnight in darkness at 37 °C. Scans were acquired using a laser confocal scanner, and images were viewed and exported with CaseViewer.
In vivo tumorigenic assay
A cohort of ten male BALB/c mice, aged five weeks, underwent subcutaneous injections on the right flank at the density of 3 × 106 YTN16 cells per mouse. The tumor volume was monitored at three-day intervals. On the 28 day of the experiment, the mice were subjected to euthanasia, following which the measurements of tumor volume and weight were performed. The experiment received ethical approval from the Animal Care and Use Ethics Committee of our institution. For the anti-PD-L1 treatment, mice were administered starting from the time tumors grew to 100 mm3. Anti-mouse PD-L1 mAb (Cat # BE0101, Bio X Cell, USA) was administered by intraperitoneal injection at a dose of 2.5 mg/kg every three days.
SDS-PAGE and protein mass spectrometry
The proteases-digested protein samples were analyzed by liquid-mass spectrometry (LC–MS/MS) to obtain a raw file of the original mass spectrometry results. Byonic software was used to analyze the raw file and search the uniprot-Homo sapiens data to obtain the identified protein results.
Analysis of glycolysis rate, lactate production, intracellular ATP production, and oxygen consumption rate
In this study, a thorough investigation into cellular metabolism was conducted, encompassing the assessment of glycolysis rate, lactate production, intracellular ATP levels, and oxygen consumption rate (OCR). For the determination of glycolysis rate and lactate production, target cells were cultured in glucose-free and pyruvate-free medium for 1 h, followed by the measurement of extracellular acidification rate (ECAR) using the Seahorse XF Glycolysis Stress Test (Cat # KA6207, Taipei, Taiwan). Simultaneously, glucose and lactate concentrations in the culture supernatant were quantified using specific assay kits (Glucose Uptake Assay Kit (Colorimetric), ab136955, Cambridge, MA, USA; l-Lactate Assay Kit (Colorimetric, ab65331). To evaluate intracellular ATP production, cells were subjected to the Seahorse XF Cell Mito Stress Test to measure OCR and assess mitochondrial function. Concurrently, intracellular ATP levels were determined by lysing cells and employing an ATP assay kit (Luminescent ATP Detection Assay Kit, ab113849). Additionally, the Seahorse XF Cell Mito Stress Test was utilized to measure OCR, providing insights into basal respiration, ATP production, and maximal respiration.
Statistical analysis
All data are displayed as mean ± standard deviation (SD). SPSS16.0 software (SPSS Inc., Chicago, IL) was used to carry out statistical comparisons. Statistical significance was analyzed by the Student's t-test or ANOVA. P < 0.05 was rendered statistically significant.
Results
Hsa_circ_0008035 is highly expressed in the serum of gastric cancer patients and positively correlates with gastric cancer progression
Utilizing bioinformatics tools, we identified hsa_circ_0008035 as a significantly upregulated circRNA in the serum samples of gastric cancer (Fig. 1A). Subsequent RT-qPCR experiments were conducted to assess the expression levels of hsa_circ_0008035 in gastric cancer patients. The results demonstrated a significant upregulation of hsa_circ_0008035 compared to the healthy control group, reaching statistical significance (P < 0.05) (Fig. 1B). To elucidate the clinical significance of hsa_circ_0008035, we performed correlation analyses with clinical features such as tumor staging and lymph node metastasis. A comprehensive analysis of clinical data revealed a positive correlation between the high expression of hsa_circ_0008035 and advanced tumor stages (Table 1), suggesting its potential involvement in different stages of gastric cancer development. Furthermore, a significant association was observed between the expression level of hsa_circ_0008035 and the presence of lymph node metastasis, indicating its potential role in the metastatic process of gastric cancer (Table 1). Consistently, hsa_circ_0008035 level was significantly increased in gastric cancer cell lines compared to that of normal gastric epithelial cells (Fig. 1C). To validate the circular structure and stability of hsa_circ_0008035, RNase R and Actinomycin D treatments were employed. Experimental results indicated that hsa_circ_0008035 maintained relative stability even under conditions of RNase R digestion (Fig. 1D and E) and Actinomycin d-induced transcriptional inhibition (Fig. 1F and G), confirming the presence of its circular RNA structure. Finally, using FISH technology, we successfully localized the position of hsa_circ_0008035 within cells, finding that hsa_circ_0008035 was mostly localized in cytoplasm of gastric cancer cells (Fig. 1H). RT-qPCR analysis obtained a consistent result (Fig. 1I). These findings robustly support the pivotal role of hsa_circ_0008035 in gastric cancer progression.
Fig. 1.
Hsa_circ_0008035 is highly expressed in the serum of gastric cancer patients and positively correlates with gastric cancer progression. (A) Hsa_circ_0008035 level was examined based on online GSE78092 dataset analysis. (B) Hsa_circ_0008035 level was detected in the serum of healthy control and gastric cancer patients through RT-qPCR assay. (C) Hsa_circ_0008035 level was detected in normal gastric epithelial and different types of gastric cancer cells. (D and E) Hsa_circ_0008035 and its parental gene level was determined gastric cancer cells with or without RNase treatment. (F and G) The stability of hsa_circ_0008035 and its parental gene mRNA level was evaluated in gastric cancer cells. (H) RNA FISH assay was constructed to assess the localization of hsa_circ_0008035 in gastric cancer cells. (I) Hsa_circ_0008035 level was measured in RNA extracted from the nucleus and cytoplasm of gastric cancer cells. **P < 0.01 vs control group.
Table 1.
Clinical pathological characteristics of the 43 gastric tumor patients.
| Frequency | % | Low hsa_circ_0008035 | High hsa_circ_0008035 | P Value | |
|---|---|---|---|---|---|
| Age | 0.736 | ||||
| Mean ± SD | 49.7 ± 11.5 | 48.1 ± 8.1 | 47.5 ± 10.5 | ||
| T grade of tumor | 0.011 | ||||
| T1 | 17 | 39.5 | 9 (52.9 %) | 8 (47.1 %) | |
| T2 | 7 | 16.3 | 3 (42.8 %) | 4 (57.2 %) | |
| T3 | 9 | 20.9 | 4 (44.4 %) | 5 (55.6 %) | |
| T4 | 10 | 23.3 | 3 (30 %) | 7 (70 %) | |
| Tumor size | 0.024 | ||||
| <6 cm | 21 | 48.8 | 11(52.3 %) | 10(47.7 %) | |
| >6 cm | 22 | 51.2 | 7(31.8 %) | 15(68.2 %) | |
| Lymph node metastasis | 0.017 | ||||
| Negative | 17 | 39.5 | 8(47.1 %) | 9(52.9 %) | |
| Positive | 26 | 60.5 | 6(23.1 %) | 20(76.9 %) |
Hsa_circ_0008035 is negatively correlated with the number and function of CD8+ T cells
GO analysis (Biological_Process section) related to circRNAs in GSE147698 data showed that genes negatively correlated with hsa_circ_0008035 were mainly enriched in signal transduction, immune response, and inflammatory response (Fig. 2A). KEGG pathway analysis also revealed that T cell receptor signaling pathway was enriched in the correlated genes (Fig. 2B). We then explored how hsa_circ_0008035 regulated tumor growth via immune pathways. Since CD8+ T cells are recognized as the main effector cells of cell immunity which kill cancer cells by releasing perforin, granzyme B, and IFN-γ. We found that hsa_circ_0008035 expression was negatively correlated with the expression of perforin, granzyme B, and IFN-γ in gastric cancer (Fig. 2C-E). Furthermore, cancer cells overexpressing hsa_circ_0008035 had an increased survival rate under coculture with isolated and reactivated CD8+ T cells, which was reversed by the knockdown of hsa_circ_0008035 (Fig. 2F and G). And the overexpression and knockdown efficiency was confirmed by RT-qPCR (Fig. 2H).
Fig. 2.
Hsa_circ_0008035 is negatively correlated with the number and function of CD8+ T cells. (A) GO analysis based on online GSE78092 dataset. (B) KEGG analysis based on online GSE78092 dataset. (C) The expression correlation between hsa_circ_0008035 and perforin was analyzed in the serum of gastric cancer patients. (D) The expression correlation between hsa_circ_0008035 and IFN-γ was analyzed in the serum of gastric cancer patients. (E) The expression correlation between hsa_circ_0008035 and Granzyme B was analyzed in the serum of gastric cancer patients. (F and G) The viability of gastric cancer cells with different treatments as indicated was determined by co-culturing with T cells. (H) The expression level of hsa_circ_0008035 was detected in the gastric cancer cells as depicted in (G). **P < 0.01 vs Vector group; ##P < 0.01 vs circRNA_OE group.
Hsa_circ_0008035 promotes gastric cancer proliferation and tumor immune escape
Then we wondered whether hsa_circ_0008035 could promote gastric cancer proliferation and tumor immune escape. Murine gastric cancer cells with or without hsa_circ_0008035 overexpression were utilized to establish the in vivo model and we found that tumors derived from hsa_circ_0008035-overexpressed cells exhibited a larger tumor weight (Fig. 3A and B) and faster tumor growth (Fig. 3C). Furthermore, it was identified that the total amount of tumor-infiltrating T cells was significantly decreased with hsa_circ_0008035 overexpression (Fig. 3D and E). In consistent, the levels of IFN-γ and IL-6, both of which were shown to suppress anti-tumor immunity by limiting T cell infiltration, were indicated to be upregulated in tumor tissues derived from hsa_circ_0008035-overexpressed cells (Fig. 3F). Importantly, gastric cancer cells with hsa_circ_0008035 knockdown displayed a stronger sensitivity to immune therapy (anti-PD-L1), as evidenced by the decrease of tumor weight (Fig. 3G and H) and tumor growth (Fig. 3I). Taken together, these results suggest that hsa_circ_0008035 can promote gastric cancer progression via facilitating the immune escape.
Fig. 3.
Hsa_circ_0008035 promotes gastric cancer proliferation and tumor immune escape. (A) Tumor image derived from murine gastric cancer cells with or without hsa_circ_0008035 overexpression. (B) Tumor weight of the tumors indicated in (A). (C) Tumor volume was measured at different time-points derived from murine gastric cancer cells with or without hsa_circ_0008035 overexpression. (D and E) The T cell ratio was determined in tumors derived from murine gastric cancer cells with or without hsa_circ_0008035 overexpression. (F) The mRNA levels of IL-6 and IFN-γ were detected in tumors derived from murine gastric cancer cells with or without hsa_circ_0008035 overexpression. (G) Tumor images derived from murine gastric cancer cells with different treatments as indicated. (H) Tumor weight was examined for the tumors described in (G). (I) Tumor volume was evaluated at different time-points as indicated derived from murine gastric cancer cells with different treatments as indicated in (G). **P < 0.01 vs Vehicle group; ##P < 0.01 vs CircRNA_kd group.
Hsa_circ_0008035 encodes a 219 amino acid (aa) novel protein, EXT1–219aa, to suppress the phosphorylation and expression of EXT1
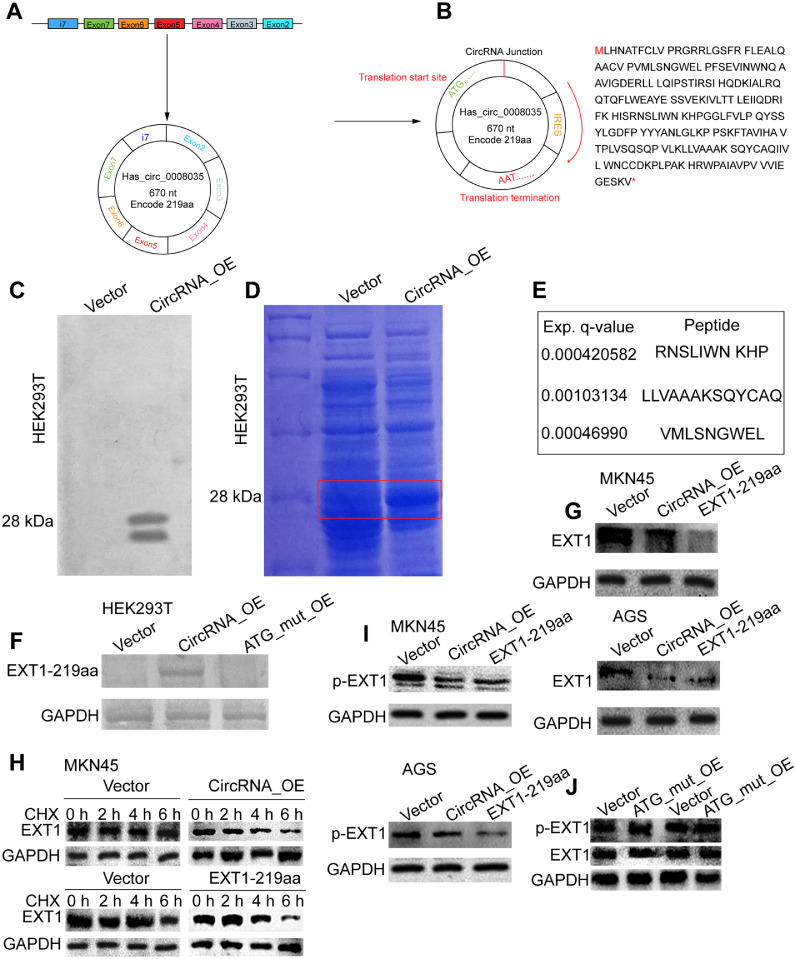
Next, we tried to reveal the underlying mechanisms by which hsa_circ_0008035 promotes the immune escape of gastric cancer. The circular RNA hsa_circ_0008035 originates from exon 2 to exon 7 and intron 7 of the EXT1 gene, with a full length of 670 nt (Fig. 4A). After analyzing the sequence of hsa_circ_0008035, we discovered the presence of a predicted ORF that may encode a 219aa protein named EXT1–219aa. The ORF starts at 'AUG' and ends at 'UAA' (Fig. 4B). To confirm the protein-encoding ability of hsa_circ_0008035, we inserted a FLAG label in the open reading frame and detected it through Western blotting. A FALG fragment was discovered near the expected molecular weight (28 kD) of EXT1–219aa, providing evidence that the open reading frame of hsa_circ_0008035 is capable of encoding protein (Fig. 4C). Once we confirmed the protein-encoding capability of hsa_circ_0008035, our primary focus shifted towards identifying the encoded product. To achieve this, we employed SDS-PAGE and protein mass spectrometry to detect the expression of EXT1–215aa. In our experimental approach, we transfected the hsa_circ_0008035 plasmid into HEK-293T cells and subsequently treated the total protein extracted from these cells. Notably, a distinct band at approximately 28 kD was observed in the experimental group, setting it apart from the control group that received an empty plasmid (Fig. 4D). The SDS-PAGE gel, where EXT1–219aa was anticipated to appear, was excised for further analysis using biological mass spectrometry. Through this technique, numerous fragments of EXT1–219aa were successfully identified and confirmed (Fig. 4E). The mutation of the start codon ATG in hsa_circ_0008035 resulted in the loss of its ability to encode EXT1–219aa (Fig. 4F). Importantly, we found that either hsa_circ_0008035 or EXT1–219aa overexpression could significantly decrease the expression of EXT1 protein (Fig. 4G) and the protein stability of EXT1 (Fig. 4H). Notably, the phosphorylation of EXT1 was significantly decreased by hsa_circ_0008035 or EXT1–219aa overexpression (Fig. 4I). However, both the expression and phosphorylation of EXT1 were unchanged in cells with or without ATG_mut_OE overexpression (Fig. 4J), this effect indicates that EXT1–219aa, but not has_circ_0008035 itself, is actually suppressing the phosphorylation and expression of EXT1. Those results verified that hsa_circ_0008035 can encode a novel protein, EXT1–219aa, which can decrease the phosphorylation of EXT1 and destabilize EXT1 protein.
Fig. 4.
Hsa_circ_0008035 encodes a 219 amino acid (aa) novel protein, EXT1–219aa, to suppress the phosphorylation and expression of EXT1. (A) The diagram depicting the encoding ability of hsa_circ_0008035. (B) The predicted amino acid sequences encoding by hsa_circ_0008035. (C) Flag expression was determined in HEK293T cells with or without hsa_circ_0008035 overexpression. (D) SDS-PAGE was subjected to detect the proteins derived from HEK293T cells with or without hsa_circ_0008035 overexpression. (E) The bands of SDS-PAGE describing in (D) were subjected to MS detection. (F) EXT1–219aa expression was detected in HEK293T cells with hsa_circ_0008035 or ATG-mut hsa_circ_0008035 overexpression, or not. (G) EXT1 protein expression was detected in gastric cancer cells with hsa_circ_0008035 or EXT1–219aa overexpression or not. (H) The protein stability of EXT1 was measured in gastric cancer cells with hsa_circ_0008035 or EXT1–219aa overexpression or not. (I) The level of p-EXT1 was determined in the cells described in (H). (J) The levels of p-EXT1 and EXT1 were examined in gastric cancer cells with or without ATG_mut_OE overexpression.
Hsa_circ_0008035 promotes the pyruvate metabolism via facilitating the nucleus localization of PKM2
Based on the online dataset analysis, it was found that the correlated genes of hsa_circ_0008035 were enriched in pyruvate metabolism (Fig. 5A). Notably, PKM2 exhibited the most correlation with hsa_circ_0008035 (Fig. 5B). Thus, we speculated that hsa_circ_0008035 could regulate the pyruvate metabolism through PKM2. As expected, overexpression of hsa_circ_0008035 or EXT1–219aa significantly promoted the glycolysis rate (Fig. 5C). Consistently, lactate production and intracellular ATP production was promoted in gastric cancer cells with hsa_circ_0008035 or EXT1–219aa overexpression (Fig. 5D and E). However, the oxygen consumption rate was unchanged with the alteration of hsa_circ_0008035 or EXT1–219aa expression (Fig. 5F). Given that the nucleus localization of PKM2 is positively correlated with its activity, we wondered whether hsa_circ_0008035 could regulate the localization of PKM2. As expected, IF analysis revealed that hsa_circ_0008035 or EXT1–219aa overexpression facilitated the nucleus localization of PKM2 (Fig. 5G). And PKM2 activity was also upregulated by hsa_circ_0008035 or EXT1–219aa overexpression in gastric cancer cells (Fig. 5H). These findings demonstrate that hsa_circ_0008035 can promote the pyruvate metabolism via facilitating the nucleus localization of PKM2.
Fig. 5.
Hsa_circ_0008035 promotes the pyruvate metabolism via facilitating the nucleus localization of PKM2. (A) The enrichment of pyruvate metabolism was identified in genes positively correlated with hsa_circ_0008035 based on online dataset analysis. (B) The genes with a positive expression correlation with hsa_circ_0008035 based on online dataset analysis. (C) Glycolysis rate was detected in gastric cancer cells with hsa_circ_0008035 or EXT1–219aa overexpression or not. (D) Lactate production was measured in gastric cancer cells with hsa_circ_0008035 or EXT1–219aa overexpression or not. (E) Intracellular ATP production was examined in gastric cancer cells with hsa_circ_0008035 or EXT1–219aa overexpression or not. (F) Oxygen consumption rate was assessed in gastric cancer cells with hsa_circ_0008035 or EXT1–219aa overexpression or not. (G) PKM2 localization was evaluated in gastric cancer cells with hsa_circ_0008035 or EXT1–219aa overexpression or not. (H) PKM2 activity was measured in gastric cancer cells with hsa_circ_0008035 or EXT1–219aa overexpression or not. **P < 0.01 vs Vector group.
Hsa_circ_0008035 regulates the nucleus localization and glycosylation level of PKM2 dependent on EXT1
EXT1 is an enzyme belonging to the Exostosin family, primarily involved in the synthesis of glycoproteins. It regulates the glycosylation process by catalyzing the addition of sugar chains. Thus, we speculated that EXT1 could regulate the glycosylation level of PKM2. As expected, we found that EXT1 overexpression significantly upshifted the PKM2 bands, which was rescued by hsa_circ_0008035 or EXT1–219aa overexpression (Fig. 6A). Interestingly, hsa_circ_0008035 or EXT1–219aa overexpression failed to affect the total level of PKM2 (Fig. 6B), implying that hsa_circ_0008035 or EXT1–219aa promotes the activity of PKM2 signaling through glycosylation of PKM2 but not its expression. Tunicamycin is widely used as a research tool to block N-linked glycosylation, to verify whether the band shift was caused by a change in the glycans, tunicamycin was added in cells with hsa_circ_0008035 knockdown. As shown in Fig. 6C, the intensity of the lower PKM2 bands was increased by hsa_circ_0008035 knockdown, this effect was attenuated by tunicamycin treatment. Additionally, the phosphorylation of PKM2 was increased by EXT1 overexpression, this effect was partially reversed by hsa_circ_0008035 overexpression or EXT1–219aa overexpression (Fig. 6D). Furthermore, the nuclear export of PKM2 led by EXT1 overexpression was rescued by hsa_circ_0008035 overexpression or EXT1–219aa overexpression (Fig. 6E). Moreover, PKM2 activity was reduced by EXT1 overexpression in gastric cancer cells, which was also abrogated by hsa_circ_0008035 overexpression or EXT1–219aa overexpression (Fig. 6F). Thus, our results indicate that hsa_circ_0008035 promotes the activity of PKM2 by facilitating the nucleus localization and attenuating the glycosylation level of PKM2 dependent on EXT1.
Fig. 6.
Hsa_circ_0008035 regulates the nucleus localization and glycosylation level of PKM2 dependent on EXT1. (A) The glycosylation level of PKM2 was determined in gastric cancer cells with EXT1 overexpression plus circRNA or EXT1–219aa overexpression or not. (B) PKM2 protein level was detected in gastric cancer cells depicted in (A). (C) The glycosylation level of PKM2 was examined in gastric cancer cells with circRNA knockdown as well as tunicamycin treatment. (D) p-PKM2 level was measured in gastric cancer cells with EXT1 overexpression plus circRNA or EXT1–219aa overexpression or not. (E) The localization of PKM2 was evaluated in MKN45 cells with EXT1 overexpression plus circRNA or EXT1–219aa overexpression or not. (F) PKM2 activity was determined in gastric cancer cells with EXT1 overexpression plus circRNA or EXT1–219aa overexpression or not.
Hsa_circ_0008035 facilitates immune evasion of gastric cancer through the EXT1/PKM2 axis
Finally, we investigated whether the EXT1/PKM2 axis was involved in the immunotherapy regulation of hsa_circ_0008035 in gastric cancer cells. Murine gastric cancer YTN16 cells with hsa_circ_0008035 overexpression were infected with the EXT1 overexpression or PKM2 knockdown lentivirus, and the stable-infected cell lines were confirmed by western blot analysis (Fig. 7A). Then, these above cell lines were utilized to establish the in vivo model and we found that the increased tumor growth derived from hsa_circ_0008035-overexpressed cells was attenuated by EXT1 overexpression or PKM2 knockdown (Fig. 7B-D). Furthermore, it was identified that the decreased total amount of tumor-infiltrating T cells led by hsa_circ_0008035 overexpression was rescued by EXT1 overexpression or PKM2 knockdown (Fig. 7E). In consistent, the increased levels of IFN-γ and IL-6 resulted by hsa_circ_0008035-overexpression were partially abrogated by EXT1 overexpression or PKM2 knockdown (Fig. 7F). Importantly, overexpression of hsa_circ_0008035-mediated increase of glycolysis rate (Fig. 7G), lactate production (Fig. 7H), and intracellular ATP production (Fig. 7I) was reduced by EXT1 overexpression or PKM2 knockdown. Notably, the decreased sensitivity of gastric cancer cells to immune therapy (anti-PD-L1) contributed by hsa_circ_0008035 overexpression was rescued by EXT1 overexpression or PKM2 knockdown (Fig. 8A–C). All over, these findings demonstrate that hsa_circ_0008035 can promote gastric cancer progression via facilitating the immune escape through the EXT1/PKM2 axis.
Fig. 7.
Hsa_circ_0008035 facilitates immune evasion of gastric cancer through the EXT1/PKM2 axis. (A) The protein levels of EXT1 and PKM2 were examined in murine gastric cancer cells with hsa_circ_0008035 overexpression infected with the EXT1 overexpression or PKM2 knockdown lentivirus. (B) Tumor images derived from cells described in (A). (C) Tumor volume was measured at different time-points as indicated derived from cells depicted in (A). (D) Tumor weight was detected for tumors derived from cells described in (A). (E) T cell ratio was measured in tumors derived from cells described in (A). (F) The mRNA levels of IL-6 and IFN-γ were examined in tumors derived from cells depicted in (A). (G) Glycolysis rate was detected in gastric cancer cells with hsa_circ_0008035 infected with the EXT1 overexpression or PKM2 knockdown lentivirus. (H) Lactate production was measured in gastric cancer cells depicted in (G). (I) Intracellular ATP production was examined in gastric cancer cells described in (G). **P < 0.01 vs Vector group; ##P < 0.01 vs circRNA_OE group.
Fig. 8.
The effects of hsa_circ_0008035 on the sensitivity of anti-PD-L1 are dependent on the EXT1/PKM2 axis. (A) Tumor images derived from murine gastric cancer cells with anti-PD-L1 treatment, anti-PD-L1 & hsa_circ_0008035 overexpression with EXT1 overexpression or PKM2 knockdown lentivirus. (B) Tumor volume was measured at different time-points as indicated derived from cells depicted in (A). (C) Tumor weight was detected for tumors derived from cells described in (A). **P < 0.01 vs control; ##P < 0.01 vs anti-PD-L1 group; $$P < 0.01 vs anti-PD-L1&circRNA_OE group.
Discussion
The presented study unravels the intricate roles of hsa_circ_0008035, EXT1, and PKM2 in shaping the landscape of gastric cancer progression, immune evasion, and metabolic reprogramming. The findings shed light on novel molecular mechanisms, providing valuable insights into potential therapeutic avenues for disrupting immune escape mechanisms in gastric cancer.
The heightened expression of hsa_circ_0008035 in the serum of gastric cancer patients underscores its potential as a diagnostic biomarker. This observation aligns with the growing recognition of circRNAs as promising candidates for non-invasive cancer detection as we previously indicated that circRNA circRPPH1 could serve as a biomarker for gastric cancer progression by promoting the stemness of gastric cancer cells [22]. The positive correlation between hsa_circ_0008035 expression and advanced tumor stages, as well as lymph node metastasis, highlights its potential as a prognostic indicator for gauging disease severity and progression. Additionally, increasing studies have revealed the immune regulatory roles of circRNAs in tumors, such as the recent study indicated that circMAPK1 promoted CD8+ T cell infiltration in lung cancer by improving the IGF2BP1 dependent CCL5 upregulation [23]; And hsa_circ_0001479 accelerates tumorigenesis of gastric cancer and mediates immune escape by upregulating DEK expression by sponge targeting miR-133a-5p [24]. An intriguing facet of hsa_circ_0008035′s regulatory network is its association with CD8+ T cells. The negative correlation observed between hsa_circ_0008035 expression and the number and function of CD8+ T cells suggests a potential role in immune evasion strategies employed by gastric cancer cells. This finding resonates with the growing understanding of circRNAs as key modulators of immune responses in the tumor microenvironment.
The identification of EXT1–219aa, a novel protein encoded by hsa_circ_0008035, adds a layer of complexity to the study. EXT1–219aa's ability to suppress the phosphorylation and expression of EXT1 unveils a regulatory axis with implications for glycoprotein synthesis and cellular processes, this is consistent with the previous study showing the roles of EXT1 in protein glycosylation [10]. The functional interplay between hsa_circ_0008035, EXT1, and PKM2 provides a unique perspective on the interconnected molecular events driving gastric cancer progression. PKM2, a central player in cellular metabolism, emerges as a key mediator of hsa_circ_0008035′s impact on glycolysis and immune escape. The facilitation of nucleus localization and glycosylation level of PKM2 by hsa_circ_0008035, dependent on EXT1, underscores the complexity of the regulatory network. This mechanism sheds light on how hsa_circ_0008035 orchestrates metabolic reprogramming, potentially contributing to the Warburg effect observed in cancer cells.
The in vivo models further substantiate the significance of the EXT1/PKM2 axis in mediating hsa_circ_0008035-induced immune escape. The attenuation of tumor growth and restoration of tumor-infiltrating T cells by EXT1 overexpression or PKM2 knockdown highlight the potential therapeutic implications of targeting this axis. Moreover, the sensitivity to immune therapy, as evidenced by decreased tumor weight and growth, further underscores the translational relevance of these findings. While this study advances our understanding of the molecular intricacies underlying gastric cancer progression and immune evasion, certain limitations should be acknowledged. Notably, we found that hsa_circ_0008035 with the mutation of the start codon ATG (ATG_mut_OE) could not affect the expression and phosphorylation of EXT1, this effect indicates that EXT1–219aa is mainly suppressing the phosphorylation and expression of EXT1 (Fig. 4). However, the complex regulatory networks involving hsa_circ_0008035, EXT1, and PKM2 warrant further investigation, and additional in vivo models and clinical validations are essential to extrapolate these findings to human populations.
In conclusion, the findings presented herein delineate a comprehensive picture of the regulatory network orchestrated by hsa_circ_0008035 in gastric cancer. The interplay of hsa_circ_0008035, EXT1, and PKM2 emerges as a critical axis in governing immune escape mechanisms and metabolic reprogramming. These insights lay the foundation for future research endeavors, pointing towards innovative therapeutic strategies aimed at disrupting immune evasion and altering the metabolic landscape in the clinical management of gastric cancer.
Declarations
Ethics approval and consent to participate
The current study was approved by the Animal Ethics Committee and was conducted in accordance to the relevant agreements with the Jinling Hospital of Nanjing University. All procedures were performed in accordance with the Guidance Suggestions for the Care and Use of Laboratory Animals, formulated by the Ministry of Science and Technology of China.
Consent for publication
Not applicable.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declared that they have no conflicts of interest regarding this work.
Funding
This work was supported by BeiJing Bethune Charitable Foundation (BCF-XD-JC-20221205–17), and Clinical ability improvement project (JSPH-MB-2020–3).
Conflict of interests
The authors declare no competing financial interest.
CRediT authorship contribution statement
Rongqi Jiang: Writing – original draft, Investigation, Formal analysis, Conceptualization. Ping Li: Investigation, Formal analysis. Enqing Meng: Methodology. Xu Cheng: Resources. Xinyi Wu: Software, Resources. Hao Wu: Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
References
- 1.Zhang F., et al. Exosomal circRNA: emerging insights into cancer progression and clinical application potential. J. Hematol. Oncol. 2023;16(1):67. doi: 10.1186/s13045-023-01452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han Y., et al. DNMT1 regulates hypermethylation and silences hsa_circ_401351 in hydroquinone-induced malignant TK6 cells. Environ. Toxicol. 2024;39(4):2092–2101. doi: 10.1002/tox.24089. [DOI] [PubMed] [Google Scholar]
- 3.Jie L., Hengyue W., Ting H. Calcitriol suppresses gastric cancer progression and cisplatin resistance by inhibiting glycolysis and M2 macrophage polarization through inhibition of mTOR activation. Environ. Toxicol. 2024;39(2):830–839. doi: 10.1002/tox.23975. [DOI] [PubMed] [Google Scholar]
- 4.Miao Z., et al. Hsa_circ_0136666 stimulates gastric cancer progression and tumor immune escape by regulating the miR-375/PRKDC Axis and PD-L1 phosphorylation. Mol. Cancer. 2023;22(1):205. doi: 10.1186/s12943-023-01883-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma X., et al. Circular RNA hsa_circ_0001846 facilitates the malignant behaviors of pancreatic cancer by sponging miR-204-3p and upregulating KRAS expression. Cell Death. Discov. 2023;9(1):448. doi: 10.1038/s41420-023-01733-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X., et al. Exostosin glycosyltransferase 1 reduces porcine reproductive and respiratory syndrome virus infection through proteasomal degradation of nsp3 and nsp5. J. Biol. Chem. 2022;298(2) doi: 10.1016/j.jbc.2021.101548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sethi S., et al. Exostosin 1/Exostosin 2-associated membranous nephropathy. J. Am. Soc. Nephrol. 2019;30(6):1123–1136. doi: 10.1681/ASN.2018080852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saez B., et al. Inhibiting stromal cell heparan sulfate synthesis improves stem cell mobilization and enables engraftment without cytotoxic conditioning. Blood. 2014;124(19):2937–2947. doi: 10.1182/blood-2014-08-593426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D., et al. Exostosin1 as a novel prognostic and predictive biomarker for squamous cell lung carcinoma: a study based on bioinformatics analysis. Cancer Med. 2021;10(8):2787–2801. doi: 10.1002/cam4.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerselidou D., et al. Alternative glycosylation controls endoplasmic reticulum dynamics and tubular extension in mammalian cells. Sci. Adv. 2021;7(19):eabe8349. doi: 10.1126/sciadv.abe8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu N.W., et al. EXT1, regulated by MiR-665, promotes cell apoptosis via ERK1/2 signaling pathway in acute lymphoblastic leukemia. Med. Sci. Monit. 2019;25:6491–6503. doi: 10.12659/MSM.918295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ajmal M., et al. Haploinsufficiency of EXT1 and Heparan Sulphate Deficiency Associated with hereditary multiple exostoses in a Pakistani Family. Medicina (Kaunas) 2022;59(1):100. doi: 10.3390/medicina59010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beltrami G., et al. Hereditary multiple exostoses: a review of clinical appearance and metabolic pattern. Clin. Cases. Miner. Bone Metab. 2016;13(2):110–118. doi: 10.11138/ccmbm/2016.13.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z., et al. PKM2, function and expression and regulation. Cell Biosci. 2019;9:52. doi: 10.1186/s13578-019-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J.Z., et al. The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun. (Lond) 2021;41(7):560–575. doi: 10.1002/cac2.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu S., et al. Pyruvate kinase M2 (PKM2) in cancer and cancer therapeutics. Cancer Lett. 2021;503:240–248. doi: 10.1016/j.canlet.2020.11.018. [DOI] [PubMed] [Google Scholar]
- 17.İlhan M. Non-metabolic functions of pyruvate kinase M2: PKM2 in tumorigenesis and therapy resistance. Neoplasma. 2022;69(4):747–754. doi: 10.4149/neo_2022_220119N77. [DOI] [PubMed] [Google Scholar]
- 18.Huang S., et al. A novel circular RNA hsa_circ_0008035 contributes to gastric cancer tumorigenesis through targeting the miR-375/YBX1 axis. Am. J. Transl. Res. 2019;11(4):2455–2462. [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Y., et al. Hsa_circ_0008035 knockdown inhibits bladder cancer progression through miR-1184/RAP2B Axis. Urol. Int. 2023;107(6):632–645. doi: 10.1159/000527873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong G., et al. Silencing hsa_circRNA_0008035 exerted repressive function on osteosarcoma cell growth and migration by upregulating microRNA-375. Cell Cycle. 2020;19(17):2139–2147. doi: 10.1080/15384101.2020.1792636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X., Wang X.L. Dexmedetomidine promotes ferroptotic cell death in gastric cancer via hsa_circ_0008035/miR-302a/E2F7 axis. Kaohsiung. J. Med. Sci. 2023;39(4):390–403. doi: 10.1002/kjm2.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J., et al. CircRPPH1 promotes the stemness of gastric cancer cells by targeting miR-375/SLC7A11 axis. Environ. Toxicol. 2023;38(1):115–125. doi: 10.1002/tox.23668. [DOI] [PubMed] [Google Scholar]
- 23.Zhao F., et al. CircMAPK1 promoted CD8 + T cell infiltration in LUAD by improving the IGF2BP1 dependent CCL5 upregulation. Int. Immunopharmacol. 2023;127 doi: 10.1016/j.intimp.2023.111267. [DOI] [PubMed] [Google Scholar]
- 24.Zang J., et al. Hsa_circ_0001479 accelerates tumorigenesis of gastric cancer and mediates immune escape. Int. Immunopharmacol. 2023;124(Pt A) doi: 10.1016/j.intimp.2023.110887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.