Abstract
Checkpoint inhibition using Fc-containing monoclonal antibodies has emerged as a powerful therapeutic approach to augment anti-tumor immunity. We recently showed that FcγRIIB, the only inhibitory IgG-Fc receptor, is expressed on a population of highly differentiated effector CD8+ T cells in the tumors of mice and humans, raising the possibility that CD8+ T cell responses may be directly modulated by checkpoint inhibitor binding to T cell-expressed FcγRIIB. Here, we show that despite exhibiting strong proliferative and cytokine responses at baseline, human FcγRIIBpos CD8+ T cells exhibited reduced responsiveness to both PD-1 and CTLA-4 checkpoint inhibition as compared to FcγRIIBneg CD8+ T cells in vitro. Moreover, frequencies of FcγRIIBpos CD8+ T cells were reduced following treatment of patients with melanoma with nivolumab in vivo. This reduced responsiveness was FcγRIIB-dependent, because conditional genetic deletion of FcγRIIB on tumor-specific CD8+ T cells improved response to checkpoint blockade in B16 and LLC mouse models of cancer. The limited responsiveness of FcγRIIBpos CD8+ T cells was also dependent on an intact Fc region of the checkpoint inhibitor, in that treatment with Fc-devoid anti-PD-1 F(ab) fragments resulted in increased proliferation of FcγRIIBpos CD8+ T cells, without altering the response of FcγRIIBneg CD8+ T cells. Finally, the addition of FcγRIIB blockade improved efficacy of PD-1 checkpoint inhibition in mouse models of melanoma, lung, and colon cancer. These results illuminate an FcγRIIB-mediated, cell-autonomous mechanism of CD8+ T-cell suppression which limits the efficacy of checkpoint inhibitors during anti-tumor immune responses in vivo.
One-sentence summary:
Due to the potential binding of Fc-containing checkpoint inhibitors, FcγRIIBpos CD8+ T cells exhibit reduced responsiveness to checkpoint inhibition.
Introduction
In the past decade, the advent of immunotherapy has altered the treatment paradigm for many cancers, including metastatic melanoma and advanced stage lung cancer (1–7). Specifically, the use of checkpoint therapies targeting programmed cell death protein 1 (PD-1) and cytotoxic T-lymphocyte associated protein 4 (CTLA-4) has led to higher response rates (8) and improved overall and progression-free survival in several cancer types, most notably advanced melanoma (9–11). Further, a single dose of PD-1 inhibitor can mediate complete pathological responses and high rates of recurrence-free survival in some patients (12). However, up to 40% of patients fail to respond to currently available checkpoint inhibitors (8). This heterogeneity in responsiveness gives rise to questions regarding mechanisms which may limit the efficacy of checkpoint inhibition in vivo.
Fcγ receptors (FcγRs) are a class of Type 1 Fc receptors for IgG, comprised of activating receptors, FcγRI, FcγRIII, and FcγRIV and the inhibitory receptor FcγRIIB (13). The ratio between activating and inhibitory FcγRs controls innate immune and B cell activation, plasma cell survival, and dendritic cell (DC) maturation and function, ultimately determining the balance between pro- and anti-inflammatory processes (13). As the only inhibitory FcγR, FcγRIIB is expressed on both innate and adaptive immune cells, including B cells (14). In the setting of cancer, the role of FcγRIIB on B cells has been studied (15). Interaction between FcγRIIB on B cells and the Fc regions of direct targeting antibodies, including anti-CD20 antibodies, has been shown to promote antibody internalization and reduced drug efficacy (16–18). Further, FcγRIIB expression on tumor cells, including melanoma, has been shown to inhibit antibody-dependent cellular cytotoxicity (ADCC) in an immunoreceptor tyrosine-based inhibition motif (ITIM)-independent manner (19). This mechanism is likely secondary to FcγRIIB binding the Fc regions of anti-tumor antibodies, thus acting as a decoy receptor. Finally, Ravetch et al. have demonstrated an improved anti-tumor response to anti-PD-1 antibody in Fcgr2−/− mice compared to WT mice, owing to the alteration of myeloid subsets within the tumor microenvironment (20).
In contrast, prevailing dogma for the last several decades is that T cells do not express any Fc receptors, including FcγRIIB (14, 21, 22). However, we and others have recently shown that a population of activated CD8+ T cells express FcγRIIB (23–27). FcγRIIB-expressing CD8+ T cells are detectable at about day 10 post tumor inoculation in the B16 murine melanoma and this expression increases with time following immune challenge, and FcγRIIB-expressing CD8+ T cells are present at later timepoints during the establishment of immunologic memory (28, 29). Consistent with this, the gene encoding FcγRIIB protein, Fcgr2b, was shown to be a differentially expressed gene in memory CD8+ T cells compared to naïve CD8+ in the seminal work by Wherry et al. (30). In our own work, we demonstrated that FcγRIIB is expressed in activated antigen-specific CD8+ T cells after ~5–6 rounds of division (23).
In the setting of cancer, inoculation of Fcgr2b−/− mice with B16-OVA melanoma cells significantly (P<0.05) reduced tumor volumes by increasing frequencies of tumor-infiltrating lymphocytes compared to wildtype (WT) mice (25). Further, adoptive transfer of Fcgr2b−/− tumor antigen-specific CD8+ T cells into B16-challenged mice resulted in a greater reduction in tumor volume compared to infusion of WT antigen-specific CD8+ T cells (25).
These findings raised important questions about the potential interaction of CD8+ T cell-expressing FcγRIIB and the Fc portion of immunoglobulin G (IgG) checkpoint inhibitors. Thus, here we sought to interrogate the impact of Fc-containing immunotherapeutics on the responsiveness of FcγRIIBpos versus FcγRIIBneg CD8+ T cells to both Fc-intact and Fc-devoid checkpoint inhibitors in tumor-bearing mice as well as in patients with melanoma. The results illuminate a previously unappreciated role for an FcγRIIB-mediated, cell-autonomous mechanism of CD8+ T-cell suppression responsible for limiting responsiveness to anti-PD-1 therapy. These findings suggest a strategy to overcome the unintended inhibitory effects of checkpoint blockade therapy on FcγRIIB-expressing anti-tumor CD8+ T cells.
Results
FcγRIIB is expressed on CD8+ T cells from human PBMCs and is a marker of activated, cytokine-producing cells
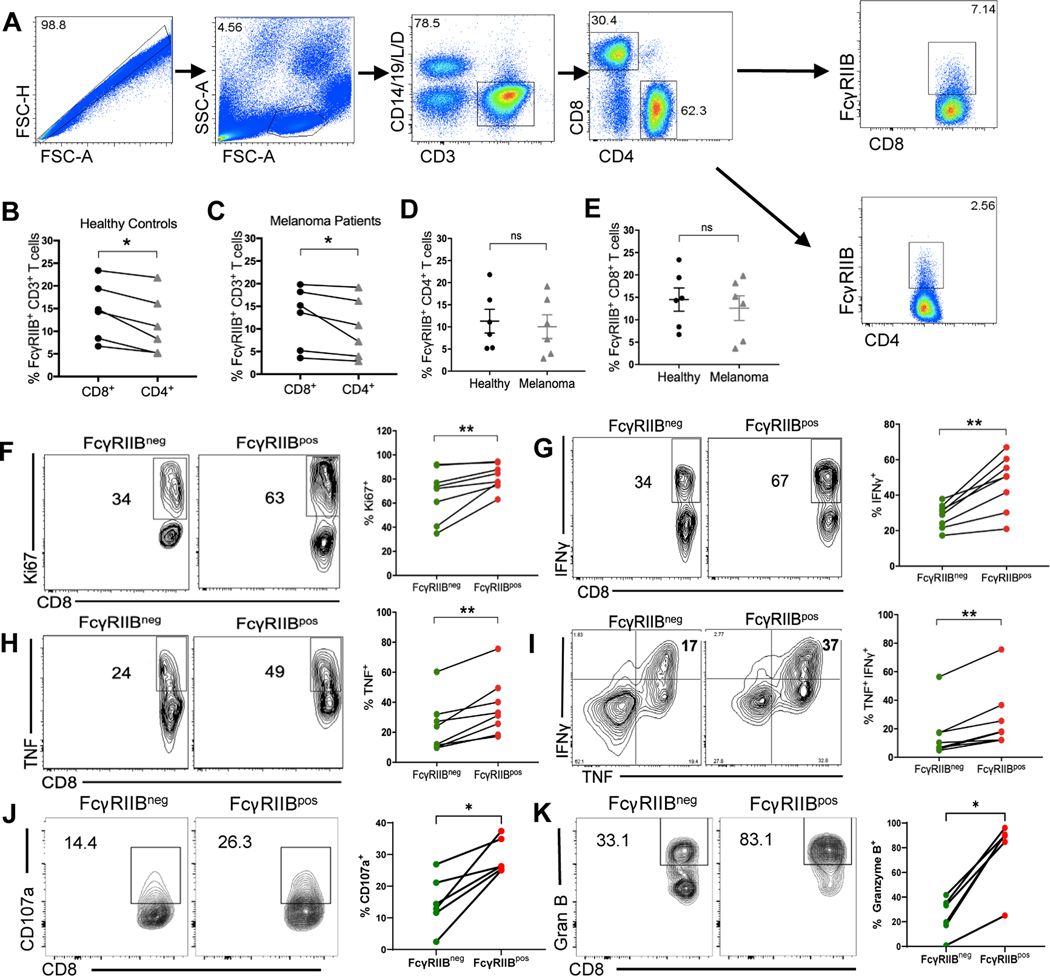
Previous work from our laboratory has shown that FcγRIIB is highly expressed on a population of activated CD8+ T cells in murine models of melanoma (25). Here, we investigated the presence of FcγRIIB+ CD8+ T cells in human healthy controls and patients with melanoma (tables S1 and S2). A stringent gating strategy was used to exclude dead cells as well as contaminating CD14+ monocytes and CD19+ B cells expressing FcγRIIB (Fig. 1A). FcγRIIB expression was detected using the anti-FcγRII clone FLI8.26 (fig. S1A), which has been reported to potentially cross-react with human FcγRIIA and FcγIIC (31). To verify that FLI8.26 was detecting only FcγRIIB on CD8+ T cells, we performed qPCR analysis on the FCGR2 genes of the population of CD8+ T cells that stained positively with FLI8.26 collected after FACS-sorting. These data show undetectable concentrations of FCGR2C n these cells (fig. S1B), making it highly unlikely that FLI8.26 could be staining FcγRIIC on CD8+ T cells. In contrast, we observed low but detectable FCGR2A mRNA in the population of FLI8.26+ CD8+ T cells, but using an FcγRIIA-specific antibody, we were unable to detect any protein expression of FcγRIIA on FLI8.26+ CD8+ T cells (fig.S1C). These data strongly suggest that FLI8.26 is staining only FcγRIIB, and not FcγRIIA or FcγRIIC, on CD8+ T cells. These findings demonstrate that in contrast to long-held dogma, but consistent with our recently published murine and human data (23, 25), FcγRIIB is expressed on a population of CD19− CD14− CD3+ CD8+ T cells, and to a lesser extent on a population of CD19− CD14− CD3+ CD4+ T cells in both healthy subjects (Fig. 1B) and patients with melanoma (Fig. 1C). The frequencies of FcγRIIBpos cells among both CD4+ and CD8+ T cells was consistent between healthy subjects and patients with melanoma (Fig. 1D–1E).
Fig. 1. FcγRIIB is expressed on CD8+ T cells from human PBMCs and is a marker of activated, cytokine-producing cells.
(A) Flow cytometry plots showing gating strategy for excluding dead cells as well as contaminating CD14+ and CD19+ cells and gating on the FcγRIIBpos population in human CD8+ and CD4+ T cell compartments of PBMCs. (B-C) Quantification comparing the frequency of FcγRIIBpos CD4+ and CD8+ T cells in (B) human healthy controls and (C) stage IV patients with melanoma. (D) Quantification comparing the frequency of FcγRIIBpos CD4+ T cells between healthy donors and stage IV melanoma patient (n=6). (E) Quantification comparing the frequency of FcγRIIBpos CD8+ T cells between healthy donors and stage IV patients with melanoma (n=6 per group). (F-K) PBMCs from healthy controls were stimulated in vitro for 5 days with anti-CD3/CD28 beads, re-stimulated with PMA/Ionomycin for 4h and stained intracellularly (n=8). Representative flow plots and quantification showing the frequency of (F) Ki-67+, of (G) IFNγ+, of (H) TNF+, of (I) TNF+ IFNγ+, of (J) CD107a+, and of (K) Granzyme B+ FcγRIIBpos and FcγRIIBneg CD8+ T cells. When comparing cell groups within the same donor, P-values were calculated using the Wilcoxon matched-pairs rank test. Mann-Whitney non-parametric, unpaired tests were used when comparing groups between donors. The error bar in summary figures denotes mean ± SEM. *P<0.05 **P<0.01, ns, not significant.
We next aimed to determine the functionality of the FcγRIIBpos CD8+ T cell population. Intracellular Ki-67 staining of stimulated peripheral blood mononuclear cells (PBMCs) revealed a significantly (P<0.01) higher frequency of proliferating cells among FcγRIIBpos CD8+ T cells as compared to FcγRIIBneg CD8+ T cells (Fig. 1F). In addition to superior proliferative potential, FcγRIIBpos CD8+ T cells also possessed significantly (P<0.01) higher frequencies of IFNγ+ (Fig. 1G), TNFα+ (TNF+) (Fig. 1H), and TNF+ IFNγ+ cells compared to the FcγRIIBneg CD8+ T cell population (Fig. 1I). The cytolytic capacity of FcγRIIBpos CD8+ T cells was also superior to that of FcγRIIBneg CD8+ T cells as demonstrated by significantly (P<0.05) increased frequencies of CD107a+ and Granzyme B+ among FcγRIIBpos versus FcγRIIBneg CD8+ T cells (Fig. 1J–1K). Similar to our previous work showing that FcγRIIB is expressed on activated and memory CD8+ T cells in mice, FcγRIIB was induced on human CD8+ T cells upon CD3/28 stimulation of healthy PBMCs ex vivo (fig. S1D).
FcγRIIB is expressed on cytokine-producing patient CD8+ tumor-infiltrating lymphocytes (TIL) that also express T cell checkpoint molecules
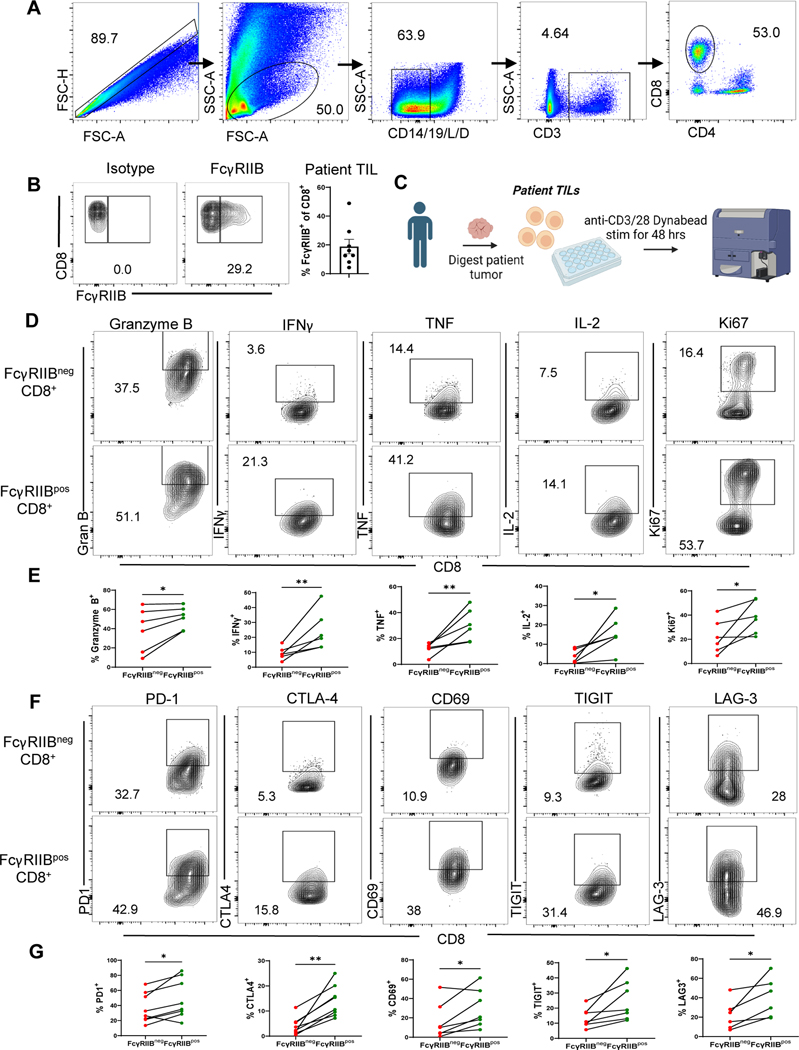
Given the finding that FcγRIIB is expressed on a population of activated human effector-like memory CD8+ T cells, we asked if FcγRIIB is expressed on CD8+ T cells at the tumor of patients with melanoma (Fig. 2A) (table S4), and found that a subset of freshly isolated patient CD8+ TIL express FcγRIIB protein (Fig. 2B). In addition, two independent publicly available single cell RNA sequencing datasets of tumors from patients with melanoma were interrogated using the single cell analysis software, BBrowser 2 (32). Results indicated that Fcgr2b was expressed in tumor-infiltrating CD8+ T cells (33) (fig. S1E-1G), and that tumor-infiltrating cytotoxic CD8+ T cells expressed significantly (P<0.01) more Fcgr2b than naïve CD8+ T cells (34) (fig. S1H-1J). Corroborating our data obtained in mouse models (25, 35) and the flow cytometric data shown above, both datasets provided external validation of Fcgr2b expression on human CD8+ T cells within tumors, and of the increased expression of Fcgr2b on effector versus naïve CD8+ T cells.
Fig. 2. FcγRIIB is expressed on activated, cytokine-producing patient CD8+ tumor-infiltrating lymphocytes (TIL) that also expressT cell checkpoint molecules.
(A) Flow cytometry plots showing gating strategy for excluding dead cells and contaminating CD14+and CD19+ cells and gating on the FcγRIIBpos population in human CD8+ TIL. (B) Representative flow cytometry plots and quantification of isotype versus FcγRIIB staining on human CD8+ TIL (n=8). (C) Schematic of experimental setup wherein TIL isolated from patients with melanoma were stimulated in vitro for 48 h with anti-CD3/CD28 beads, then Golgi Stop was added 4h prior to harvest and cells were stained for extracellular and intracellular markers (D) Representative flow plots and (E) quantification showing frequencies of Granzyme B+, IFNγ+, TNF+, IL-2+, and Ki67+ of FcγRIIBpos as compared to FcγRIIBneg CD8+ patient TIL (n=6). (F) Representative flow plots and (G) quantification showing frequencies of PD-1+, CTLA-4+, CD69+, TIGIT+, and LAG-3+ on FcγRIIBpos and FcγRIIBneg CD8+ patient TIL (n=7–8). The error bar in summary figures denotes mean ± SEM. *P<0.05, **P<0.01, Wilcoxon matched-pairs rank test.
We next sought to characterize the immune phenotype of FcγRIIB+ versus FcγRIIB− CD8+ T cells at the tumor of patients with melanoma. Flow cytometric analysis revealed that FcγRIIBpos CD8+ TIL contained increased frequencies of Granzyme B+, IFNγ+, TNF+, IL-2+, and Ki67+ cells as compared to FcγRIIBneg CD8+ TIL (Fig. 2C–2E). Further, FcγRIIBposCD8+ TIL exhibited significantly (P<0.05) higher frequencies of PD-1-, CTLA-4-, CD69-, TIGIT-, and LAG-3- expressing cells as compared to the FcγRIIBneg CD8+ TIL (Fig. 2F–2G). The phenotype and function of FcγRIIBpos CD8+ T cells was similar in both healthy control PBMCs (fig. S2A-2C) and PBMCs from patients with melanoma (fig. S2D-2F). These data show that, despite the fact that they express putative exhaustion markes such as PD-1, TIGIT, and LAG-3, FcγRIIBpos CD8+ T cells are not functionally exhausted but instead possess increased proliferative and cytokine effector functions relative to FcγRIIBneg CD8+ T cells.
FcγRIIBpos CD8+ T cells are resistant to immune checkpoint blockade in an Fc-dependent manner
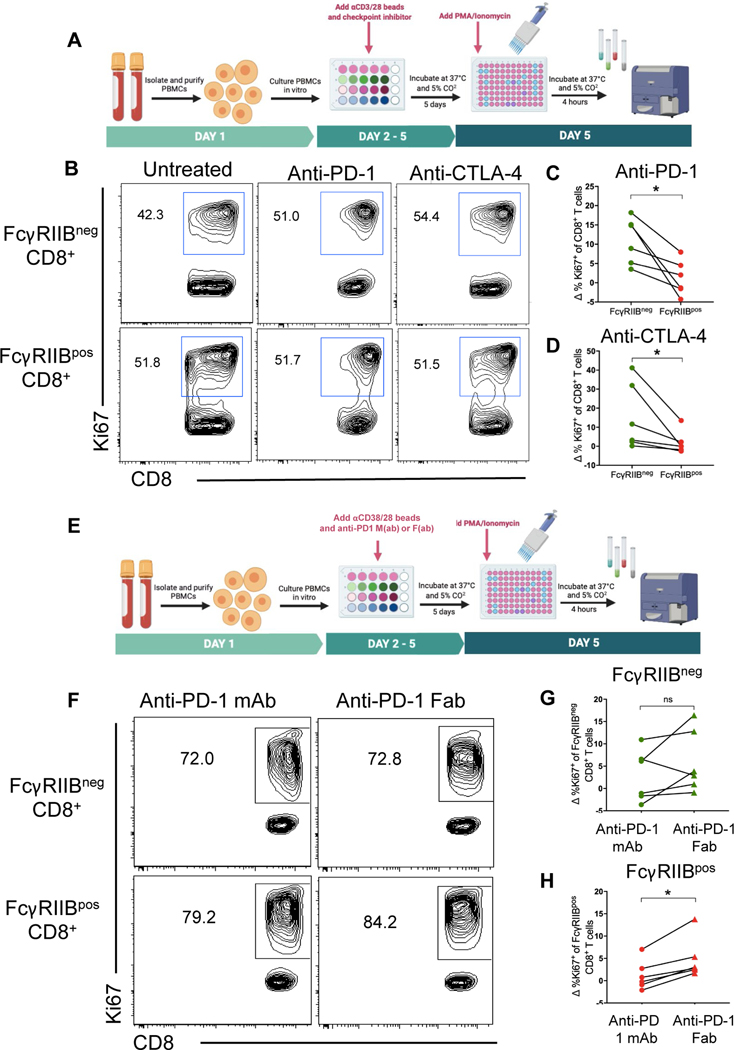
Because FcγRIIBpos CD8+ T cells expressed elevated amounts of T cell checkpoint molecules and these molecules are also targets of immune checkpoint blockade, we hypothesized that the IgG domains of checkpoint inhibitors could function as an FcγRIIB ligand. In this case, the binding of the Fc region of checkpoint inhibitors to FcγRIIB on the surface of CD8+ T cells could transduce an inhibitory signal to limit the ability of cells to proliferate or that induces apoptosis. To test if FcγRIIBpos cells were negatively impacted by immune checkpoint blockade antibodies, an in vitro stimulation assay was used to determine if the presence of FcγRIIB on the surface of CD8+ T cells affected their responsiveness to checkpoint inhibition, specifically anti-PD-1 and anti-CTLA-4 antibody blockade (Fig. 3A–3B). Results indicated that relative to untreated controls, FcγRIIBneg CD8+ T cells exhibited an increase in the frequency of Ki-67+ proliferating T cells in the presence of PD-1 blockade. When comparing the increase in Ki-67+ cells from the untreated controls to the anti-PD-1 treated group, the FcγRIIBpos CD8+ T cells exhibited significantly (P<0.05) fewer Ki-67+ proliferating cells relative to FcγRIIBneg CD8+ T cells in response to PD-1 blockade (Fig. 3C). The differential proliferative response to checkpoint inhibition evident in FcγRIIBneg CD8+ T cells after PD-1 blockade was also observed in cultures treated with anti-CTLA-4 (Fig. 3D). In contrast, no differences in the frequencies of caspase 3/7+ 7AAD+ apoptotic cells, IFNγ+ or TNF+ cells were observed when comparing FcγRIIBpos to FcγRIIBneg CD8+ T cells treated with PD-1 blockade (fig. S3A-3D). Because cells were stained with detection antibody (anti-FcγRIIB) after an incubation with therapeutic antibodies (anti-PD-1 or anti-CTLA4), we hypothesized that the therapeutic antibody could be masking detection of FcγRIIB. However, this was not the case because a similar amount of FcγRIIB was detected with or without the addition of checkpoint inhibitor (fig. S3E-3F). These data show that the therapeutic antibody did not compete with the detection antibody and support the hypothesis that the decrease in frequency of FcγRIIB+ CD8+ T cells in anti-PD-1 treated patients is due to a true reduction and not an artifact of the therapeutic antibody masking FcγRIIB detection. Taken together, these results demonstrate that FcγRIIBpos CD8+ T cells fail to accumulate following checkpoint inhibition due to reduced proliferation, and not increased cell death, relative to FcγRIIBneg CD8+ T cells. We reasoned that the reduced responsiveness of FcγRIIBpos CD8+ T cells to immune checkpoint blockade could be due to the impact of FcγRIIB-mediated inhibitory signaling following ligation by these Fc-containing monoclonal antibodies.
Fig. 3. FcγRIIBpos CD8+ T cells are resistant to immune checkpoint blockade in an Fc-dependent manner.
(A) Schematic of experimental setup where PBMCs from human healthy controls were cultured with anti-PD-1-mAb or anti-CTLA4 mAb for 5 days and were re-stimulated with PMA/Ionomycin and stained intracellularly. Responses were measured by frequency of Ki-67+ of FcγRIIBpos and FcγRIIBneg CD8+ T cells (n=6). (B) Representative flow cytometry plots showing the frequency of Ki-67+ of FcγRIIBpos and FcγRIIBneg CD8+ T cells in untreated human controls and after incubation with anti-PD-1 or anti-CTLA4 mAbs. Quantification showing the change in frequency of Ki-67+ of FcγRIIBneg and FcγRIIBpos CD8+ T cells after incubation with anti-PD-1 mAb (C) and anti-CTLA-4 mAb (D) in vitro compared to untreated controls. (E) Schematic of experimental setup wherein PBMCs from healthy controls were cultured with anti-PD-1-mAb or anti-PD-1-F(ab) for 5 days and were re-stimulated with PMA/Ionomycin and stained intracellularly. Responses were measured by frequency of Ki-67+ of FcγRIIBpos and FcγRIIBneg CD8+ T cells (n=6).(F) Representative flow plots showing the frequency of Ki-67+ of FcγRIIBpos and FcγRIIBneg CD8+ T cells after incubation with anti-PD-1-mAb or anti-PD-1-F(ab). (G) Quantification comparing the frequency of FcγRIIBneg CD8+ T cells after incubation with anti-PD-1 mAb or antI-PD-1 F(ab). (H) Quantification comparing the change in frequency of Ki-67+ of FcγRIIBpos CD8+ T cells after incubation with anti-PD-1-mAb or anti-PD-1-F(ab) compared to untreated controls. *P<0.05, ns, not significant. Wilcoxon matched-pairs rank test.
To determine if the interaction between Fc regions of checkpoint inhibitors and FcγRIIB was critical to the differential response of FcγRIIBneg CD8+ T cells following checkpoint blockade, F(ab) fragments from the commercially available anti-PD-1 mAb nivolumab (fig. S4) were generated, and their ability to augment proliferation of both FcγRIIBpos and FcγRIIBneg CD8+ T cells following in vitro stimulation was assessed relative to that of intact anti-PD-1 mAb (Fig. 3E–3F). Results indicated that the FcγRIIBneg CD8+ T cell population showed a similar increase in Ki-67 staining in response to both the intact anti-PD-1 mAb and the anti-PD-1 F(ab) (Fig. 3G). In contrast, the FcγRIIBpos CD8+ T cell population was more responsive to the anti-PD-1 F(ab), in that the frequency of Ki-67+ cells was significantly (P<0.05) elevated in the anti-PD-1 F(ab)-treated cultures as compared to the anti-PD-1 mAb-treated cultures (Fig. 3H). Additionally, FcγRIIB+ CD8+ T cells did not fit the gene expression profile of anti-PD-1 resistant CXCR5− TIM-3+ cells as defined by Im et al. (36) (fig. S3I-3J). These data suggest that FcγRIIB+ CD8+ T cells were not resistant to PD-1 therapy because of their differentiation state, but because of FcγRIIB-mediated inhibitory signals generated following ligation with the Fc portion of the checkpoint inhibitor.
PD-1 checkpoint blockade results in a loss of FcγRIIBpos CD8+ T cells in patients with melanoma and in B16 and LLC mouse models
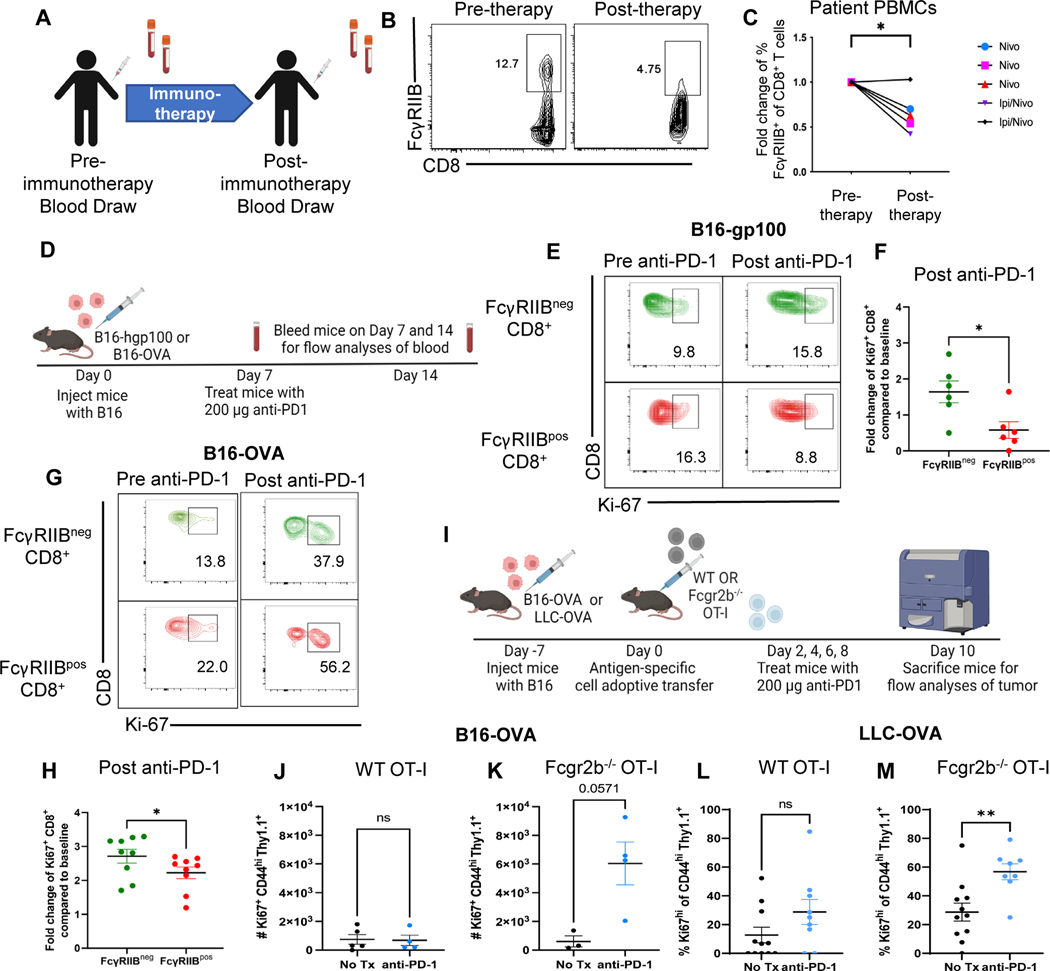
We next sought to determine the responsiveness of FcγRIIBpos versus FcγRIIBneg CD8+ T cells to PD-1 checkpoint blockade in patients with melanoma in vivo. PBMCs from patients with advanced stage melanoma (table S3) were obtained both at baseline and following administration of anti-PD-1 (nivolumab) or a combination of anti-PD-1 and anti-CTLA-4 (nivolumab + ipilumimab) (Fig. 4A). The frequency of FcγRIIBpos among CD8+ T cells significantly (P<0.05) decreased following administration of checkpoint inhibitors (Figures 4B–4C), suggesting a reduced response of FcγRIIBpos as compared to FcγRIIBneg CD8+ T cells to checkpoint inhibition in vivo.
Fig. 4. PD-1 checkpoint blockade results in a loss of FcγRIIBpos CD8+ T cells in patients with melanoma and in B16 and LLC mouse models.
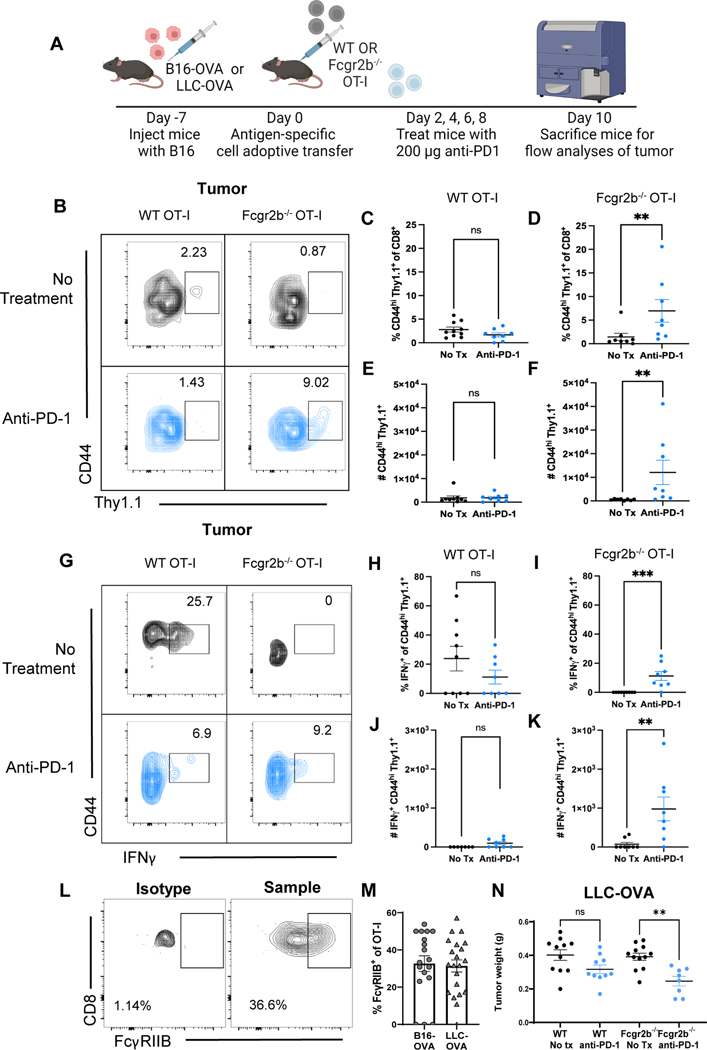
(A) Blood samples were drawn at baseline and following one dose of anti-PD-1 mAb. Fresh PBMCs were processed directly after collection and stained for extracellular surface markers (B) Representative flow cytometry plots of the frequency of FcγRIIBpos CD8+ T cells at baseline (pre-anti-PD-1) and after one cycle of anti-PD-1 mAb (post-anti-PD-1). (C) Quantification of the frequency of FcγRIIBpos of CD8+ T cells at baseline and one cycle of anti-PD-1 mAb, frequency of FcγRIIBpos of CD8+ T cells were normalized to the frequency of the patient’s FcγRIIBpos CD8+ T cells prior to initiation of anti- PD-1 therapy. (D) Schematic of the mouse model and treatment used wherein C57BL/6J mice were challenged with B16-gp100 or B16-OVA melanoma and blood was drawn from mice before (day 7) and after PD-1 blockade (day 14) to measure proliferation of FcγRIIBpos and FcγRIIBneg CD8+ T cells during PD-1 blockade (n=6). (E) Representative flow plots showing the frequency of Ki-67+ FcγRIIBpos and FcγRIIBneg CD8+ T cells before and after PD-1 blockade in the B16-gp100 model. (F) Quantification of fold change values shown from the B16-gp100 model. The blood was lysed and underwent extracellular and intracellular staining for flow cytometric analyses (n=6). (G) Representative flow plots showing the frequency of Ki-67+ FcγRIIBpos and FcγRIIBneg CD8+ T cells before and after PD-1 blockade in the B16-OVA mouse model. (H) Quantification of the frequency of Ki-67+ CD8+ T cells within FcγRIIBpos and FcγRIIBneg CD8+ T cells (n=6). Fold change compared to baseline is the frequency of Ki-67+ cells after PD-1 over the frequency of Ki-67+ cells before PD-1 blockade. (I) Schematic shown wherein C57BL/6J mice were challenged with B16-OVA or LLC-OVA. One week later, one million WT or Fcgr2b−/− OT-I transgenic CD8+ T cells were harvested from donor spleen and adoptively transferred into challenged B6 mice. Mice were then treated with 200 μg of PD-1 antibody or PBS on days 2, 4, 6, and 8 and then sacrificed for flow analyses of tumor. Quantification of the number of Ki67+ OT-Is isolated from the tumors of anti-PD1 treated versus untreated mice that were given (J) WT OT-Is or (K) Fcgr2b−/− OT-Is in the B16-OVA model (n=4 per group). Quantification of frequency of Ki67+ OT-Is isolated from the tumors of anti-PD1 treated versus untreated mice that were given (L) WT OT-Is or (M) Fcgr2b−/− OT-Is in the LLC-OVA model (n=11 per group, pooled data from two experiments). The error bar in summary figures denotes mean ± SEM.Quantification, *P<0.05, **P<0.01, ns, not significant. Wilcoxon matched-pairs rank test.
Because these data in human patients with melanoma suggested an association between immune checkpoint therapy and a decrease in FcγRIIBpos CD8+ T cells, a mouse model of melanoma was employed to interrogate the causal relationship between T cell expression of FcγRIIB and responsiveness to checkpoint inhibition. To begin to address this, we sought to corroborate the association between T cell expression of FcγRIIB and responsiveness to checkpoint inhibition in the B16-melanoma mouse model. Naïve B6 recipients of B16-hgp100 melanoma cells were treated with anti-PD-1 on days 7 and 14 post-tumor inoculation (Fig. 4D) and blood was obtained on days 7 and 14 for the pre- and post-PD-1 timepoints. Results indicated that FcγRIIBneg CD8+ T cells in the blood of mice treated with anti-PD-1 exhibited significantly (P<0.05) more proliferation as measured by the fold change in Ki-67+ proliferating cells as compared to FcγRIIBpos CD8+ T cells (Fig. 4E–4F). A similarly significant (P<0.05) increase in the fold change of Ki-67+ proliferating cells among FcγRIIBneg as compared to FcγRIIBpos CD8+ T cells following anti-PD-1 administration was also observed in a B16-OVA mouse model (Fig. 4G–4H). Thus, consistent with data from patients with melanoma, these results from two mouse models of melanoma demonstrate that FcγRIIBpos cells exhibit reduced responsiveness to anti-PD-1 therapy.
Given this association between FcγRIIB expression on CD8+ T cells and reduced responsiveness to PD-1 blockade, we next questioned whether genetic deletion of the Fcgr2b gene from tumor-specific CD8+ T cells would augment their response to PD-1 blockade. Naïve B6 mice were inoculated with B16-OVA cells and the tumor was allowed to establish for seven days. After seven days, WT or Fcgr2b−/− OT-I transgenic CD8+ T cells were adoptively transferred into tumor-challenged mice. Mice were then treated with anti-PD-1 antibody on days 9, 11, 13, and 15 until day 17 post tumor inoculation (Fig. 4I). We observed that the absolute number of proliferating WT OT-Is in the tumors of mice treated with anti-PD-1 was not significantly different from proliferation of OT-Is in untreated mice as measured by Ki67+ of CD44hi OT-Is (Fig. 4J). In contrast, Fcgr2b−/− OT-I from tumors of mice treated with anti-PD1 exhibited increased (p=0.057) proliferation compared to untreated mice (Fig. 4K). To verify our results in another murine cancer model, we repeated the WT versus Fcgr2b−/− OT-I experiments in the Lewis lung carcinoma (LLC-OVA) model. Naïve B6 animals were inoculated with LLC tumor cells that express the OVA cognate antigen and then WTor Fcgr2b−/− OT-I T cells were adoptively transferred into the mice on day 7. Animals were treated with anti-PD-1 on days 9, 11, 13, and 15 following inoculation and sacrificed on day 17 for analysis of tumor-specific cells in the tumor. Results indicated that, as was observed in mice inoculated with B16-OVA, anti-PD-1 failed to elicit an increase in the frequency of proliferating Ki67+ WT Thy1.1+ CD8+ OT-I T cells (Fig. 4L). In contrast, anti-PD-1 did elicit a significant (P<0.01) increase in the frequency of Ki67+ Fcgr2b−/− Thy1.1+ CD8+ OT-I T cells (Fig. 4M). This increase in proliferation of the Fcgr2b−/− CD8+ OT-I T cells in both models shows that the lack of proliferative response by OT-Is to anti-PD-1 is indeed due to the expression of FcγRIIB on the antigen-specific CD8+ T cell itself and not due to FcγRIIB expressed on other immune cell types. These data further support the conclusion that Fcgr2b on CD8+ T cells negatively impacts proliferation in response to anti-PD-1 therapy in mice and humans.
T cell-expressed FcγRIIB restrains CD8+ T cell responsiveness to PD-1 blockade B16 melanoma in vivo
Using the B16-OVA model aforementioned (Fig. 5A), we further investigated the response of antigen-specific cells to anti-PD-1 in the absence of Fcgr2b. Frequencies and absolute numbers of WT OT-I T cells from tumors of anti-PD-1-treated mice were not increased relative to WT OT-I T cells isolated from tumors of untreated mice (Fig. 5B–5D). In contrast, frequencies and absolute numbers of Fcgr2b−/− OT-I T cells isolated from tumors of anti-PD-1-treated mice were significantly (P<0.01) increased relative to Fcgr2b−/− OT-I T cells from tumors of untreated mice (Fig. 5B, 5E and 5F).
Fig. 5. T cell-expressed FcγRIIB restrains CD8+ T cell responsiveness to PD-1 blockade B16 and LLC mouse models in vivo.
(A) Schematic shown wherein C57BL/6J mice were inoculated with the B16-OVA cancer cell line. One week later, one million WT or Fcgr2b−/− OT-I transgenic CD8+ T cells were harvested from donor spleen and adoptively transferred into B16-challenged B6 mice. Animals were treated with 200 μg of PD-1 antibody or PBS on days 2, 4, 6, 8 post adoptive transfer (days 9, 11, 13, and 15 post tumor inoculation). Spleen and tumor tissues were harvested on day 10 post adoptive transfer (day 17 post tumor) for flow analyses. (B) Representative flow plot showing CD44hi Thy1.1+ WT and Fcgr2b−/− OT-I T cells in the tumor comparing the no treatment group and the anti-PD-1 treated group, n=8–10 per group, pooled data from two independent experiments. (C-F) Summary figures comparing the frequencies (C) and absolute cell numbers of (D) CD44hi WT versus the frequencies (E) and absolute cell numbers (F) Fcgr2b−/− OT-I T cells in the tumor, (G) Representative flow plot showing IFNγ+ CD44hi Thy1.1+ WT and Fcgr2b−/− OT-I T cells in the tumor comparing the no treatment and anti-PD-1 treatment groups, n=8–10 per group, pooled data from two independent experiments. (H-K) Summary figures comparing the frequencies (H) and absolute cell numbers (I) of IFNγ+ CD44hi WT versus the frequencies (J) and absolute cell numbers (K) of IFNγ+ Fcgr2b−/− OT-I T cells in the tumor. (L) Representative flow cytometry plots showing FcγRIIB staining on Thy1.1+ OT-Is compared to isotype control and (M) quantification showing FcγRIIB staining on Thy1.1+ OT-Is from the tumors of mice challenged with B16-OVA or LLC-OVA. (N) Summary figurecomparing tumor weight of mice challenged with LLC-OVA, timeline and experimental design as mentioned above, using LLC-OVA instead of B16-OVA cell line, n=10–12 per group. Mann-Whitney test or multiple comparisons in one-way ANOVA was used. The error bar in summary figures denotes mean ± SEM. *P<0.05, **P < 0.01, ***P < 0.001, ns, not significant.
In addition to tumor infiltration, IFNγ production by OT-Is at the tumor was measured. No significant differences in frequencies or cell numbers of activated CD44hi IFNγ-producing WT OT-I cells isolated from tumors of untreated versus anti-PD-1 treated mice were observed (Fig. 5G–5I). In contrast, significantly increased (P<0.001) frequencies and cell numbers (P<0.01) of activated CD44hi IFNγ-producing Fcgr2b−/− OT-I were isolated from tumors of anti-PD-1 treated mice (Fig. 5G, 5J and 5K) compared to Fcgr2b−/− OT-I T cells isolated from tumors of untreated mice. Corroborating our previous data in murine and human TIL , FcγRIIB was expressed on 20–50% of OT-I CD8+ T cells during B16-OVA and LLC-OVA challenge (Fig. 5L–5M). Although genetic deletion of Fcgr2b from CD8+ T cells did increase OT-I frequency and cell number as well as the frequency and cell number of IFNγ+ OT-Is at the tumor of anti-PD-1 treated mice, this was not sufficient to decrease tumor weight in mice inoculated with B16-OVA (fig. S5B). In the LLC-OVA model, treatment with anti-PD-1 alone was insufficient to significantly reduce tumor weight when mice contained WT tumor-specific OT-I T cells. However, when mice were given Fcgr2b−/− tumor-specific OT-I CD8+ T cells, a significant (P<0.01) reduction in tumor weight was observed following PD-1 blockade (Fig. 5N). Anti-PD1 therapy did not delay tumor growth in the B16-OVA model (fig. S5C), but was able to do so in the LLC-OVA model (fig. S5D). These data show that FcγRIIB expression on CD8+ T cells is a factor governing anti-PD-1 response to some murine models of cancer.
In the spleens of anti-PD-1 treated mice, there were no significant differences in the frequencies or cell number of WT versus Fcgr2b−/− OT-I cells (fig. S5E-5H). Additionally, the frequencies and cell numbers of activated CD44hi IFNγ-producing WT vs. Fcgr2b−/− OT-I T cells in the spleen were not different (fig. S5I-5L. These data suggest that the impact of T cell-expressed FcγRIIB functions at the tumor.
Dual blockade of anti-FcγRIIB and anti-PD-1 promotes infiltration of CD8+ T cells into the tumor to delay tumor progression in a B16 melanoma mouse model
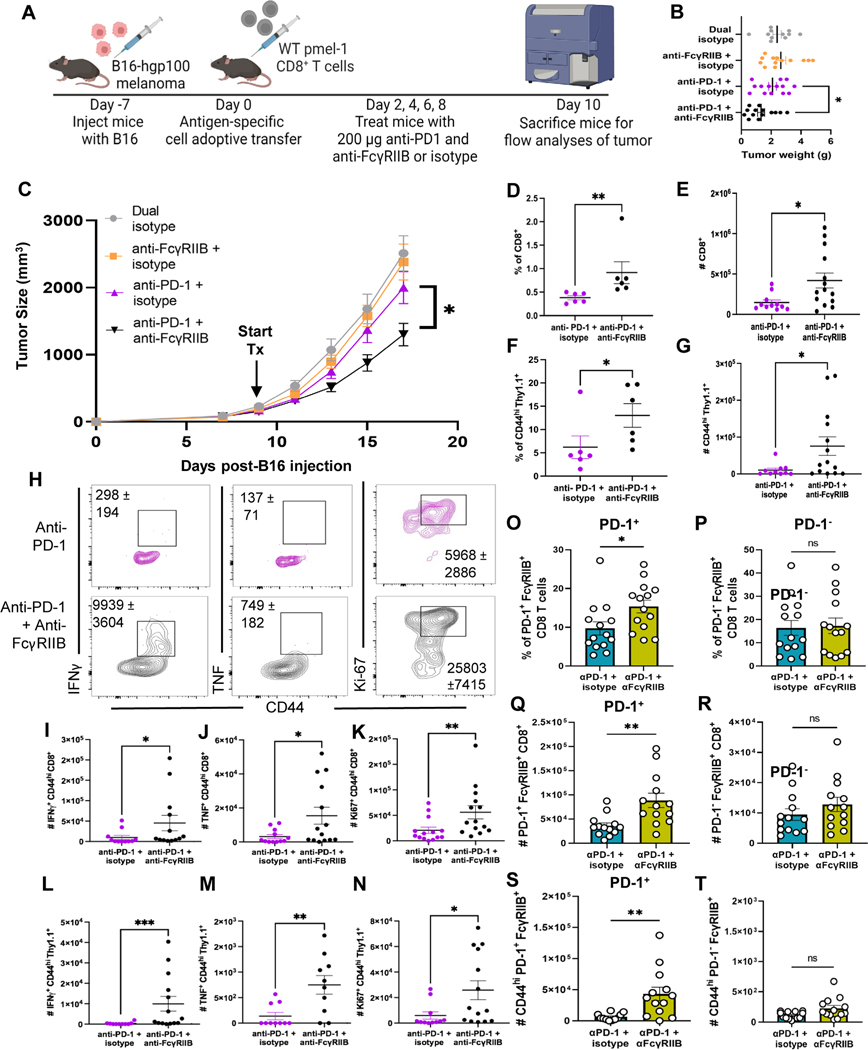
Because FcγRIIBpos CD8+ T cells are more proliferative and produce more cytokines than their FcγRIIBneg CD8+ T cell counterparts but were not effectively augmented by PD-1 blockade, we asked if FcγRIIB blockade in the context of PD-1 blockade would enhance therapeutic efficacy. Here, we utilized a B16-hgp100 mouse model and gp100-specific CD8+ pmel-1 T cells (Fig. 6A). In mice with B16-hgp100 tumors established for seven days (~5–6 mm) and adoptively transferred Thy1.1+ WT pmel-1 CD8+ TCR transgenic T cells, dual blockade of anti-FcγRIIB (clone 2.4G2) and anti-PD-1 (RMP1–14) significantly decreased tumor weight (P<0.05) and delayed tumor progression (P<0.05) relative to mice treated with anti-PD-1 blockade alone (Fig. 6B–6C). The dual blockade also significantly increased tumor infiltration by host CD8+ T cells as evidenced by elevated frequencies (P<0.01) and cell number (P<0.05) of CD8+ T cells in dual blockade mice compared to PD-1 treated mice (Fig. 6D and 6E). The frequencies and cell number of activated donor CD44hi Thy1.1+ tumor-specific pmel-1 CD8+ T cells were also significantly (P<0.05) increased in the tumors in animals treated with dual blockade (Fig. 6F–6G) compared to anti-PD-1 blockade alone.
Fig. 6. Dual blockade of anti-FcγRIIB and anti-PD-1 promotes infiltration of CD8+ T cells into the tumor to delay tumor progression in a B16 melanoma mouse model.
(A) Schematic shown wherein C57BL/6J mice were inoculated with B16-hgp100 melanoma. One week later (tumors ~5mm), WT pmel-1 CD8+ T cells were isolated from mesenteric lymph node and spleen of donor pmel-1 mice and adoptively transferred into B16-hgp100-challenged mice. On days 2, 4, 6, and 8 post adoptive transfer (days 9, 11, 13, and 15 post tumor inoculation), mice were treated in four groups: 200 μg of IgG2a and IgG2b isotypes, 200 μg of anti-FcγRIIB + IgG2a isotype, 200 μg anti-PD-1 + 200 μg IgG2b, or 200 μg anti-PD-1 + 200 μg anti-FcγRIIB. On day 10 post adoptive transfer, (day 17 post tumor), the mice were sacrificed and draining lymph node, spleens, and tumor were harvested for flow cytometric analyses. (B) Tumor weight was measured after excision of each mouse and (C) tumor volume was measured with calipers for each mouse every other day after adoptive transfer to compose the tumor progression plot. Mice were sacrificed at humane endpoint defined by >2cm in either dimension, necrotic, or affected animal health/ mobility as approved by the Emory University IACUC, n=13–18 per group, pooled data from three independent experiments. (D) Quantification for the frequencies (data shown are representative of three independent repeats, n=4–6 per group) and (E) absolute cell number of bulk CD8+ T cells in PD-1 single blockade or PD-1 + FcγRIIB blockade treated animals. (F) Quantification for the frequencies (data shown are representative of three independent repeats, n=4–6 per group) and (G) absolute cell number of CD44hi Thy1.1+ pmel-1 CD8+ T cells in PD-1 single blockade or dual blockade treated animals. (H) Representative flow plots of CD44hi Thy1.1+ cells in both treatment groups, cell numbers ± SEM are shown on the flow plot as well. (I-K) Quantification for the absolute cell number of (I) CD44hi IFNγ+ and (J) CD44hi TNF+ CD8+ T cells as well as (K) CD44hi Ki67+ CD8+ T cells, pooled data from three independent experiments, n=11–14 per group. (L-N) Quantification for the absolute cell number of (L) CD44hi IFNγ+ and (M) CD44hi TNF+ Thy1.1+ pmel-1 CD8+ T cells as well as (N) CD44hi Ki67+ Thy1.1+ pmel-1 CD8+ T cells, pooled data from three independent experiments, n=11–14 per group. (O-R) Quantification for the (O) frequency and (P) absolute cell number of PD-1+FcγRIIB+ CD8+ T cells at the tumor and the (Q) frequency and (R) absolute cell number of PD-1−FcγRIIB+ CD8+ T cells as well as absolute cell number of (S) CD44hiPD-1+FcγRIIB+ CD8+ T cells and (T) CD44hiPD-1−FcγRIIB+ CD8+ T cells, pooled data from three independent experiments, n=14–15 per group. The error bar in summary figures denotes mean ± SEM. *P<0.05, **P < 0.01, ***P< 0.001, ns, not significant, Mann-Whitney Test.
Blocking FcγRIIB alongside PD-1 also increased the quality of these cells at the tumor (Fig. 6H), significantly increasing the number of bulk IFNγ+ (P<0.05) (Fig. 6I), TNF+ (P<0.05) (Fig. 6J), Ki-67+ (P<0.01) (Fig. 6K).The number of tumor-specific CD8+ T cells (CD44hi Thy1.1+) producing IFNγ (P<0.001) (Fig. 6L), TNF (P<0.01) (Fig. 6M), and expressing Ki-67 (P<0.05) (Fig. 6N) was also significantly increased in dual blockade treated mice compared to mice given anti-PD-1 blockade alone. Overall, these data show that blocking FcγRIIB augments activation and cytokine production of activated tumor-specific pmel-1 CD8+ T cells during PD-1 blockade, likely allowing these high-quality cells to elicit enhanced anti-tumor functions to delay B16 tumor progression in vivo.
We posited that because of the co-expression of PD-1 and FcγRIIB on activated CD8+ T cells (25), when anti-PD-1 antibody binds its target it could be co-engaging FcγRIIB and eliciting a negative regulatory signal to activated FcγRIIB+ CD8+ T cells. If this were the case, one would predict that FcγRIIB blockade would impact only PD-1+ FcγRIIB+ CD8+ T cells and not PD-1− FcγRIIB+ CD8+ T cells. Thus, we compared the effect of blocking FcγRIIB on PD-1+ FcγRIIB+ versus PD-1− FcγRIIB+ CD8+ T cells. Results indicated that the addition of FcγRIIB blockade significantly increased frequencies (P<0.05) and cell number (P<0.01) of PD-1+ FcγRIIB+ cells as compared to anti-PD-1 alone (Fig. 6O and 6P). In contrast, it did not impact the PD-1− FcγRIIB+ CD8+ T cell population (Fig. 6Q and 6R). Focusing on activated CD8+ T cells, the addition of FcγRIIB blockade resulted in a significant (P<0.01) increase in cell number of PD-1+ CD44hi FcγRIIB+ CD8+ T cells at the tumor as compared to anti-PD-1 alone (Fig. 6S). In contrast, it did not impact the number of PD-1− CD44hi FcγRIIB+ CD8+ T cells (Fig. 6T). These data demonstrate that PD-1+ FcγRIIB+ CD8+ T cells, and not all FcγRIIB+ CD8+ T cells, are preferentially “rescued” by the addition of FcγRIIB blockade.
Dual blockade of FcγRIIB and PD-1 elicits enhanced CD8+ T cell antitumor efficacy to regress tumors in an LLC lung carcinoma mouse model
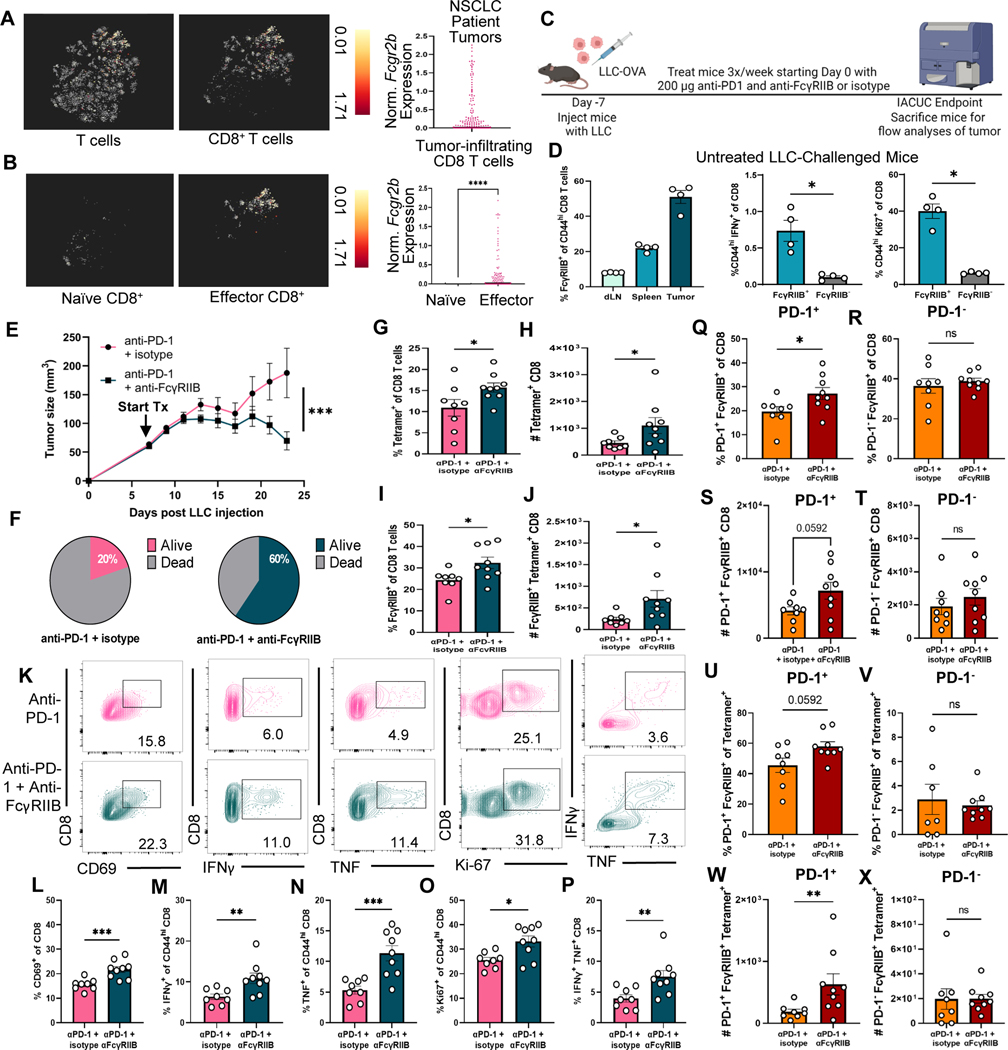
We next investigated whether the ability of FcγRIIB blockade to augment anti-tumor immunity in the context of PD-1 blockade was generalizable to other cancers. Interrogation of an existing single cell RNA-sequencing dataset of 9,055 T cells from 14 patients with non-small cell lung cancer (NSCLC) deposited by Guo et al. (37) revealed that Fcgr2b is expressed on tumor-infiltrating CD8+ T cells (Fig. 7A). The Guo et al. dataset also stratified CD8+ T cells into functional states based on canonical cell markers. Thus, we queried the expression of Fcgr2b on cytotoxic versus naïve CD8+ T cells and found that cytotoxic CD8+ T cells expressed significantly (P<0.0001) more Fcgr2b than naïve CD8+ T cells at the tumor of patients with NSCLC (Fig. 7B). To further investigate the presence and phenotype of FcγRIIB+ CD8+ T cells in the tumor of lung cancer, we used a Lewis lung cancer (LLC) mouse model wherein LLC cells were engineered to express the OVA epitope (LLC-OVA) (Fig. 7C). Upon phenotyping FcγRIIB+ CD8+ T cells in the LLC model, we found that FcγRIIB is most expressed on activated cells at the tumor, then in the spleen, with the least expression in the draining lymph node (Fig. 7D), similar to our published findings in a melanoma model (25). Similar to our previous studies, FcγRIIB+ CD8+ cells exhibited superior effector function compared to FcγRIIB− cells as demonstrated by an increased production of IFNγ as well as increased proliferation as measured by Ki67 (Fig. 7D).
Fig. 7. Dual blockade of FcγRIIB and PD-1 elicits enhanced CD8+ T cell antitumor efficacy to regress tumors in an LLC lung carcinoma mouse model.
(A-B) Reanalysis of lung cancer TIL single cell datasets. (A) t-SNE plot from a publicly available single cell dataset visualizing FCGR2B expression on 9,005 single T cells isolated from NSCLC patient tumors (n=14) (left), followed by a t-SNE visualization of FCGR2B expression CD8+ T cells (middle), summary of FCGR2B expression within tumor-infiltrating CD8+ T cells (right), data normalized in log2 format and centered to each patient. (B) t-SNE plot projecting FCGR2B expression in naïve (left) versus dataset-defined effector (middle) CD8+ T cells at the tumor from the same publicly available single cell dataset of lung cancer samples with a summary plot of FCGR2B normalized in log2 format and centered to each patient (right). (C) Mice were inoculated with LLC-OVA cells. One week later (tumors ~5mm), treatment of 200 μg anti-PD-1 + 200 μg IgG2b or 200 μg anti-PD-1 + 200 μg anti-FcγRIIB three times a week until IACUC endpoint was reached. The mice were sacrificed and draining lymph nodes, spleens, and tumor were harvested for flow cytometric analyses. (D) Quantification measuring the frequency of FcγRIIB+ of CD44hi CD8+ T cells at the draining lymph node, spleen, and tumor (left) and the frequency of CD44hi IFNγ+ CD8+ T cells (middle) and of CD44hi Ki67+ CD8+ T cells (right) in cells stimulated ex vivo isolated from untreated LLC-challenged mice, n=4 per group. (E) Tumor progression of mice treated accordingly is displayed, tumor volume was measured with calipers for each mouse every other day after start of treatment. (F) Survival data were plotted on Kaplan-Meier curves, and a log-rank (Mantel-Cox) test was performed. (G-J) Quantification for the (G) frequency and (H) absolute cell number of tetramer+ CD8+ as well as the (I) frequency of FcγRIIB+ CD8+ and (J) absolute cell number of FcγRIIB+ tetramer+ CD8+ T cells in the tumor of treated mice. (K-P) Quantification for the frequency of (L) CD69+ CD8+, (M) IFNγ+ of CD44hi CD8+, (N) TNF+ of CD44hi CD8+, (O) Ki67+ of CD44hi CD8+ T cells, and (P) IFNγ+ TNF+ CD8+ T cells stimulated ex vivo from the tumors of treated mice, n=8–9 per group. Representative flow plots from both treatment groups are shown with the frequency shown on the flow plot as well. (Q-T) Quantification for the frequency (Q) and absolute cell number (R) of PD-1+FcγRIIB+ CD8+ T cells vs. the frequency (S) and absolute cell number (T) of PD-1−FcγRIIB+ CD8+. (U-X) Quantification for the frequency (U) and absolute cell number (V) of PD-1+FcγRIIB+ tetramer+ cells versus the frequency (W) and absolute cell number (X) of PD-1−FcγRIIB+ tetramer+ cellsat the tumor, n=8–9 per group. Mann-Whitney non-parametric, unpaired tests were used when comparing groups of mice. Area under curve was calculated and then total area was compared using an unpaired T test. The error bar in summary figures denotes mean ± SEM. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05, ns, not significant.
To assess the effect of the addition of FcγRIIB blockade to PD-1 checkpoint inhibition on the endogenous CD8+ T cell response to tumors, mice with LLC-OVA tumors (established for seven days ~5 mm) were treated three times a week with dual blockade or with anti-PD-1 + isotype control. Strikingly, the addition of FcγRIIB blockade resulted in significantly (P<0.001) decreased tumor size compared to mice treated with anti-PD-1 alone (Fig. 7E). Moreover, anti-PD-1 and anti-FcγRIIB significantly (P<0.05) increased survival wherein 60% of mice in the dual blockade-treated group were alive at day 50 compared to only 20% of mice in the PD-1 blockade treated group (Fig. 7F). Mice treated with dual blockade exhibited significantly increased (P<0.05) frequencies and (P<0.05) numbers of tumor-infiltrating OVA-specific tetramer+ CD8+ T cells as compared to anti-PD-1 treated mice (Fig. 7G and 7H). The addition of FcγRIIB blockade resulted in increased frequency and number of FcγRIIB+ cells among bulk CD8+ T cells (Fig. 7I) and among tetramer+ cells (Fig. 7J) compared to treatment with anti-PD-1 alone. When stimulated with cognate antigen ex vivo, tumor-infiltrating CD8+ T cells isolated from dual blockade-treated mice were more activated and functional (Fig. 7K) as measured by significantly increased frequencies of CD69+ (P<0.001) (Fig. 7L), IFNγ+ (P<0.01) (Fig. 7M), TNF+ (P<0.001) (Fig. 7N), Ki67+ (P<0.05) (Fig. 7O), and double-producer IFNγ+TNF+ CD8+ T cells (P<0.01) (Fig. 7P) compared to mice treated with anti-PD-1 alone. We next assessed whether FcγRIIB blockade preferentially impacts PD-1+ FcγRIIB+ cells, as compared to PD-1− FcγRIIB+ cells, and found that the addition of FcγRIIB blockade resulted in a significant (P<0.05) increase in frequency (Fig. 7Q) and number (p=0.0592) (Fig. 7R) of PD-1+ FcγRIIB+ CD8+ cells as compared to PD-1 blockade alone. In contrast, the addition of FcγRIIB blockade did not increase the frequency (Fig. 7S) and number (Fig. 7T) of PD-1− FcγRIIB+ cells as compared to PD-1 blockade alone. These findings were also true for PD-1+ FcγRIIB+ cells among the tetramer+ population (Fig. 7U–7X).
Blocking FcγRIIB with clone 2.4G2 or non-cross-reactive FcγRIIB-specific antibody AT-128 increases response to anti-PD-1 therapy in an MC38 mouse colon cancer model
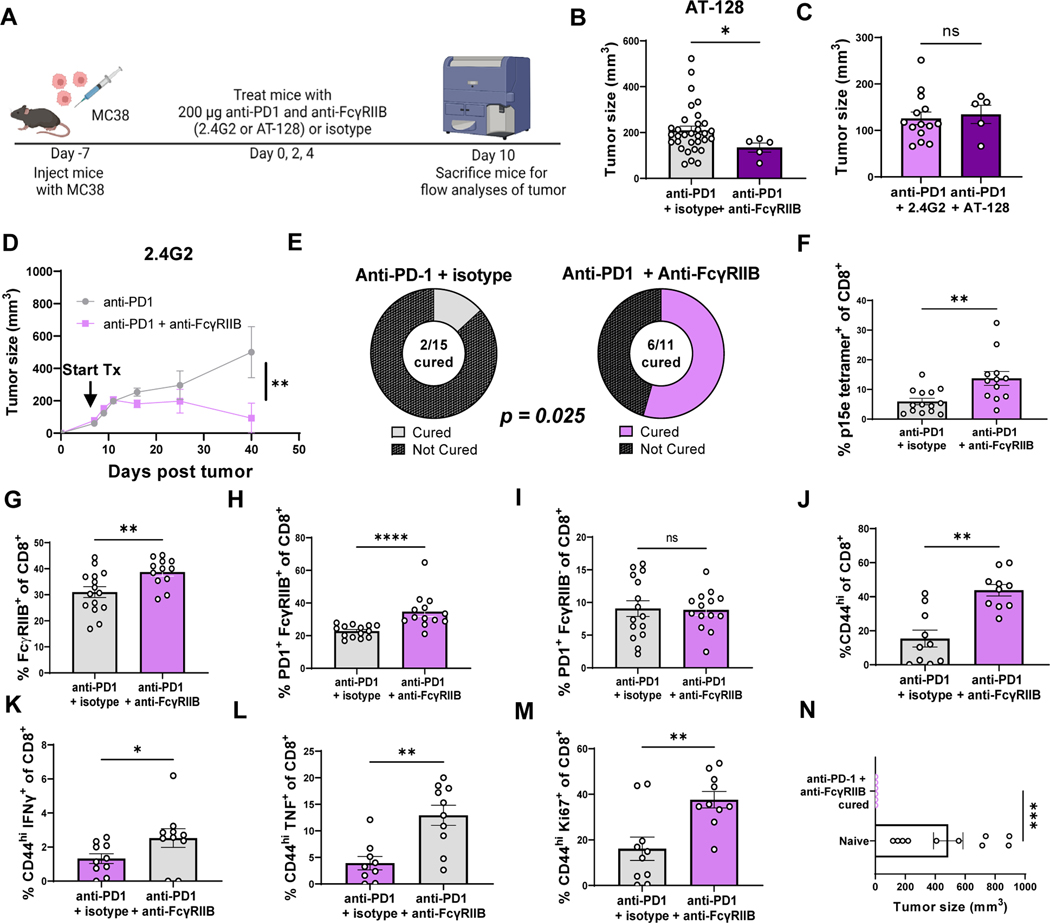
Anti-FcγRIIB clone 2.4G2 cross reacts with FcγRIII. Although we did not observe expression of FcγRIII on CD8+ T cells, to definitively evaluate the impact of blocking FcγRIIB on CD8+ T cells and rule out any potential effect of anti-FcγRIII binding, we utilized a distinct antibody which is FcγRIIB-specific and non-cross-reactive. Animals were inoculated with MC38 tumors and treated with anti-PD-1 along with either isotype control or the FcγRIIB-specific clone AT-128 starting 7 days after inoculation. Tumor-specific CD8+ T cells were identified using p15e/Kb tetramer, a retroviral epitope expressed by MC38 cells (Fig. 8A). In mice with MC38 tumors established for seven days (~5–6 mm), dual blockade of anti-FcγRIIB (clone AT-128) and anti-PD-1 (RMP1–14) significantly (P<0.05) decreased final tumor size at day 17 post tumor inoculation relative to mice treated with anti-PD-1 blockade alone (Fig. 8B). These data using a non cross-reactive, FcγRIIB-specific antibody definitively demonstrate that blocking FcγRIIB improves the efficacy of anti-PD-1. Moreover, no differences were observed in the tumor sizes of mice treated with clone 2.4G2 and anti-PD-1 versus clone AT-128 and anti-PD-1, suggesting that the clones are similar in antitumor efficacy (Fig. 8C). In the MC38 mouse model, we showed that blocking FcγRIIB alongside PD-1 significantly (P<0.01) delayed tumor progression (Fig. 8D) and resulted in significantly (P<0.05) more curative responses (Fig. 8E, 54.5% vs. 13.3%) than anti-PD-1 treatment alone. Analysis of the TIL within these animals revealed, as in the two other cancer models, more tetramer+ (Fig. 8F) and FcγRIIB+ CD8+ T cells (Fig. 8G) in dual blockade treated animals compared to animals treated with anti-PD-1 alone. In addition, double positive PD-1+FcγRIB+ CD8+ T cells were significantly (P<0.0001) increased following dual blockade (Fig. 8H) whereas PD-1−FcγRIB+ CD8+ T cells (Fig. 8I) were not. In tumors of the dual blockade-treated mice, there were significantly higher frequencies of CD44hi CD8+ T cells (P<0.01) (Fig. 8J) as well as higher frequencies of IFNγ+ (P<0.05) (Fig. 8K), TNF+ (P<0.01) (Fig. 8L) and Ki67+ CD44hi CD8+ T cells (P<0.01) (Fig. 8M) compared to mice treated with anti-PD-1 alone. To assess the durability of the anti-tumor response elicited in recipients treated with PD-1 blockade in the presence or absence of concomitant FcγRIIB blockade, we rechallenged mice that had experienced tumor cure by anti-PD-1 + anti-FcγRIIB dual blockade with a new inoculation of MC38 cancer cells 14 weeks after initial inoculation and 10–12 weeks after tumor clearance. Results indicated that all mice that had been cured by the combination of anti-PD-1 + anti-FcγRIIB (n=6) remained tumor-free (Fig. 8N) whereas large tumors formed in naïve mouse controls (n=10) (Fig. 8N). The low (13%) cure rate in the anti-PD-1 group precluded the ability to generate enough animals in this group for statistical comparison. These data highlight that blocking FcγRIIB in another mouse model of cancer augments activation and cytokine production of activated tumor-specific CD8+ T cells during PD-1 blockade, resulting in delayed tumor progression, and curative responses in some mice. The use of AT-128, a non-cross-reactive FcγRIIB-specific antibody, supports the claim that the antitumor efficacy observed in these models is likely due to blocking FcγRIIB and not blocking FcγRIII.
Fig. 8. Blocking FcγRIIB with clone 2.4G2 or non-cross-reactive FcγRIIB-specific antibody AT-128 increases response to anti-PD-1 therapy in an MC38 mouse colon cancer model.
(A) Schematic shown wherein C57BL/6J mice were inoculated with MC38 cells. One week later (tumors ~5mm), mice were treated with 200 μg anti-PD-1 + 200 μg IgG2b or 200 μg anti-PD-1 + 200 μg anti-FcγRIIB (clone 2.4G2 or clone AT-128) on days 0, 2, and 4. Mice were sacrificed and draining lymph nodes, spleens, and tumor were harvested for flow cytometric analyses on day 10 or IACUC endpoint. (B) Tumor size of mice treated with anti-PD1 and isotype or anti-PD1 and anti-FcγRIIB (clone AT-128) is displayed at sacrifice on day 10 (n=14–15 per group), pooled data from two experiments. (C) Tumor size of mice treated with anti-PD1 and anti-FcγRIIB (clone 2.4G2) or anti-PD1 and anti-FcγRIIB (clone AT-128) is displayed at sacrifice (n=32 or 5 per group). (D) Tumor time course is displayed with tumor volume as measured with calipers for each mouse every other day after start of treatment until IACUC endpoint (n=11 or 15 per group). (E) Pie graphs depicting proportion of MC38-challenged mice that were cured after anti-PD1 and isotype treatment vs anti-PD1 and anti-FcγRIIB (clone 2.4G2) (n=11 or 15 per group). Quantification for the (F) frequency of p15e tetramer+ CD8+ T cells, (G) frequency of FcγRIIB+ CD8+ T cells, and frequency of (H) PD-1+FcγRIIB+ CD8+ T cells versus (I) PD-1−FcγRIIB+ CD8+. Quantification for the frequency of (J) CD44hi CD8+, (K) CD44hi IFNγ+ CD8+, (L) CD44hi TNF+ of CD8+, (M) CD44hi Ki67+ of CD8+ T cells stimulated with p15e peptide ex vivo from the tumors of treated mice, n=9–10 per group, pooled data from two experiments. (N) Tumor size of mice rechallenged with MC38 tumors 10–12 weeks after tumor cure with anti-PD-1 + anti-FcγRIIB (n=6) or naïve mice controls challenged for first time. Mann-Whitney non-parametric, unpaired tests were used when comparing groups of mice. The error bar in summary figures denotes mean ± SEM. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05, ns, not signf.
Discussion
Here we report the presence of the inhibitory Fc receptor FcγRIIB on a population of activated memory CD8+ T cells isolated from both healthy humans and patients with melanoma. Although FcγRIIBpos CD8+ T cells exhibit high functionality, they possess limited responsiveness to checkpoint inhibition in vitro as well as in in vivo mouse melanoma models and in human patients with melanoma. This reduced responsiveness of FcγRIIBpos relative to FcγRIIBneg CD8+ cell population was Fc-dependent since removal of the Fc region from anti-PD-1 antibody improved the response of FcγRIIBpos CD8+ T cells without affecting FcγRIIBneg CD8+ T cells. Moreover, when Fcgr2b was genetically deleted from tumor-specific OT-I CD8+ T cells in B16-OVA and LLC-OVA mouse models, the anti-tumor efficacy of these cells was improved during PD-1 blockade. Taken together, these data suggest a model wherein the use of Fc-containing checkpoint inhibitors provides the ligand for FcγRIIB by binding in the reverse orientation to FcγRIIB expressed by multi-potent effector tumor-specific CD8+ T cells, resulting in the transmission of a negative signal that functions to counteract the stimulatory effect of checkpoint inhibition.
This work corroborates recent reports from our laboratory and others demonstrating expression of FcγRIIB at the RNA and protein level in CD8+ T cells in mouse models of infection, transplant, and cancer (23–27, 29, 30, 38–40). Of particular relevance to this work, Fcgr2b is a differentially expressed gene in several datasets in the settings of chronic viral infection and cancer in memory and effector CD8+ T cells versus naïve CD8+ T cells (27, 29, 30, 41). Our own analysis of patient TIL and reanalysis of three independent datasets of single cell RNA sequencing of immune cells isolated from patient tumor confirm that Fcgr2b transcript is present in CD8+ T cells, and that Fcgr2b expression is increased in cytotoxic CD8+ compared to naïve T cells. Together these studies challenge the long-held notion that T cells do not express Fc receptors (23). Consistent with our results reported here, published studies in murine models of transplantation and melanoma also showed that FcγRIIBpos CD8+ T cells exhibit potent cytokine production and proliferation, along with higher expression of activation markers, compared to FcγRIIBneg CD8+ T cells (23, 25). These functional characteristics suggest that FcγRIIBpos CD8+ T cells are not exhausted, but instead are highly functional effectors that can be regulated through ligation of FcγRIIB. Our results demonstrating the impact of Fc-containing immunotherapeutics on T cells expressing FcγRIIB are also consistent with previous reports demonstrating clinically- relevant interactions of Fc-containing biologics with FcγRIIB on other cell types including B cells and myeloid cells (15, 17–20, 42, 43). Further, the anti-tumor effect of anti-PD-1 mAb was greater in Fcgr2−/− vs. WT mice (20), and FcγRIIB inhibition improved PD-1 therapy by blocking FcγRIIB-mediated macrophage uptake of PD-1 antibody from T cells (44). Although these studies demonstrated that PD-1 antibodies interact with FcγRIIB, the main mechanism of action in both studies was attributed to the expression of FcγRIIB on antigen-presenting cells or B cells, but not on CD8+ T cells. This study therefore highlights a novel mechanism by which the Fc portion of checkpoint inhibitors can directly modulate immune function of activated CD8+ T cells in the setting of cancer immunotherapy.
Our work has identified a potential immunosuppressive effect of Fc-containing biologics on a population of CD8+ T cells in the context of anti-tumor immunity. Due to the ability of FcγRIIB to bind the subclasses of IgG with different affinity, it could therefore be advantageous to generate immunotherapeutic agents using IgG isotypes with lower affinity for FcγRIIB. Although FcγRIIB is a low-affinity receptor, IgG4 binds FcγRIIB with similar affinity as high-affinity activating receptors (45, 46). Of clinical import, FDA-approved anti-PD-1 drugs nivolumab and pembrolizumab are both IgG4 isotypes. Given this potential interaction, expression of FcγRIIB on CD8+ T cells in patients with cancer could be a determinant of response to PD-1 blockade.
Our study has some limitations. First, analysis of human PBMC and TIL were conducted on cells isolated from a small cohort of patients with a single tumor type (melonma), thus analysis of additional tumor types and in a larger patient population is warranted. However, the fact that FcγRIIB blockade synergized with anti-PD-1 in three different cancer types in murine models, two with a self-antigen and one with a surrogate neoantigen, as well as in settings of both endogenous and adoptively transferred CD8+ T cells, expand the generalizability of these findings. In addition, the enhanced antitumor efficacy of FcγRIIB blockade in combination with anti-PD-1 in all three models of varying sensitivities to PD-1 blockade highlights the potential clinical utility of blocking FcγRIIB together with PD-1 in cancer models with varying sensitivities to PD-1 blockade, thus expanding the patient population to which these findings could potentially apply. An additional limitation of our study is the inability to generalize findings across racially diverse patient populations, as our patient population with melanoma did not include any non-white patients.
In conclusion, the findings herein describe a previously unappreciated mechanism by which immune checkpoint antibodies may impede anti-tumor CD8+ T cell responses through FcγRIIB, and suggest strategies to overcome this unintended effect. In an era of immune checkpoint blockade, identifying nuances and unintended drug interactions is of paramount importance to increase patient response, and ultimately, improve patient survival.
Materials and Methods
Study design
The primary objective of this study was to investigate the impact of CD8+ T-cell expressed FcγRIIB on the efficacy of Fc-containing immune checkpoint antibodies. Using in vitro stimulation in the presence or absence of anti-PD-1 and anti-CTLA-4 and flow cytometric readouts, we compared the efficacy of Fc-containing checkpoint inhibitors to improve the proliferation and effector function of FcγRIIB-expressing vs non-FcγRIIB-expressing CD8+ T cells. We then sought to determine how the genetic deletion of FcγRIIB on tumor-specific CD8+ T cells impacted their function and ability to clear the tumor in the setting of checkpoint blockade in murine cancer models. Animals were randomized to groups; treatments were not blinded. Outliers were detected using the ROUT method. In each experiment, at least 4–5 mice were used per group and the experiment was repeated 2–3 times; mice of similar sex and age were used across groups. ARRIVE guidelines (Animal Research: Reporting of In Vivo Experiments) were utilized in the presentation of data in the figures and figure legends.
Patient population
Healthy controls were enrolled after informed consent (table S1, n=8). Patients undergoing treatment at Emory University Hospital for advanced stage II-IV melanoma between 2009 and 2019 were enrolled in a protocol approved by Emory University’s Institutional Review Board (IRB #00046593). For PBMC immunophenotyping, melanoma patient demographic data are shown in table S2 (n=6). For FcγRIIB+ T cell analyses, patient blood samples were drawn at baseline (pre-therapy) and after one dose of immunotherapy with anti-PD-1 alone or in combination with anti-CTLA4 (demographic data shown in table S3 (n=5)). For TIL studies, melanoma tumor tissues were collected (IRB #00095411), deidentified, and distributed by the Cancer Tissue and Pathology shared resource of Winship Cancer Institute of Emory University (demographic data shown in table S4 (n=8)).
FLI8.26 Antibody Specificity Verification
FcγRIIB staining was performed using PE-conjugated anti-CD32 clone FLI8.26 (BD Biosciences). FLI8.26 has been shown to be cross-reactive with human FcγRIIA, FcγRIIB, and FcγIIC (31). To verify that FLI8.26 was detecting only FcγRIIB on CD8+ T cells, we performed qPCR analysis of the FCGR2 genes of the population of CD8+ T cells that stained positively with FLI8.26 (by FACS-sorting). These data show undetectable amounts of FCGR2C on these cells (fig. S1B), making it unlikely that that FLI8.26 could be staining FcγRIIC on CD8+ T cells. In contrast, we observed low but detectable FCGR2A expression on the population of FLI8.26+ CD8+ T cells, but using an FcγRIIA-specific antibody, we were unable to detect any protein expression of FcγRIIA on FLI8.26+ CD8+ T cells (fig. S1C).
PBMC isolation and processing
PBMCs from patients with melanoma pre- and post-PD-1 mAb therapy (Fig. 4) were purified using BD Vacutainer CPT tubes (BD Biosciences) and were processed and stained for extracellular surface markers directly.
Human flow cytometry staining
Staining for lineage and intracellular markers was performed using antibodies listed in Data File S2. Prior to intracellular staining, cells were fixed and permeabilized using the Foxp3 Staining Kit (Thermofisher). Live/Dead Aqua Dead Cell Stain (Thermofisher) was used according to manufacturer instructions. All flow cytometry samples were acquired on a Fortessa flow cytometer (BD Biosciences) and data were analyzed using FlowJo (v9.9.6) and Prism 9 (GraphPad Software). Absolute numbers were calculated using Countbright bead analysis according to the manufacturer’s instructions (Thermofisher).
Patient tumor preparation and in vitro stimulation
For TIL experiments, patient tumor tissue was collected and dissociated using the Human Tumor Dissociation Kit (Miltenyi) and GentleMACS Octo Dissociator (Miltenyi). Cells were activated in vitro using 15 μl/ml anti-CD3/28-coated Dynabeads (Thermofisher) in R10 for 48h. Four hours prior to harvest, Golgi Stop was added at 1 μl/ml and flow cytometry staining was performed as described above.
Patient tumor-derived single cell data analyses
Three independent single-cell RNA-sequencing data of human melanoma and human lung carcinoma TIL were reanalyzed using BBrowser2 (BioTuring) (32). The third dataset deposited by Guo et al. (37) consists of 9,055 single T cells from 14 non-small cell lung cancer (NSCLC) patients formed into sixteen clusters through the SMART-Seq2 protocol described by Picelli et al. (47). Utilizing Bioturing’s single cell analysis workflow (version 2.10.30) described by Le et al. (32), we queried FCGR2B within cell types at the tumor. Statistical analysis was performed with GraphPad Prism 9. Datasets are available on the GEO database under accession numbers: GSE120575, GSE123139, and GSE99254 respectively.
In vitro stimulation of human PBMCs in presence of checkpoint inhibitor antibodies
PBMCs were cultured ex vivo at a concentration of 106 cells/ml containing 15 μl/ml anti-CD3/CD28-coated Dynabeads (Thermofisher) for 5 days in R10 media with different concentrations (5 mg/ml, 10 mg/ml, 15 mg/ml, 20 mg/ml) of anti-PD-1 (BioXcell, clone J116) or anti-CTLA4 (BioXcell, clone BN13) monoclonal antibodies. Cells were then re-stimulated with PMA (1 mg/ml) and Ionomycin (0.7 mg/ml) (MilliporeSigma) and Golgi Stop (1 μl/ml) for 4h. After staining for FcγRIIB and cell lineage markers, cells were stained intracellularly for cytokines (TNF, IFNγ) and Ki-67. Apoptosis and cell death were also assessed through caspase 3/7 and 7-AAD staining using the caspase 3/7 kit (Thermofisher). Isotype controls were initially used and no differences between isotype-treated wells and untreated wells were observed. Thus, untreated controls were used in subsequent experiments. Samples without PMA/Ionomycin were analyzed as unstimulated controls, stimulated samples without anti-PD-1 or anti-CTLA4 were analyzed as untreated controls. For samples treated with anti-PD-1 or anti-CTLA4, the best response dose was tabulated as defined by Kamphorst et al. (48) and used for analysis.
Anti-PD-1 F(ab) generation
F(ab) fragments were generated from the anti-PD-1 monoclonal antibody Nivolumab (McKesson) using the Pierce F(ab) Preparation Kit (Thermofisher). F(ab) fragments were purified and concentrated to 1 mg/ml. F(ab) quantification was assessed using absorbance at 280 nm by NanoDrop (Thermofisher) and validated through gel imaging and western blotting using an anti-IgG4 rabbit Fc antibody conjugated to HRP (ab99823) (Abcam) at a 1:10,000 dilution. The anti-IgG4 rabbit Fc antibody was used to control for contamination by Fc fragments. Intact PD-1 mAb (Nivolumab) and elution fractions with undigested mAb and Fc fragments were used for comparison.
Ex vivo culture with nivolumab mAb and F(ab) fragments
Following F(ab) digestion and purification, PBMCs were cultured in the presence of anti-PD-1 mAb (Nivolumab) or anti-PD-1-F(ab) ex vivo as described above. After extracellular staining, cells were stained for cytokines, Ki-67, or caspase 3/7 and 7AAD. IgG4 isotype controls were initially used and no differences between isotype-treated wells and untreated wells were observed. Thus, untreated controls were used in subsequent experiments. Samples without PMA/Ionomycin were analyzed as unstimulated controls, while stimulated samples untreated with anti-PD-1 mAb or anti-PD-1 F(ab) were analyzed as untreated controls. For samples treated with anti-PD-1 mAb or anti-PD-1 F(ab), the best response dose was tabulated according to Kamphorst et al. (48) and used for analysis.
Mice
8–12wk C57BL/6J male mice were acquired from The Jackson Laboratory. OT-I TCR transgenic mice were purchased from The Jackson Laboratory and crossed with Thy1.1 animals (from Jackson Laboratory). B6.Cg-Thy1a/CyTg(TcraTcrb)8Rest/J transgenic mice possessing TCRs specific for premelanosome (pmel-1, gp100) were acquired from The Jackson Laboratory. The protocol (PROTO201700558) was approved by the Emory University IACUC. All surgery was performed under general anesthesia with maximum efforts made to minimize suffering. All animals were housed in specific pathogen-free animal facilities at Emory University.
Murine cancer cell lines
B16 melanoma cell line engineered to express the OVA epitope was provided by Dr. Y. Fu, University of Texas Southwest, Dallas, TX (49). B16-F10 melanoma cells engineered to express the hgp100 protein were kindly provided by Dr. N. Restifo (NIH) (17). The MC38 cell line was a gift from Dr. H. Kissick (Emory University). The Lewis lung carcinoma (LLC) cell line expressing OVA was provided by Dr. Gabrilovich, Wistar Institute (50). B16-hgp100 and B16-OVA cells were cultured in RPMI1640; LLC-OVA, and MC38 were cultured in DMEM for LLC and MC38 (Sigma). Guidelines for LLC-OVA and MC38 culture and cryopreservation outlined by the American Type Culture Collection were followed. Guidelines for B16 culture and cryopreservation outlined by the Restifo laboratory were followed (51).
Syngeneic mouse melanoma tumor challenge
Cancer cells were trypsinized, washed with cold PBS, and filtered prior to cancer cell inoculation (51). 3×105 B16-hgp100, B16-OVA, 4×105 LLC-OVA, or 5×105 MC38 cells were injected in PBS subcutaneously into the right flank. Tumor volume was monitored using electronic calipers and tumor size was calculated using the formula: tumor volume (mm3) = (L × W2)/2. After one week, tumor-bearing mice were randomized to experimental groups. Animals were sacrificed when tumors reached IACUC endpoint (2 cm in either dimension).
Adoptive transfers
WT OT-I, Fcgr2b−/− OT-I, or WT pmel-1 splenocytes were harvested from 8–12wk old mice. Cells were counted using a Nexcelom Cellometer(Nexcelom Bioscience) and stained with CD8- BV785, CD4-PacBlue, Thy1.1- PerCP, Vα2- FITC, Vβ5- PE, and Vβ13-PE (Biolegend). OT-I frequency was determined by Vα2/Vβ5 co-expression,pmel-1 frequency was determined by Vα2/Vβ13 co-expression. 106 OT-I T cells or pmel-1 T cells were transferred intravenously into C57BL/6J mice seven days post tumor challenge.
PD-1 and FcγRIIB blockade
For single PD-1 blockade experiments, C57BL/6J mice were injected as indicated with 200 μg of anti-PD-1 ip (RMP1–14, BioXCell). For dual blockade experiments, mice given B16-gp100 were treated on days 2, 4, 6, and 8 post pmel-1 cell adoptive transfer. Mice given LLC-OVA were treated three times a week starting one week after tumor inoculation until mice reached endpoint. Mice given MC38 were treated on days 7, 9, and 11 post tumor challenge. PD-1 antibody or the IgG2a isotype control (2A3, BioXCell) was administered with 200 μg of the FcγRIIB blocking antibody (2.4G2, BioXCell) or the respective isotype, IgG2b (LTF-2, BioXCell). Mouse FcγRIIB antibody specificity was previously verified as described by Morris et al. wherein the anti-mouse clone 2.4G2 (which is known to bind to FcγRIIB and FcγRIII) does not stain Fcgr2b−/− T cells, demonstrating that the staining observed on WT CD8+ T cells is indicative of the presence of FcγRIIB (35). 2.4G2 specificity was also verified through RNA-sequencing in that sorted 2.4G2+ CD8+ T cells expressed more Fcgr2b transcripts than 2.4G2− CD8+ T cells. To confirm specificity of FcγRIIB blockade (Fig. 8), the non-crossreactive FcγRIIB antibody clone, AT-128, was used as well at the same dose and timing as clone 2.4G2. The antagonistic FcγRIIB-specific antibody clone AT-128 was produced using hybridoma technology and identifying clones that bound to FcγRIIB but not FcγRI or FcγRIII as described by Williams et al. (52), this antibody or the respective isotype was used in the dual blockade experiments in the MC38 model. The AT-128 antibody was a kind gift from Dr. M. Cragg (University of Southhampton).
Murine flow cytometry staining
Spleen, draining lymph node (right inguinal proximal to tumor), and tumor cells from mice were processed to cell suspensions, blood underwent red blood cell (RBC) lysis prior to staining. The samples were then stained with extracellular and intracellular markers with antibodies listed in Data File S2. For cytokine staining, splenocytes were ex vivo stimulated at 37°C for 4h with 10 nM OVA257–264 (SIINFEKL) peptide (B16-OVA),1 μM SIINFEKL peptide (LLC-OVA), or 10 μM p15e KSPWFTTL (MC38) and 10 μg/mL GolgiPlug (BD Biosciences). For pmel-1 experiments, cells were incubated at 37°C for 4h in the presence of 30 ng/mL PMA, 400 ng/mL Ionomycin, and 10 μg/mL GolgiPlug (BD Biosciences). After 4h, cells were processed, fixed, and stained for flow cytometry analyses. Samples without peptide were analyzed for unstimulated controls. Pre-incubation experiments with the anti-PD-1 therapeutic antibody clone used in mice in vivo (RMP1–14) were performed to ensure that the staining anti-PD1 and the therapeutic anti-PD-1 did not compete for the same epitope (fig. S3G-3H).
Statistics
Non-parametric, unpaired t-test (Mann-Whitney) was used to compare cell populations between groups, Wilcoxon matched-pairs rank tests were performed to compare subsets within the same donor. One-way ANOVA with multiple comparisons (Kruskal-Wallis) was performed when comparing multiple groups. For normalization, the baseline value for each mouse or patient sample served as the denominator and fold change was calculated relative to baseline. Survival data was plotted on Kaplan-Meier curves and a log-rank (Mantel-Cox) test was performed. For pie graphs, chi-square test was used. For tumor growth curves, area under the curve was calculated and then an unpaired T test was used to calculate accompanying p-value. Tumor size and weight were calculated using a Mann-Whitney t-test. The ROUT outliers test was used to determine any outliers. All analyses were done using Prism 9 (GraphPad Software). In all legends and figures, mean ± SEM is shown, and *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Dr. M. Cragg for the kind gift of the AT-128 antibody as well as Dr. H. Kissick for the contribution of the MC-38 cell line. Drs. K. Delman, C. Farley, A. Morris, and Ford lab members for helpful discussions on this work.
Funding
This work was supported by awards AI073707 (to M.L.F.) and Society of Surgical Oncology Young Investigator Award (to M.C.L.). K.B.B. was supported by NIH fellowship F31CA271764. M.T. was supported by a Surgical Oncology Melanoma Research Fellowship. Research reported in this publication was supported in part by the Cancer Tissue and Pathology shared resource of Winship Cancer Institute of Emory University and NIH/NCI under award number P30AC138292. This study was supported in part by the Emory Flow Cytometry Core (EFCC), one of the Emory Integrated Core Facilities (EICF), and is subsidized by the Emory University School of Medicine. Additional support was provided by the National Center for Georgia Clinical & Translational Science Alliance of the National Institutes of Health under Award Number UL1TR002378.
Footnotes
Competing interests
K.B.B. and M.L.F are coinventors on a provisional patent (63/423,098, Emory University) entitled: Methods to Block FcgRIIB and PD-1 on CD8 T Cells related to this work. M.L.F has consulted for Veloxis Pharmaceuticals and Sanofi. M.C.L. has consulted for Merck. R.R.K. is a current employee of Daiichi Sankyo. C.M.P. has received research funding from Obsidian, Lycera, and Thermo Fisher Scientific, and is the cofounder of Ares Immunotherapy. At the time of the study, all authors were employed at Emory University. The other authors declare that they have no competing interests.
Conflict of interest: All authors have declared that no conflict of interest exists.
Data and materials availability
All data associated with this study are present in the paper or supplementary materials.
References
- 1.Yun S, Vincelette ND, Green MR, Wahner Hendrickson AE, Abraham I, Targeting immune checkpoints in unresectable metastatic cutaneous melanoma: a systematic review and meta-analysis of anti-CTLA-4 and anti-PD-1 agents trials. Cancer medicine 5, 1481–1491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gettinger SN, Horn L, Gandhi L, Spigel DR, Antonia SJ, Rizvi NA, Powderly JD, Heist RS, Carvajal RD, Jackman DM, Sequist LV, Smith DC, Leming P, Carbone DP, Pinder-Schenck MC, Topalian SL, Hodi FS, Sosman JA, Sznol M, McDermott DF, Pardoll DM, Sankar V, Ahlers CM, Salvati M, Wigginton JM, Hellmann MD, Kollia GD, Gupta AK, Brahmer JR, Overall Survival and Long-Term Safety of Nivolumab (Anti-Programmed Death 1 Antibody, BMS-936558, ONO-4538) in Patients With Previously Treated Advanced Non-Small-Cell Lung Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 33, 2004–2012 (2015) 10.1200/JCO.2014.58.3708). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gettinger S, Rizvi NA, Chow LQ, Borghaei H, Brahmer J, Ready N, Gerber DE, Shepherd FA, Antonia S, Goldman JW, Juergens RA, Laurie SA, Nathan FE, Shen Y, Harbison CT, Hellmann MD, Nivolumab Monotherapy for First-Line Treatment of Advanced Non-Small-Cell Lung Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 34, 2980–2987 (2016) 10.1200/JCO.2016.66.9929). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lugowska I, Teterycz P, Rutkowski P, Immunotherapy of melanoma. Contemp Oncol (Pozn) 22, 61–67 (2018) 10.5114/wo.2018.73889). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J, Dummer R, Ferrucci PF, Smylie M, Hogg D, Hill A, Márquez-Rodas I, Haanen J, Guidoboni M, Maio M, Schöffski P, Carlino MS, Lebbé C, McArthur G, Ascierto PA, Daniels GA, Long GV, Bastholt L, Rizzo JI, Balogh A, Moshyk A, Hodi FS, Wolchok JD, Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 381, 1535–1546 (2019); published online Epub2019/10/17 ( 10.1056/NEJMoa1910836). [DOI] [PubMed] [Google Scholar]
- 6.Mansfield AS, Markovic SN, Novel therapeutics for the treatment of metastatic melanoma. (2009). [DOI] [PubMed] [Google Scholar]
- 7.Wasif N, Bagaria SP, Ray P, Morton DL, Does metastasectomy improve survival in patients with Stage IV melanoma? A cancer registry analysis of outcomes. Journal of surgical oncology 104, 111–115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS, Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. New England Journal of Medicine 372, 2006–2017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodi FS, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Cowey CL, Lao CD, Schadendorf D, Wagstaff J, Dummer R, Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. The Lancet Oncology 19, 1480–1492 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Carlino MS, Larkin J, Long GV, Immune checkpoint inhibitors in melanoma. The Lancet 398, 1002–1014 (2021); published online Epub2021/09/11/ ( 10.1016/S0140-6736(21)01206-X). [DOI] [PubMed] [Google Scholar]
- 11.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J, Dummer R, Ferrucci PF, Smylie M, Butler MO, Hill A, Márquez-Rodas I, Haanen J, Guidoboni M, Maio M, Schöffski P, Carlino MS, Lebbé C, McArthur G, Ascierto PA, Daniels GA, Long GV, Bas T, Ritchings C, Larkin J, Hodi FS, Long-Term Outcomes With Nivolumab Plus Ipilimumab or Nivolumab Alone Versus Ipilimumab in Patients With Advanced Melanoma. J. Clin. Oncol. 40, 127–137 (2022); published online EpubJan 10 ( 10.1200/jco.21.02229). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang AC, Orlowski RJ, Xu X, Mick R, George SM, Yan PK, Manne S, Kraya AA, Wubbenhorst B, Dorfman L, A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nature medicine 25, 454–461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nimmerjahn F, Ravetch JV, Fcγ receptors as regulators of immune responses. Nature Reviews Immunology 8, 34–47 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Smith KG, Clatworthy MR, FcγRIIB in autoimmunity and infection: evolutionary and therapeutic implications. Nature Reviews Immunology 10, 328–343 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roghanian A, Cragg MS, Frendéus B, Resistance is futile: Targeting the inhibitory FcγRIIB (CD32B) to maximize immunotherapy. Oncoimmunology 5, e1069939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim SH, Vaughan AT, Ashton-Key M, Williams EL, Dixon SV, Chan HC, Beers SA, French RR, Cox KL, Davies AJ, Fc gamma receptor IIb on target B cells promotes rituximab internalization and reduces clinical efficacy. Blood, The Journal of the American Society of Hematology 118, 2530–2540 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Vaughan AT, Iriyama C, Beers SA, Chan CH, Lim SH, Williams EL, Shah V, Roghanian A, Frendéus B, Glennie MJ, Inhibitory FcγRIIb (CD32b) becomes activated by therapeutic mAb in both cis and trans and drives internalization according to antibody specificity. Blood, The Journal of the American Society of Hematology 123, 669–677 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Clynes RA, Towers TL, Presta LG, Ravetch JV, Inhibitory Fc receptors modulate in vivo cytoxicity against tumor targets. Nat. Med. 6, 443–446 (2000); published online Epub2000/04/01 ( 10.1038/74704). [DOI] [PubMed] [Google Scholar]
- 19.Cohen-Solal JF, Cassard L, Fournier EM, Loncar SM, Fridman WH, Sautès-Fridman C, Metastatic melanomas express inhibitory low affinity fc gamma receptor and escape humoral immunity. Dermatology research and practice 2010, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahan R, Sega E, Engelhardt J, Selby M, Korman AJ, Ravetch JV, FcγRs modulate the anti-tumor activity of antibodies targeting the PD-1/PD-L1 axis. Cancer cell 28, 285–295 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Leclerc J, Plater C, Fridman W, The role of the Fc receptor (FcR) of thymus-derived lymphocytes I. Presence of FcR on cytotoxic lymphocytes and absence of direct role in cytotoxicity. European journal of immunology 7, 543–548 (1977). [DOI] [PubMed] [Google Scholar]
- 22.Anderson CL, Grey HM, Receptors for aggregated IgG on mouse lymphocytes: their presence on thymocytes, thymus-derived, and bone marrow-derived lymphocytes. The Journal of experimental medicine 139, 1175–1188 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris AB, Farley CR, Pinelli DF, Adams LE, Cragg MS, Boss JM, Scharer CD, Fribourg M, Cravedi P, Heeger PS, Signaling through the Inhibitory Fc Receptor FcγRIIB Induces CD8+ T Cell Apoptosis to Limit T Cell Immunity. Immunity 52, 136–150. e136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris AB, Pinelli DF, Liu D, Wagener M, Ford ML, Memory T cell-mediated rejection is mitigated by FcγRIIB expression on CD8(+) T cells. Am. J. Transplant. 20, 2206–2215 (2020); published online EpubAug ( 10.1111/ajt.15837). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farley CR, Morris AB, Tariq M, Bennion KB, Potdar S, Kudchadkar R, Lowe MC, Ford ML, FcγRIIB is a T cell checkpoint in antitumor immunity. JCI Insight 6, (2021); published online Epub02/22/ ( 10.1172/jci.insight.135623). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starbeck-Miller GR, Badovinac VP, Barber DL, Harty JT, Cutting edge: Expression of FcγRIIB tempers memory CD8 T cell function in vivo. The Journal of Immunology 192, 35–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin J-X, Konieczny BT, Im SJ, Freeman GJ, Leonard WJ, Kissick HT, Ahmed R, Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1+ Stem-like CD8+ T Cells during Chronic Infection. Immunity 51, 1043–1058.e1044 (2019) 10.1016/j.immuni.2019.11.002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starbeck-Miller GR, Badovinac VP, Barber DL, Harty JT, Cutting edge: Expression of FcgammaRIIB tempers memory CD8 T cell function in vivo. J Immunol 192, 35–39 (2014); published online EpubJan 1 ( 10.4049/jimmunol.1302232). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D, Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. U. S. A. 103, 3304–3309 (2006); published online EpubFeb 28 ( 10.1073/pnas.0511137103). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wherry EJ, Ha S-J, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R, Molecular Signature of CD8+ T Cell Exhaustion during Chronic Viral Infection. Immunity 27, 670–684 (2007); published online Epub2007/10/26/ ( 10.1016/j.immuni.2007.09.006). [DOI] [PubMed] [Google Scholar]
- 31.Veri MC, Gorlatov S, Li H, Burke S, Johnson S, Stavenhagen J, Stein KE, Bonvini E, Koenig S, Monoclonal antibodies capable of discriminating the human inhibitory Fcgamma-receptor IIB (CD32B) from the activating Fcgamma-receptor IIA (CD32A): biochemical, biological and functional characterization. Immunology 121, 392–404 (2007); published online EpubJul ( 10.1111/j.1365-2567.2007.02588.x). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le T, Phan T, Pham M, Tran D, Lam L, Nguyen T, Truong T, Vuong H, Luu T, Phung N, Pham N, Nguyen T, Pham O, Nguyen A, Nguyen H, Tran H, Tran L, Nguyen HA, Tran T, Nguyen N, Tran N, Boysen C, Nguyen U, Pham V, Kim T, Pham N, Gill T, Pham S, BBrowser: Making single-cell data easily accessible. bioRxiv, 2020.2012.2011.414136 (2020) 10.1101/2020.12.11.414136). [DOI] [Google Scholar]
- 33.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, Freeman SS, Reuben A, Hoover PJ, Villani A-C, Ivanova E, Portell A, Lizotte PH, Aref AR, Eliane J-P, Hammond MR, Vitzthum H, Blackmon SM, Li B, Gopalakrishnan V, Reddy SM, Cooper ZA, Paweletz CP, Barbie DA, Stemmer-Rachamimov A, Flaherty KT, Wargo JA, Boland GM, Sullivan RJ, Getz G, Hacohen N, Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013.e1020 (2018) 10.1016/j.cell.2018.10.038). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, van den Braber M, Rozeman EA, Haanen JBAG, Blank CU, Horlings HM, David E, Baran Y, Bercovich A, Lifshitz A, Schumacher TN, Tanay A, Amit I, Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789.e718 (2019) 10.1016/j.cell.2018.11.043). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris AB, Farley CR, Pinelli DF, Adams LE, Cragg MS, Boss JM, Scharer CD, Fribourg M, Cravedi P, Heeger PS, Ford ML, Signaling through the Inhibitory Fc Receptor FcgammaRIIB Induces CD8(+) T Cell Apoptosis to Limit T Cell Immunity. Immunity 52, 136–150 e136 (2020); published online EpubJan 14 ( 10.1016/j.immuni.2019.12.006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, Sharpe AH, Freeman GJ, Germain RN, Nakaya HI, Xue H-H, Ahmed R, Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016); published online Epub2016/09/01 ( 10.1038/nature19330). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R, Gao R, Zhang L, Dong M, Hu X, Ren X, Kirchhoff D, Roider HG, Yan T, Zhang Z, Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24, 978–985 (2018); published online Epub2018/07/01 ( 10.1038/s41591-018-0045-3). [DOI] [PubMed] [Google Scholar]
- 38.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, Manos M, Gjini E, Kuchroo JR, Ishizuka JJ, Collier JL, Griffin GK, Maleri S, Comstock DE, Weiss SA, Brown FD, Panda A, Zimmer MD, Manguso RT, Hodi FS, Rodig SJ, Sharpe AH, Haining WN, Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nature Immunology 20, 326–336 (2019); published online Epub2019/03/01 ( 10.1038/s41590-019-0312-6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D, Xia J, Kwon JYH, Nevin J, Herbst RH, Yanai I, Rozenblatt-Rosen O, Kuchroo VK, Regev A, Anderson AC, A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell 166, 1500–1511.e1509 (2016); published online EpubSep 8 ( 10.1016/j.cell.2016.08.052). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, Utzschneider DT, von Hoesslin M, Cullen JG, Fan Y, Eisenberg V, Wohlleber D, Steiger K, Merkler D, Delorenzi M, Knolle PA, Cohen CJ, Thimme R, Youngblood B, Zehn D, TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019); published online EpubJul ( 10.1038/s41586-019-1326-9). [DOI] [PubMed] [Google Scholar]
- 41.Yao C, Sun H-W, Lacey NE, Ji Y, Moseman EA, Shih H-Y, Heuston EF, Kirby M, Anderson S, Cheng J, Khan O, Handon R, Reilley J, Fioravanti J, Hu J, Gossa S, Wherry EJ, Gattinoni L, McGavern DB, O’Shea JJ, Schwartzberg PL, Wu T, Single-cell RNA-seq reveals TOX as a key regulator of CD8(+) T cell persistence in chronic infection. Nature immunology 20, 890–901 (2019) 10.1038/s41590-019-0403-4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim SH, Vaughan AT, Ashton-Key M, Williams EL, Dixon SV, Chan HTC, Beers SA, French RR, Cox KL, Davies AJ, Potter KN, Mockridge CI, Oscier DG, Johnson PWM, Cragg MS, Glennie MJ, Fc gamma receptor IIb on target B cells promotes rituximab internalization and reduces clinical efficacy. Blood 118, 2530–2540 (2011) 10.1182/blood-2011-01-330357). [DOI] [PubMed] [Google Scholar]
- 43.Simpson AP, Roghanian A, Oldham RJ, Chan HTC, Penfold CA, Kim HJ, Inzhelevskaya T, Mockridge CI, Cox KL, Bogdanov YD, James S, Tutt AL, Rycroft D, Morley P, Dahal LN, Teige I, Frendeus B, Beers SA, Cragg MS, FcγRIIB controls antibody-mediated target cell depletion by ITIM-independent mechanisms. Cell Rep. 40, 111099 (2022); published online EpubJul 19 ( 10.1016/j.celrep.2022.111099). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arlauckas SP, Garris CS, Kohler RH, Kitaoka M, Cuccarese MF, Yang KS, Miller MA, Carlson JC, Freeman GJ, Anthony RM, Weissleder R, Pittet MJ, In vivo imaging reveals a tumor-associated macrophage-mediated resistance pathway in anti-PD-1 therapy. Sci. Transl. Med. 9, eaal3604 (2017) 10.1126/scitranslmed.aal3604). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, Daëron M, Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725 (2009); published online Epub2009/04/16/ ( 10.1182/blood-2008-09-179754). [DOI] [PubMed] [Google Scholar]
- 46.Yu J, Song Y, Tian W, How to select IgG subclasses in developing anti-tumor therapeutic antibodies. J. Hematol. Oncol. 13, 45 (2020); published online Epub2020/05/05 ( 10.1186/s13045-020-00876-4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R, Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014); published online Epub2014/01/01 ( 10.1038/nprot.2014.006). [DOI] [PubMed] [Google Scholar]
- 48.Kamphorst AO, Pillai RN, Yang S, Nasti TH, Akondy RS, Wieland A, Sica GL, Yu K, Koenig L, Patel NT, Behera M, Wu H, McCausland M, Chen Z, Zhang C, Khuri FR, Owonikoko TK, Ahmed R, Ramalingam SS, Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1–targeted therapy in lung cancer patients. Proceedings of the National Academy of Sciences 114, 4993 (2017) 10.1073/pnas.1705327114). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown DM, Fisher TL, Wei C, Frelinger JG, Lord EM, Tumours can act as adjuvants for humoral immunity. Immunology 102, 486–497 (2001); published online Epub2001/04/01 ( 10.1046/j.1365-2567.2001.01213.x). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu T, Ramakrishnan R, Altiok S, Youn JI, Cheng P, Celis E, Pisarev V, Sherman S, Sporn MB, Gabrilovich D, Tumor-infiltrating myeloid cells induce tumor cell resistance to cytotoxic T cells in mice. J. Clin. Invest. 121, 4015–4029 (2011); published online EpubOct ( 10.1172/jci45862). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Overwijk WW, Restifo NP, B16 as a mouse model for human melanoma. Curr Protoc Immunol Chapter 20, Unit 20 21 (2001); published online EpubMay ( 10.1002/0471142735.im2001s39). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams EL, Tutt AL, French RR, Chan HTC, Lau B, Penfold CA, Mockridge CI, Roghanian A, Cox KL, Verbeek JS, Glennie MJ, Cragg MS, Development and characterisation of monoclonal antibodies specific for the murine inhibitory FcγRIIB (CD32B). European Journal of Immunology 42, 2109–2120 (2012) 10.1002/eji.201142302). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or supplementary materials.