Alterations leading to unconstrained protein kinase A signaling, regardless of the presence or absence of PRKACA fusions, drive the phenotypes of fibrolamellar hepatocellular carcinoma, reshaping understanding of the pathogenesis of this rare liver cancer.
Abstract
Fibrolamellar hepatocellular carcinoma (FLC) is a rare liver cancer that is driven by the fusion of DNAJB1 and PRKACA, the catalytic subunit of protein kinase A (PKA). PKA activity is controlled through regulatory proteins that both inhibit catalytic activity and control localization, and an excess of regulatory subunits ensures PRKACA activity is inhibited. Here, we found an increase in the ratio of catalytic to regulatory units in FLC patient tumors driven by DNAJB1::PRKACA using mass spectrometry, biochemistry, and immunofluorescence, with increased nuclear localization of the kinase. Overexpression of DNAJB1::PRKACA, ATP1B1::PRKACA, or PRKACA, but not catalytically inactive kinase, caused similar transcriptomic changes in primary human hepatocytes, recapitulating the changes observed in FLC. Consistently, tumors in patients missing a regulatory subunit or harboring an ATP1B1::PRKACA fusion were indistinguishable from FLC based on the histopathological, transcriptomic, and drug–response profiles. Together, these findings indicate that the DNAJB1 domain of DNAJB1::PRKACA is not required for FLC. Instead, changes in PKA activity and localization determine the FLC phenotype.
Significance: Alterations leading to unconstrained protein kinase A signaling, regardless of the presence or absence of PRKACA fusions, drive the phenotypes of fibrolamellar hepatocellular carcinoma, reshaping understanding of the pathogenesis of this rare liver cancer.
Introduction
Many cancers are driven by kinases that are mutated, overexpressed, or expressed as fusion proteins. For most of these, the pathways of pathogenesis are unknown or conjectural. Fibrolamellar hepatocellular carcinoma (FLC), a primary cancer of the liver, is driven by an oncokinase in the hepatocytes that is the fusion of the first exon of DNAJB1, a heat shock protein, to PRKACA, a catalytic subunit of protein kinase A (PKA; ref. 1). DNAJB1::PRKACA is the oncogenic driver, and the expression of catalytically active DNAJB1::PRKACA initiates FLC tumors in mice (2, 3), with tumors dying upon the elimination of DNAJB1::PRKACA (4, 5). FLC affects adolescents and young adults (6–8), and as a result, patients have a low mutational burden of 1.85 to 2.8 mutations/Mb (9, 10), with the deletion that creates DNAJB1::PRKACA as the only recurrent mutation. These findings make FLC a powerful system for studying the role of dysregulation of kinase activity in tumorigenesis.
It is unresolved whether transformation is the result of increased kinase expression or is instead the result of a structurally altered PKA catalytic subunit with novel properties. The activity of PKA is regulated by the formation of a holoenzyme consisting of two catalytic subunits, C, and two regulatory subunits, R (11, 12). There are four known isoforms of the R subunits, RIα, RIIα, RIβ, and RIIβ, with RIα and RIIα being the most abundant in the mammalian liver (13). The amino-terminus of R occludes the substrate binding site of C, thereby inhibiting its activity—inhibition that is relieved by cAMP-mediated dissociation of R from C (14, 15). In normal tissues, catalytic activity is inhibited in the basal state because there is an excess of R over C subunits (13, 16). Each R interacts with different A kinase–associated proteins, which serve to localize the holoenzyme (17). Free C that enters the nucleus is then bound by protein kinase inhibitor peptide (PKI), with sub-nanomolar affinity for the same substrate binding site as the regulatory subunits (18). The PKI-catalytic subunit complex is exported back to the cytosol, where C re-engages R (19–21).
In FLC, DNAJB1::PRKACA is expressed from one copy of chromosome 19 and native PRKACA from the other copy. As measured by Western blots, the expression of DNAJB1::PRKACA is usually greater than that of PRKACA (1, 22), consistent with the reads in the transcriptome (23). Additionally, there are decreased reads for the R subunit RIIβ transcript (23), and an increase of RNA and protein of RIα has been reported (22). It is unknown whether there is a net change in the ratio of expression level of the R to C subunits in FLC. Tumors with histological features that are the same as those of FLC are seen with alternative forms of dysregulation of PKA signaling, and among these are the loss of a regulatory component of the holoenzyme (24), fusions of the first exon of ATP1B1 to the PKA catalytic subunits PRKACA or PRKACB (25), and deletions of BAP1, with duplication of PRKACA (26).
Here, we analyze the dysregulation of PKA in FLC, using mass spectrometry, biochemistry, quantitative immunofluorescence, spatial transcriptional analysis and drug–response profiles. We determine the levels and subcellular distribution of C and R of PKA in FLC tumor and in adjacent nontransformed tissue. We find an increase in the ratio of C to R, with some of the RI sequestered into cytoplasmic condensates, most of which are devoid of C. This leads both to an increase of basal unconstrained catalytic activity and increased localization of catalytic subunit to the nucleus. In primary human hepatocytes, augmenting expression of active catalytic subunit with PRKACA, DNAJB1::PRKACA, or ATP1B1::PRKACA all produced the same redistribution of C, as assessed by microscopy, and transcriptomic changes as determined by bulk and single-cell spatial transcriptional analysis, as observed in patient FLC tumor tissue. We conclude that unconstrained kinase and its re-localization, regardless of the presence or absence of a structural adduct to PRKACA, drives the transformation phenotype of FLC.
Materials and Methods
Patient tissue processing
Under supervision of our Institutional Review Board approval (Rockefeller IRB #SSI 0797, SSI 0798) written informed consent was obtained from patients scheduled for tumor resection, and studies were conducted in accordance with recognized ethical guidelines (e.g., the Declaration of Helsinki, Council for International Organizations of Medical Sciences, Belmont Report, U.S. Common Rule). Since this is a rare disease, we accepted samples from all patients, without regard to age or sex. For each patient, the diagnosis of FLC was confirmed both by the demonstration of the DNAJB1::PRKACA fusion transcript by using RT-PCR and DNAJB1::PRKACA fusion protein by using Western blot (1). Tissue preparation protocols for the study were modified from Mertin and colleagues (27). Liver samples for proteome analysis were placed on dry ice immediately after the resection and stored at −80°C. For tissue lysis, small pieces (2–5 mm in diameter) were cut from the specimen and placed in microfuge tubes. The tube was placed in a liquid-nitrogen-cooled mini mortar and pestle set that accommodates microfuge tubes (Bel-Art; catalog # H37260-0100). The tissue was pulverized over liquid nitrogen. Ice-cold lysis buffer [8 mol/L urea, 75 mmol/L NaCl, 50 mmol/L Tris (pH 8.0), 1 mmol/L EDTA] supplemented with cOmplete EDTA-free protease inhibitor (Roche), and PhosSTOP phosphatase inhibitor (Roche) tablets were added to the pulverized tissue. The tube was then vortexed at maximum speed for 15 seconds, incubated on ice for 15 minutes, and then vortexed again at maximum speed for 15 seconds. The sample was then spun at 30,000 × relative centrifugal field (RCF) for 30 minutes. To avoid contamination by lipids, the microfuge tube was punctured near the bottom (above the pellet) and the supernatant decanted into another microfuge tube until the lipid layer reached the level of the puncture. All spins were performed at 4°C. The lysate was assayed for protein concentration using the BCA protein assay (Pierce).
Protein digestion
Proteins were precipitated using a chloroform/water/methanol method (28) to remove contaminant lipids. Pellets were dissolved in 8 mol/L urea (GE Healthcare), 50 mmol/L ammonium bicarbonate (AMBIC, Fluka Chemicals), and 10 mmol/L dithiothreitol (EMD Chemicals) in water. After dissolution in 8 mol/L urea, the samples were vigorously shaken at room temperature for 1 hour to reduce disulfide bonds. Iodoaceamide (Sigma) was added to 20 mmol/L and alkylation proceeded for 1 hour at room temperature in the dark. The samples were diluted with 50 mmol/L AMBIC and digested with lysyl endopeptidase (Wako) overnight at room temperature. The concentration of urea was adjusted to 1.5 mol/L using 50 mmol/L AMBIC, and the samples were further digested for 6 hours at room temperature, using Sequencing Grade Modified Trypsin (Promega). Digestion was stopped by acidification using either formic acid (Fluka) or trifluoroacetic acid (Thermo Fisher Scientific), and peptides were purified using high-capacity 30 mg Oasis HLB Cartridges (Waters), according to the manufacturer's specifications. The peptides were then tagged with tandem mass tags from the TMTpro 16plex Label Reagent Set (Thermo Fisher Scientific) following the manufacturer’s instructions.
LC-MS/MS-SPS-MS analysis
Peptides were fractionated using a Dionex 3000 UltiMate loading pump equipped with a 2.1 × 150 mm, 3.5-µm Xbridge C18 column (Waters). Solvent A consisted of 10 mmol/L ammonium hydroxide (Sigma-Aldrich) in water at pH 10, and solvent B consisted of 10 mmol/L ammonium hydroxide, 90% acetonitrile in water at pH 10. Peptides were separated across a 60-minute gradient, and 96 fractions were collected and pooled for a total of 24 fractions. The fractions were analyzed using a Fusion Lumos mass spectrometer with synchronous precursor population (SPS)-MS3 acquisition. Data were analyzed using Proteome Discoverer v.2.3. Spectra were queried against the human proteome with a 1% false discovery rate. An 80% SPS match was required for a result to be recorded.
Shotgun proteomics
In a previous study of this tumor, we performed Shotgun proteomics (34). The raw data from this study was used here both to quantify the relative levels of the catalytic and regulatory subunits (Supplementary Table S1) and to help select peptides for absolute quantification (AQUA). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (1) partner repository with the dataset identifier PXD046905.
Absolute quantification
The chosen peptides were synthesized (Thermo Fisher Scientific) with lysine at the carboxyl terminus labeled with a stable isotope (15N and 13C) and quantified by amino acid analysis to provide accurate quantitation of the synthesized peptides. The mass difference between the heavy lysine peptide and the endogenous cleaved peptide was 8 Da. Proteins were precipitated with ice-cold acetone. Pellets were dissolved in 8 mol/L urea, 50 mmol/L triethylamonium bicarbonate, and 10 mmol/L DTT. Reduction and alkylation with iodoacetamide were carried out at room temperature for 1 hour for each process. Alkylation was carried out in the dark. Proteins were digested with Lys-C and spiked with isotopically labeled AQUA peptides to a concentration of 11 fmol peptide/microgram of lysate. Samples were purified by reverse-phase microcolumns and analyzed by LC-MS/MS using a Q-Exactive HF mass spectrometer operating in positive ion PRM mode (the PRM method designed from injections of AQUA peptide without background). Peak area quantitation was performed using the Skyline software platform.
The peptides that were used to quantify the proteins are REIFDRYGEEVK (DNAJB1::PRKACA junction), GNAAAAK (PRKACA), ATEQYYAMK (PRKACB), GIVSLSDILQALVLTGGEK (PRKACG), HNIQALLK and VSILESLDK (RIα), VSILESLEK (RIβ), RNISHYEEQLVK and SLEVSERMK (RIIα), and TDDQRNRLQEACK (RIIβ). For RIα and RIIα, in which more than one peptide was used, the average of two peptides was used for calculating the sum of subunits.
Data were analyzed using Proteome Discoverer v. 2.5. Spectra were queried against the human proteome concatenated with the chimeric fusion protein along with common contaminants. Peptides used for quantitation were selected based on having no missed cleavages, oxidized residues, or missing values throughout the dataset.
Protein kinase A activity assay
PKA activity was measured using a calorimetric PKA kinase activity assay (Invitrogen, catalog #EIAPKA) based upon a solid-phase ELISA method using an antibody against phosphorylated Kemptide. The measurements were performed at dilution series of tissue extract lysed in Tris pH 7.4 with no detergent. The catalytic activity was measured as the initial slope of absorbances at different dilutions, and the slope for the standard in various dilutions was used to determine units of activity. Total activity is reported as PKA activity after stimulation with 20 µmol/L cAMP, while free PKA activity represents basal activity in the absence of cAMP stimulation. PKI5-24 at a concentration of 5 µmol/L was used to assure all calculated activity was specific to PKA. All activity measurements were normalized for the concentration of protein extracted.
cAMP assay
The level of cAMP was measured using a calorimetric cAMP assay kit ab290713 (Abcam, Cambridge, UK) based on a competitive ELISA method. Measurements were performed following the kit’s manual on cryo-ground issue.
Separation of regulatory and catalytic subunits on DEAE-Sepharose columns
Flash frozen tissues were cryoground and lysed in room temperature lysis buffer (25 mmol/L Tris pH 7.4 and 5 mmol/L β-mercaptoethanol) supplemented with cOmplete EDTA-free protease inhibitor (Roche), and PhosSTOP phosphatase inhibitor (Roche). The samples were then spun at 20,000 × RCF for 10 minutes at 4°C, and the supernatant was collected without disturbing the lipid solution layer. The clarified cell extract was assayed for protein concentration using BCA. One milligram to 2 mg of total protein was mixed with 400 μL of a 1:1 DEAE-Sepharose slurry (GE Healthcare Life Sciences) in lysis buffer incubated with continuous rotation at 4°C for 2 hours. Subsequently, the lysate and DEAE-Sepharose mix were packed into mini spin columns. The flow-through was collected by centrifuging at 7,600 × RCF for 1 minute. After three washes with the lysis buffer, the catalytic subunit was eluted with increasing concentrations of cAMP (2 µmol/L, 20 µmol/L, and 200 µmol/L) in the lysis buffer. A final elution was performed using 1 mol/L NaCl. The PKA catalytic subunits were visualized and quantified by Western blot probing with an anti-PRKACA (D38C6) rabbit monoclonal antibody (Cell Signaling Technology) at 1:3,000 dilution and a goat anti-rabbit IgG A0545 (Sigma) antibody at 1:25,000 dilution.
Use of GST-PKIA to measure free catalytic subunit
Cleared lysate obtained from BL21 bacterial cells overexpressing GST-PKIA was incubated with Cytiva Glutathione Sepharose beads and purified as described above. Cleared supernatant from cryo-ground tumor and adjacent normal tissue, lysed in tris buffer (containing NP-40, 100 µmol/L ATP, and 1 mmol/L MgCl2, supplemented with protease and phosphatase inhibitors), was divided into halves. cAMP, 15 µmol/L, was added to one half of the treated supernatant and incubated with GST-PKIA-Gluseph beads for 30 minutes with rotation at 4°C. Flow-through was collected by centrifuging at 500 × RCF at 4°C for 5 minutes. The beads were washed four times with 50 mmol/L Tris pH 7.4, 100 µmol/L ATP, and 1 mmol/L MgCl2. Unbound PKA catalytic subunits were eluted by heating the samples in 1.1 × lithium dodecyl sulfate sample buffer at 75°C for 10 minutes. The catalytic subunits were visualized and quantified using Western blot as described above.
Immunofluorescence and proximity ligation assay
Staining
Frozen, optimal cutting temperature–embedded tissue was sectioned at −20°C to a thickness of 14 μm and mounted onto glass microscope slides (Fisherbrand Superfrost Plus). The mounted tissue was then fixed with 4% (wt/vol) paraformaldehyde in PBS for 15 minutes at room temperature. After washing with PBS three times each for 5 minutes, the tissue was permeabilized and blocked for 1 hour in blocking buffer: 0.1% (vol/vol) Triton X-100, 2.5% normal donkey serum, 2.5% normal goat serum, and 1% BSA (all from Sigma). Primary antibodies were then added in blocking buffer and incubated overnight at 4°C in a humid chamber. The tissue was then washed three times each for 5 minutes in PBS and then incubated with a secondary antibody diluted in blocking buffer for 1 hour at room temperature. The secondary antibody was washed three times each for 5 minutes. For the proximity ligation assay (PLA), Duolink In Situ Orange Kit (Sigma, DUO92007) was used in accordance with the manufacturer's instructions. Phalloidin Atto 488 (Sigma, 49409) was used to define the cell boundaries when needed. After staining the nuclei with either Hoechst (Thermo Fisher Scientific, H3569) or DAPI (Sigma, DUO82040), the tissue was mounted with ProLong Gold Antifade Mountant (Invitrogen).
Antibodies used in this study are monoclonal rabbit anti-PKA catalytic subunit as raised against the sequence surrounding Ser326 mapped to the C-terminus (PKA C-α, 1:200; #5842, Cell Signaling Technology), monoclonal mouse anti PKA regulatory subunit II α raised against amino acids 21 to 100 mapped near the N-terminus of RII α [PKA IIα reg (H-12), 1:200, sc-137220; Santa Cruz Biotechnology], monoclonal mouse anti-PKA regulatory subunit RI raised against amino acids 225 to 381 (PKA RI, 1:200, 610165, BD Biosciences), Goat anti-Rabbit IgG (H&L) secondary antibody, Alexa Fluor 568 (1:1,000, Invitrogen), Goat anti-Mouse IgG (H&L) highly cross-adsorbed secondary antibody, Alexa Fluor 488 (1:1,000, Invitrogen).
Image quantification
DAPI and immunostained (catalytic and regulatory subunits) tissue samples were imaged via confocal microscopy (Olympus FluoView FV3000, UPLXAPO 60× oil immersion objective, numerical aperture: 1.42). From DAPI images, z-stack planes containing full-size nuclei were identified, and the regions of interest (ROI) were drawn around cell peripheries and the nuclei. Between 50 and 250 ROIs were identified in each patient’s normal and tumor tissue samples. The average fluorescence intensity in each ROI was quantified in cellSens (Olympus Life Science), by using pixel offset subtraction by measuring the average pixel intensity in a dark region away from the tissue samples. For each patient sample, the average fluorescence intensity of each ROI was normalized by the ROI in the tumor sample with the maximum average fluorescence. The tumor and normal samples for each patient were stained and imaged on the same slide, with the same microscope illumination and collection parameters.
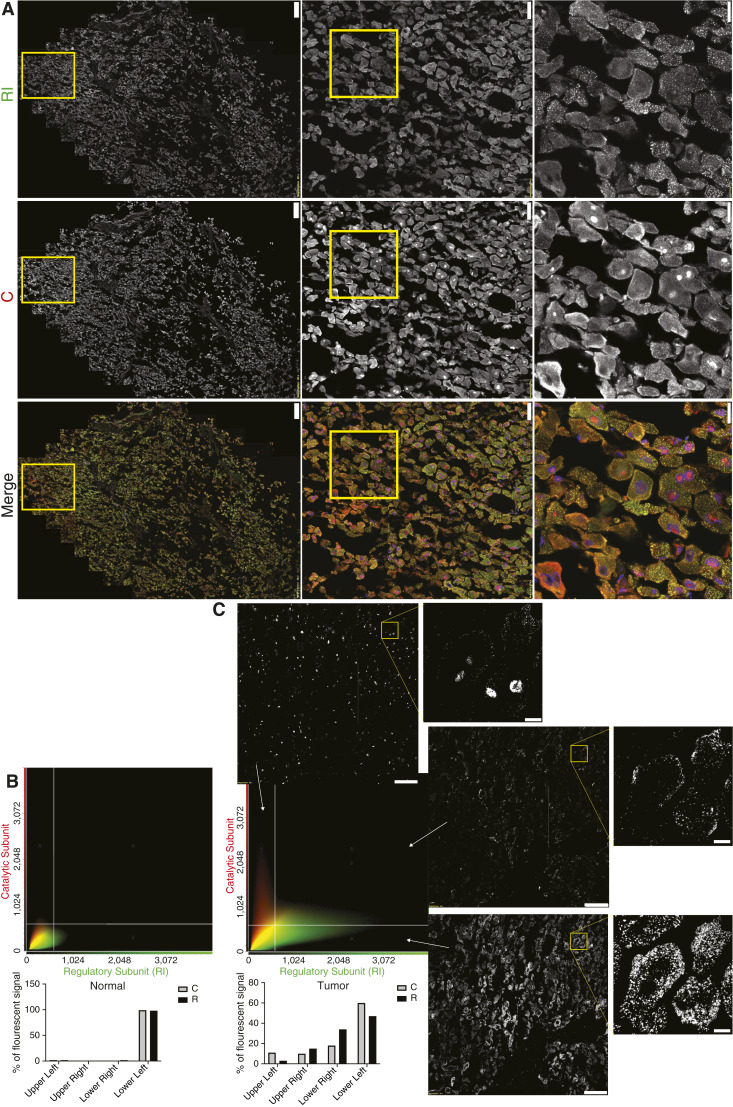
Scatter plots comparing pixel-by-pixel catalytic and regulatory subunit fluorescence intensity were generated in cellSens. A threshold intensity of 600 was used to segment the scatter plot into four different regions, with the upper right region representing individual pixels with both catalytic and regulatory subunit fluorescence above 600, the upper left with only catalytic subunit fluorescence greater than 600, and the lower right with only regulatory subunit fluorescence greater than 600. Masked images were generated from the pixels that satisfied the 600-threshold in each of these three regions. The total fluorescence in each threshold region was determined for both the catalytic and regulatory subunits by summing the intensity of all the pixels in the region. During this summation, the detector offset for each pixel was first removed. The percentage of fluorescence in each region for either catalytic or regulatory subunit was then determined by dividing the total fluorescence for each region by the total fluorescence of all four regions.
Proximity of C and R was quantified from the PLA images by first identifying spots with the segmentation function in cellSens. An automatic threshold with the object size of two or more pixels was used for segmentation. The total area of the identified PLA spots was then normalized to the total area of the cells in which the spots were identified.
MERFISH
The design of the gene panel, tissue preparation, and imaging were done according to manufacturer’ instructions (29). Frozen, optimal cutting temperature–embedded tissue was sectioned using a cryostat set to 10 μm thickness and mounted onto a functionalized 20-mm coverslip (MERSCOPE Slide Box, Vizgen, 10500001). The tissue was then fixed with 4% paraformaldehyde in PBS, washed with PBS, and stored in 70% ethanol at 4°C for at least 1 day. The cell boundary was stained using the MERSCOPE Cell Boundary Staining Kit (Vizgen, 10400009). The designed gene panel was synthesized by Vizgen. The RNA integrity values of the tissues were measured in the Agilent BioAnalyzer and TapeStation to confirm RNA quality. The samples were processed for spatial transcriptional analysis as previously described (29). The images were collected using a 60× oil immersion objective from multiple rounds of three-color imaging using 749, 638, and 546 nm from seven focal planes.
Primary human hepatocytes transduction
Plasmid construction
Genes expressing PRKACA, DNAJB1::PRKACA, DNAJB1::PRKACAK128H, and ATP1B1-PRKACA were amplified using Q5 high-fidelity DNA polymerase (New England Biolabs, NEB) and cloned into SCRPSY vector (accession KT368137) using HiFi DNA Assembly (NEB). The plasmids were grown in NEB Stable Competent E. coli under ampicillin (100 μg/mL). DNA was isolated using Plasmid Miniprep Kit (Zymo Research) or NucleoBond Xtra Midi EF (Takara Bio). Oligonucleotide primers were chemically synthesized by Integrated DNA Technologies. DNA sequences were confirmed using Genewiz.
Lentiviral production
HEK293 cells were seeded in a 150-mm dish in DMEM supplemented with 1% nonessential amino acids (Gibco) and 3% FBS (Sigma). Virus-containing media was collected at 24 hours (and at 48 and 72 hours, as needed) post transfection, filtered through a 0.45-μm filter (Millipore) and concentrated 50- to 100-fold overnight at 4°C using Lenti-X concentrator (Takara, cat. #631232) following the manufacturer’s instructions. Aliquots were stored at −80°C.
Lentiviral transduction of primary human hepatocytes (PHH; ref. 30) was done in hepatocyte culture medium (Lonza, CC-3199) supplemented with HCM SingleQuot (CC-4182) and 4 to 10 μg/mL polybrene for 6 to 16 hours, after which, the media was changed to fresh HCM. For the titration of lentiviruses, the PHHs were plated in 96-well plates at 50 thousand cells/well. The following morning, serial dilutions of lentivirus stocks were made in HCM, and 50 µL of each dilution was used to infect individual wells. The media was changed the following day. At 72 hours post infection, the media was changed to HCM with 4 mg/mL puromycin. After 3 days of puromycin selection, cell viability was measured using CellTiter-Glo (Promega). For the transcriptome studies, PHHs were plated and transduced in 6-well plates at a density of 1.5 million cells/well. Per construct, we used three wells as three replicates. We extracted the RNA 9 days after transduction. For the mouse studies, the transduced PHHs were implanted back into FNRG mice, as previously described (30). All mouse studies were performed with the approval of the Institutional Animal Care and Use Committee at the Rockefeller University.
RNA isolation, RNA sequencing, and bioinformatics
Total RNA from PHHs was extracted using the RNeasy Mini Kit (Qiagen). RNA concentration levels were measured using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific) in the 260/280 nm absorbance ratio. RNA integrity values were measured using an Agilent BioAnalyzer and TapeStation. Paired-end 2 × 150 nt RNA sequencing libraries were prepared using the TruSeq Stranded Total RNA sample preparation kit with the Ribo-Zero Gold Kit for rRNA and mt-rRNA depletion (Illumina). These libraries were sequenced in an Illumina NovaSeq SP instrument at 60 million reads per sample. Quality assessment and trimming were performed using FastQC v0.11.9, MultiQC v1.11 (31) and BBDuk (included in BBMap v38.95). Reads per transcript were quantified using Salmon (32) with the human reference genome hg38 and the EMSEMBL GRCh38.103 gene annotations as the template. Differential gene expression analysis was conducted in R v4.2.0 and RStudio 2022.07.1+554 using DESeq2 v1.36.0, excluding genes corresponding to rRNAs and mt-rRNAs. The results were analyzed in detail through PCA, tSNE, and UMAP plots, as well as heat maps, and box-scatter plots.
The expression level of each kinase was measured as the ratio of the expression of PRKACA in each condition over the expression of PRKACA in cells expressing the red fluorescent protein (RFP), that is, cells that were not transduced with additional C. If we normalize the level of expression of each kinase fusion to the level of PRKACA in the cells transduced with RFP, then the expression of ATP1B1::PRKACA was 315, DNAJB1::PRKACA was 46, DNAJB1::PRKACAK128H was 271, and PRKACA was 166.
Methods for drug response
The tissue samples obtained fresh from surgical resections on patients were processed as previously described (33).
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD043816, and the full transcriptomics is available via GEO accession number: GSE237697. All other raw data generated in this study are available upon request from the corresponding author.
Results
Catalytic subunit is increased relative to regulatory subunit in FLC
To quantify PKA subunits, we analyzed Tandem Mass Tag–based shotgun proteomics from eight pairs of FLC tumor and the adjacent normal tissue (34) and found increased C, RIα, and RIβ proteins in the FLC tumor and decreased RIIα and RIIβ subunits (Supplementary Table S1). AQUA (35) allows calibrated quantification of proteins in a complex mix. Peptides chosen for calibration were those detected in shotgun experiments, unique to the protein of interest and composed of Lys-C peptides (i.e., a lysine at the C terminus, but no internal lysines). The unique fusion junction peptide of DNAJB1::PRKACA proved detectable when digested with Lys-C in AQUA experiments. We used multiple peptides when possible, but due to homology, some proteins had a single unique identifying peptide. Calibration peptides were chosen for the PRKACA, PRKACB, PRKACG, DNAJB1::PRKACA, RIα, RIβ, RIIα, and RIIβ proteins. They bore isotopic labels and were synthesized with lysine at the carboxyl terminus and quantified by amino acid analysis.
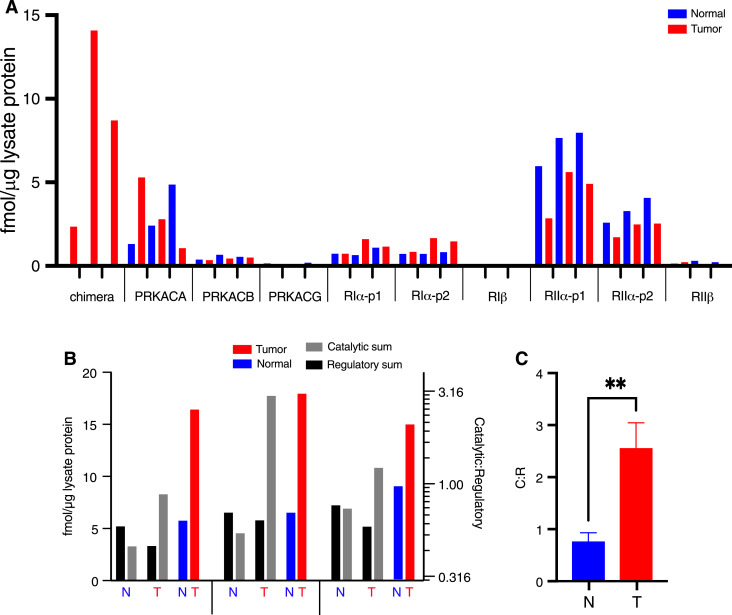
For the AQUA experiment, three FLC tumors and their matched normal liver samples were analyzed. The chimeric fusion peptide was not detected in normal liver and was detected in all FLC tumor samples. RIα was detected and increased in some patients, whereas RIβ was not detected, implying low abundance. Consistent with previous work (13), RIα and RIIα were more abundant than RIβ and RIIβ in liver tissue (Fig. 1A).
Figure 1.
Quantification of PKA subunits in FLC tumor and adjacent nontransformed tissues for three patients. A, AQUA of peptides resulting from the PKA catalytic and regulatory subunits. p1 and p2 refer to two different peptides for the same protein (see Materials and Methods). B, Comparing sum of catalytic subunits (PRKACA, DNAJB1::PRKACA, PRKACB, and PRKACG; gray) to sum of regulatory subunits (RIα, RIβ, RIIα, and RIIβ; black), as derived from the data presented in A (right log scale is the ratio of C:R). C, Mean of three patients with SD. Multiple unpaired t test analysis on the ratio of catalytic subunits to regulatory subunits measured using AQUA peptides in normal and tumor tissues. In A and B, the red bars are from tumor tissue and blue are from adjacent nontransformed tissue. **, P < 0.005.
To assess the absolute and relative amounts of global R and C, total PKA C subunit quantities were compared to total quantities of R subunits. The ratio of R:C subunits shifted from R > C in normal tissue to C > R in FLC tissue (Fig. 1B and C).
Unbound catalytic subunit is increased in FLC
The MS data led us to hypothesize that the increased ratio of catalytic to regulatory subunits leads to a higher amount of unbound C subunit. We tested this with two different biochemical assays, comparing FLC tumor and adjacent normal tissue.
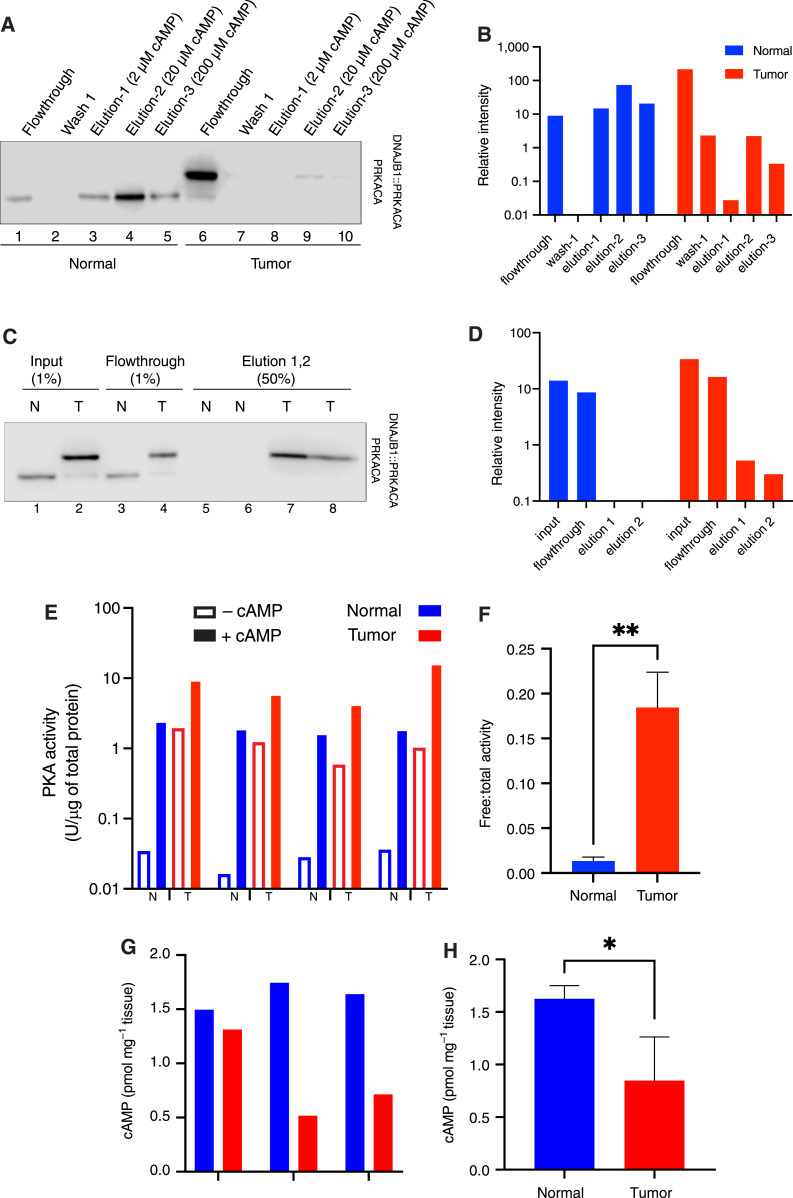
The first assay utilized DEAE-Sepharose to selectively bind holoenzyme R, thus capturing C bound to R (36). Unbound C is not captured; free C is quantified by the flowthrough of the column (37, 38). From normal tissue, most of the C was in the elution and was barely detectable in the flowthrough (Fig. 2A, lane 1). By contrast, in FLC tissue, there was much more C in the flowthrough relative to C eluted with cAMP (lane 6). This indicates that there is little free C in the normal tissue relative to free C in the FLC tissue (Fig. 2B). The addition of cAMP eluted additional C from the column in normal tissue (lanes 3–5), but barely detectable amounts of C were eluted by cAMP from the tumor tissue sample (lanes 8–10). This demonstrates a 24-fold greater free catalytic subunit in the tumor tissue than in the normal tissue (Fig. 2B).
Figure 2.
Free PKA catalytic subunit and activity level in tumor vs. normal liver. A, DEAE-Sepharose resin pulldown. Human FLC normal and tumor liver lysate were incubated with DEAE resin. Unbound PKA C subunits (non-holoenzyme) flow through the column. Three washes were performed, followed by elution with increasing concentrations of cAMP. The flowthroughs, first wash, and elution were visualized by immunoblotting using an antibody to the carboxyl terminus of the C subunit, which recognizes both PRKACA (bottom band) and DNAJB1::PRKACA (top band). The same percent of each fraction was loaded on the gel. B, Quantification of PRKACA and DNAJB1::PRKACA bands from the Western blot in A. Blue, normal; red, tumor. C, GST-PKIα pulldown. Lysate from tumor and normal tissue was incubated with GST-PKI immobilized on glutathione Sepharose beads. Flowthrough was collected, and after three washes, catalytic subunits captured by GST-PKI were eluted with excess arginine. The percent of each fraction loaded on the gel is shown on the top of each well. D, Quantification of both PRKACA and DNAJB1::PRKACA from the Western blot on C. The loading percent in each well was counted for the quantification. Blue, normal; red, tumor. E, Free PKA (cAMP-independent, basal) activity is higher in tumor (T) tissue lysate than in normal (N) liver. FLC tumor has higher total PKA activity (in the presence of cAMP) than normal liver tissue. Results of paired tumor and normal tissue from four different patients. The first three are flash frozen and lysed by cryogrinding; the sample on the right is fresh and homogenized tissue. F, The ratio of free-to-total PKA (cAMP-dependent) in the flash-frozen tissue in tumor (red) compared to normal liver (blue). G and H, cAMP levels in tissue samples from three different patients (G) and the average of the three patients (H). t test was done using PRISM. *, P < 0.05; **, P < 0.005. Note the log scale in vertical axes of B, D, and E.
The second assay relied on the binding of the endogenous PKI to unbound PKA C subunits. PKI shares a binding site on C with R but cannot competitively displace R (39). Thus, free C is determined by the amount retained on a PKI-resin. In the normal sample, ∼60% of the C subunit flowed through the PKI resin (Fig. 2C, lane 3, and D) relative to the input (lane 1), whereas for the FLC sample, ∼50% of C flowed through the resin (lane 4) relative to the input (lane 2). The addition of arginine, which competes for binding of C to PKI, did not elute additional C in the normal sample (lanes 5 and 6), but it did elute C in the FLC sample (lanes 7 and 8). These two assays demonstrate a substantial pool of free C in tumor that is much lower, or not detectable, in normal tissue.
Basal PKA activity is increased in FLC
To test if the increased ratio of C:R protein also increases basal PKA catalytic activity, we assessed samples from four FLC tumors and adjacent normal tissue. Baseline PKA activity without cAMP stimulation was ∼50-fold higher in FLC tumor tissue, regardless of the tissue lysis method or preservation state (Fig. 2E). Additionally, the ratio of free:total PKA activity was 10 times higher in the tumor tissue, indicating an elevated basal PKA activity in the FLC tumor (Fig. 2F). We quantified the levels of cAMP in the normal and tumor tissues. The levels of cAMP were lower in the tumor sample of each patient (Fig. 2G), and its average level in the tumors was significantly lower (Fig. 2H). Thus, the increased level of basal activity and the increased levels of free catalytic subunit in the tumor were not the result of an increase in basal cAMP in the tumor-driving kinase activity.
Catalytic subunit increases more than holoenzyme by proximity ligation assay
The previous measurements have demonstrated an increase of C relative to R, as well as an increase in the amount of free C that is not bound in the holoenzyme, in the total tumor tissue relative to the adjacent tissue. To test for potential heterogeneous distribution of R and C within the tissue or within individual cells that could impact kinase activity, we used confocal immunofluorescence.
Microscopy, as a complement to biochemistry and mass spectrometry, has a few advantages. First, measurements are made in intact cells rather than in lysates. Second, it allows measurements at the single-cell level and to selectively distinguish FLC tumor cells from normal hepatocytes, stromal cells, or immune infiltrates while making the measurements. Third, it provides data on subcellular localization. The degree to which C activity is constrained by R depends not only on their amounts but also on their proximity; R cannot regulate C if it is not colocalized. We studied R and C localization by confocal immunofluorescence and the holoenzyme by Proximity Ligation Assay (PLA), which measures the proximity of R and C antibodies in intact cells (40, 41).
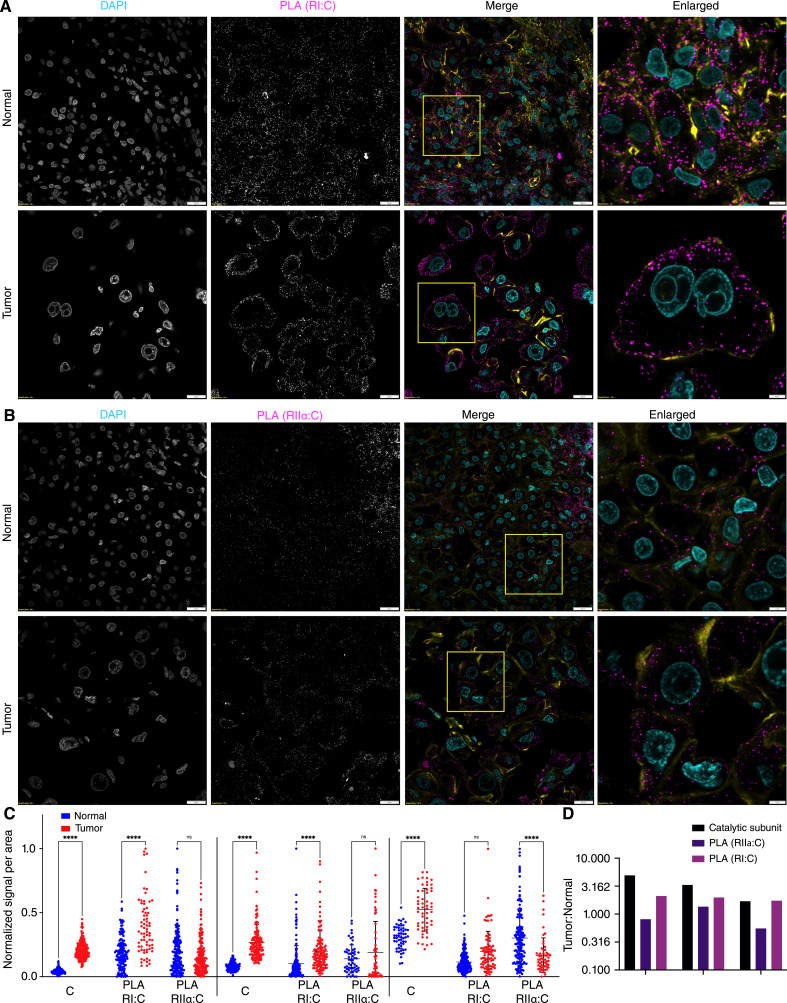
The PLA association of regulatory and catalytic subunits was observed as discrete fluorescence puncta with RI isoforms and RIIα (Fig. 3A and B). We did not assess the RIIβ isoform at the RNA and protein levels due to its low abundance in normal liver and tumor tissues, according to transcriptome (23), mass spectrometry (Fig. 1), and previous studies (13). The interaction of C with either RI and RIIα was restricted to the cytoplasm, and no intranuclear PLA was observed. PLA signals for RI were increased near the cytoplasmic membrane (Fig. 3A and B). The quantification of PLA signals showed comparable or greater intensity in tumor tissues than in the normal tissue for C:RI interactions (Fig. 3C). However, the increase in C:RI PLA was not as large as the increase of C in the tumor tissue (Fig. 3D). The number of PLA events per area between C and RIIα did not increase in the tumor tissue (Fig. 3C). These results are consistent with the upregulation of RIα observed by mass spectrometry and are validated using immunofluorescence. These results demonstrate that in FLC tumor tissue, there is a greater increase of C than of holoenzyme.
Figure 3.
Proximity ligation assay to detect binding of PKA catalytic and regulatory subunits. A and B, PLA using antibody against catalytic subunit and antibody against regulatory subunit RI (A) and RIIα (B). C, Analysis of PLA signals per cell reported as the total area of dots per area of cells for tumor and adjacent normal tissues from three different patients. Statistics were done using two-way ANOVA in PRISM. ****, P < 0.0001. D, Ratio of PKA catalytic subunit signal, PLA of C:RI, and PLA of C:RIIα in tumor over normal tissue in three different patients. Scale bars, 20 μm, except for the right column, which is 5 μm. In the merged images, DAPI is in cyan, PLA in maroon, and phalloidin (actin) in yellow. Note the use of log scale in D.
Catalytic subunit in the nucleus and regulatory subunit in cytosolic condensates in FLC
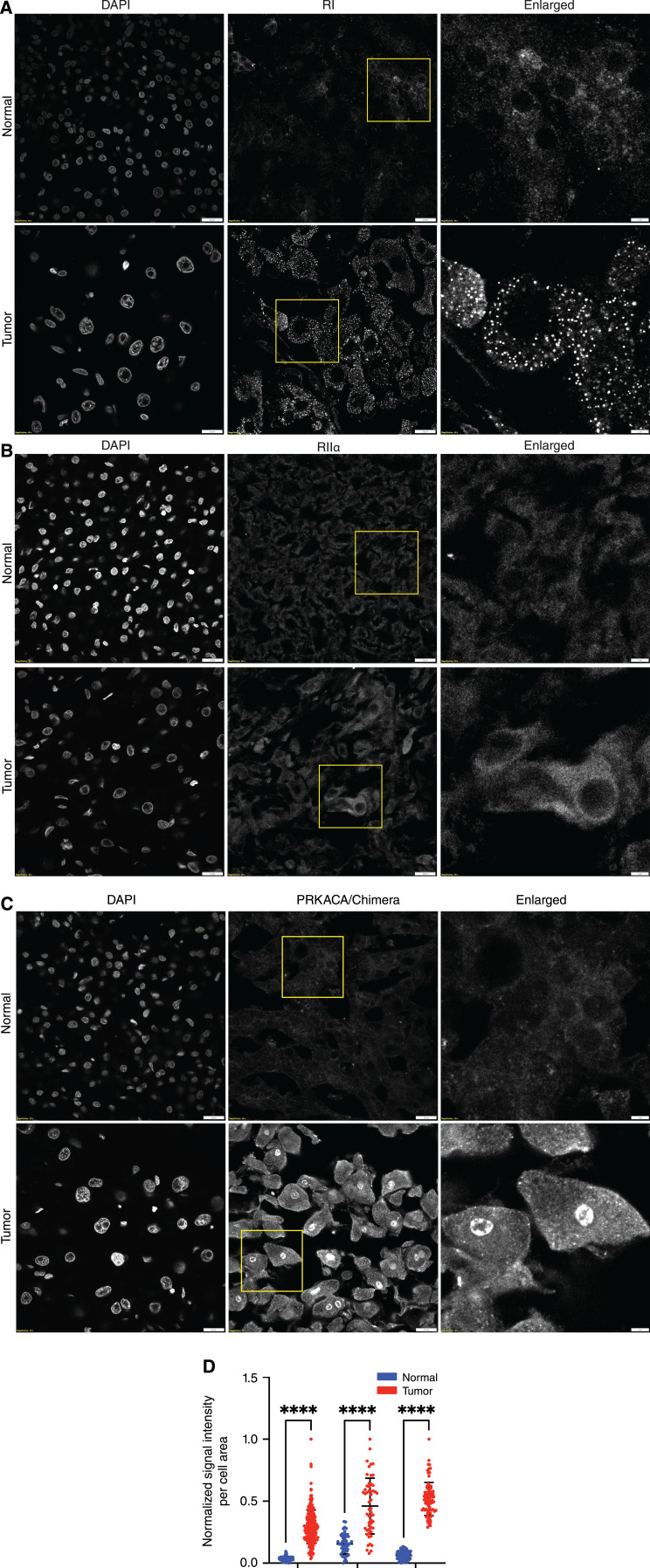
The results from MS and biochemistry have demonstrated an increase in C relative to R and an increase of free catalytic activity. Confocal immunofluorescence was used to examine the spatial distribution of R and C in FLC and the adjacent normal tissue. In FLC tumor tissue, the distribution pattern of RI subunits showed distinct clusters throughout the cytosol, whereas normal tissue showed a more diffuse pattern (Fig. 4A). The distribution of RIIα did not differ between tumor and normal tissue (Fig. 4B).
Figure 4.
Distribution of RI(α/β), RIIα, and C. A–C, Representative immunofluorescence images of FLC tumor and adjacent normal liver incubated with DAPI and antibody against RI (α/β) subunits (A), antibody against RIIα subunit (B), antibody against PRKACA/DNAJB::PRKACA (C). Scale bar in the left and middle column, 20 μm; right column, 5 μm. D, Quantification of PRKACA/DNAJB1::PRKACA signal in the nuclei of FLC cells (red) and adjacent normal liver tissue (blue) from three different patients. Statistics were done using two-way ANOVA in PRISM. ****, P < 0.0001.
In the normal tissue, C was only cytosolic, whereas in FLC tumor tissue, it was both cytosolic and nuclear (Fig. 4C and D). When we previously examined the localization of C in the nuclei of FLC tumor cells, the results were not as distinct (1). The duration between the resection of tumor and its freezing may account for the variation. For the experiments in this work, all the samples were frozen while in the operating room.
The distribution of R and C in the FLC tumor tissue was further analyzed by costaining C and RI (Fig. 5A). The intensity of C in each pixel of the field was plotted as a function of the intensity of R for both tumor and normal tissue (Fig. 5B and C). Scatter plot analysis revealed distinct quadrants representing diffuse R and C in the cytoplasm, predominantly C in the nucleus, R and C with similar amounts near the plasma membrane, and puncta consisting mainly of R dispersed in the cytosol. These results demonstrate that in FLC tumor, much of the R subunit is in cytosolic condensates with minimal C (lower right quadrant), with the excess of C in the nucleus (upper left quadrant).
Figure 5.
Distribution and colocalization of RI and C. A, Immunofluorescent image of FLC tumor, showing the localization of RI (top), C (middle), and the merged (bottom) images. Scale bars, left, 200 μm; middle, 50 μm; right, 20 μm. B and C, Colocalization analysis of PRKACA/DNAJB1::PRKACA and RI in nontransformed (B) and adjacent FLC tumor (C) from (A). Scatterplot and quantification of PRKACA/ DNAJB1::PRKACA (red) and RI (green) pixel intensities of the FLC cells (right) and adjacent normal (left). A representative field from the tumor was selected where the intensity of C and RI were plotted for each pixel in each of the three quadrants (cellSens software, Olympus). Scale bars, left images, 100 μm; right enlarged image, 10 μm.
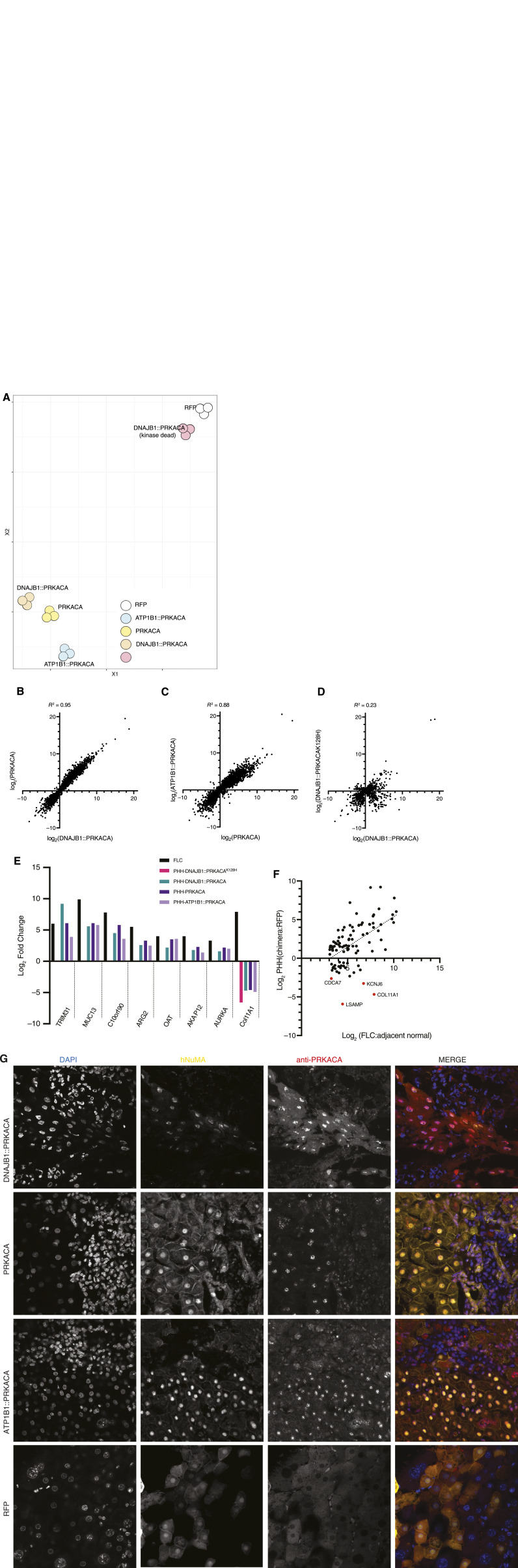
Overexpression of chimeric or wild-type kinase similarly engenders an FLC-like transcriptome
These observations demonstrate that in the FLC tumor, there is an increase of C relative to R, an increase of free C, an increase of basal catalytic activity, a segregation of R into condensates, and an increase of C in the nucleus. We next tested whether these robust observations are just correlations or are causal of the distinct transcriptome of FLC compared to normal liver (23). We transduced PHHs, isolated from the human liver and propagated in mice (30), with RFP (as the negative control for transduction effects), with native PRKACA, DNAJB1::PRKACA, or a kinase-inactive version of DNAJB1::PRKACA (made catalytically inactive by a K128H mutation) or with ATP1B1::PRKACA. The last of these was reported in a few patients with intraductal oncocytic papillary neoplasms (25, 42). Transcriptome analysis revealed that cells expressing active kinases clustered near each other, whereas cells expressing an inactive kinase (DNAJB1::PRKACAK128H) clustered near the RFP control group, as assessed by tSNE (Fig. 6A). The |log2| fold change of genes in PHHs expressing DNAJB1::PRKACA had a strong correlation, R2 = 0.95, with PHHs overexpressing PRKACA (Fig. 6B) and for PHHs expressing ATP1B1::PRKACA, and a correlation of R2 = 0.88 with PHHs expressing PRKACA (Fig. 6C). The only comparison that showed a weak correlation was that of the active to inactive kinase. PHHs expressing DNAJB1::PRKACAK128H had a correlation of R2 = 0.23 with PHHs expressing active DNAJB1::PRKACA (Fig. 6D). Some of the inactive DNAJB1::PRKACAK128H may be displacing active catalytic subunit from the holoenzyme, which could explain why there is any correlation at all.
Figure 6.
Transcriptome in PHHs overexpressing PKA catalytic subunits. A, tSNE plot from whole transcriptomes of PHHs expressing PRKACA, DNAJB1::PRKACA, ATP1B1-PRKACA, DNAJB1::PRKACAK128H(inactive kinase), and RFP. B, Scatter plot comparing the |log2| fold change of the transcripts in PHHs expressing PRKACA (y-axis) to cells expressing DNAJB1::PRKACA (x-axis); transcripts = 7,406. C, Scatter plot comparing the |log2| fold change of the transcripts in PHHs expressing ATP1B1::PRKACA (y-axis) to cells expressing PRKACA (x-axis); transcripts = 7,986. D, Scatter plot comparing the |log2| fold change of the transcripts in PHHs expressing DNAJB1::PRKACAK128H (inactive kinase; y-axis) to cells expressing DNAJB1::PRKACA (x-axis); transcripts = 2,513; the number of transcripts are lower because there are fewer significant changes with the DNAJB1::PRKACAK128H. The R2 in B, C, and D was calculated by using the Pearson method using PRISM. E, Comparison of some of the known overexpressed transcripts in FLC with PHHs overexpressing DNAJB1::PRKACA, PRKACA, ATP1B1::PRKACA, and DNAJB1::PRKACAK128H. F, Scatter plot comparing the |log2| fold change of the transcripts in PHHs expressing DNAJB1::PRKACA with the |log2| fold change of the same transcripts in FLC for some of the most overexpressed transcripts in FLC tumors. G, Immunofluorescence on PHH implanted into mouse liver. The PHHs were transduced with top row, DNAJB1::PRKACA; second row, PRKACA; third row, ATP1B1::PRKACA; bottom row, RFP. All samples were probed with DAPI (left column); antibody to human nuclear mitotic apparatus (NuMA; second column); and antibody to the carboxyl terminus of PRKACA (third column). Merged image, fourth column. Each image is 210 μm by 210 μm.
We examined eight transcripts that we most frequently find increased in FLC: TRIM31, MUC13, C10orf90, ARG2, OAT, AKAP12, AURKA, and Col11A1 (23). All of these, except for Col11A1, increased in expression in PHHs transduced with any of the active kinases. Importantly, they increased to an extent similar to their increase in patient FLC (Fig. 6E). None of them increased with the inactive kinase or a control RFP. By contrast, the expression of Col11A1 was decreased in FLC. We then expanded our set to 40 transcripts that are highly overexpressed (log2 > 3) in FLC tumor. Most of these increased in PHHs expressing DNAJB1::PRKACA to a similar extent as their increase in FLC tumor tissue (Fig. 6E). But again, there were four striking exceptions, one of which was Col11A1. A test of 700 transcripts assessed whether they increased or decreased in PHHs with active kinase relative to whether they increase or decrease in FLC tumor and found a correlation of 0.69. When we examined the points that deviated the most in the analysis, these, besides Col11A1, included KCNJ6 that is known to be expressed only in stellate cells of the liver, CDCA7 that is expressed only in blood cells in the liver (B cells, T cells, plasma cells), and LSAMB that expresses in plasma cells but not in hepatocytes (Fig. 6F, outliers marked by red dots).
The subcellular localization of DNAJB1::PRKACA observed in FLC patients (Fig. 4) could be the consequence of novel structural properties of a chimeric fusion protein or, alternatively, simply the result of kinase overexpression. To test these conjectures, we expressed DNAJB1::PRKACA, wild-type PRKACA ,or the alternative oncogenic fusion ATP1B1::PRKACA in PHHs (Fig. 6G). The distribution of the kinase for each of the three kinases tested emulated what was observed in FLC, a marked nuclear localization. By contrast, a negative control, red fluorescent protein expression, preserved the diffuse distribution of the kinase characteristic of PHHs. These results further confirm the validity of transduced PHHs as a model of FLC. They also confirm that the localization of C in the nucleus of tumor cells can be effected just by overexpression and does not require fusion adducts to PRKACA.
Spatial transcriptional analysis reveals variability of cell types in tumor tissue
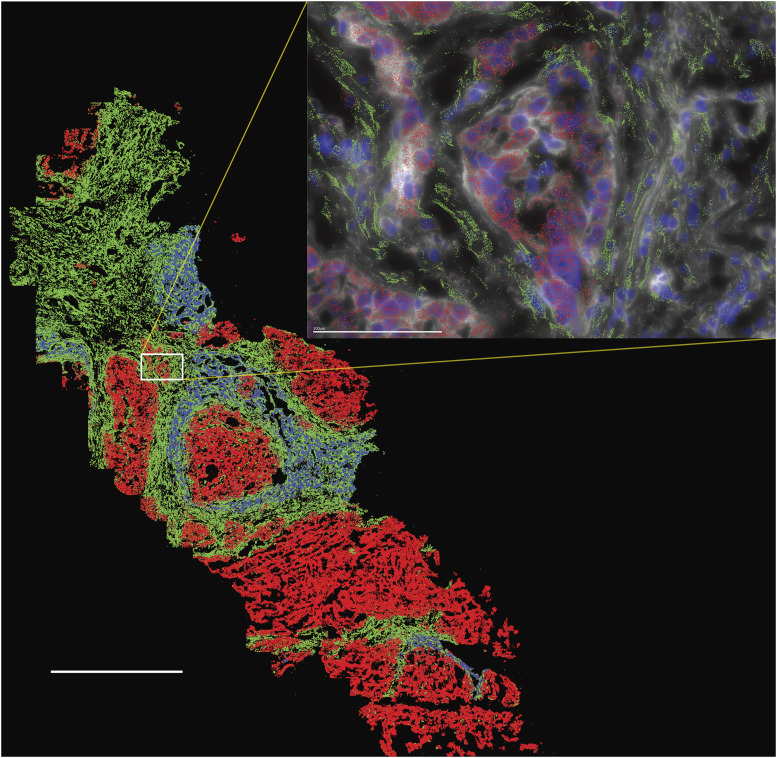
FLC tumor is a mixture of tumor cells, benign hepatocytes, benign cholangiocytes, stromal cells, and reticular endothelial and immune cells (43). To determine the expression pattern of transcripts in individual cells of the FLC tumor tissue, we used single-cell spatial transcriptional analysis. Normal, tumor, and stromal cells can be easily distinguished based on cell morphology: stromal cells are elongated, and FLC tumor cells are larger than normal cells (Fig. 7). As expected, Cyp3A (blue) showed higher expression in normal hepatocytes than in tumor cells, and Muc13 (red) was much higher in the tumor cells (Fig. 7; Supplementary Data S3). While the expression of Col11A1 (green) was increased in the FLC tumor tissue, the expression in the FLC tumor cells was decreased relative to the adjacent nontransformed hepatocytes (Fig. 7; Supplementary Data S3). However, the expression of Col11A1 was greatly increased in the stromal cells. Imaging revealed not only the diversity of cells in the tumor tissue but also the variability of the relative populations of different cell types in different sections, even within the same tumor tissue. Tissue inhomogeneity and sampling variation could contribute to the variability in the measurements of, for example, the relative amounts of DNAJB1::PRKACA to PRKACA or ratios of free to total PKA activity.
Figure 7.
Spatial transcriptional analysis of an FLC tumor tissue. Three transcripts are shown: Cyp3A (blue) that is high in normal tissue and downregulated in FLC, Muc13 (red) that is high in FLC and low in normal tissue, and Col11A1 (green) that is high in stromal cells. The distinction between the tumor and stromal (stellate) cells can be seen in the inset where the nuclei are also labeled with DAPI (dark blue) and the Vizgen Cell Boundary Staining Kit (white). Inset:, the Col11A1 was only seen above the wispy stellate cells and not above the FLC cells. Scale bar, large image, 1 mm; inset, 100 μm.
Tumors engendered by DNAJB1::PRKACA and ATP1B1::PRKACA are similar
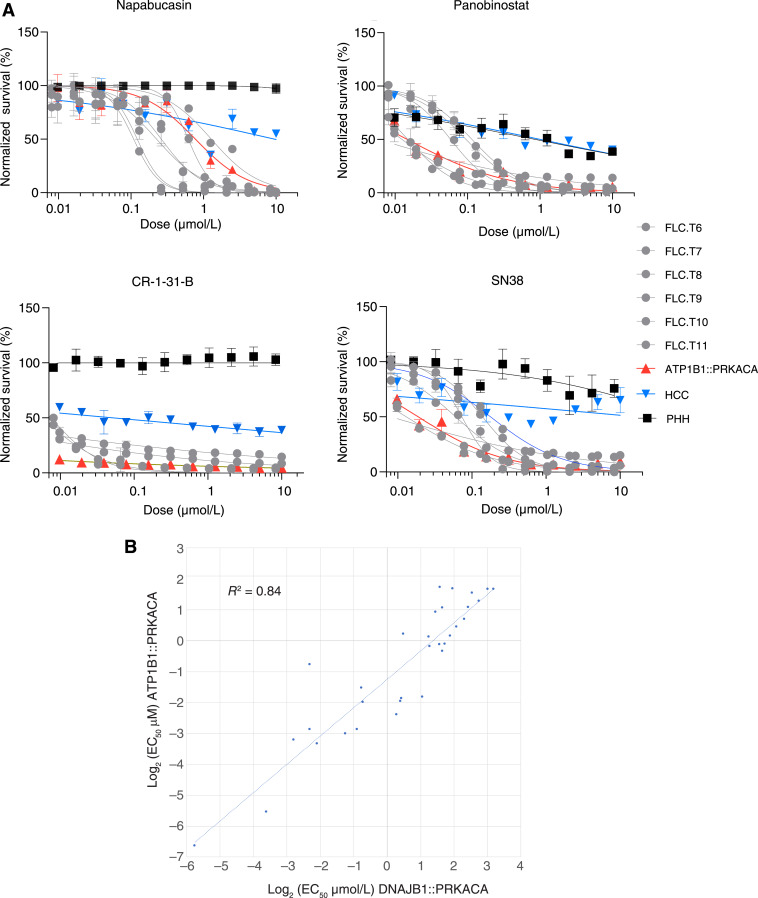
Each of the fusion kinases, ATP1B1::PRKACA, PRKACA, or DNAJB1::PRKACA, if active, produce a similar transcriptome when expressed in PHHs. It has previously been reported that tumors in ductal cells of the liver (cholangiocytes) and pancreas that express these fusion kinases have tumors that are similar to FLC based on histology (abundant eosinophil cytoplasm and a prominent nucleus) and transcriptome (25). To further explore the extent to which these tumors are similar, we obtained a freshly resected tumor from a patient expressing ATP1B1::PRKACA in the ductal cells of the liver. With a high-throughput assay, we assessed the drug–response properties of dissociated tumor cells in the first 72 hours after the resection (Fig. 8; Supplementary Table S2; ref. 33). Shown in the figure are the dose–response relationship for what we previously reported were four of the most efficacious agents (33). More globally, the EC50 values for the cells from the cholangiocarcinoma expressing ATP1B1::PRKACA strongly correlated (R2 = 0.84) with the freshly resected samples from FLC patients with DNAJB1::PRKACA, but clearly distinct from PHHs and other liver cancers (Fig. 8B). These results demonstrate that human liver tumors that express ATP1B1::PRKACA and DNAJB1::PRKACA not only have a similar histopathology and transcriptome but also a similar drug–response profile.
Figure 8.
Drug–response profile of cells freshly resected from patient tumors. A, Dose–response curve of four effective drugs against FLC against cells dissociated directly from patients’ tumors with DNAJB1::PRKACA (gray), HCC (blue), ATP1B1::PRKACA (red), or PHH (black). Each patient sample was tested in triplicate. Error bars, SD. Drugs were tested at 10 μM to 10 nM with twofold serial dilution. The y-axis shows normalized percent survival calculated as 100 − [(positive control − drug response)/(positive control − negative control) × 100]. Cells were treated for 72 hours, and normalized percent survival was quantified using CellTiter-Glo. B, Comparison of the EC50 in cells dissociated from patient tumors with DNAJB1::PRKACA mutation vs. cells dissociated from patient tumors with ATP1B1::PRKACA mutation.
Discussion
The Precision Medicine Initiative was launched with the goal of precisely defining disease not by organ or histopathology but by molecular etiology (44, 45). Here, we show that three cancers, that are FLC or FLC-like, have different somatic mutations but a common disruption in the ecology of PKA signaling. There is an increased amount of the catalytic subunit (C) of PKA relative to the amount of inhibitory regulatory subunit (R) in the tumor. Consequently, these tumors share histopathology, transcriptome, and drug–response profiles. Whether they are called three separate cancers, or three subvariants of the same cancer, they all respond the same to therapeutics, and this should be paramount when considering patient care.
PKA signaling is regulated by the inhibition of the catalytic subunit PRKACA, C, when in a holoenzyme with one of the regulatory subunits, R. In tumors considered here, there is the loss of the dominant R of the liver, RIα (24), or fusions to the catalytic subunit C: DNAJB1::PRKACA, ATP1B1::PRKACA, or ATP1B1::PRKACB (25, 42). In the most common form of FLC, that is driven by DNAJB1::PRKACA, changes were observed at the levels of both C and R in patient tumors. First, there was an increase of C protein, likely due to increased transcription (22, 23). Second, we found an increase of RIα and a decrease of RIIβ proteins. RIIβ is decreased in the transcriptome of both FLC (23) and PHHs overexpressing C (Supplementary Table S1). RIα was not altered in the transcriptome. The increase in RIα may be influenced by increased expression of C, which can stabilize R protein by promoting its greater inclusion in the R2C2 holoenzyme (46, 47). In the tumors, much of the R pool is sequestered in the cytosolic puncta, potential condensates, which may also contribute to the increased RIα level. These condensates are qualitatively similar to findings in brain neurons (48, 49). These condensates, primarily positioned more centrally in the cytoplasm, show neither robust PLA nor detectable immunofluorescence of C. This suggests a decreased accessibility of RI for C binding within them. RI may be in the interior of these condensates, leaving only RI on the surface available to bind C. Therefore, the increase in RI does not mean increased R available for binding to C. By contrast, the puncta of R1 in the periphery of the cytoplasm had robust PLA between C and RI. Recent reports have shown that the expression of DNAJB1::PRKACA in HEK293 cells disassembled RIα condensates (50), although the overexpression of PRKACA has been associated with the absence of R puncta (51, 52). By contrast, FLC cells expressing DNAJB1::PRKACA exhibit numerous RIα puncta throughout the cytosol with none observed in adjacent normal cells. The difference may be attributable to different levels of R and C expressions in the HEK293 experiment and the FLC cells of patients.
Multiple techniques have confirmed that the ratio of C to R increases in FLC tumors compared to the adjacent normal tissue. This increased C:R ratio contributes to increasing the amount of free uninhibited C outside the holoenzyme. The increase of free C was seen by two different biochemical assays, as well as by confocal microscopy and proximity ligation. Consistent with this observation, the PKA activity and the fraction of active C were higher in FLC tumor extracts than in normal tissue, indicating an increase in free C and its active state. A previous study on PKA activity in FLC showed increased free catalytic activity in one patient sample but not in three others (22). The reasons for the difference in these results could be attributed to variations in tissue sample composition (the ratio of tumor:normal:stromal cells), tissue processing, or other factors. In the present data, the consistent finding across multiple different experimental techniques using tissue that was freshly frozen upon resection gives confidence in the conclusion that there is an increase in free C.
With the increase of free C in FLC tumor cells, there was a shift in its subcellular localization into the nucleus. This is likely a consequence of both the increased ratio of C:R and the extranuclear abundance of RI in puncta devoid of C. Myristoylation of PRKACA, but not of DNAJB1::PRKACA, may also affect its localization and movement (53). The increased basal PKA activity, particularly in the nucleus, can have many physiological effects, including transcriptional and splicing perturbations (54). Overexpressing active C, either native or fusion, in PHHs resulted in nuclear localization of PKA and transcriptomic changes similar to those observed in FLC (see Fig. 6G; Supplementary Table S1). These changes include most transcripts that are consistently dysregulated in FLC (Fig 6E and F; ref. 23). The changes are attributable to the kinase activity of the proteins; the expression of inactive kinase did not result in comparable changes. These results demonstrate that an increase in the amount of C, of any enzymatically active sort, is sufficient to produce a substantial fraction of the transcriptomic changes observed in FLC.
There are a few genes consistently upregulated in FLC tumor that instead show decreased expression in PHHs expressing an active kinase. A key example is collagen Col11A1 (23). Collagen is known to be expressed in stromal cells, often in response to inflammation of the liver (55). Stromal cells are absent in the PHH model. Indeed, an examination of gene expression by spatial transcriptional analysis of FLC tumors showed that expression of Col11A1 was decreased in tumor cells relative to the adjacent normal cells but was greatly increased in stromal cells. Thus, the population transcriptomics that report an increased collagen expression in FLC tumor reflects the presence of stromal cells within the tumor and is not due to expression changes in tumor hepatocytes themselves. Another example is VCAN, which also did not increase in the PHH model. An examination of each of these exceptions helps resolve whether they are the result of expression in non-hepatocytes or whether these changes in FLC are observed only in the context of cells growing as a three-dimensional tumor and not in cells grown in culture in two dimensions. Thus, variations of transcriptomics or proteomics from patient to patient and even variations within a patient between different metastases, or even different samples within a tumor, may reflect different ratios of the populations of different cells (tumor, normal, stromal, immune), and not necessarily variations in the tumor cells.
These results reshape our understanding of the pathogenesis of FLC. More PRKACA, regardless of its specific structural form, native or fusion, creates the FLC phenotype. In FLC, increased catalytic subunit, increased basal PKA activity, and changes in its subcellular localization with a large increase of catalytic subunit in the nucleus all affect the accessibility of kinase to its substrates. Such a change is consistent with the generation of FLC tumors that are associated with the loss of RIα expression, without structural changes to the catalytic subunit (24). This conclusion is also consistent with the existence of tumors in cholangiocytes that have an alternate oncogenic kinase, one with the first exon of ATP1B1 fused to the catalytic subunit. These liver tumors show a histology and transcriptome similar to that of DNAJB1::PRKACA (25). Here, we show that they also have a response profile to therapeutics similar to that of tumors driven by DNAJB1::PRKACA in the hepatocytes (Fig. 8). These rare but informative cases show that FLC transformation does not require a specific role for the DNAJB1 moiety. As shown here, transcriptional changes associated with FLC are similarly caused by increased expression of PRKACA, DNAJB1::PRKACA, or ATP1B1::PRKACA in primary human hepatocytes. This implies that a key hallmark of the disease, its invariant transcriptome, is similarly generated by these diverse kinases. Increased catalytic activity can be responsible for changes observed in hepatocyte histology when PRKACA is overexpressed using a Sleeping Beauty transposon (2) in the livers of mice. In that system, overexpression of native PRKACA triggered the formation of tumors that by histopathology looked like FLC. Over the course of a few years, a small fraction of mice expressing DNAJB1::PRKACA died, but the number of mice expressing PRKACA was too small to determine whether there was a statistical difference in their survival (2). Our present results, together with patient and experimental genetic evidence, demonstrate that, contrary to widespread expectations (2, 50, 56, 57), DNAJB1 is not required for the FLC phenotype. Instead, increased activity and altered localization of PKA are sufficient to transform liver cells into FLC.
Supplementary Material
Supplemental Table 1 Levels of PKA subunits between FLC and adjacent normal tissue as assessed by mass spectrometry (peptides) or transcriptome. With trypsin digestion a unique peptide was not detected at the fusion junction between DNAJB1 and PRKACA
Supplemental Table 2: EC50 (µm) of the drugs tested on cells dissociated from patients tumors expressing (middle column) DNAJB1::PRKACA or (right column) ATP1B1::PRKACA.
The Spatial transcriptomics data from figure 7 with the cell by cell data for the transcripts of Cyp3A, Muc13 and Col11A1
Acknowledgments
We would like to thank all of the fibrolamellar patients, and their caregivers, through their contributions to The Fibrolamellar Registry, to our Fibrolamellar Tissue Repository, through work at the bench, and contributions in too many ways to enumerate. We would also like to thank for their financial support critical seed funds from private foundations and NIH/NCI P50CA210964 and NIH/NCI U54CA243126 (S.M. Simon); Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust (S.M. Simon); The Sohn Conference Foundation (S.M. Simon); The Rally Foundation (S.M. Simon); The Bear Necessities (S.M. Simon); The Truth365 (S.M. Simon). Supported in part by grant NIH/NCATS #UL1 TR001866 from a Clinical and Translational Science Award (CTSA).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors’ Disclosures
S. Levin reports grants from NIH during the conduct of the study. G. Lalazar reports a patent for Novel Therapeutic Targets in Fibrolamellar Hepatocellular Carcinoma identified in high throughput screens of patient-derived xenografts and direct from patient tissue pending. M.S. Torbenson reports grants from NIH during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
M. Shirani: Conceptualization, resources, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. S. Levin: Data curation, investigation, methodology. B. Shebl: Data curation, formal analysis, investigation, methodology. D. Requena: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. T.M. Finkelstein: Investigation, methodology. D.S. Johnson: Conceptualization, investigation, visualization, methodology, writing–review and editing. D. Ng: Investigation, methodology. G. Lalazar: Investigation, methodology. S. Heissel: Investigation, methodology, writing–review and editing. P. Hojrup: Investigation. H. Molina: Investigation, project administration, writing–review and editing. Y.P. de Jong: Investigation, methodology. C.M. Rice: Investigation, methodology, project administration. A.D. Singhi: Data curation, investigation. M.S. Torbenson: Formal analysis, investigation. P. Coffino: Formal analysis, supervision, investigation, project administration, writing–review and editing. B. Lyons: Supervision, investigation, project administration. S.M. Simon: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Honeyman JN, Simon EP, Robine N, Chiaroni-Clarke R, Darcy DG, Lim IIP, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 2014;343:1010–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kastenhuber ER, Lalazar G, Houlihan SL, Tschaharganeh DF, Baslan T, Chen C-C, et al. DNAJB1-PRKACA fusion kinase interacts with β-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc Natl Acad Sci U S A 2017;114:13076–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Engelholm LH, Riaz A, Serra D, Dagnaes-Hansen F, Johansen JV, Santoni-Rugiu E, et al. CRISPR/Cas9 engineering of adult mouse liver demonstrates that the Dnajb1-Prkaca gene fusion is sufficient to induce tumors resembling fibrolamellar hepatocellular carcinoma. Gastroenterology 2017;153:1662–73.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neumayer C, Ng D, Jiang CS, Qureshi A, Lalazar G, Vaughan R, et al. Oncogenic addiction of fibrolamellar hepatocellular carcinoma to the fusion kinase DNAJB1-PRKACA. Clin Cancer Res 2022;29:271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neumayer C, Ng D, Requena D, Jiang CS, Qureshi A, Vaughan R, et al. GalNAc-conjugated siRNA targeting the DNAJB1-PRKACA fusion junction in fibrolamellar hepatocellular carcinoma. Mol Ther 2024;32:140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Torbenson M. Fibrolamellar carcinoma: 2012 update. Scientifica Cairo 2012;2012:743790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lalazar G, Simon SM. Fibrolamellar carcinoma: recent advances and unresolved questions on the molecular mechanisms. Semin Liver Dis 2018;38:51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O’Neill AF, Church AJ, Perez-Atayde AR, Shaikh R, Marcus KJ, Vakili K. Fibrolamellar carcinoma: an entity all its own. Curr Probl Cancer 2021;45:100770. [DOI] [PubMed] [Google Scholar]
- 9. Darcy DG, Chiaroni-Clarke R, Murphy JM, Honeyman JN, Bhanot U, LaQuaglia MP, et al. The genomic landscape of fibrolamellar hepatocellular carcinoma: whole genome sequencing of ten patients. Oncotarget 2015;6:755–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen KY, Popovic A, Hsiehchen D, Baretti M, Griffith P, Bista R, et al. Clinical outcomes in fibrolamellar hepatocellular carcinoma treated with immune checkpoint inhibitors. Cancers Basel 2022;14:5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Potter RL, Taylor SS. Correlation of the cAMP binding domain with a site of autophosphorylation on the regulatory subunit of cAMP-dependent protein kinase II from porcine skeletal muscle. J Biol Chem 1979;254:9000–5. [PubMed] [Google Scholar]
- 12. Corbin JD, Soderling TR, Park CR. Regulation of adenosine 3′,5′-monophosphate-dependent protein kinase. I. Preliminary characterization of the adipose tissue enzyme in crude extracts. J Biol Chem 1973;248:1813–21. [PubMed] [Google Scholar]
- 13. Walker-Gray R, Stengel F, Gold MG. Mechanisms for restraining cAMP-dependent protein kinase revealed by subunit quantitation and cross-linking approaches. Proc Natl Acad Sci U S A 2017;114:10414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corbin JD, Sugden PH, Lincoln TM, Keely SL. Compartmentalization of adenosine 3′:5′-monophosphate and adenosine 3′:5′-monophosphate-dependent protein kinase in heart tissue. J Biol Chem 1977;252:3854–61. [PubMed] [Google Scholar]
- 15. Walsh DA, Perkins JP, Krebs EG. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem 1968;243:3763–5. [PubMed] [Google Scholar]
- 16. Aye TT, Scholten A, Taouatas N, Varro A, Van Veen TAB, Vos MA, et al. Proteome-wide protein concentrations in the human heart. Mol Biosyst 2010;6:1917–27. [DOI] [PubMed] [Google Scholar]
- 17. Scott JD, Dessauer CW, Tasken K. Creating order from chaos: cellular regulation by kinase anchoring. Annu Rev Pharmacol Toxicol 2013;53:187–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zimmermann B, Schweinsberg S, Drewianka S, Herberg FW. Effect of metal ions on high-affinity binding of pseudosubstrate inhibitors to PKA. Biochem J 2008;413:93–101. [DOI] [PubMed] [Google Scholar]
- 19. Dalton GD, Dewey WL. Protein kinase inhibitor peptide (PKI): a family of endogenous neuropeptides that modulate neuronal cAMP-dependent protein kinase function. Neuropeptides 2006;40:23–34. [DOI] [PubMed] [Google Scholar]
- 20. Fantozzi DA, Taylor SS, Howard PW, Maurer RA, Feramisco JR, Meinkoth JL. Effect of the thermostable protein kinase inhibitor on intracellular localization of the catalytic subunit of cAMP-dependent protein kinase. J Biol Chem 1992;267:16824–8. [PubMed] [Google Scholar]
- 21. Wiley JC, Wailes LA, Idzerda RL, McKnight GS. Role of regulatory subunits and protein kinase inhibitor (PKI) in determining nuclear localization and activity of the catalytic subunit of protein kinase A. J Biol Chem 1999;274:6381–7. [DOI] [PubMed] [Google Scholar]
- 22. Riggle KM, Riehle KJ, Kenerson HL, Turnham R, Homma MK, Kazami M, et al. Enhanced cAMP-stimulated protein kinase A activity in human fibrolamellar hepatocellular carcinoma. Pediatr Res 2016;80:110–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simon EP, Freije CA, Farber BA, Lalazar G, Darcy DG, Honeyman JN, et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proc Natl Acad Sci U S A 2015;112:E5916–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graham RP, Lackner C, Terracciano L, Gonzalez-Cantu Y, Maleszewski JJ, Greipp PT, et al. Fibrolamellar carcinoma in the Carney complex: PRKAR1A loss instead of the classic DNAJB1-PRKACA fusion. Hepatology 2018;68:1441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Singhi AD, Wood LD, Parks E, Torbenson MS, Felsenstein M, Hruban RH, et al. Recurrent rearrangements in PRKACA and PRKACB in intraductal oncocytic papillary neoplasms of the pancreas and bile duct. Gastroenterology 2020;158:573–82.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hirsch TZ, Negulescu A, Gupta B, Caruso S, Noblet B, Couchy G, et al. BAP1 mutations define a homogeneous subgroup of hepatocellular carcinoma with fibrolamellar-like features and activated PKA. J Hepatol 2020;72:924–36. [DOI] [PubMed] [Google Scholar]
- 27. Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc 2018;13:1632–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957;226:497–509. [PubMed] [Google Scholar]
- 29. Liu J, Tran V, Vemuri VNP, Byrne A, Borja M, Kim YJ, et al. Concordance of MERFISH spatial transcriptomics with bulk and single-cell RNA sequencing. Life Sci Alliance 2023;6:e202201701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michailidis E, Vercauteren K, Mancio-Silva L, Andrus L, Jahan C, Ricardo-Lax I, et al. Expansion, in vivo-ex vivo cycling, and genetic manipulation of primary human hepatocytes. Proc Natl Acad Sci U S A 2020;117:1678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016;32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017;14:417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lalazar G, Requena D, Ramos-Espiritu L, Ng D, Bhola PD, de Jong YP, et al. Identification of novel therapeutic targets for fibrolamellar carcinoma using patient-derived xenografts and direct-from-patient screening. Cancer Discov 2021;11:2544–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Levin SN, Tomasini MD, Knox J, Shirani M, Shebl B, Requena D, et al. Disruption of proteome by an oncogenic fusion kinase alters metabolism in fibrolamellar hepatocellular carcinoma. Sci Adv 2023;9:eadg7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A 2003;100:6940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Calebiro D, Hannawacker A, Lyga S, Bathon K, Zabel U, Ronchi C, et al. PKA catalytic subunit mutations in adrenocortical Cushing’s adenoma impair association with the regulatory subunit. Nat Commun 2014;5:5680. [DOI] [PubMed] [Google Scholar]
- 37. Kirchhefer U, Schmitz W, Scholz H, Neumann J. Activity of cAMP-dependent protein kinase and Ca2+/calmodulin-dependent protein kinase in failing and nonfailing human hearts. Cardiovasc Res 1999;42:254–61. [DOI] [PubMed] [Google Scholar]
- 38. Nesterova M, Yokozaki H, McDuffie E, Cho-Chung YS. Overexpression of RII beta regulatory subunit of protein kinase A in human colon carcinoma cell induces growth arrest and phenotypic changes that are abolished by site-directed mutation of RII beta. Eur J Biochem 1996;235:486–94. [DOI] [PubMed] [Google Scholar]
- 39. Paulucci-Holthauzen AA, O’Connor KL. Use of pseudosubstrate affinity to measure active protein kinase A. Anal Biochem 2006;355:175–82. [DOI] [PubMed] [Google Scholar]
- 40. Hegazy M, Cohen-Barak E, Koetsier JL, Najor NA, Arvanitis C, Sprecher E, et al. Proximity ligation assay for detecting protein-protein interactions and protein modifications in cells and tissues in Situ. Curr Protoc Cell Biol 2020;89:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ooki T, Hatakeyama M. Protocol for visualizing conditional interaction between transmembrane and cytoplasmic proteins. STAR Protoc 2021;2:100430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vyas M, Hechtman JF, Zhang Y, Benayed R, Yavas A, Askan G, et al. DNAJB1-PRKACA fusions occur in oncocytic pancreatic and biliary neoplasms and are not specific for fibrolamellar hepatocellular carcinoma. Mod Pathol 2020;33:648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Torbenson M, Zen Y, Yeh MM. Tumors of the liver. Rockville (MD): American Registry of Pathology; 2018. [Google Scholar]
- 44. Simon EP, Obama BH. Launch of the precision medicine initiative; 2016. Available from:https://youtu.be/qHD-_NYOcVA?si=yanlhWV-dn-jZiyU.
- 45. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Steinberg RA, Agard DA. Turnover of regulatory subunit of cyclic AMP-dependent protein kinase in S49 mouse lymphoma cells. Regulation by catalytic subunit and analogs of cyclic AMP. J Biol Chem 1981;256:10731–4. [PubMed] [Google Scholar]
- 47. Uhler MD, McKnight GS. Expression of cDNAs for two isoforms of the catalytic subunit of cAMP-dependent protein kinase. J Biol Chem 1987;262:15202–7. [PubMed] [Google Scholar]
- 48. Mucignat-Caretta C, Caretta A. Protein kinase A catalytic and regulatory subunits interact differently in various areas of mouse brain. Int J Mol Sci 2020;21:3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mucignat-Caretta C. Aggregates of cAMP-dependent kinase RIalpha characterize a type of cholinergic neurons in the rat brain. Brain Res Mol Brain Res 2000;80:233–6. [DOI] [PubMed] [Google Scholar]
- 50. Zhang JZ, Lu T-W, Stolerman LM, Tenner B, Yang JR, Zhang J-F, et al. Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell 2020;182:1531–44.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Day ME, Gaietta GM, Sastri M, Koller A, Mackey MR, Scott JD, et al. Isoform-specific targeting of PKA to multivesicular bodies. J Cell Biol 2011;193:347–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ahn S-H, Qin S, Zhang JZ, McCammon JA, Zhang J, Zhou H-X. Characterizing protein kinase A (PKA) subunits as macromolecular regulators of PKA RIα liquid-liquid phase separation. J Chem Phys 2021;154:221101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tillo SE, Xiong W-H, Takahashi M, Miao S, Andrade AL, Fortin DA, et al. Liberated PKA catalytic subunits associate with the membrane via myristoylation to preferentially phosphorylate membrane substrates. Cell Rep 2017;19:617–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang L, Zambon AC, Vranizan K, Pothula K, Conklin BR, Insel PA. Gene expression signatures of cAMP/protein kinase A (PKA)-promoted, mitochondrial-dependent apoptosis. Comparative analysis of wild-type and cAMP-deathless S49 lymphoma cells. J Biol Chem 2008;283:4304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang S, Friedman SL. Hepatic fibrosis: a convergent response to liver injury that is reversible. J Hepatol 2020;73:210–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Turnham RE, Smith FD, Kenerson HL, Omar MH, Golkowski M, Garcia I, et al. An acquired scaffolding function of the DNAJ-PKAc fusion contributes to oncogenic signaling in fibrolamellar carcinoma. eLife 2019;8:e44187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simon SM. Fighting rare cancers: lessons from fibrolamellar hepatocellular carcinoma. Nat Rev Cancer 2023;23:335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1 Levels of PKA subunits between FLC and adjacent normal tissue as assessed by mass spectrometry (peptides) or transcriptome. With trypsin digestion a unique peptide was not detected at the fusion junction between DNAJB1 and PRKACA
Supplemental Table 2: EC50 (µm) of the drugs tested on cells dissociated from patients tumors expressing (middle column) DNAJB1::PRKACA or (right column) ATP1B1::PRKACA.
The Spatial transcriptomics data from figure 7 with the cell by cell data for the transcripts of Cyp3A, Muc13 and Col11A1
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD043816, and the full transcriptomics is available via GEO accession number: GSE237697. All other raw data generated in this study are available upon request from the corresponding author.