Abstract
Background:
Pretreatment HIV drug resistance (PDR) has the potential to affect treatment outcome and mortality. We present here the first nationally representative PDR study conducted in Cameroon.
Methods:
From February to July 2015, HIV-infected ART initiators were recruited from 24 randomly selected clinics situated in both urban and rural regions. Dried blood spot specimens were collected from study participants at these clinics and centralized in a reference laboratory in Yaoundé, Cameroon, for drug resistance testing. HIV drug resistance mutations were identified using the Stanford algorithm.
Results:
Overall, from the 379 participants recruited, 321 pol sequences were successfully interpreted. Two hundred and five sequences were from patients attending urban ART clinics and 116 from patients seen at rural facilities. Nine percent of sequences (29/321) were from participants reporting previous exposure to antiretrovirals. PDR prevalence among all initiators was 10.4% (95% CI 5.4%–19.1%), with 14.2% (95% CI 6.6%–27.9%) reported in urban areas and 4.3% (95% CI 1.2%–14.3%) in rural areas. Among participants with no prior exposure to antiretrovirals, PDR prevalence was 10.4% (95% CI 4.7%–21.5%) overall, with 13.5% (95% CI 5.1%–31.5%) and 5.3% (95% CI 1.4%–17.5%) reported in urban and rural areas, respectively.
Conclusions:
Our findings indicate that at least 10% of patients initiating ART in Cameroon carry viruses with PDR and may be at risk of premature ART failure. The high level of NNRTI-associated resistance is of particular concern and supports introduction of drugs with a higher genetic barrier to resistance.
Introduction
HIV drug resistance (HIVDR) can significantly compromise ART outcome. Investigating HIVDR is essential to identify the most effective antiretroviral (ARV) regimens for patients initiating first-line ART and for those changing regimen after ART failure. Understanding HIVDR is also useful to select optimal options for preventing mother-to-child transmission of HIV (PMTCT) and for adequate definition of pre- and post-exposure prophylaxis. Significant attention is generally paid to monitoring HIVDR emergence during ART, but pretreatment HIVDR (PDR) also represents a significant threat, potentially decreasing first-line regimen efficacy and increasing mortality.1-3 In many resource-rich countries, guidelines therefore recommend baseline genotypic testing prior to ART initiation to guide the choice of the first-line regimens. In resource-limited countries, mostly in sub-Saharan Africa (SSA), similar guidelines are currently not in place because of financial and operational constraints. In these countries, strategies to expand access to ART rely on standard first-line regimens and not individualized regimens as in developed countries.4
The WHO initially recommended periodical assessment of the transmission of HIVDR (TDR) in recently infected individuals in order to generate strategic information for policymaking and guidance, aiming to minimize the impact of PDR on the efficacy of first-line ARVs in countries scaling up ART.5 Between 2006 and 2010, in African countries adopting the strategy, the majority of results indicated low (≤5%) to moderate (5%–15%) levels of TDR, but a few urban sites had begun to show a significant increase.6-10 Monitoring TDR was relevant, but hard to implement in the majority of sub-Saharan countries owing to difficulties in reaching target populations of newly HIV-infected individuals, therefore making regular implementation almost impossible.6 Since 2014, the WHO has recommended a new methodology to monitor HIVDR in populations initiating ART.11 This new approach aims to assess HIVDR just before ART initiation and is called PDR. With this information, national ART programmes may anticipate first-line ART efficacy and develop adequate treatment policies. In addition, contrary to previous recommendations to conduct studies in selected cities, which were generally major cities where the first ART programmes were implemented,5 the new approach strongly recommended nationally representative studies.
In this study, we conducted a nationally representative assessment of PDR in ART initiators in Cameroon.
Methods
Study sites and participants
This was a cross-sectional study, initiated in Cameroon in February 2015 and the methodology was adapted from the 2014 WHO concept note to estimate HIV PDR.11 Twenty-nine ART clinics (19 urban clinics, with one clinic sampled twice, and 10 rural clinics) were sampled using the probability proportional to proxy size (PPPS) approach with urban/rural stratification, from a countrywide listing of all 154 clinics providing ART. The respective number of patients on ART at the end of 2014 in each clinic was provided to estimate clinic size. In PPPS sampling, clinics are sampled proportionally to the total number of patients on ART in each clinic. Thus, clinics with a larger number of patients on ART are more likely to be sampled than smaller clinics.11 The required sample size (n = 300, 32 participants per clinic) was calculated to obtain an estimate of drug resistance with a CI of ±5%, assuming a drug resistance prevalence of 10% and genotyping failure rate of 10%. To minimize the loss of effectiveness due to anticipated intraclass correlation, we assumed an intraclass correlation of 0.01 and the design effect due to imperfect weighting information was set to 1.5.11
Participants were recruited if they were HIV-1 positive, aged ≥18 years, initiating ART irrespective of their prior exposure to ARVs and provided written informed consent. Minimal sociodemographic and clinical data on participant age, gender, planned ART regimen and prior exposure to ARVs were collected using a questionnaire.
Laboratory methods
For each eligible study participant, 5 mL of whole blood was collected using EDTA tubes. Freshly collected whole blood was used to prepare dried blood spots (DBSs) according to previously published standards.12 Briefly, 50 μL of whole blood was spotted onto each of the five circles of a 903 Whatman filter paper and dried at ambient temperature for 3 h. Three DBS cards were prepared for each participant and individually packed into zipper-closure plastic bags containing two silica desiccants. Bagged DBSs were then stored at room temperature, in air-free plastic boxes in the ART clinics for a maximum of two weeks before shipment to the reference national HIVDR genotyping laboratory. Upon arrival at the genotyping laboratory, DBSs were stored between −20°C and −30°C until use for HIV-1 drug resistance genotyping.
Viral RNA was extracted from DBSs using the Abbott m2000rt RealTime HIV-1 kit according to manufacturer recommendations (Abbott Park, IL, USA). Briefly, two punched out DBS spots were incubated in 1.7 mL of m2000rt lysis buffer for 30 min under constant shaking. The recovered eluate was transferred into 2 mL collection tubes and centrifuged for 10 min at 2000 rpm to eliminate paper particles. Once cleared, the eluate was used for RNA extraction. The purified nucleic acids were then used for HIV-1 drug resistance genotyping using an in-house protocol optimized for DBS testing. The viral protease (PR) and reverse transcriptase (RT) regions were separately amplified, as previously published.12 A one-step RT–PCR was performed with primers PR2 (5′-CCTAGRAAAARGGGCTGTTGGAAATGT-3′, forward) and TR2as (5′-AATYTGACTTGCCCARTTTARTTTTCC-3′, reverse). Separate nested PCRs were performed in the PR region (amino acids 1–99) using PR3 (5′-GARGGACAYCAAATGAAAGAYTGYAC-3′) and PR3as (5′-GCCATTGTTTAACYTTTGGDCCATCCATT-3′), and in the RT region (amino acids 1–260) with TR3 (5′-TGATAGGRGGAATTGGAGGTTTTATCAA-3′) and TR3as (5′-CTAAYTTYTGTATRTCATTGACAGTCCA-3′). RT-PCRs were carried out with 10–15 μL of RNA extracts using the QIAGEN OneStep RT–PCR kit (QIAGEN, Courtaboeuf, France). Five microlitres of the RT–PCR product were used for nested PCRs using the HotStartTaq master mix kit (QIAGEN). PCR products were purified and directly sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Carlsbad, CA). HIV-1 subtyping was generated with MEGA 6 (https://www.megasoftware.net/).13 Relevant drug resistance mutations (DRMs) in PR and RT were identified using the Stanford interpretation algorithms (https://hivdb.stanford.edu/). Sequences classified as susceptible and potential low-level resistant were considered to have no drug resistance.
Statistical analysis
The outcomes generated included: (i) the overall PDR among all ART initiators; (ii) HIVDR among treatment initiators without prior ARV exposure; and (iii) HIVDR among individuals initiating NNRTI-based ART with prior ARV exposure. Prevalence of each outcome was calculated as a ratio, where the denominator was an estimate of the number of eligible individuals in the country during the survey period and the numerator was an estimate of such individuals with the outcome of interest. Proportions and means were estimated using ‘svy’ Stata commands (Stata 14, StataCorp, College Station, TX, USA) to take into account the stratified two-stage cluster design. Intraclass correlation, i.e. the proportion of an outcome’s total variance that is shared within clinics, was estimated for PDR prevalence outcome using analysis of variance. Sampling weights accounting for probability of selection at each stage (clinic, patient) and non-response were defined for all outcomes. A finite population correction was applied. Ninety-five percent CIs were calculated using a logit transformation.
Sequence accession numbers
The newly reported PR and RT sequences are available in GenBank under the following accession numbers: MF797286 to MF797622.
Ethics
All study participants provided written informed consent and anonymous identifiers were used to avoid any identification of the participants by the research team. The study protocol was approved by the Cameroon National Ethics Committee for Health Research (Yaoundé, Cameroon). This project was reviewed according to the CDC human research protection procedures and was determined to be research but the CDC was not engaged.
Results
Participant characteristics
Three hundred and eighty-two participants were recruited in 24 clinics out of 29. Five clinics, located in the northern region, were excluded owing to Boko Haram attacks and the related security issues and additional patients were recruited in the remaining 24 clinics; 379 participants were ultimately considered for the assessment after 3 were excluded because they did not meet eligibility criteria. Two-thirds (n = 249) of the study participants were from urban clinics and the remaining one-third (n = 130) were from rural clinics. The mean age of the study participants was 37.4 (95% CI 36.2–38.7) years; women represented the predominant population (67.7%; 95% CI 61.4%–73.5%). ARV regimens planned for ART initiation were primarily tenofovir + lamivudine + efavirenz/nevirapine (86.1%; 95% CI 75.1%–92.7%) and zidovudine + lamivudine + efavirenz/nevirapine (13.7%; 95% CI 7.1%–24.7%) (Table 1). Two hundred and sixty (78.3%; 95% CI 69.0%–85.4%) patients were ARV naive, 42 (10.5%; 95% CI 6.2%–17.2%) declared prior exposure to ARVs and 77 (11.2%; 95% CI 5.6%–21.3%) had unknown status regarding prior exposure to ARVs. Amongst those exposed to ARVs, 23 (64.6%; 95% CI 34.0%–86.6%) were exposed through PMTCT intervention and 18 (33.7%; 95% CI 13.0%–63.3%) were exposed through ART. The mean time delay between DBS collection in the clinics and their shipment and storage at −20°C to −30°C in the central laboratory was 10 days overall, 8 days for urban sites and 13 days for rural sites (Table 1).
Table 1.
Study participants and characteristics
| Overall |
Urban sites |
Rural sites |
||||
|---|---|---|---|---|---|---|
| n | estimatea | n | estimatea | n | estimatea | |
| Total participants recruited | 382 | 252 | 130 | |||
| not eligible (excluded) | 3 | 3 | 0 | |||
| Total eligible for the study | 379 | 249 | 130 | |||
| Female | 247 | 67.7% (61.4–73.5) | 159 | 67.6% (58.4–75.6) | 88 | 67.9% (58.1–76.4) |
| Age (years) | 38 | 37.4 (36.2–38.7) | 38 | 37.3 (35.6–39.1) | 37 | 37.6 (35.7–39.4) |
| ARV regimen (planned) | ||||||
| TDF+3TC+EFV/NVP | 318 | 86.1% (75.1–92.7) | 206 | 84.4% (67.6–93.3) | 112 | 88.8% (71.1–96.2) |
| ZDV+3TC+EFV/NVP | 56 | 13.7% (7.1–24.7) | 39 | 15.5% (6.6–32.2) | 17 | 10.9% (3.6–28.7) |
| TDF+FTC+EFV | 3 | 0.2% (0.0–0.9) | 2 | 0.1% (0.0–0.8) | 1 | <0.5% |
| 2 NRTIs+LPV/r | 1 | <0.1% (0.0–0.3) | 1 | 0.1% (0.0–0.5) | 0 | −/− |
| Prior exposure status to ARVs | ||||||
| no prior exposure | 260 | 78.3% (69.0–85.4) | 169 | 77.5% (67.4–85.2) | 91 | 79.6% (55.9–92.3) |
| prior exposure declared | 42 | 10.5% (6.2–17.2) | 36 | 14.5% (8.5–23.5) | 6 | 4.0% (0.9–16.7) |
| unknown | 77 | 11.2% (5.6–21.3) | 44 | 8.0% (3.3–18.2) | 33 | 16.4% (4.9–42.6) |
| Type of exposure | 42 | 36 | 6 | |||
| PMTCT | 23 | 64.6% (34.0–86.6) | 17 | −/− | 6 | −/− |
| ART | 18 | 33.7% (13.0–63.3) | 18 | −/− | 0 | −/− |
| other | 1 | 1.7% (0.2–13.9) | 1 | −/− | 0 | −/− |
| Time DBS spent on site (days) | 10.1 (7.8–12.4) | 8.2 (4.8–11.7) | 13.2 (11.5–14.9) | |||
TDF, tenofovir; 3TC, lamivudine; EFV, efavirenz; NVP, nevirapine; ZDV, zidovudine; FTC, emtricitabine; LPV/r, boosted lopinavir.
−/−, not displayed because of small sample size.
Estimates are study design-weighted proportion [% (95% CI)] or study design-weighted mean (95% CI).
Prevalence of PDR
Three hundred and twenty-one viral sequences representing 85% (321/379) of all samples tested were successfully genotyped and interpreted. HIV subtypes identified included CRF02_AG (56%), CRF36_cpx (9%), CRF22_01A1 (8%), A1 (5%), CRF11_cpx (4%), F2 (4%), CRF37_cpx (3%), CRF43_02G (3%), D (2%), H (1%) CRF18_cpx (1%), CRF14_BG (1%) and other subtypes (2%).
Overall, PDR prevalence among all initiators was 10.4% (95% CI 5.4%–19.1%) for any DRM. The estimated intraclass correlation, providing an indication of the similarity of participants within the sites for the PDR prevalence outcome, was 0.11 (95% CI 0.02–0.21). PI DRMs represented 0.3% (95% CI 0.1%–1.5%), NRTI DRMs represented 2.4% (95% CI 0.4%–12.9%) and NNRTI DRMs represented 10.0% (95% CI 5.1%–18.8%). In urban clinics, PDR prevalence was 14.2% (95% CI 6.6%–27.9%) overall, 0.5% (95% CI 0.1%–2.6%) for PI DRMs, 3.9% (95% CI 0.6%–20.9%) for NRTI DRMs and 13.7% (95% CI 6.2%–27.5%) for NNRTI DRMs (Table 2). In rural regions, the PDR frequency was 4.3% (95% CI 1.2%–14.3%) and included NNRTI DRMs only. Amongst ART initiators with no prior exposure to ARVs, PDR frequency was 10.4% (95% CI 4.7%–21.5%) overall. PI DRMs represented 0.3% (95% CI 0.0%–2.1%), NRTI DRMs represented 2.8% (95% CI 0.4%–16.3%) and NNRTI DRMs represented 10.1% (95% CI 4.4%–21.3%). In urban clinics, the frequency of PDR amongst initiators with no prior exposure to ARVs was 13.5% (95% CI 5.1%–31.5%) for any DRM; 0.5% (95% CI 0.1%–3.7%) were PI DRMs, 4.6% (95% CI 0.7%–26.2%) were NRTI DRMs and 13.1% (95% CI 4.7%–31.2%) were NNRTI DRMs. In rural clinics, PDR prevalence in initiators with no prior exposure was 5.3% (95% CI 1.4%–17.5%) and all DRMs were associated with NNRTIs. In ART initiators with history of prior exposure to ARVs, the PDR frequency was 14.7% (95% CI 4.6%–38.2%) for any DRM. PI DRMs represented 1.1% (95% CI 0.1%–8.8%), NRTI DRMs represented 1.6% (95% CI 0.2%–9.9%) and NNRTI DRMs represented 13.6% (95% CI 3.9%–37.9%). Sequences carrying DRMs in this group were from urban clinics only (Table 2).
Table 2.
Frequency of PDR
| Variable | Overall | Urban sites | Rural sites |
|---|---|---|---|
| Total specimens eligible for genotyping, n | 379 | 249 | 130 |
| Total successfully genotyped, n | 321 | 205 | 116 |
| Genotyping rate, % | 85 | 82 | 89 |
| HIVDR in all initiators, n | 321 | 205 | 116 |
| any DRM, % (95% CI) | 10.4 (5.4–19.1) | 14.2 (6.6–27.9) | 4.3 (1.2–14.3) |
| PI DRMs, % (95% CI) | 0.3 (0.1–1.5) | 0.5 (0.1–2.6) | 0 |
| NRTI DRMs, % (95% CI) | 2.4 (0.4–12.9) | 3.9 (0.6–20.9) | 0 |
| NNRTI DRMs, % (95% CI) | 10.0 (5.1–18.8) | 13.7 (6.2–27.5) | 4.3 (1.2–14.3) |
| HIVDR in initiators with no prior exposure to ARVs | 223 | 141 | 82 |
| any DRM, % (95% CI) | 10.4 (4.7–21.5) | 13.5 (5.1–31.5) | 5.3 (1.4–17.5) |
| PI DRMs, % (95% CI) | 0.3 (0.0–2.1) | 0.5 (0.1–3.7) | 0 |
| NRTI DRMs, % (95% CI) | 2.8 (0.4–16.3) | 4.6 (0.7–26.2) | 0 |
| NNRTI DRMs, % (95% CI) | 10.1 (4.4–21.3) | 13.1 (4.7–31.2) | 5.3 (1.4–17.5) |
| HIVDR in initiators with prior exposure to ARVs, n | 29 | 23 | 6 |
| any DRM | 14.7 (4.6–38.2) | 18.8 (5.5–48.0) | 0 |
| PI DRMs | 1.1 (0.1–8.8) | — | — |
| NRTI DRMs | 1.6 (0.2–9.9) | — | — |
| NNRTI DRMs | 13.6 (3.9–37.9) | — | — |
—, not displayed.
Data are presented as n, % or study design-weighted proportion [% (95% CI)].
DRMs and baseline susceptibility
Mutations associated with resistance to PIs included L33F (0.7%), M46IV (0.3%), I47R (0.2%) and L90M (0.2%). Mutations associated with resistance to NRTIs were D67N (1.1%), M184V (1.2%), T215FV (2.3%) and K219Q (1.1%). For NNRTI drugs, DRMs identified included K103N (4.7%), V108I (1.3%), E138AGK (3.7%), V179ADET (1.8%), Y181C (1.9%) and G190A (0.7%).
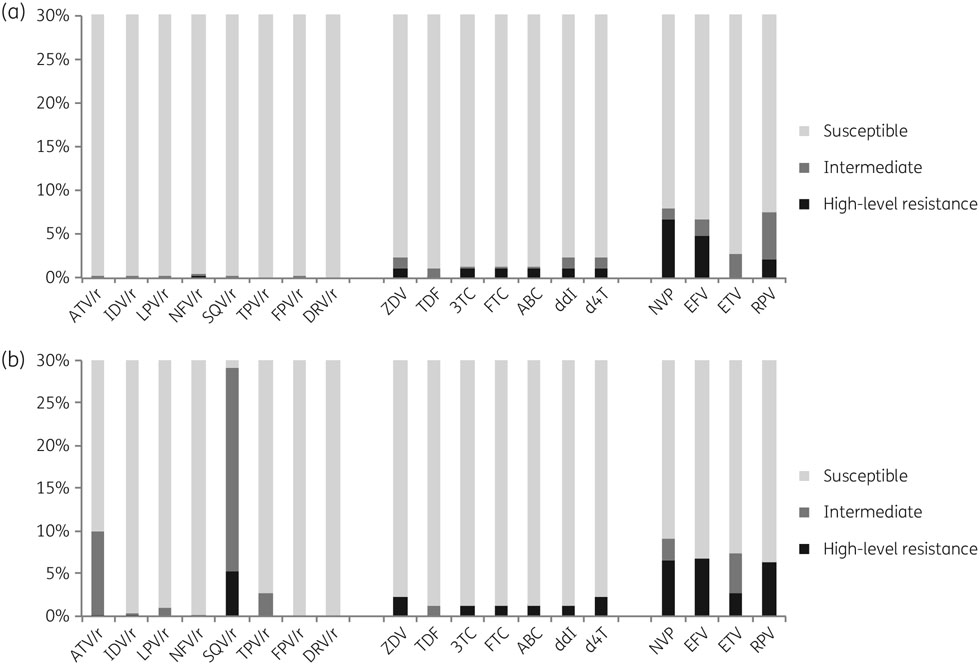
Results generated with the Stanford algorithm showed effects of identified DRMs on susceptibility to all NRTIs, for which 1.2%–2.3% of viruses were predicted to have some resistance. The highest effects were observed for zidovudine, didanosine and stavudine (2.3%), followed by lamivudine, emtricitabine and abacavir (1.3%) (Figure 1a). For NNRTIs, 6.7% and 8.1% of viruses were predicted to be resistant to efavirenz and nevirapine, respectively, and 2.8% and 7.6% of viruses were predicted to have some resistance to etravirine and rilpivirine, respectively. Less than 1% of viruses were predicted to be resistant to any PI. Results using the Agence Nationale de Recherche sur le SIDA (ANRS) algorithm (http://www.hivfrenchresistance.org/) were similar for NRTIs (Figure 1b). For NNRTIs, important discordance was observed for etravirine, for which the ANRS algorithm predicted 7.5% of viruses would have some resistance. In addition, significant discordances were observed also for PIs, especially for atazanavir, nelfinavir and saquinavir, for which 9.9%–29% of viruses were predicted to have some resistance (Figure 1b). Mutations associated with this interpretation of resistance to PIs were naturally occurring polymorphisms in the PR region that included L10I/V, I15A/V, G16E, K20I/MIR, D60E and V77I.
Figure 1.
Predicted efficacy of ARV drugs using the Stanford HIVdb (a) and the ANRS (b) algorithms. 3TC, lamivudine; ABC, abacavir; ATV, atazanavir; ZDV, zidovudine; d4T, stavudine; ddI, didanosine; DRV, darunavir; EFV, efavirenz; ETV, etravirine; FPV, fosamprenavir; FTC, emtricitabine; IDV, indinavir; LPV, lopinavir; NFV, nelfinavir; NVP, nevirapine; /r, ritonavir-boosted; RPV, rilpivirine; SQV, saquinavir; TDF, tenofovir; TPV, tipranavir.
Discussion
In this study, we estimated that 10.4% of patients in a nationally representative sample of ART initiators in Cameroon had PDR. All previous studies conducted in the country to assess HIVDR among ART-naive individuals were not designed to provide nationally representative estimates, having focused on populations at a few or only one specific clinic in a particular city or region of the country. Also, the use of differing methodologies makes it hard to compare results from one study with those from another. A 2008 study involving 79 newly diagnosed individuals from Yaoundé reported up to 9.3% harboured viruses with resistance mutations to NRTIs and NNRTIs and 2.6% had resistance to PIs.14 We conducted a survey in 2009 using the WHO methodology to estimate TDR among newly infected individuals and reported TDR prevalence of <5% in Yaoundé and 5%–15% in Douala, the two major cities of the country.6 A 2012 study of 49 ART-naive individuals in Yaoundé reported a TDR rate of 8.2%.15 Although not standardized, these studies suggest an increase in HIVDR prevalence in the ART-naive population following ART scale-up in Cameroon. Indeed, we have shown in a study conducted amongst the ART-naive population in Yaoundé, that HIVDR prevalence increased from 0% in 1996–99 to 12.3% in 2007.16 Because Yaoundé is the city where the first ART programmes were implemented in the country in early 2000, it is reasonable to expect a higher rate of HIVDR than in areas with a shorter history of ART. Consistent with this expectation, we found that rates of PDR in urban regions, where ART clinics were implemented around 2000, were four times higher than rates in rural areas, where ART clinics were not made fully operational until 2007–10. Thus, although PDR is currently low in rural areas of the country, in the absence of effective preventative actions this may increase rapidly. Indeed, a higher rate of HIVDR, up to 9.8%, has been already reported in rural Cameroon.17
Outside Cameroon, published reports on national estimates of PDR using the new WHO methodology are still scarce, although several investigations are ongoing in Eastern and Southern Africa. Two similar studies recently conducted in 2016, in Mexico and Argentina, reported high levels of PDR, with overall rates of 15.5% and 14%, respectively; and 10.6% and 11% resistance to NNRTIs, respectively.18,19 Additional investigations in other resource-limited countries can help support and guide decision making at the country and global levels.
In our study, resistance to NNRTIs was most common and was found at very high rates compared with other drug classes. Mutations associated with resistance to NRTIs were found at a low rate and PI mutations were close to absent. This was particularly noticeable in rural areas where only NNRTI resistance was found, probably because of the limited use of PI regimens. Several studies assessing TDR inside and outside SSA have reported a high prevalence of NNRTI-associated resistance, generally with K103N predominating.20-23 Reasons for the high prevalence of NNRTI-associated mutations may be the previous large-scale use of nevirapine monotherapy or nevirapine tail in short-course triple ART for PMTCT in Cameroon and in many other countries, and also long-term use of nevirapine as the NNRTI backbone in first lines of ART. The ease of transmission of K103N compared with NRTI- and PI-associated PDR mutations, coupled with its tendency to persist for long periods of time could also explain its high prevalence.24,25 Indeed, the high rate of NNRTI-associated mutations directly affects viral susceptibility to this drug class, especially nevirapine and efavirenz, which are rendered less effective. Rilpivirine was also affected, with a high level of intermediate resistance observed, mostly due to E138AGK mutations, the second most prevalent NNRTI resistance mutations observed. E138A is a polymorphic mutation, which depending on subtype, occurs in 0.5%–5% of viruses from ARV-naive patients;26,27 E138G/K are non-polymorphic mutations and all these mutations at position E138 are associated with 2- to 3-fold reduced susceptibility to etravirine and/or rilpivirine.28 These second-generation NNRTIs have not been widely used in SSA to date, but reports of their low genetic barrier to resistance in ART-experienced patients,29 coupled with their high frequency of PDR mutations, make them less suitable for salvage therapy.
HIV subtype distribution in this study correlated with previous findings, with the co-circulation of several HIV-1 subtypes and recombinant forms, including the predominating CRF02_AG strain.30 We identified no HIV strain known to naturally carry mutations capable of affecting the interpretation of HIVDR.31
The WHO recommends the Stanford algorithm for the interpretation of PDR, which ensures standardization in the interpretation of results, but many other interpretation rules exist, including the ANRS rule, the REGA rule (https://rega.kuleuven.be/cev/regadb) and the International AIDS Society (IAS) DRM list (https://www.iasusa.org/content/drug-resistance-mutations-in-HIV). These rules are used in various settings for routine interpretation of HIVDR. In our report, an additional analysis performed with the ANRS rule showed concordant results for NRTI and NNRTI resistance, with only minor differences. The differences occurred mostly for etravirine and rilpivirine and were associated with the intermediate and high-level resistance interpretations as previously reported.32,33 Major differences were associated with the interpretation of resistance to PIs, mostly for atazanavir and saquinavir for which intermediate and high-level resistance, up to 29%, were found with the ANRS rule. Such large differences will significantly affect reported levels of PDR and call into question the necessity of a standard mutation list as previously developed for TDR studies.34
A potential limitation of this study is the exclusion of a few clinics where access was not possible and that may have introduced some bias on the representativeness at the national level.
In conclusion, the national rate of PDR is high in Cameroon and potentially represents an important threat affecting the efficacy of ongoing ART programmes. This study is the first nationally representative evaluation of PDR in Cameroon and future studies will be essential to assess the evolution of HIVDR in ART initiators. In the meantime, strategies to prevent PDR should be reinforced, including efforts to prevent HIVDR in those on ART in order to reduce TDR. The high level of NNRTI-associated PDR in this study, and in many other previous reports, suggests a review of the current NNRTI backbone in the recommended first line of ART and consideration of drugs with higher genetic barrier. With the ongoing scale-up of the WHO ‘treat all’ approach in the region, assessing and preventing PDR should be considered a priority.
Acknowledgements
We thank all patients who participated in this study, medical staff, the CREMER laboratory staff and the Cameroon national health authorities.
Funding
This work was supported by the President’s Emergency Plan for AIDS Relief (PEPFAR) through the US CDC under the terms of 1U01GH000755.
Members of the EHRICA Study Group
Sylvie Abia, Avelin Fobang Aghokeng, Silvia Bertagnolio, Dorothée Bessala, Christelle Butel, Corneluis Chebo, Oumarou Chifen, Amandine Cournil, John E. Ebonloe, Sabrina Eymard-Duvernay, Gaspary Fodjeu, Suzanne Izard, Brigitte Kamtie, Emmanuel Chia Kiawi, Charles Kouam, Charles Kouanfack, Nadine Lamare, Emilienne Mamang, Nadia Mandeng, Eyongetah Mbu, Bouba Mfokue, Jembia Joseph Mosoko, Bernard Nandjou, Mireille Mpoudi, Eitel Mpoudi-Ngole, Mariama Ndam, Anne Njom Nlend, Batam Nlend, Cecile Nouboué, Pierrette Omgba, Thierry Owono, Florant Oyono, Ida Penda, Elliot Raizes, Laetitia Serrano, Xavier Tchetnya, Christian Tchinou and Gaëlle Francine Tchouwa.
Footnotes
Members are listed in the Acknowledgments section.
Transparency declarations
None to declare.
Disclaimer
The findings and conclusions in this paper are those of the authors and do not necessarily represent the official position of the funding agency.
References
- 1.Deeks SG. Treatment of antiretroviral-drug-resistant HIV-1 infection. Lancet 2003; 362: 2002–11. [DOI] [PubMed] [Google Scholar]
- 2.Hofstra LM, Sauvageot N, Albert J et al. Transmission of HIV drug resistance and the predicted effect on current first-line regimens in Europe. Clin Infect Dis 2016; 62: 655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cambiano V, Bertagnolio S, Jordan MR et al. Transmission of drug resistant HIV and its potential impact on mortality and treatment outcomes in resource-limited settings. J Infect Dis 2013; 207: S57–62. [DOI] [PubMed] [Google Scholar]
- 4.WHO. Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection—Recommendations for a Public Health Approach—Second Edition. 2016. http://apps.who.int/iris/bitstream/10665/208825/1/9789241549684_eng.pdf. [PubMed]
- 5.Bennett DE, Myatt M, Bertagnolio S et al. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther 2008; 13 Suppl 2: 25–36. [PubMed] [Google Scholar]
- 6.Aghokeng AF, Vergne L, Mpoudi-Ngole E et al. Evaluation of transmitted HIV drug resistance among recently-infected antenatal clinic attendees in four Central African countries. Antivir Ther 2009; 14: 401–11. [DOI] [PubMed] [Google Scholar]
- 7.Ayouba A, Lien TT, Nouhin J et al. Low prevalence of HIV type 1 drug resistance mutations in untreated, recently infected patients from Burkina Faso, Côte d’Ivoire, Senegal, Thailand, and Vietnam: the ANRS 12134 study. AIDS Res Hum Retroviruses 2009; 25: 1193–6. [DOI] [PubMed] [Google Scholar]
- 8.Ndembi N, Hamers RL, Sigaloff KC et al. Transmitted antiretroviral drug resistance among newly HIV-1 diagnosed young individuals in Kampala. AIDS 2011; 25: 905–10. [DOI] [PubMed] [Google Scholar]
- 9.Nouhin J, Ngin S, Martin PR et al. Low prevalence of drug resistance transmitted virus in HIV Type 1-infected ARV-naive patients in Cambodia. AIDS Res Hum Retroviruses 2009; 25: 543. [DOI] [PubMed] [Google Scholar]
- 10.Price MA, Wallis CL, Lakhi S et al. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses 2011; 27: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO. Surveillance of HIV Drug Resistance in Populations Initiating Antiretroviral Therapy (Pre-Treatment HIV Drug Resistance). 2014. http://apps.who.int/iris/bitstream/10665/112802/1/9789241507196_eng.pdf.
- 12.Monleau M, Aghokeng AF, Eymard-Duvernay S et al. Field evaluation of dried blood spots for routine HIV-1 viral load and drug resistance monitoring in patients receiving antiretroviral therapy in Africa and Asia. J Clin Microbiol 2014; 52: 578–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 2013; 30: 1229–35. [DOI] [PubMed] [Google Scholar]
- 14.Ndembi N, Abraha A, Pilch H et al. Molecular characterization of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in Yaounde, Cameroon: evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B. J Clin Microbiol 2008; 46: 177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceccarelli L, Salpini R, Moudourou S et al. Characterization of drug resistance mutations in naïve and ART-treated patients infected with HIV-1 in Yaounde, Cameroon. J Med Virol 2012; 84: 721–7. [DOI] [PubMed] [Google Scholar]
- 16.Aghokeng AF, Kouanfack C, Laurent C et al. Scale-up of antiretroviral treatment in sub-Saharan Africa is accompanied by increasing HIV-1 drug resistance mutations in drug-naive patients. AIDS 2011; 25: 2183–8. [DOI] [PubMed] [Google Scholar]
- 17.Koizumi Y, Ndembi N, Miyashita M et al. Emergence of antiretroviral therapy resistance-associated primary mutations among drug-naive HIV-1-infected individuals in rural western Cameroon. J Acquir Immune Defic Syndr 2006; 43: 15–22. [DOI] [PubMed] [Google Scholar]
- 18.Ávila-Rios S, García-Morales C, Matías-Florentino M et al. Pretreatment HIV-drug resistance in Mexico and its impact on the effectiveness of first-line antiretroviral therapy: a nationally representative 2015 WHO survey. Lancet HIV 2016; 3: e579–91. [DOI] [PubMed] [Google Scholar]
- 19.Bissio E, Barbás MG, Bouzas MB et al. Pretreatment HIV-1 drug resistance in Argentina: results from a surveillance study performed according to WHO-proposed new methodology in 2014–15. J Antimicrob Chemother 2017; 72: 504–10. [DOI] [PubMed] [Google Scholar]
- 20.Castor D, Low A, Evering T et al. Transmitted drug resistance and phylogenetic relationships among acute and early HIV-1-infected individuals in New York City. J Acquir Immune Defic Syndr 2012; 61: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamers RL, Wallis CL, Kityo C et al. HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: a multicentre observational study. Lancet Infect Dis 2011; 11: 750–9. [DOI] [PubMed] [Google Scholar]
- 22.Nichols BE, Sigaloff KC, Kityo C et al. Averted HIV infections due to expanded antiretroviral treatment eligibility offsets risk of transmitted drug resistance: a modeling study. AIDS 2014; 28: 73–83. [DOI] [PubMed] [Google Scholar]
- 23.Rhee SY, Blanco JL, Jordan MR et al. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med 2015; 12: e1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little SJ, Frost SD, Wong JK et al. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol 2008; 82: 5510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruelle J, Ingels MG, Jnaoui K et al. Transmission network of an HIV type 1 strain with K103N in young Belgian patients from different risk groups. AIDS Res Hum Retroviruses 2013; 29: 1306–9. [DOI] [PubMed] [Google Scholar]
- 26.Sluis-Cremer N, Jordan MR, Huber K et al. E138A in HIV-1 reverse transcriptase is more common in subtype C than B: implications for rilpivirine use in resource-limited settings. Antiviral Res 2014; 107: 31–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Theys K, Van Laethem K, Gomes P et al. Sub-epidemics explain localized high prevalence of reduced susceptibility to rilpivirine in treatment-naive HIV-1-infected patients: subtype and geographic compartmentalization of baseline resistance mutations. AIDS Res Hum Retroviruses 2016; 32: 427–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tambuyzer L, Nijs S, Daems B et al. Effect of mutations at position E138 in HIV-1 reverse transcriptase on phenotypic susceptibility and virologic response to etravirine. J Acquir Immune Defic Syndr 2011; 58: 18–22. [DOI] [PubMed] [Google Scholar]
- 29.Fofana DB, Soulié C, Baldé A et al. High level of HIV-1 resistance in patients failing long-term first-line antiretroviral therapy in Mali. J Antimicrob Chemother 2014; 69: 2531–5. [DOI] [PubMed] [Google Scholar]
- 30.Courtney CR, Agyingi L, Fokou A et al. Monitoring HIV-1 group M subtypes in Yaoundé, Cameroon reveals broad genetic diversity and a novel CRF02_AG/F2 infection. AIDS Res Hum Retroviruses 2016; 32: 381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villabona-Arenas CJ, Domyeum J, Mouacha F et al. HIV-1 group O infection in Cameroon from 2006 to 2013: prevalence, genetic diversity, evolution and public health challenges. Infect Genet Evol 2015; 36: 210–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cotte L, Trabaud MA, Tardy JC et al. Prediction of the virological response to etravirine in clinical practice: comparison of three genotype algorithms. J Med Virol 2009; 81: 672–7. [DOI] [PubMed] [Google Scholar]
- 33.Vergne L, Snoeck J, Aghokeng A et al. Genotypic drug resistance interpretation algorithms display high levels of discordance when applied to non-B strains from HIV-1 naive and treated patients. FEMS Immunol Med Microbiol 2006; 46: 53–62. [DOI] [PubMed] [Google Scholar]
- 34.Bennett DE, Camacho RJ, Otelea D et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009; 4: e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]